Abstract
How plants adapt to low temperature is not well understood. To identify components involved in low-temperature signaling, we characterized the previously isolated chilling-sensitive2 mutant (chs2) of Arabidopsis (Arabidopsis thaliana). This mutant grew normally at 22°C but showed phenotypes similar to activation of defense responses when shifted to temperatures below 16°C. These phenotypes include yellowish and collapsed leaves, increased electrolyte leakage, up-regulation of PATHOGENESIS RELATED genes, and accumulation of excess hydrogen peroxide and salicylic acid (SA). Moreover, the chs2 mutant was seedling lethal when germinated at or shifted for more than 3 d to low temperatures of 4°C to 12°C. Map-based cloning revealed that a single amino acid substitution occurred in the TIR-NB-LRR (for Toll/Interleukin-1 receptor- nucleotide-binding Leucine-rich repeat)-type resistance (R) protein RPP4 (for Recognition of Peronospora parasitica4), which causes a deregulation of the R protein in a temperature-dependent manner. The chs2 mutation led to an increase in the mutated RPP4 mRNA transcript, activation of defense responses, and an induction of cell death at low temperatures. In addition, a chs2 intragenic suppressor, in which the mutation occurs in the conserved NB domain, abolished defense responses at lower temperatures. Genetic analyses of chs2 in combination with known SA pathway and immune signaling mutants indicate that the chs2-conferred temperature sensitivity requires ENHANCED DISEASE SUSCEPTIBILITY1, REQUIRED FOR Mla12 RESISTANCE, and SUPPRESSOR OF G2 ALLELE OF skp1 but does not require PHYTOALEXIN DEFICIENT4, NONEXPRESSOR OF PR GENES1, or SA. This study reveals that an activated TIR-NB-LRR protein has a large impact on temperature sensitivity in plant growth and survival.
For optimal growth and survival, plants have evolved unique and sophisticated defense mechanisms against multiple stresses, both abiotic and biotic. Cold stress has a significant limiting effect on the geographic location of plants and on crop productivity (Guy, 1990). It can disrupt cellular homeostasis by altering the fatty acid composition of membrane lipids, which can deactivate membrane proteins and uncouple major physiological processes (Los and Murata, 2004). Plants respond and adapt to cold stress in many biochemical and physiological processes. A number of genes are involved in the DREB/CBF (for DRE-binding protein/C-repeat-binding factor)-dependent pathway to control cold acclimation (Gilmour et al., 1992, 2004), and DREB/CBF-independent pathways have been identified as important for cold responses as well (Xin and Browse, 1998; Dong et al., 2006; Lee et al., 2006; Xin et al., 2007; Zhu et al., 2008).
Plants have evolved at least two layers of defense mechanisms against pathogens. One of them is mediated by resistance (R) proteins. Interaction of an R protein with a specific pathogen avirulence protein triggers the hypersensitive response (HR), which is a form of programmed cell death that limits pathogen growth and spread (Scheel, 1998). Most of the characterized R proteins encode proteins with nucleotide-binding Leu-rich repeat (NB-LRR) domains. A well-conserved ARC (for Apaf-1, R protein, and CED4) domain is found just after the NB domain, and these two domains are often referred to as the NB-ARC domain. The NB-LRR proteins can be grouped into two main classes based on their N-terminal structure, which has either a Toll/Interleukin-1 receptor (TIR) domain or a coiled-coil domain (Meyers et al., 2003).
