Abstract
Systemic acquired resistance (SAR) is a form of defense that provides resistance against a broad spectrum of pathogens in plants. Previous work indicates a role for plastidial glycerolipid biosynthesis in SAR. Specifically, mutations in FATTY ACID DESATURASE7 (FAD7), which lead to reduced trienoic fatty acid levels and compromised plastidial lipid biosynthesis, have been associated with defective SAR. We show that the defective SAR in Arabidopsis (Arabidopsis thaliana) fad7-1 plants is not associated with a mutation in FAD7 but rather with a second-site mutation in GLABRA1 (GL1), a gene well known for its role in trichome formation. The compromised SAR in gl1 plants is associated with impairment in their cuticles. Furthermore, mutations in two other components of trichome development, GL3 and TRANSPARENT TESTA GLABRA1, also impaired cuticle development and SAR. This suggests an overlap in the biochemical pathways leading to cuticle and trichome development. Interestingly, exogenous application of gibberellic acid (GA) not only enhanced SAR in wild-type plants but also restored SAR in gl1 plants. In contrast to GA, the defense phytohoromes salicylic acid and jasmonic acid were unable to restore SAR in gl1 plants. GA application increased levels of cuticular components but not trichome formation on gl1 plants, thus implicating cuticle, but not trichomes, as an important component of SAR. Our findings question the prudence of using mutant backgrounds for genetic screens and underscore a need to reevaluate phenotypes previously studied in the gl1 background.
Plants have evolved a large array of defense mechanisms to resist infection by pathogens. Upon recognition, the host plant initiates one or more signal transduction pathways that activate various plant defenses and thereby prevent pathogen colonization. In many cases, resistance is associated with increased expression of defense genes, including the pathogenesis-related (PR) genes and the accumulation of salicylic acid (SA) in the inoculated leaf. Induction of these responses is accompanied by localized cell death at the site of pathogen entry, which can often restrict the spread of pathogen to cells within and immediately surrounding the lesions. This phenomenon, known as the hypersensitive response, is one of the earliest visible manifestations of induced defense responses and resembles programmed cell death in animals (Dangl et al., 1996; Gray, 2002; Glazebrook, 2005; Kachroo and Kachroo, 2006). Concurrent with hypersensitive response development, defense reactions are triggered in sites both local and distal from the primary infection. This phenomenon, known as systemic acquired resistance (SAR), is accompanied by a local and systemic increase in SA and jasmonic acid (JA) and a concomitant up-regulation of a large set of defense genes (Durrant and Dong, 2004; Truman et al., 2007; Vlot et al., 2009).
SAR involves the generation of a mobile signal in the primary leaves that, upon translocation to the distal tissues, activates defense responses resulting in broad-spectrum resistance. The production of the mobile signal takes places within 3 to 6 h of avirulent pathogen inoculation in the primary leaves (Smith-Becker et al., 1998), and the inoculated leaf must remain attached for at least 4 h after inoculation for immunity to be induced in the systemic tissues (Rasmussen et al., 1991). Mutations compromising SA synthesis or impairing SA, JA, or auxin signaling abolish SAR (Durrant and Dong, 2004; Truman et al., 2007, 2010). SAR is also dependent on the SALICYLIC ACID-BINDING PROTEIN2 (SABP2)-catalyzed conversion of methyl SA to SA in the distal tissues (Kumar and Klessig, 2003). Recent studies have suggested that methyl SA is the mobile signal required to initiate SAR in distal tissues in tobacco (Nicotiana tabacum; Park et al., 2007) and Arabidopsis (Arabidopsis thaliana; Liu et al., 2010), although another group reported a disparity in their findings related to the role of methyl SA in Arabidopsis (Attaran et al., 2009). Notably, the time point of requirement of SABP2 activity (between 48 and 72 h post inoculation; Park et al., 2009) does not coincide with the early generation and/or translocation of the mobile signal into distal tissues (within 6 h post inoculation).
The mutations acyl carrier protein4 (acp4), long-chain acyl-CoA synthetase2 (lacs2), and lacs9, which are impaired in fatty acid (FA)/lipid flux (Schnurr et al., 2004; Xia et al., 2009), also compromise SAR (Xia et al., 2009). Detailed characterization has shown that the SAR defect in acp4, lacs2, and lacs9 mutants correlates with their defective cuticles. Analysis of the SAR response in acp4 plants has shown that these plants can generate the mobile signal required for inducing SAR but are unable to respond to it. It is likely that the defective cuticle in these plants impairs their ability to perceive the SAR signal, because mechanical abrasion of cuticles disrupts SAR in wild-type plants (Xia et al., 2009). This SAR-disruptive effect of cuticle abrasion is highly specific, because it does not alter local defenses and hinders SAR only during the time frame during which the mobile signal is translocated to distal tissues.
SAR is also compromised in plants that contain a mutation in glycerol-3-phosphate dehydrogenase (Nandi et al., 2004). The glycerol-3-phosphate dehydrogenase (GLY1) reduces dihydroxyacetone phosphate to generate glycerol-3-phosphate, an obligatory component and precursor for the biosynthesis of all plant glycerolipids. Consequently, a mutation in GLY1 results in reduced carbon flux through the prokaryotic pathway of lipid biosynthesis, which leads to a reduction in the hexadecatrienoic (16:3) FAs (Miquel et al., 1998; Kachroo et al., 2004). Carbon flux and SAR are also impaired in plants containing mutations in FATTY ACID DESATURASE7 (FAD7; Chaturvedi et al., 2008). The FAD7 enzyme desaturates 16:2 and 18:2 FA species present on plastidial lipids to 16:3 and 18:3, respectively. Consequently, the fad7 mutant plants accumulate significantly reduced levels of trienoic FAs (16:3 and 18:3). Compromised SAR in mutants affected in certain plastidial FA/lipid pathways has prompted the suggestion that plastidial FA/lipids participate in SAR (Chaturvedi et al., 2008). Such a tempting conclusion is also favored by the fact that SAR requires the DIR1-encoded nonspecific lipid transfer protein, which is required for the generation and/or translocation of the mobile signal (Maldonado et al., 2002). In addition, azelaic acid, a dicarboxylic acid, was recently shown to prime SA biosynthesis and thereby SAR (Jung et al., 2009). The fact that azelaic acid is derived from oleic acid, a FA well known for its role in defense (Kachroo et al., 2003, 2004, 2005, 2007, 2008; Chandra-Shekara et al., 2007; Jiang et al., 2009; Venugopal et al., 2009; Xia et al., 2009), further suggests that FA/lipids might participate in SAR.
This study was undertaken to reexamine the role of the FA/lipid pathways in SAR and to determine the nature of the FA/lipid species mediating SAR in fad7-1 plants. Our results show that impaired FA/lipid flux is not associated with compromised SAR in fad7-1 plants but, rather, with an abnormal cuticle, which is the result of a nonallelic mutation in the GLABRA1 (GL1) gene. Besides GL1, other mutations affecting trichome formation also compromised cuticle and thereby SAR. A compensatory effect of exogenous GA on gl1 plants suggests that GA might participate in resistance to bacterial pathogens by restoring cuticle formation.
RESULTS
SAR Is Compromised in the fad7-1 Plants But Not in Other fad Mutants
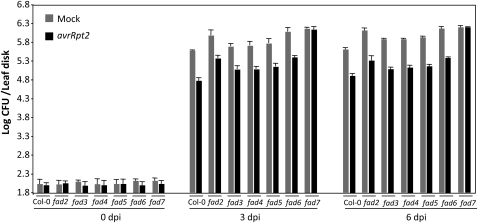
To determine which, if any, of the desaturated FA species contributed to SAR, we first evaluated SAR in all fad mutants that are defective in the desaturation of various FA species present on plastidial or extraplastidial membrane lipids (for the respective FAD-catalyzed activities, see Supplemental Fig. S1; for review, see Kachroo and Kachroo, 2009). The fad2-1, fad3-2, fad4-1, fad5-1, fad6-1, and fad7-1 mutants were first inoculated with MgCl2 or avirulent bacteria (avrRpt2) followed by a second inoculation with virulent bacteria on distal tissues at 48 h post primary inoculation. The growth of the virulent bacteria was monitored at 0, 3, and 6 d post inoculation (dpi; Fig. 1). As expected, MgCl2-infiltrated leaves of wild-type (ecotype Columbia [Col-0]) plants supported more growth of the secondary virulent pathogen than the plants that were preinfected with the avrRpt2 strain, indicating appropriate induction of SAR (Fig. 1). Similar to wild-type plants, the fad2-1, fad3-2, fad4-1, fad5-1, and fad6-1 mutants also showed proper induction of SAR. In agreement with a previous report (Chaturvedi et al., 2008), the fad7-1 mutant plants showed compromised SAR; plants infiltrated with MgCl2 or avrRpt2 bacteria were equally susceptible to the secondary virulent bacteria in the distal tissues. Together, these data suggested that the FAD7-catalyzed desaturation of 18:2 to 18:3 on plastidial membrane lipids might be important for the proper induction of SAR.
Figure 1.