The Arabidopsis (Arabidopsis thaliana) RPP5 (for Recognition of Peronospora parasitica5) locus in Columbia-0 (Col) is composed of seven TIR-NB-LRR class R genes, including RPP4 and SNC1 (for Suppressor of npr1-1, constitutive 1) genes (Noel et al., 1999). RPP4 plays an important role in resistance to Hyaloperonospora parasitica through multiple signaling components, including DETACHMENT 9 (DTH9; Mayda et al., 2000), ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1; Aarts et al., 1998), PHYTOALEXIN DEFICIENT4 (PAD4; Glazebrook et al., 1996), NONEXPRESSOR OF PR GENES1 (NPR1; Cao et al., 1997), NON-RACE-SPECIFIC DISEASE RESISTANCE1 (NDR1; Century et al., 1995), Phenylalanine Ammonium Lyase (PAL; Mauch-Mani and Slusarenko, 1996), avrPphB SUSCEPTIBLE2 (PBS2) and PBS3 (Warren et al., 1999), SUPPRESSOR OF G2 ALLELE OF skp1 (SGT1b) and REQUIRED FOR Mla12 RESISTANCE (RAR1; Austin et al., 2002), RPS5 (Warren et al., 1998), and SALICYLIC ACID INDUCTION-DEFICIENT1 (SID1), SID2, and salicylic acid (SA; McDowell et al., 2000; van der Biezen et al., 2002). In addition, RPP4 mediates disease resistance and basal defense against H. parasitica through the transcription factor AtWRKY70 (Knoth et al., 2007). SNC1 confers disease resistance and suppresses plant growth in a temperature-dependent manner when activated (Stokes and Richards, 2002; Zhang et al., 2003; Yang and Hua, 2004; Zhu et al., 2010). The RPP5 locus R genes are coordinately regulated by transcriptional activation and RNA silencing (Yi and Richards, 2007).
Although the initial stimuli of cold and biotic stresses are obviously different, in many cases these signals are integrated into a unified scheme and trigger a common set of responses. For instance, cold and defense responses are shown to share common targets, such as PATHOGENESIS-RELATED (PR) genes, which not only play a role in pathogen resistance but also are induced by cold stress and promote freezing tolerance (Snider et al., 2000; Seo et al., 2008). Furthermore, cold and defense responses share common regulators, such as the SUMO E3 ligase SIZ1 (for SAP and Miz1; Lee et al., 2007; Miura et al., 2007), AtSR1/CAMTA3 (for Arabidopsis signal responsive/Calmodulin-binding transcription activator 3; Doherty et al., 2009; Du et al., 2009), and the transcriptional repressor of DREB protein DEAR1 (for DREB and EAR protein 1; Tsutsui et al., 2009). In addition, defense responses induced by a number of R genes are modulated by temperature, including Mi in tomato (Solanum lycopersicum; Hwang et al., 2000), N in tobacco (Nicotiana tabacum; Someya et al., 2004), and RESISTANCE TO POWDERY MILDEW8, SUPPRESSOR OF SALICYLIC ACID INSENSITIVE4, SNC1, and the RPP1-like TIR-NB-LRR cluster in Arabidopsis (Xiao et al., 2003; Yang and Hua, 2004; Zhou et al., 2008; Alcazar et al., 2009). A recent study revealed that the NB-LRR proteins function as temperature-sensitive components in plant immune responses (Zhu et al., 2010). Some of the defense signaling components, such as PAD4, EDS1, and SA, are also regulated by temperature (Clarke et al., 2004; Yang and Hua, 2004). Moreover, the plasma membrane-bound NAC transcription factor NTL6 is proteolytically activated by cold and in turn enters the nucleus, thereby inducing defense responses by binding to the promoter of PR genes (Seo et al., 2010). All of these findings support an extensive signaling network between cold stress and defense responses.
Here, we report the investigation of a cold-sensitive mechanism of chilling-sensitive2 (chs2) in Arabidopsis. The chs2 mutant exhibits HR-like cell death and consequent lethality under cold stress. Map-based cloning revealed that CHS2 encodes the TIR-NB-LRR-type R protein RPP4. An amino acid substitution in the NB-ARC region leads to a temperature-dependent gain-of-function phenotype. This study reveals the involvement of an activated R gene in cold response, suggesting a contribution of defense responses to temperature sensitivity.
RESULTS
Morphological Phenotypes of the Chilling-Sensitive Mutant chs2
The chs2-1 and chs2-2 mutants were isolated as chilling sensitive from an ethane methyl sulfonate (EMS)-mutagenized pool of Arabidopsis (Schneider et al., 1995). We further characterized the mutant phenotypes of these two alleles. They resembled the wild type when grown in soil at 22°C; however, the leaves of these two mutants turned yellow and wilted 3 d after being shifted to low temperature of 4°C to 12°C, and they eventually died (Fig. 1A). When planted on Murashige and Skoog (MS) plates directly at 4°C, the chs2 seedlings died shortly after germination (Fig. 1C). Given that these two alleles showed similar phenotypes, we chose chs2-2 (referred as chs2 hereafter) for further studies.
Figure 1.
Cold sensitivity of chs2 mutant plants. A, Phenotypes of wild-type Col and chs2 plants grown in soil at 22°C for 4 weeks (top row), cold treated at 4°C for 10 d (middle row), followed by 22°C for 3 d (bottom row). B, Phenotypes of wild-type Col and chs2 plants grown in soil at 22°C for 6 weeks (top) followed by cold treatment at 4°C for 10 d (bottom). C and D, Phenotypes of Col and chs2 plants directly geminated on MS plates and grown at 4°C for 3 months (C) or grown at 22°C for 3 weeks and then transferred to 4°C for an additional 10 d (D). E, Phenotypes of wild-type Col and chs2 plants grown in soil at 16°C for 7 weeks. F, Phenotypes of chs2 plants grown on MS plates at the indicated temperatures for 4 weeks. Images are of representative plants.
To get a better understanding of chs2 in response to chilling, we examined the phenotypes of chs2 plants by shifting them to cold conditions at different growth stages either in soil or on agar plates. The 22°C-grown chs2 plants at every developmental stage tested were hypersensitive to cold stress both in soil and on MS plates (Fig. 1, B–D). All parts of the chs2 plants including the rosette leaves, cauline leaves, stems, flowers, and siliques became yellow, collapsed, and then died quickly after cold exposure (Fig. 1B). It is noteworthy that the mutant grown at 16°C to 18°C showed dwarf stature with curly chlorotic leaves and short inflorescence internodes (Fig. 1E). With decreased temperature, the chs2 mutant plants showed more severe growth defects, and they were lethal when temperature was below 12°C (Fig. 1F). Therefore, the chs2 mutant is sensitive to low temperature throughout plant development, with lower temperature causing more severe growth defects.
Physiological Characteristics of chs2 at Low Temperatures
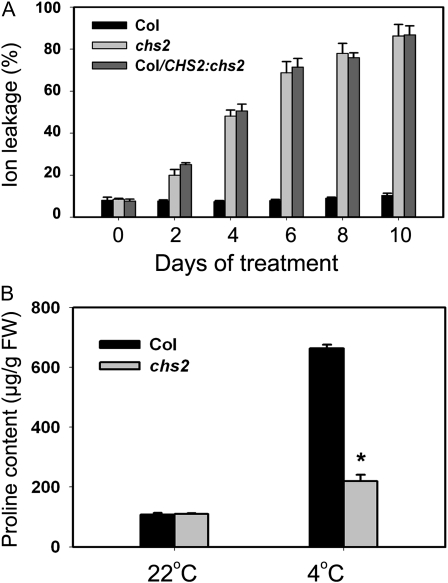
Leakage of ions from cell membranes is a good index to measure chilling sensitivity in plants (Lyons, 1973). We carried out ion leakage assays to determine the extent of chilling injury to chs2 plants. No obvious changes in ion leakage were detected in wild-type leaves during cold treatment. However, ion leakage of chs2 plants increased drastically following cold treatment (Fig. 2A). This result indicates that the cell membranes of chs2 leaves are severely injured under cold stress, which is in agreement with the cold-sensitive phenotype of chs2.
Figure 2.
Physiological analysis of chs2 mutant plants. A, Ion leakage assay in chs2 plants. Plants grown at 22°C for 3 weeks were then treated at 4°C for the indicated times. The data represent means of three replicates ± sd. Similar results were observed in three independent experiments. B, Pro content in chs2 plants. Plants grown at 22°C for 3 weeks were treated at 4°C for 6 d. The data represent means of four replicates ± sd. * P < 0.01 (t test), significant difference from Col. Similar results were observed in three independent experiments. FW, Fresh weight.