SAR response in fad mutants. Primary leaves were inoculated with MgCl2 (gray bars) or P. syringae expressing avrRpt2 (black bars), and the systemic leaves were inoculated 48 h later with a virulent strain of P. syringae.
fad7-1 Plants Are Compromised in the Pathogen-Induced Accumulation of SA
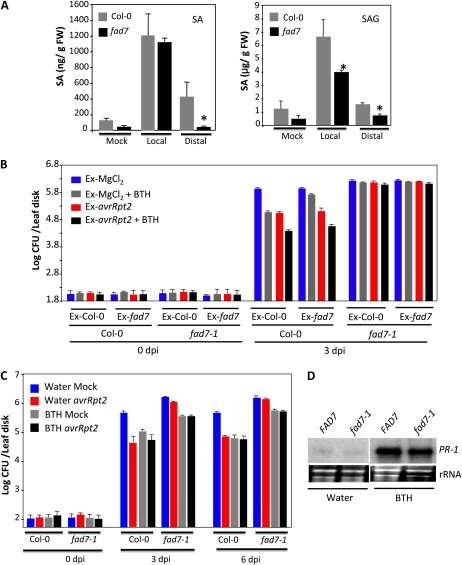
Since SA plays a critical role in SAR, we first tested if the fad7-1 mutant plants were competent in pathogen-responsive accumulation of SA. SA levels in wild-type and fad7-1 plants were determined before and after inoculation of Pseudomonas syringae containing avrRpt2. As expected, wild-type plants inoculated with avirulent pathogen showed a significant increase in both free SA and salicylic acid glucoside (SAG) in their primary as well as systemic tissues. Although the fad7-1 plants also showed an increase in SA and SAG levels in the primary tissues, levels of SA/SAG were significantly lower in their distal leaves in comparison with wild-type plants (Fig. 2A). Thus, impaired SAR in fad7-1 plants correlated with their inability to accumulate SA in the distal tissues. To confirm this, we tested the effect of exogenously supplied SA on SAR in fad7-1 plants. The wild-type and fad7-1 leaves were infiltrated with MgCl2 (mock) or avrRpt2 bacteria. Vascular exudates collected from these infiltrated leaves were mixed with water or the SA analog benzo(1,2,3)thiadiazole-7-carbothioic acid (BTH) and injected into the leaves of a fresh set of wild-type and fad7-1 plants. The distal leaves of this second set of plants were then inoculated with virulent bacteria, and proliferation of the virulent bacteria was monitored at 0 and 3 dpi (Fig. 2B). Notably, BTH-containing exudate from MgCl2-infiltrated Col-0 plants conferred SAR on Col-0 plants but not on fad7-1 plants. In comparison, the BTH-containing exudate from MgCl2-infiltrated fad7-1 was unable to confer SAR on either Col-0 or fad7-1 plants. Interestingly, the exudate from avrRpt2-infiltrated fad7-1 plants conferred SAR on Col-0 plants but not on fad7-1 plants. The BTH-containing exudate from avrRpt2-infiltrated Col-0 or fad7-1 plants produced better SAR on Col-0 but not on fad7-1, suggesting that exogenous BTH had an additive effect on SAR in Col-0. Together, these results suggested that exogenously supplied BTH was unable to restore the defective SAR in fad7-1 plants.
Figure 2.
SA levels and pathogen response of Col-0 and fad7-1 plants after exogenous application of BTH. A, SA and SAG levels in Col-0 and fad7-1 plants inoculated with MgCl2 or P. syringae expressing avrRpt2. FW, Fresh weight. B, SAR response in Col-0 and fad7-1 plants infiltrated with exudates collected from wild-type or fad7-1 plants that were treated either with MgCl2 or P. syringae expressing avrRpt2. Exudates (Ex) were mixed with water or 100 μm BTH prior to infiltration into a fresh set of plants. C, SAR in Col-0 or fad7-1 plants treated with water or BTH for 48 h prior to inoculation. D, RNA gel blot showing transcript levels of PR-1 gene in plants treated with water or BTH for 48 h. [See online article for color version of this figure.]
To determine if pretreatment of the whole plant with BTH restored SAR in fad7-1 plants, we treated Col-0 and fad7-1 plants with BTH for two consecutive days followed by inoculation of primary leaves with MgCl2 (mock) or avrRpt2 bacteria. The virulent bacteria were then infiltrated into the distal leaves 48 h after primary infiltration (MgCl2/avrRpt2), and growth of virulent bacteria was monitored at 3 and 6 dpi (Fig. 2C). Whole plant application of BTH conferred enhanced resistance to virulent pathogen in both wild-type and fad7-1 plants. However, the BTH-treated fad7-1 plants continued to support higher growth of virulent bacteria compared with BTH-treated Col-0, suggesting that BTH application on fad7-1 enhanced resistance but not to wild-type-like levels. To determine if this was due to a partial insensitivity to SA, we compared the levels of the SA-inducible marker, PR-1, in fad7-1 plants. Similar levels of PR-1 transcript were induced in response to BTH in both Col-0 and fad7 plants (Fig. 2D), suggesting that fad7-1 plants are able to induce SA-derived marker gene expression.
fad7-1 Plants Are Compromised in Pathogen-Induced Accumulation of JA
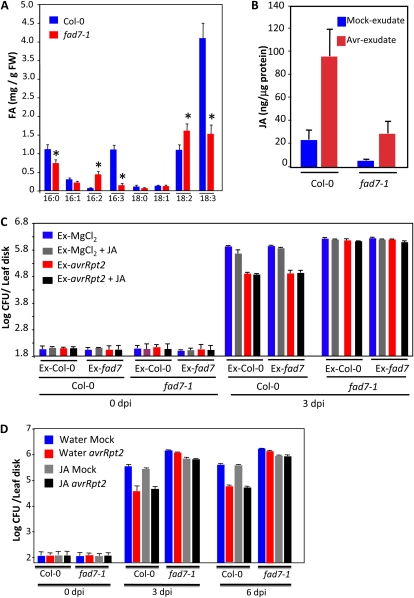
The FAD7 enzyme catalyzes the desaturation of 18:2 to 18:3 FA on membranous lipids, and a mutation in FAD7 reduces trienoic FA levels in the plant (Fig. 3A). Notably, the fad7-1 plants accumulated reduced levels of basal and pathogen-induced JA (Fig. 3B), which correlated well with reduced levels of the JA precursor 16:3 and 18:3 FAs. To determine if reduced accumulation of JA was responsible for compromised SAR, we tested if exogenously supplied JA restored SAR in fad7 plants. The wild-type and fad7-1 leaves were infiltrated with MgCl2 (mock) or avrRpt2 bacteria. Vascular exudates collected from the MgCl2- or avrRpt2-infiltrated leaves were mixed with water or JA and injected into the leaves of a fresh set of wild-type and fad7-1 plants. The distal leaves of this second set of plants were then inoculated with virulent bacteria, and their proliferation was monitored at 0 and 3 dpi (Fig. 3C). Unlike BTH, addition of JA to exudates from MgCl2-infiltrated Col-0 or fad7-1 plants did not reduce the growth of virulent bacteria. Furthermore, addition of JA to exudate from avrRpt2-infiltrated Col-0 plants also did not increase the potency of SAR in Col-0. Application of exudate on the fad7-1 plants did not induce SAR, regardless of the source of the exudate (Col-0 or fad7-1 plants). Together, these data suggested that the inability to accumulate JA likely did not contribute to the compromised SAR in fad7-1 plants. This was further confirmed by whole plant application of JA (Fig. 3D). The Col-0 and fad7-1 plants were treated with JA for 2 d followed by inoculation of primary leaves with MgCl2 (mock) or avrRpt2 bacteria. The virulent bacteria were inoculated on distal leaves 48 h after primary infiltrations, and their growth was monitored at 3 and 6 dpi. JA-treated fad7-1 plants showed a marginal reduction in the growth of the virulent pathogen but they did not show SAR. Additionally, similar to results obtained in the exudate-related experiment (Fig. 3C), whole plant pretreatment with JA did not improve SAR in either mock- or avrRpt2-inoculated wild-type plants. Together, these results argue against reduced JA as the cause of defective SAR in fad7-1 plants.
Figure 3.
FA and JA levels and pathogen response of Col-0 and fad7-1 plants after exogenous application of JA. A, Levels of FAs in 4-week-old Col-0 and fad7-1 leaves. The error bars represent sd. Asterisks denote a significant difference with Col-0 (t test, P < 0.05). FW, Fresh weight. B, JA levels in Col-0 and fad7-1 plants inoculated with MgCl2 or P. syringae expressing avrRpt2. C, SAR response in Col-0 and fad7-1 plants infiltrated with exudates collected from wild-type or fad7-1 plants that were treated either with MgCl2 or P. syringae expressing avrRpt2. Exudates (Ex) were mixed with water or 50 μm JA prior to infiltration into a fresh set of plants. D, SAR in Col-0 or fad7-1 plants treated with water or JA for 48 h prior to inoculation. [See online article for color version of this figure.]
A gl1 Mutation in the fad7-1 Plants Is Responsible for Their Defective SAR
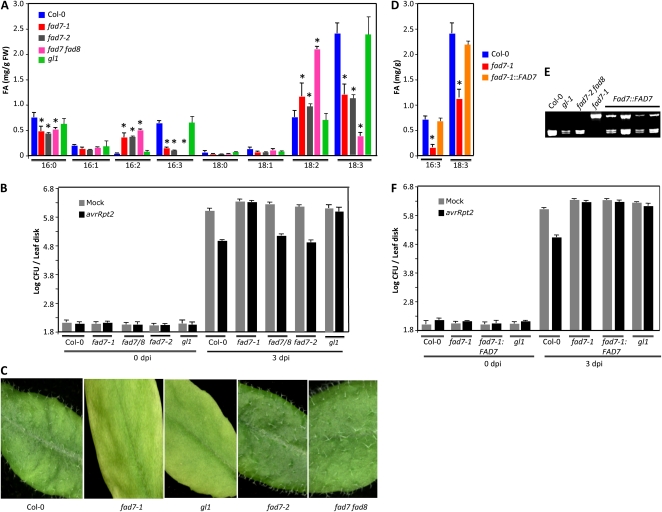
In addition to FAD7, the Arabidopsis FAD8 enzyme also catalyzes the desaturation of 18:2 to 18:3 on plastidial lipids. This is evident in the fad7-2 fad8-1 double mutant plants, which are more severely reduced in their trienoic FA levels. To test if trienoic FAs contribute to SAR, we assayed SAR in the fad7-2 fad8-1 double mutant plants. As expected, the fad7-2 fad8-1 plants showed negligible levels of 16:3 and a significant reduction in 18:3 levels (Fig. 4A). Surprisingly, the fad7-2 fad8-1 plants showed normal SAR (Fig. 4B). One possibility was that the fad8 mutation restored SAR in the fad7-2 fad8 double mutant. However, this was rather unlikely, given the fact that both FAD7 and FAD8 catalyze the same desaturation event (18:2–18:3). It was also not easily testable, since the fad8 single mutation has no detectable effect on overall FA composition (McConn et al., 1994). Therefore, we retested the requirement of FAD7 in SAR induction by evaluating SAR in the fad7-1 allelic mutant, fad7-2. Both fad7-1 and fad7-2 showed similar profile of FAs and resulted in a similar decrease in the levels of trienoic FAs (Fig. 4A). However, unlike fad7-1, the fad7-2 plants showed normal SAR (Fig. 4B). Together, these data suggested that compromised SAR in fad7-1 plants was not associated with their reduced trienoic FA levels or their altered FA/lipid profile.
Figure 4.
FA levels, SAR response, and trichome phenotypes in fad7 and fad7 fad8 plants. A, Levels of FAs in 4-week-old leaves. The error bars represent sd. Asterisks denote a significant difference with Col-0 (t test, P < 0.05). FW, Fresh weight. B, SAR response in the indicated genotypes. C, Leaves from the indicated genotypes showing the presence or absence of trichomes. D, Levels of FAs in the indicated genotypes. At least 10 different T2 transgenic plants expressing a genomic copy of FAD7 in the fad7-1 background were analyzed, and all showed a similar profile. The error bars represent sd. Asterisks denote a significant difference with Col-0 (t test, P < 0.05). E, Cleaved amplified polymorphic sequence analysis of the indicated genotypes for fad7-1 mutation. F, SAR response in the indicated genotypes. Two independent transgenic lines were analyzed, and both showed compromised SAR.
A comparison of the genetic backgrounds used for isolating fad7-1 and fad7-2 mutants revealed that fad7-1 was isolated in the gl1 background whereas fad7-2 was isolated in the wild-type background. The GL1 gene encodes the R2R2-MYB transcription factor, which is required for trichome differentiation in Arabidopsis (Oppenheimer et al., 1991). A mutation in the GL1 gene results in a glabrous phenotype due to a lack of trichomes on leaves. Indeed, similar to leaves from gl1 plants, fad7-1 gl1 leaves were devoid of any trichomes (Fig. 4C). On the other hand, wild-type, fad7-2, and fad7-2 fad8-1 leaves showed normal trichomes (Fig. 4C). Quantitative FA profiling showed a wild-type-like profile of FAs in gl1 plants (Fig. 4A), suggesting that, unlike fad7-1, the gl1 mutation was not associated with an altered FA profile. However, similar to fad7-1, the gl1 plants were compromised in SAR (Fig. 4B). Together, these data suggested that the gl1 mutation was responsible for compromised SAR in fad7-1 plants.
To confirm this further, we expressed a genomic copy of the FAD7 gene in fad7-1 gl1 plants. All the T1 plants showed wild-type-like levels of trienoic acid but were still devoid of trichomes, suggesting that these transgenic plants were complemented for fad7-1 but not for the gl1 mutation. Three independent T1 lines were analyzed in the T2 generation, and these segregated for the fad7-1 mutation in an approximately 3:1 wild-type-like:fad7-like manner; the fad7-1 plants were scored based on their genotype as well as FA profile (Fig. 4, D and E). However, all the T2 plants lacked trichomes (Supplemental Fig. S2), further confirming that the FAD7 transgene complemented only the fad7-1 and not the gl1 mutation. The fad7:FAD7 transgenic plants showed compromised SAR, like fad7-1 and gl1 plants (Fig. 4F), further reiterating the fact that the fad7-1 mutation was not responsible for compromised SAR.
gl1 Plants Are Impaired in the Perception, But Not the Generation, of the Mobile Signal
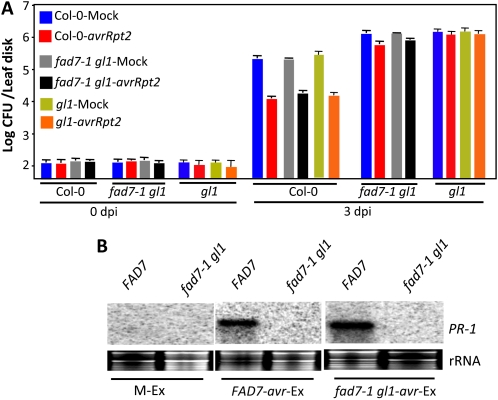
We next assessed whether GL1 contributed to mobile signal production or perception in the SAR response. We evaluated the response of wild-type, gl1, and fad7-1 gl1 plants to vascular exudates collected from pathogen-inoculated wild-type, gl1, or fad7-1 gl1 leaves. The wild-type, gl1, or fad7-1 gl1 leaves were infiltrated with MgCl2 or avrRpt2 bacteria, and vascular exudates collected from the inoculated leaves were injected into the leaves of a fresh set of wild-type, gl1, or fad7-1 gl1 plants. Distal leaves of the exudate-infiltrated plants were then inoculated with virulent bacteria, and proliferation of virulent bacteria was monitored at 0 and 3 dpi (Fig. 5A). As expected, exudates from avrRpt2-infected wild-type plants conferred protection against virulent pathogen in wild-type plants. Similar to the wild type, exudates from avrRpt2-infected gl1 or fad7-1 gl1 plants also conferred resistance against virulent pathogen in wild-type plants. In comparison, the same exudates (wild-type, gl1, or fad7-1 gl1 plants) conferred only marginal SAR on gl1 or fad7-1 gl1 plants. These data also correlated with PR-1 gene expression; the fad7-1 gl1 plants showed basal levels of PR-1 transcript in response to exudate application compared with wild-type plants (Fig. 5B). Together, these results suggested that gl1 plants are competent in generating the mobile SAR signal(s) but are defective in its perception.
Figure 5.
Responsiveness of gl1 plants to petiole exudates collected from pathogen-inoculated plants. A, SAR response in Col-0, fad7-1 gl1, and gl1 plants infiltrated with exudates collected from wild-type or acp4 plants that were treated either with MgCl2 or avrRpt2. B, RNA gel blot showing transcript levels of PR-1 in Col-0 and fad7-1 gl1 leaves infiltrated with petiole exudates (Ex). PR-1 transcript levels were analyzed 48 h after treatments. M and avr indicate petiole exudates collected from leaves infiltrated with MgCl2 or P. syringae containing avrRpt2. [See online article for color version of this figure.]
The gl1 Plants Are Defective in Their Cuticle
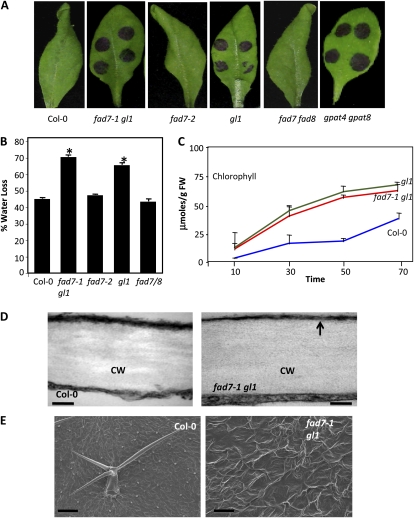
The inability of the fad7-1 gl1 plants to perceive SAR signal(s) was reminiscent of phenotypes associated with acp4 plants (Fig. 5A; Xia et al., 2009), which are defective in cuticle formation. These similarities prompted us to investigate whether the fad7-1 gl1 and gl1 plants also contained defective cuticles, similar to acp4 plants. The fad2-1, fad3-2, fad4-1, fad5-1, fad6-1, fad7-2, and fad7-2 fad8-1 mutants were also included in this analysis. The known cuticle-impaired mutant gpat4 gpat8 was included as a positive control (Fig. 6A; Li et al., 2007). Toluidine blue, a hydrophilic dye that only penetrates leaves with permeable cuticles (Tanaka et al., 2004), stained leaves in fad7-1 gl1, gl1, and gpat4 gpat8 mutants. Furthermore, fad7-1 gl1 and gl1 also showed increased water loss (Fig. 6B) and chlorophyll leaching (Fig. 6C), further supporting their defective cuticle phenotype.