Free Pro is an osmolyte considered to protect plants against cold stress (Xin and Browse, 1998; Nanjo et al., 1999). We investigated if the cold sensitivity of chs2 is accompanied by reduced Pro levels. Indeed, the Pro content in chs2 was much lower than in the wild type when treated at 4°C for 6 d (Fig. 2B), suggesting that less Pro accumulation in chs2 might at least partly account for its cold sensitivity.
Chloroplasts Are Damaged in chs2 Plants under Cold Stress
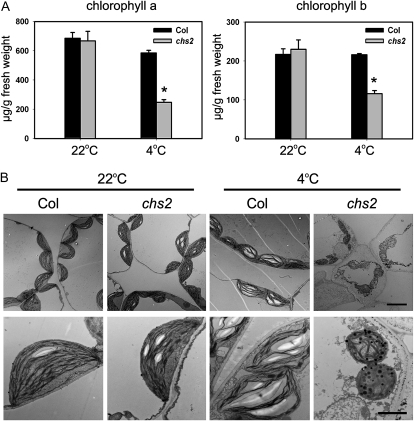
Because the chs2 plants exhibited yellow leaves under cold stress (Fig. 1), we measured the chlorophyll content in the chs2 mutant. The levels of chlorophyll a and chlorophyll b in cold-treated chs2 plants were approximately 42% and 50% of those in the wild-type plants, respectively (Fig. 3A), implying that the chloroplasts in chs2 are severely damaged under cold conditions.
Figure 3.
The effect of the chs2 mutation on chloroplast development under cold stress. Wild-type Col and chs2 plants were grown at 22°C for 3 weeks and then treated at 4°C for 10 d. A, Chlorophyll content of Col and chs2 seedlings. The data represent means of four replicates ± sd. * P < 0.01 (t test), significant difference from Col. Similar results were observed in three independent experiments. B, Transmission electron microscopy of plastids from chs2 plants. Bar = 5 μm (top row) and 2 μm (bottom row). Images are of representative plants.
The chloroplast morphology in cold-treated chs2 plants was further examined using transmission electron microscopy. The mature chloroplasts of the chs2 and wild-type plants at 22°C exhibited crescent-shaped and well-developed thylakoid membranes. Chloroplasts in cold-treated wild-type plants were similar to those in plants without cold treatment, but with larger starch granules, which is a normal response to cold stress. In contrast, cold-treated chs2 chloroplasts were smaller and more spherical than those in the wild-type plants, and they contained fewer internal thylakoid membranes. Moreover, the starch grains in cold-treated chs2 chloroplasts were either absent or reduced in size and number. The mutant chloroplasts also appeared to contain more plastoglobuli than wild-type chloroplasts (Fig. 3B). Thus, cold stress causes serious damage to the chloroplasts in chs2 plants.
We then determined whether light had an effect on cold-induced phenotypes of chs2. Although the cold-induced phenotype of chs2 was significantly delayed in the dark (Supplemental Fig. S1A), the plants eventually died. Accordingly, the degradation of chlorophyll a and b was also delayed in the dark (Supplemental Fig. S1B). These results demonstrate that light accelerates the chs2-conferred phenotype, but low temperature triggers this phenotype in the absence of light.