Figure 6.
Evaluation of cuticle-associated phenotypes in fad7-1 gl1 plants. A, Toluidine blue-stained leaves from 4-week-old plants of the indicated genotypes. B, Measurement of water lost from leaves subjected to drought conditions for 4 d. C, A time-course measurement of chlorophyll leaching in the indicated genotypes. FW, Fresh weight. D, Transmission electron micrographs showing the cuticle layer on the adaxial surface of leaves from the indicated genotypes. The arrow indicates electron-opaque regions. CW, Cell wall. Bars = 50 nm. E, Scanning electron micrographs showing adaxial surface of leaves from the indicated genotypes. Bars = 200 μm. [See online article for color version of this figure.]
We also analyzed the outermost cell wall of the epidermis of fad7-1 gl1 and gl1 leaves by transmission electron microscopy (TEM). As expected, the cuticle of the wild-type leaf appeared as a continuous and regular electron-dense osmiophilic layer outside the cell wall (Fig. 6D). In comparison, fad7-1 gl1 mutants showed electron-opaque cuticles (Fig. 6D, arrow). Comparison of the scanning electron micrography (SEM) results of wild-type, fad7-1 gl1, and gl1 leaf surfaces showed that the mutants each had an uneven surface, which was greatly folded compared with that of wild-type leaves (Fig. 6E; data not shown for gl1).
To determine if this defect in cuticle structure was associated with alterations in the content and/or composition of cuticular waxes or cutin polyester monomers, we compared levels of waxes and cutin monomers between wild-type, fad7-1 gl1, and gl1 leaves (Supplemental Fig. S3). The fad7-1 gl1 and gl1 leaves showed reduction in several FAs, alkane and primary alcohol species, compared with wild-type plants (Supplemental Fig. S3A). The gl1 leaves showed an approximately 32% overall decrease in cutin aliphatic monomer content. In the case of gl1, the decrease was more pronounced in two major monomers (16:0-, 18:1-, and 18:2-dicarboxylic acid and 16-OH-16:0 [Supplemental Fig. S3B]). Taken together, these results show that GL1 is required for the biosynthesis of the cuticular wax and cutin polymers in leaves.
The gl1 Plants Show Compromised Resistance to Fungal Pathogens
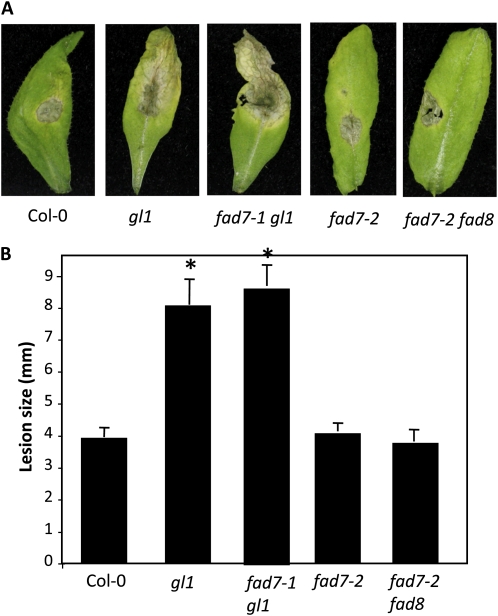
Since cuticle plays an important role in defense against fungal pathogens (Kurdyukov et al., 2006; Bessire et al., 2007; Chassot et al., 2007; Li et al., 2007; Mang et al., 2009), we next evaluated the response of fad7-1 gl1 and gl1 plants to the necrotrophic pathogen Botrytis cinerea and the hemibiotrophic fungal pathogen Colletotrichum higginsianum. Interestingly, both fad7-1 gl1 and gl1 plants showed enhanced susceptibility to B. cinerea and C. higginsianum; spray and spot inoculations showed significantly larger lesions on fad7-1 gl1 and gl1 leaves (Fig. 7; Supplemental Fig. S4). Unlike fad7-1 gl1 and gl1 plants, the fad7-2 and fad7-2 fad8 plants showed wild-type-like response to B. cinerea and C. higginsianum, arguing against a role for trienoic FAs in resistance to necrotrophic or hemibiotrophic fungal pathogens.
Figure 7.
Response of gl1 to the hemibiotrophic pathogen C. higginsianum. A, Disease symptoms on the indicated genotypes spot inoculated with 106 spores mL−1 C. higginsianum. B, Lesion size in spot-inoculated genotypes. The plants were spot inoculated with 106 spores mL−1 C. higginsianum, and the lesion size was measured from 20 to 30 independent leaves at 6 dpi. Statistical significance was determined using Student’s t test. Asterisks indicate data significantly different from those of the control (Col-0; P < 0.05). Error bars indicate sd. [See online article for color version of this figure.]
Mutations in GL3 and TTG1 Impair the Cuticle and Compromise SAR
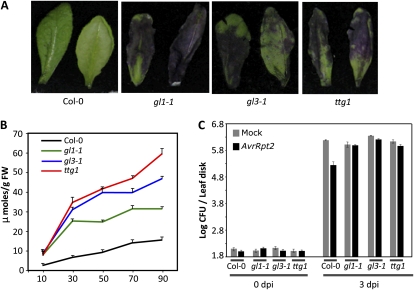
Since the GL1 gene is involved in trichome formation, we next tested if trichome formation was generally associated with a defective cuticle and thereby impaired SAR. The cuticular phenotypes were assessed in two known mutants, gl3 and transparent testa glabra1 (ttg1), which are affected in the differentiation of trichomes (Supplemental Fig. S5). The GL3 and TTG1 genes encode a basic helix-loop-helix protein and WD40 repeat protein, respectively. GL3 and TTG1 along with GL1 are thought to form a combinatorial regulatory complex during trichome differentiation (Walker et al., 1999; Payne et al., 2000; Zhang et al., 2003; Morohashi et al., 2007). Indeed, similar to gl1, both gl3-1 and ttg1 mutants do not form any trichomes on their leaves (Supplemental Fig. S5). Interestingly, the gl3 and ttg1 leaves rapidly stained with toluidine blue (Fig. 8A) and also showed increased chlorophyll leaching (Fig. 8B). These results suggest that gl3 and ttg1 mutants contain a defective cuticle. Consistent with the proposed role for cuticle in SAR, both gl3 and ttg1 plants were unable to induce proper SAR (Fig. 8C).
Figure 8.
Evaluation of cuticle-associated phenotypes and SAR response in gl3 and ttg1 plants. A, Toluidine blue-stained leaves from 4-week-old plants of the indicated genotypes. B, A time-course measurement of chlorophyll leaching in the indicated genotypes. FW, Fresh weight. C, SAR response in the indicated genotypes. [See online article for color version of this figure.]
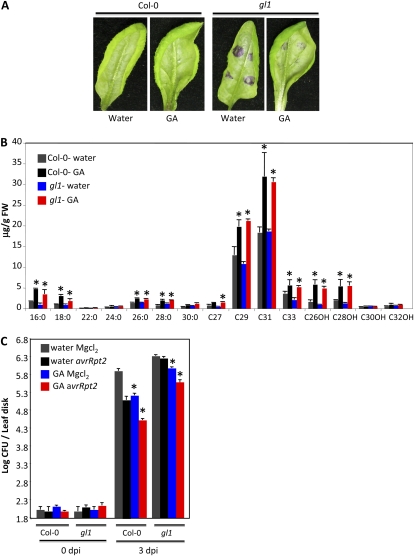
GA application is known to stimulate trichome formation (Chien and Sussex, 1996; Perazza et al., 1998). Since our results indicated that trichome differentiation was associated with cuticle formation, we next tested whether GA can restore trichome and/or cuticle formation on gl1 leaves. Exogenous application of GA to wild-type and gl1 plants caused a significant enhancement in their growth (Supplemental Fig. S6A). Although GA application was unable to induce trichome formation on gl1 leaves (Supplemental Fig. S6A), the GA-treated gl1 plants showed a significant reduction in the toluidine blue-stained areas (Fig. 9A). Furthermore, GA-treated wild-type and gl1 plants showed a significant reduction in chlorophyll leaching from these leaves (Supplemental Fig. 6B). These data suggested that GA application led to enhancement of the cuticle in both wild-type and gl1 plants. Consistent with toluidine blue-staining and chlorophyll-leaching phenotypes, the GA-treated wild-type and gl1 plants showed a pronounced increase in the levels of cuticular components (Fig. 9B). Interestingly, GA-treated wild-type plants showed significantly better SAR compared with water-treated plants (Fig. 9C). Exogenous application of GA also improved SAR on gl1 plants, but the effect was less pronounced compared with wild-type plants, likely because the cuticle might not be completely restored. Nonetheless, unlike SA or JA, GA was able to partially restore defective SAR in gl1 plants.