The chs2 Mutation Causes Reactive Oxidative Species Accumulation and Imbalanced Reactive Oxidative Species-Scavenging Network under Cold Stress
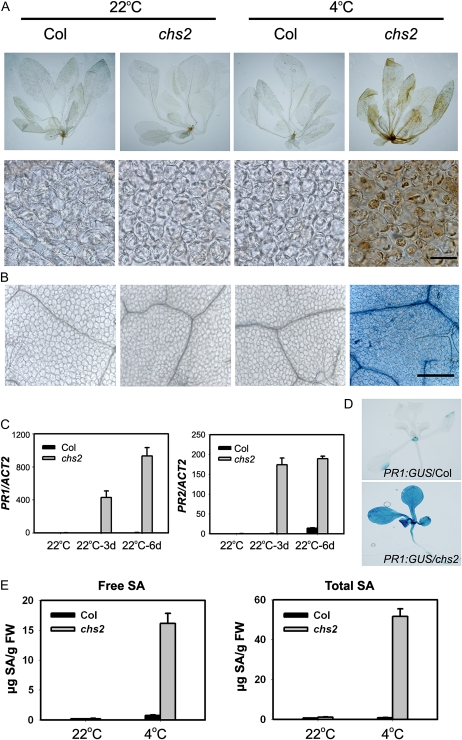
Low temperature can perturb electron transport chains and cause the production of reactive oxidative species (ROS; Fryer et al., 2002; Hideg et al., 2002; Pfannschmidt et al., 2003). Therefore, we examined hydrogen peroxide (H2O2) accumulation in chs2 plants under cold conditions by 3,3′-diaminobenzidine (DAB) staining. Strong staining was detected in cold-treated chs2 plants but not in wild-type plants (Fig. 4A), indicating that the mutant plants accumulated more H2O2 than the wild-type plants. Under light, chloroplast is the main site of ROS generation; consistently, DAB precipitates were mostly present in the chloroplasts. Therefore, the chs2-induced phenotypes under cold stress might be caused by the impairment of normal chloroplast function and by the overgeneration of ROS in the chloroplasts.
Figure 4.
chs2 constitutively activates defense responses to cold. Wild-type Col and chs2 plants were grown at 22°C for 3 weeks and then treated at 4°C for 6 d. For A, B, and E, 20 plants were tested for each genotype. Images are of representative plants from one of three independent experiments. A, H2O2 accumulation in chs2 plants stained by DAB. Bar = 20 μm. B, Cold-induced cell death in chs2 plants. Detached leaves were stained with trypan blue. Bar = 100 μm. Images are of representative plants. C, Expression of PR1 and PR2 in wild-type and chs2 plants by real-time RT-PCR. The data represent means of three replicates ± sd. Similar results were observed in three independent experiments. D, GUS analysis of PR1 in chs2 plants. PR1:GUS transgenic plants were crossed with chs2 plants. The F2 homozygous lines were used for GUS staining analysis. Images are of representative plants. E, SA accumulation in chs2 under cold conditions. Three-week-old 22°C-grown plants were treated at 4°C for 6 d. The data represent means of three replicates ± sd. Similar results were observed in three independent experiments. FW, Fresh weight.
When subjected to low temperature, plants accumulate excess H2O2 (O’Kane et al., 1996), which in turn induces the expression of genes associated with oxidative stress (Iba, 2002; Mittler et al., 2004; Rizhsky et al., 2004). More H2O2 accumulation in chs2 was observed under cold conditions (Fig. 4A). Therefore, we examined the expression of several genes encoding ROS-detoxification enzymes, including copper/zinc superoxide dismutase (CSD), ascorbate peroxidase (APX), and catalase (CAT), in cold-treated chs2 plants. No obvious differences in expression of CSD1, APX1, or CAT1 were detected between wild-type and chs2 plants at 22°C. In contrast, the expression of these genes was substantially elevated in chs2 plants relative to wild-type plants under cold stress (Supplemental Fig. S2B). The zinc-finger protein ZAT12 plays a crucial role in oxidative and abiotic stress signaling (Rizhsky et al., 2004; Davletova et al., 2005). In addition, ferritin protein nanocages are essential for protecting cells against oxidative damage (Ravet et al., 2009). We found that ZAT12 and FER1 were also significantly up-regulated in cold-treated chs2 plants relative to wild-type plants (Supplemental Fig. S2B). Therefore, the chilling sensitivity of chs2 might result from an imbalance of ROS detoxification and consequent impairment of oxidative signaling.
The Expression of Cold-Regulated Genes Is Not Affected in chs2
We further examined whether the chs2 mutation affects the induction of cold-regulated genes. The CBF1 to CBF3 genes were rapidly induced in chs2 and wild-type plants 3 h after exposure to cold, and their target genes RD29A and COR47 were substantially induced at 6 to 12 h after cold treatment. No significant difference in expression of these genes was observed between chs2 and wild-type plants (Supplemental Fig. S3). Therefore, the chs2 gene appears not to affect the CBF pathway.
chs2 Constitutively Activates Defense Responses under Cold Conditions
Leaves in cold-treated chs2 plants turned yellow, lost turgor pressure, and collapsed (Fig. 1), resembling the pathogen-induced HR cell death response. Extensive cell death did occur in cold-treated chs2 plants but not in wild-type plants, as revealed by trypan blue staining (Fig. 4B). Furthermore, PR1 and PR2 were highly expressed in chs2 plants under cold stress (Fig. 4C). Consistently, cold-treated chs2 plants harboring a PR1:GUS construct showed stronger staining of GUS than wild-type PR1:GUS transgenic plants (Fig. 4D).