Figure 9.
Evaluation of cuticle-associated phenotypes and SAR response in plants treated with GA. A, Toluidine blue-stained leaves from 4-week-old plants of the indicated genotypes. B, Analysis of wax components from leaves of 4-week old Col-0 and gl1 plants treated with water or GA. 16:0 to 30:0 are FAs, C27 to C33 are alkanes, and C26OH to C32OH are primary alcohols. FW, Fresh weight. C, SAR response in Col-0 and gl1 plants pretreated with water or GA. Asterisks denote significant differences with respect to water-treated and MgCl2-infiltrated plants (t test, P < 0.05). [See online article for color version of this figure.]
DISCUSSION
The gl1 mutant was first isolated based on its glabrous phenotype (Koornneef et al., 1982), and this phenotype has served as a useful marker for numerous mutant isolations and/or screenings. A cursory database search identified over 200 articles in which the gl1 background has been used to study molecular, genetic, and biochemical aspects of diverse aspects of plant physiology (Liu and Zhu, 1997; Aarts et al., 1998; Bender and Fink, 1998; Cecchini et al., 1998; Xie et al., 1998; Yi and Jack, 1998; Bieza and Lois, 2001; Ellis and Turner, 2001; Laby et al., 2001; Devoto et al., 2002; He et al., 2002; Collins et al., 2003; Quiel and Bender, 2003; Barkan et al., 2006; Zhu et al., 2007; Speth et al., 2009; Jung and Niyogi, 2010; Rowe et al., 2010). Notably, many of the mutants that were isolated or crossed into the gl1 background have been studied for the defense physiologies. Interestingly, most studies were conducted with the assumption that the gl1 mutant itself does not affect any phenotype but the loss of trichomes. Our work shows that this is clearly not the case and that gl1 plants are compromised in defense to both bacterial and fungal pathogens. Additionally, the gl1 mutation affects cuticular development. Our findings question the prudence of using mutant backgrounds to design genetic screens and underscore the need to reevaluate phenotypes that were previously studied in the gl1 background.
This is clearly the case for the fad7-1 mutant, which was originally isolated in the gl1 background (Browse et al., 1986). Subsequent studies suggested a role for glycerolipid synthesis in SAR due to the inability of the fad7-1 (fad7-1 gl1) plants to induce SAR (Chaturvedi et al., 2008). However, normal SAR phenotype in allelic fad7-2 and the fad7-2 fad8 double mutant argue against a role for trienoic FA/lipid metabolism in SAR. This is further reiterated by the fact that trienoic FA levels but not the ability to induce SAR were restored in fad7-1 transgenic lines expressing a wild-type copy of FAD7. Characterization of the fad7-1 mutant showed that its impaired SAR is associated with a second-site mutation in the GL1 gene. Notably, a mutation in the GL1 gene impaired cuticle development, thereby compromising the plant’s ability to induce SAR. Interestingly, besides GL1, two other mutations affecting trichome formation (gl3 and ttg1) also contained deformed cuticles and were defective in inducing SAR. These data suggest that trichome differentiation and cuticle development may involve an overlapping biochemical basis. However, a correlation between the lack of trichomes and the presence of a defective cuticle was not observed in transgenic plants expressing the FATTY ACID ELONGATION1 gene specifically in the epidermis; these plants do not contain trichomes but have a normal cuticle (Reina-Pinto et al., 2009).
Is there a connection between trichome formation and cuticle development? To begin with, epidermal cells are involved in both of these processes. Trichomes differentiate from the pluripotent epidermal cells in a patterned manner, which is determined by genetic interactions between trichome-promoting and trichome-repressing factors (for review, see Schellmann and Hülskamp, 2005; Ishida et al., 2008). Proteins encoded by GL1, GL3, and TTG1 act as positive regulators, and mutations in these genes affect trichome development. Both GL1 and GL3 act as transcriptional regulators and are known to regulate genes involved in various other processes besides trichome development (Morohashi and Grotewold, 2009). Interestingly, one of the direct targets of GL3/GL1 includes FIDDLEHEAD (FDH), which encodes a protein related to β-ketoacyl-CoA synthase involved in cuticle development (Pruitt et al., 2000; Voisin et al., 2009). Notably, the fdh mutant shows a significant reduction in the number of trichomes (Yephremov et al., 1999). Similarly, several other mutants, including lcr (for LACERATA), bdg (for BODYGUARD), yre (for YORE-YORE, allelic to WAX2/CER3), and dso (for DESPERADO/AtWBC11), which are defective in cuticle formation, exhibit abnormal differentiation of trichomes and reduced trichome numbers (Wellesen et al., 2001; Kurata et al., 2003; Kurdyukov et al., 2006; Panikashvili et al., 2007). Suppression of ketoacyl reductase activity, which catalyzes the first reduction step leading to the synthesis of very-long-chain FAs, also results in abnormal trichomes and correlates with reduced cuticular wax levels (Beaudoin et al., 2009). These findings, together with our results, support the notion that overlapping pathways involving GL1, GL3, and TTG1 might regulate cuticle development as well as trichome formation. This is further supported by the fact the defective trichome phenotypes of the yre gl2 and yre ttg1 double mutant plants are more severe than the yre, ttg1, and gl2 single mutants (Kurata et al., 2003).
The ability of exogenous GA to complement the SAR defect in gl1 plants but not restore trichome formation suggests that trichomes might not play a role in SAR. This is further supported by the fact that compromised SAR in acp4 plants is not associated with trichome formation; acp4 plants show wild-type-like trichomes on their leaves (Supplemental Fig. S7). It appears that GA might participate in systemic immunity by affecting cuticle development. This is based on the fact that GA application reduced chlorophyll leaching by increasing the levels of cuticular components. Increased incorporation of palmitic acid and oleic acid into cutin monomers was also seen in GA-treated rice (Oryza sativa) and pea (Pisum sativum) plants (Bowen and Walton, 1988; Hoffmann-Benning and Kende, 1994), suggesting that GA-mediated increased carbon flux into cuticle might be a common phenomenon in plants. GA application also induced stronger SAR in wild-type plants. GA application is known to derepress its signaling pathway by inducing the degradation of DELLA proteins (Achard and Genschik, 2009), which negatively regulates the accumulation of reactive oxygen species (ROS; Achard et al., 2008). Notably, GA treatment, in the absence of pathogen inoculation, did not alter ROS levels; ROS levels in uninoculated, GA-treated wild-type or gl1 plants were similar to those in water-treated plants. This suggests that the GA-triggered increased carbon flux into cuticle is not mediated via ROS. However, it does not rule out a role for ROS in GA-triggered SAR. The fact that exogenous application of hydrogen peroxide is unable to improve the resistance response of wild-type plants to Pseudomonas discounts a role for ROS in GA-triggered gl1 plants. This is further supported by the result that exogenous application of SA, which potentiated a defense response leading to increased accumulation of ROS (Shirasu et al., 1997), was unable to trigger SAR in gl1 plants. Similarly, SA and JA treatments were also unable to complement defective SAR in another cuticle-deficient mutant, acp4 (Xia et al., 2009). Together, these observations support the notion that GA-mediated restoration of cuticle might be responsible for the restored SAR in gl1 plants. More importantly, these and our earlier results (Xia et al., 2009) reconfirm the fact that cuticle-deficient mutants are impaired in their ability to perceive mobile signal.
MATERIALS AND METHODS
Plant Growth Conditions and Complementation Analysis
Arabidopsis (Arabidopsis thaliana) plants were grown in MTPS 144 Conviron walk-in chambers at 22°C, 65% relative humidity, and a 14-h photoperiod. For complementation, an approximately 3.5-kb region encompassing the FAD7 coding region was amplified from Col-0 genomic DNA using BamHI and XbaI linkered primers, 5′-AAATTCATGCGGAATCAGAGAACG-3′ and 5′-TCTATGTTTCCGATACTGAAGC-3′, respectively, and cloned into the pBAR1 binary vector. After confirmation of the DNA sequence, the binary vector was transformed into fad7-1 plants using the floral dip method. The transgenic plants were selected on soil sprayed with the herbicide Basta. The complementation was confirmed by FA and genotype analysis of the T1 plants and by analyzing the segregation of FA phenotypes in the T2 generation. The fad7-1 mutation was identified as a C-to-T transition that converted the amino acid at position 253 from Pro to Leu. The coding region of FAD7 was amplified using 5′-ATGGCGAACTTGGTCTTATCAGAA-3′ and 5′-GAGGTCAAAGTAAGAGCAGATTGA-3′ primers. Genomic cleaved-amplified polymorphic sequence analysis for the fad7-1 was performed by amplifying a 481-bp fragment using primers 5′-GAGGAGTCTCCATTGGAGGAA-3′ and 5′-CATGTTGCTAGTAGACCAACCC-3′, which was digested with MspI.
RNA Extraction and Northern Analyses
Small-scale extraction of RNA from one or two leaves was performed in the TRIzol reagent (Invitrogen) following the manufacturer’s instructions. Northern analyses and synthesis of random primed probes was carried out as described before (Kachroo et al., 2005).
SA, FA, and JA Quantifications
SA and SAG were extracted and measured from approximately 0.3 g fresh weight of leaf tissue as described before (Chandra-Shekara et al., 2006). Measurements were performed in triplicate, and the experiment was repeated twice.
FA extraction was carried out by placing leaf tissue (n = 6) in 2 mL of 3% H2SO4 in methanol. After 30 min of incubation at 80°C, 1 mL of hexane with 0.001% butylated hydroxytoluene was added. The hexane phase was then transferred to vials for gas chromatography (GC) analysis. One-microliter samples were analyzed by GC on a Varian FAME 0.25-mm × 50-m column and quantified with flame ionization detection. For quantification of FAs, leaves (50 mg) were extracted together with an internal standard, 19:0, and the FA levels were calculated based on the detected peak areas corresponding to the FA retention time relative to the areas of the internal standard. FA quantifications were repeated three times.
For JA estimations, leaves (1 g) were ground in liquid nitrogen and extracted in 100% methanol using dihydro-JA as an internal standard. The extract was acidified to pH ≤ 4 with 1 m HCl and passed through C-18 Sep-Pak columns (Waters; 500 mg, 3 mL), which were preequilibrated with 75% methanol containing 0.2% acetic acid. The column-purified extract was saturated with sodium chloride and reextracted with diethyl ether. The ether extract was completely dried under a stream of nitrogen gas and methylated with diazomethane. The oxylipins were solubilized in 0.5 mL of hexane and dried to approximately 10 μL under a gentle stream of nitrogen gas, and 1 μL was injected for GC-mass spectrometry (MS) as described above. The JA peaks were identified using mass spectrometry. For quantitative analysis of JA, the mass spectrometer was run in selective ion monitoring mode for mass-to-charge ratio (m/z) 224 and 226. The ratio of m/z 224 to 226 in the peak area (retention time between 9.5 and 11.0 min) was used to calculate the JA levels based on the abundance of m/z 224 and 226 generated by standard JA, dihydro-JA, and their equal-level mixture. Measurements were performed in triplicate, and the experiment was repeated twice.
For JA levels in vascular exudates, samples were extracted using a solution containing glacial acetic acid, methanol, chloroform, and potassium chloride (0.9%; 1:4:8:8, v/v) and 17:0 as an internal standard. The lower phase was removed and dried under a stream of nitrogen gas, and samples were derivatized with diazomethane, dried and reconstituted in methyl tert-butyl ether, transferred to a glass insert, dried again under a stream of nitrogen gas, and reconstituted in a minimum volume of acetonitrile. Samples (1 μL) were analyzed with GC-MS as described above. Measurements were performed in triplicate, and the experiment was repeated twice.
SA, JA, and GA Treatments
SA, JA, and GA (GA4 and GA7) treatments were carried out by spraying 500, 50, or 100 μm solutions, respectively. JA-treated plants were covered with a transparent plastic dome to maximize exposure to JA.
Pathogen Infections
Inoculations with the bacterial pathogen Pseudomonas syringae were conducted as described before (Kachroo et al., 2005). The bacterial cultures were grown overnight in King’s B medium containing rifampicin and/or kanamycin. The cells were washed and suspended in 10 mm MgCl2. The bacterial suspension was injected into the abaxial surface of the leaf using a needleless syringe. Three discs from the inoculated leaves were collected and homogenized in 10 mm MgCl2. The extract was diluted, and appropriate dilutions were plated on King’s B medium. For analysis of SAR, the primary leaves were inoculated with MgCl2 or the avirulent bacteria (107 colony-forming units [CFU] mL−1), and 48 h later the systemic leaves were inoculated with virulent bacteria (105 CFU mL−1). Unless noted otherwise, samples from the systemic leaves were harvested at 3 dpi. All SAR experiments were repeated at least three times.
Colletotrichum higginsianum (IMI 349063) and Botrytis cinerea were maintained on potato dextrose agar and V8 medium, respectively. Four-week-old Arabidopsis plants were used for both spray and spot inoculations. Spore suspensions at concentrations of 104 to 106 spores mL−1 were used for various experiments. For spot inoculations, 10 μL of spore suspension was used to inoculate Arabidopsis leaves. After inoculations, the plants were transferred to a PGV36 Conviron walk-in chamber and covered with a plastic dome to maintain high humidity. Disease symptoms were scored between 4 and 11 dpi. A digital Vernier caliper was used to measure lesion size in spot-inoculated leaves. Each experiment was repeated at least twice, and each included 30 to 50 individual plants. Statistical significance was determined using Student’s t test.
Collection of Phloem Exudate
Leaf exudate was collected as described earlier (Maldonado et al., 2002). In brief, plants were induced for SAR by inoculation with P. syringae containing avrRpt2 (106 CFU mL−1). Twelve to 24 h later, petioles were excised, surface sterilized in 50% ethanol and 0.0006% bleach, rinsed in sterile 1 mm EDTA, and submerged in approximately 1.9 mL of 1 mm EDTA and 100 μg mL−1 ampicillin. Exudates were collected over 48 h and infiltrated into healthy plants. Infiltrated leaves were harvested after 2 d for PR-1 gene expression studies. For SAR studies, virulent pathogen was inoculated in the distal leaves 2 d after infiltration of exudate.
Microscopy, Chlorophyll Leaching, and Water Loss
For SEM analysis, both abaxial and adaxial surfaces of the leaf samples were mounted on sample holders with 12-mm conductive carbon tabs (Ted Pella, Inc.), sputter coated with gold-palladium, and observed on a Hitachi S-3200 SEM device with and without a backscatter detector at 5 and 20 kV.
For TEM analysis, leaves were fixed in paraformaldehyde and embedded in Epon-Araldite. Leaves were sectioned on a Reichert-Jung Ultracut E microtome with a Diatome diamond knife and observed with a Philips Tecnai Biotwin 12 TEM device.
For chlorophyll leaching assays, 100 mg of leaves was weighed and gently agitated, in the dark at room temperature, in tubes containing 80% ethanol. Absorbance of each sample was measured at 664 and 647 nm, and micromolar concentrations of total chlorophyll per gram fresh weight were calculated using the following formula: total micromoles of chlorophyll = 7.93 (A664) + 19.3 (A647).
For water loss measurements, 4-week-old plants were either subjected to drought or kept moist. The leaf weight was measured from approximately 50 leaves.
Analysis of Wax and Cutin Components
For analysis of the wax component, 500 mg of 4-week-old leaves was immersed in 10 mL of chloroform for 10 s. The leaves were rinsed once more with 10 mL of chloroform. An internal standard (100 μg of n-tetracosane) was added, and the sample volume was evaporated under a gentle steam of nitrogen. The samples were dried under a stream of nitrogen gas and methylated with diazomethane, dried again, and derivatized with 100 μL of acetic anhydride in 100 μL of pyridine, and the sealed tubes were incubated for 60 min at 60°C. The samples were again dried under a stream of nitrogen and dissolved in 1 mL of acetonitrile. Samples (1 μL) were injected into an HP-5 column of the GC device equipped with a flame ionization detector. The same samples were also run on an HP-5 column (30-m × 0.25-mm × 0.25-mm film thickness) on a GC device equipped with a mass spectrometer. Various components were identified based on their retention times as compared with standards and by MS analysis. Quantification was based on flame ionization detector peak areas as compared with the peak areas of the internal standard n-tetracosane added prior to derivatization. Measurements were performed in triplicate, and the experiment was repeated twice.
Cutin monomer composition and content were determined using the sodium methoxide-catalyzed transmethylation method followed by acetylation of the hydroxyl groups with acetic anhydride and GC-MS slightly modified from a previous description (Bonaventure and Ohlrogge, 2002; Molina et al., 2006). After methanolysis, the methylene dichloride extract of cutin monomers was washed with 0.9% potassium chloride instead of 0.5 m sodium chloride. For GC-MS analysis, the Varian FAME capillary column used was as described in wax analysis with helium carrier gas at 1 mL min−1. The mass spectrometer was run in scan mode over 35 to 450 atomic mass units (electron impact ionization). Measurements were performed in triplicate, and the experiment was repeated twice.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. An abbreviated scheme for fatty acid and lipid biosynthesis.
Supplemental Figure S2. Leaves from the indicated genotypes showing the presence or absence of trichomes.
Supplemental Figure S3. Cuticular wax and cutin monomer profiles.
Supplemental Figure S4. Response of gl1 to the necrotrophic pathogen B. cinerea.
Supplemental Figure S5. Leaves of the indicated genotypes showing the absence of trichomes on gl3 and ttg1 plants.
Supplemental Figure S6. Effects of GA treatment on trichome formation, leaf size, and chlorophyll leaching.
Supplemental Figure S7. Scanning electron micrographs of leaves from Nö and acp4 plants.
Supplementary Material
Acknowledgments
We thank Ljerka Kunst, David Smith, and David Hildebrand for useful comments. We thank John Johnson for help with GC, Ludmila Lapchyk for technical help, Larry Rice for help with SEM, and Mary Gail Engle for help with TEM. We thank Bruce Downie for GA, John Ohlrogge for gpat4 gpat8 seeds, and the Arabidopsis Biological Resource Center for fad7-1 gl1, gl1, gl3, and ttg1 seeds.
References
- Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE. (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci USA 95: 10306–10311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P, Genschik P. (2009) Releasing the brakes of plant growth: how GAs shutdown DELLA proteins. J Exp Bot 60: 1085–1092 [DOI] [PubMed] [Google Scholar]
- Achard P, Renou JP, Berthome R, Harberd NP, Genschik P. (2008) Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr Biol 18: 656–660 [DOI] [PubMed] [Google Scholar]
- Attaran E, Zeier TE, Griebel T, Zeier J. (2009) Methyl salicylate production and jasmonate signaling are not essential for systemic acquired resistance in Arabidopsis. Plant Cell 21: 954–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan L, Vijayan P, Carlsson AS, Mekhedov S, Browse J. (2006) A suppressor of fab1 challenges hypotheses on the role of thylakoid unsaturation in photosynthetic function. Plant Physiol 141: 1012–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin F, Wu X, Li F, Haslam RP, Markham JE, Zheng H, Napier JA, Kunst L. (2009) Functional characterization of the Arabidopsis B-ketoacyl-coenzyme A reductase candidates of the fatty acid elongase. Plant Physiol 150: 1174–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender J, Fink GR. (1998) A Myb homologue, ATR1, activates tryptophan gene expression in Arabidopsis. Proc Natl Acad Sci USA 95: 5655–5660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessire MCC, Jacquat AC, Humphry M, Borel S, Petétot JMC, Métraux JP, Nawrath C. (2007) A permeable cuticle in Arabidopsis leads to a strong resistance to Botrytis cinerea. EMBO J 26: 2158–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieza K, Lois R. (2001) An Arabidopsis mutant tolerant to lethal ultraviolet-B levels shows constitutively elevated accumulation of flavonoids and other phenolics. Plant Physiol 126: 1105–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaventure G, Ohlrogge JB. (2002) Differential regulation of mRNA levels of acyl carrier protein isoforms in Arabidopsis. Plant Physiol 128: 223–235 [PMC free article] [PubMed] [Google Scholar]
- Bowen DJ, Walton TJ. (1988) Cutin composition and biosynthesis during gibberellic acid-induced stem extension of Pisum sativum var Meteor. Plant Sci 55: 115–127 [Google Scholar]
- Browse J, McCourt P, Somerville C. (1986) A mutant of Arabidopsis deficient in C18:3 and C16:3 leaf lipids. Plant Physiol 81: 859–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini E, Al-Kaff N, Bannister A, Giannakou M, McCallum D, Maule A, Milner J, Covey S. (1998) Pathogenic interactions between variants of cauliflower mosaic virus and Arabidopsis thaliana. J Exp Bot 49: 731–737 [Google Scholar]
- Chandra-Shekara AC, Gupte M, Navarre D, Raina S, Raina R, Klessig D, Kachroo P. (2006) Light-dependent hypersensitive response and resistance signaling to turnip crinkle virus in Arabidopsis. Plant J 45: 320–335 [DOI] [PubMed] [Google Scholar]
- Chandra-Shekara AC, Venugopal SC, Barman SR, Kachroo A, Kachroo P. (2007) Plastidial fatty acid levels regulate resistance gene-dependent defense signaling in Arabidopsis. Proc Natl Acad Sci USA 104: 7277–7282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassot C, Nawrath C, Metraux JP. (2007) Cuticular defects lead to full immunity to a major plant pathogen. Plant J 49: 972–980 [DOI] [PubMed] [Google Scholar]
- Chaturvedi R, Krothapalli K, Makandar R, Nandi A, Sparks AA, Roth M, Welti R, Shah J. (2008) Plastid ω-3 desaturase-dependent accumulation of a systemic acquired resistance inducing activity in petiole exudates of Arabidopsis thaliana is independent of jasmonic acid. Plant J 54: 106–117 [DOI] [PubMed] [Google Scholar]
- Chien JC, Sussex IM. (1996) Differential regulation of trichome formation on the adaxial and abaxial surfaces by gibberellins and photoperiod in Arabidopsis thaliana (L) Heynh. Plant Physiol 111: 1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins NC, Thordal-Christense H, Lipka V, Bau S, Kombrink E, Qiu JL, Hückelhoven R, Stein M, Freialdenhoven A, Somerville SC, et al. (2003) SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425: 973–977 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Dietrich RA, Richberg MH. (1996) Death don’t have no mercy: cell death programs in plant-microbe interactions. Plant Cell 8: 1793–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto A, Nieto-Rostro M, Xie D, Ellis C, Harmston R, Patrick E, Davis J, Sherratt L, Coleman M, Turner JG. (2002) COI1 links jasmonate signaling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J 32: 457–466 [DOI] [PubMed] [Google Scholar]
- Durrant WE, Dong X. (2004) Systemic acquired resistance. Annu Rev Phytopathol 42: 185–209 [DOI] [PubMed] [Google Scholar]
- Ellis C, Turner JG. (2001) The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13: 1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Gray WM. (2002) Plant defense: a new weapon in the arsenal. Curr Biol 12: R352–R354 [DOI] [PubMed] [Google Scholar]
- He Y, Fukushige H, Hildebrand DF, Gan S. (2002) Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol 128: 876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann-Benning S, Kende H. (1994) Cuticle biosynthesis in rapidly growing internodes of deepwater rice. Plant Physiol 104: 719–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Kurata T, Okada K, Wada T. (2008) A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol 59: 365–386 [DOI] [PubMed] [Google Scholar]
- Jiang CJ, Shimono M, Maeda S, Inoue H, Mori M, Hasegawa M, Sugano S, Takatsuji H. (2009) Suppression of the rice fatty-acid desaturase gene OsSSI2 enhances resistance to blast and leaf blight diseases in rice. Mol Plant Microbe Interact 22: 820–829 [DOI] [PubMed] [Google Scholar]
- Jung HS, Niyogi KK. (2010) Mutations in Arabidopsis YCF20-like genes affect thermal dissipation of excess absorbed light energy. Planta 231: 923–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HW, Tschaplinksi TJ, Wang L, Glazebrook J, Greenberg J. (2009) Priming in systemic plant immunity. Science 324: 89–91 [DOI] [PubMed] [Google Scholar]
- Kachroo A, Daqi F, Havens W, Navarre D, Kachroo P, Ghabrial S. (2008) An oleic acid-mediated pathway induces constitutive defense signaling and enhanced resistance to multiple pathogens in soybean. Mol Plant Microbe Interact 21: 564–575 [DOI] [PubMed] [Google Scholar]
- Kachroo A, Kachroo P. (2006) Salicylic acid-, jasmonic acid- and ethylene-mediated regulation of plant defense signaling. Setlow J, , Genetic Regulation of Plant Defense Mechanisms, Vol 28. Springer, New York, pp 55–83 [DOI] [PubMed] [Google Scholar]
- Kachroo A, Kachroo P. (2009) Fatty acid derived signals in plant defense. Annu Rev Phytopathol 47: 153–176 [DOI] [PubMed] [Google Scholar]
- Kachroo A, Lapchyk L, Fukushigae H, Hildebrand D, Klessig D, Kachroo P. (2003) Plastidial fatty acid signaling modulates salicylic acid- and jasmonic acid-mediated defense pathways in the Arabidopsis ssi2 mutant. Plant Cell 12: 2952–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo A, Shanklin J, Lapchyk L, Whittle E, Hildebrand D, Kachroo P. (2007) The Arabidopsis stearoyl-acyl carrier protein-desaturase family and the contribution of leaf isoforms to oleic acid synthesis. Plant Mol Biol 63: 257–271 [DOI] [PubMed] [Google Scholar]
- Kachroo A, Venugopal SC, Lapchyk L, Falcone D, Hildebrand D, Kachroo P. (2004) Oleic acid levels regulated by glycerolipid metabolism modulate defense gene expression in Arabidopsis. Proc Natl Acad Sci USA 101: 5152–5157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo P, Venugopal SC, Navarre DA, Lapchyk L, Kachroo A. (2005) Role of salicylic acid and fatty acid desaturation pathways in ssi2-mediated signaling. Plant Physiol 139: 1717–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Dellaert LWM, van der Veen JH. (1982) EMS- and radiation-induced mutation frequencies at individual loci in Arabidopsis thaliana (L) Heynh. Mutat Res 93: 109–123 [DOI] [PubMed] [Google Scholar]
- Kumar D, Klessig DF. (2003) The high-affinity salicylic acid-binding protein 2 is required for plant innate immunity and has salicylic acid-stimulated lipase activity. Proc Natl Acad Sci USA 100: 16101–16106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata T, Kawabata-Awai C, Sakuradani E, Shimizu S, Okada K, Wada T. (2003) The YORE-YORE gene regulates multiple aspects of epidermal cell differentiation in Arabidopsis. Plant J 36: 55–56 [DOI] [PubMed] [Google Scholar]
- Kurdyukov S, Faust A, Nawrath C, Bär S, Voisin D, Efremova N, Franke R, Schreiber L, Saedler H, Métraux JP, et al. (2006) The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis. Plant Cell 18: 321–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laby RJ, Kim D, Gibson SI. (2001) The ram1 mutant of Arabidopsis exhibits severely decreased β-amylase activity. Plant Physiol 127: 1798–1807 [PMC free article] [PubMed] [Google Scholar]
- Li Y, Beisson F, Koo AJK, Molina I, Pollard M, Ohlrogge J. (2007) Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proc Natl Acad Sci USA 104: 18339–18344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu JK. (1997) An Arabidopsis mutant that requires increased calcium for potassium nutrition and salt tolerance. Proc Natl Acad Sci USA 94: 14960–14964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PP, Yang Y, Pichersky E, Klessig DF. (2010) Altering expression of Benzoic acid/salicylic acid carboxyl methyltransferase 1 compromises systemic acquired resistance and PAMP-triggered immunity in Arabidopsis. Mol Plant Microbe Interact 23: 82–90 [DOI] [PubMed] [Google Scholar]
- Maldonado AM, Doerner P, Dixon RA, Lamb CJ, Cameron RK. (2002) A putative lipid transfer protein involved in systemic resistance signaling in Arabidopsis. Nature 419: 399–403 [DOI] [PubMed] [Google Scholar]
- Mang HG, Laluk KA, Parsons EP, Kosma DK, Cooper BR, Park HC, AbuQamar S, Boccongelli C, Miyazaki S, Consiglio F, et al. (2009) The Arabidopsis RESURRECTION1 gene regulates a novel antagonistic interaction in plant defense to biotrophs and necrotrophs. Plant Physiol 151: 290–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn M, Hugly S, Browse J, Somerville C. (1994) A mutation at the fad8 locus of Arabidopsis identifies a second chloroplast ω-3 desaturase. Plant Physiol 106: 1609–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel M, Cassagne C, Browse J. (1998) A new class of Arabidopsis mutants with reduced hexadecatrienoic acid fatty acid levels. Plant Physiol 117: 923–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina I, Bonaventure G, Ohlrogge J, Pollard M. (2006) The lipid polyester composition of Arabidopsis thaliana and Brassica napus seeds. Phytochemistry 67: 2597–2610 [DOI] [PubMed] [Google Scholar]
- Morohashi K, Grotewold E. (2009) A system approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet 5: e1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K, Zhao M, Yang M, Read B, Lloyd A. (2007) Participation of the Arabidopsis bHLH factor GL3 in trichome initiation regulatory events. Plant Physiol 145: 736–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi A, Welti R, Shah J. (2004) The Arabidopsis thaliana dihydroxyacetone phosphate reductase gene SUPPRESSOR OF FATTY ACID DESATURASE DEFICIENCY1 is required for glycerolipid metabolism and for the activation of systemic acquired resistance. Plant Cell 16: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD. (1991) A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67: 483–493 [DOI] [PubMed] [Google Scholar]
- Panikashvili D, Savaldi-Goldstein S, Mandel T, Yifhar T, Franke RB, Höfer R, Schreiber L, Chory J, Aharoni A. (2007) The Arabidopsis DESPERADO/AtWBC11 transporter is required for cutin and wax secretion. Plant Physiol 145: 1345–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SW, Kaimoyo E, Kumar D, Mosher S, Klessig DF. (2007) Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science 318: 113–116 [DOI] [PubMed] [Google Scholar]
- Park SW, Liu PP, Forouhar F, Vlot AC, Tong L, Tietjen K, Klessig DF. (2009) Use of a synthetic salicylic acid analog to investigate the roles of methyl salicylate and its esterases in plant disease resistance. J Biol Chem 284: 7307–7317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne CT, Zhang F, Llyod AM. (2000) GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazza D, Vachon G, Herzog M. (1998) Gibberellins promote trichome formation by up-regulating GLABROUS1 in Arabidopsis. Plant Physiol 117: 375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt RE, Vielle-Calzada JP, Ploense SE, Grossniklaus U, Lolle SJ. (2000) FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proc Natl Acad Sci USA 97: 1311–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiel JA, Bender J. (2003) Glucose conjugation of anthranilate by the Arabidopsis UGT74F2 glucosyltransferase is required for tryptophan mutant blue fluorescence. J Biol Chem 278: 6275–6281 [DOI] [PubMed] [Google Scholar]
- Rasmussen JB, Hammerschmidt R, Zook MN. (1991) Systemic induction of salicylic acid accumulation in cucumber after inoculation with Pseudomonas syringae pv syringae. Plant Physiol 97: 1342–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-Pinto JJ, Voisin D, Kurdyukov S, Faust A, Haslam RP, Michaelson LV, Efremova N, Franke B, Schreiber L, Napier JA, et al. (2009) Misexpression of FATTY ACID ELONGATION1 in the Arabidopsis epidermis induces cell death and suggests a critical role for phospholipase A2 in this process. Plant Cell 21: 1252–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HC, Walley JW, Corwin J, Chan EKF, Dehesh K, Kliebenstein DJ. (2010) Deficiencies in jasmonate-mediated plant defense reveal quantitative variation in Botrytis cinerea pathogenesis. PLoS Pathog 6: e1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellmann S, Hülskamp M. (2005) Epidermal differentiation: trichomes in Arabidopsis as a model system. Int J Dev Biol 49: 579–584 [DOI] [PubMed] [Google Scholar]
- Schnurr J, Shockey J, Browse J. (2004) The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell 16: 629–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K, Nakajima H, Rajasekhar VK, Dixon RA, Lamb C. (1997) Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Plant Cell 9: 261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Becker J, Marois E, Huguet EJ, Midland SL, Sims JJ, Keen NT. (1998) Accumulation of salicylic acid and 4-hydroxybenzoic acid in phloem fluids of cucumber during systemic acquired resistance is preceded by a transient increase in phenylalanine ammonia-lyase activity in petioles and stems. Plant Physiol 116: 231–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speth EB, Imboden L, Hauck P, He SY. (2009) Subcellular localization and functional analysis of the Arabidopsis GTPase RabE. Plant Physiol 149: 1824–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Tanaka H, Machida C, Watanabe M, Machida Y. (2004) A new method for rapid visualization of defects in leaf cuticle reveals five intrinsic patterns of surface defects in Arabidopsis. Plant J 37: 139–146 [DOI] [PubMed] [Google Scholar]
- Truman W, Bennett MH, Kubigsteltig I, Turnbull C, Grant M. (2007) Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proc Natl Acad Sci USA 104: 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman W, Bennett MH, Turnbull CGN, Grant MR. (2010) Arabidopsis auxin mutants are compromised in systemic acquired resistance and exhibit aberrant accumulation of various indolic compounds. Plant Physiol 152: 1562–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal SC, Jeong RD, Mandal M, Zhu S, Chandra-Shekara AC, Xia Y, Hersh M, Stromberg AJ, Navarre D, Kachroo A, et al. (2009) ENHANCED DISEASE SUSCEPTIBILITY 1 and SALICYLIC ACID act redundantly to regulate resistance gene expression and low OLEATE-induced defense signaling. PLoS Genet 5: e1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF. (2009) Salicylic acid, a multifaceted horomone to combat disease. Annu Rev Phytopathol 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Voisin D, Nawrath C, Kurdyukov S, Franke RB, Reina-Pinto JJ, Efremova N, Will I, Schreiber L, Yephremov A. (2009) Dissection of the complex phenotype in cuticular mutants of Arabidopsis reveals a role of SERRATE as a mediator. PLoS Genet 5: e1000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC. (1999) The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11: 1337–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellesen K, Durst F, Pinot F, Benveniste I, Nettesheim K, Wisman E, Steiner-Lange S, Saedler H, Yephremov A. (2001) Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different roles of fatty acid ω-hydroxylation in development. Proc Natl Acad Sci USA 98: 9694–9699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Gao QM, Yu K, Lapchyk L, Navarre D, Hildebrand D, Kachroo A, Kachroo P. (2009) An intact cuticle in distal tissues is essential for the induction of systemic acquired resistance in plants. Cell Host Microbe 5: 151–165 [DOI] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Yephremov A, Wisman E, Huijser P, Huijser C, Wellesen K, Saedler H. (1999) Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell 11: 2187–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y, Jack T. (1998) An intragenic suppressor of the Arabidopsis floral organ identity mutant apetala3-1 functions by suppressing defects in splicing. Plant Cell 10: 1465–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869 [DOI] [PubMed] [Google Scholar]
- Zhu J, Fu Z, Koo YD, Zhu JK, Jenny FE, Jr, Adams MWW, Zhu Y, Shi H, Yun DJ, Hasegawa PM, et al. (2007) An enhancer mutant of Arabidopsis salt overly sensitive 3 mediates both ion homeostasis and the oxidative stress response. Mol Cell Biol 27: 5214–5224 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.