Because high PR gene expression is often associated with elevated levels of SA, the endogenous SA levels in chs2 were examined. The levels of both free SA and total SA in chs2 were comparable to those in wild-type plants grown at 22°C. However, cold-treated chs2 plants accumulated approximately 22- and 65-fold higher levels of SA and total SA, respectively, than wild-type plants (Fig. 4E). Thus, chs2 plants constitutively activate defense responses under cold stress.
A Mutation in RPP4 Is Responsible for the Chilling-Sensitive Phenotype
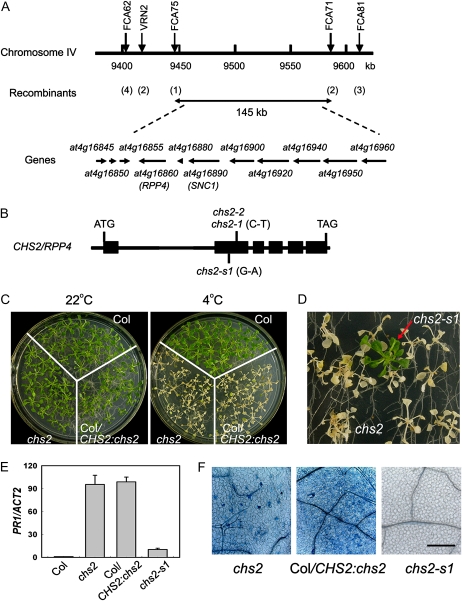
The chs2 mutant was previously shown to contain a dominant mutation in a single nuclear locus (Schneider et al., 1995). To identify the chs2 mutation, chs2-2 was crossed with Landsberg erecta (Ler) to generate a mapping population. Given that the chs2 mutation is dominant, wild-type-looking seedlings were chosen for mapping from the segregating F2 population after cold treatment. The chs2 mutation was initially mapped to the middle of chromosome IV. Approximately 3,000 plants were then selected for fine mapping. The chs2 mutation was narrowed to a 145-kb region containing the RPP5 cluster region (Fig. 5A). To identify the molecular lesion in chs2-2, all of the annotated genes in this region were amplified from chs2-2 and sequenced. Only one nucleotide substitution of C to T was found in the second exon of At4g16860 (RPP4 or ColA) in chs2-2, resulting in a Ser-to-Phe change at residue 389 (Fig. 5B). The same mutation was found in chs2-1.
Figure 5.
Map-based cloning of CHS2. A, A genetic map of the CHS2 locus on chromosome IV. Positions of the markers used for mapping are indicated above the line. The corresponding nucleotide positions are numbered in kilobases below the line. The number of recombinants is indicated in parentheses. Predicted genes are shown by arrows indicating the direction of transcription. B, A schematic diagram of the genomic structure of the CHS2 gene. Boxes and lines indicate exons and introns, respectively. The nucleotide substitutions in chs2 and chs2-s1 are shown. C, Complementation of the chs2 mutant. Wild-type Col, chs2, and Col transformed with a genomic clone containing the mutated chs2 (Col/CHS2:chs2) were grown at 22°C for 2 weeks (left) and then treated at 4°C for 10 d (right). D, Screening of the chs2 suppressor chs2-s1. EMS-mutagenized chs2 plants were grown at 22°C for 2 weeks and then treated at 4°C for 10 d. E, PR1 gene expression in Col, chs2, Col/CHS2:chs2, and chs2-s1 plants treated at 4°C for 6 d by real-time RT-PCR. The data represent means of three replicates ± sd. Similar results were observed in three independent experiments. F, Trypan blue staining of the leaves from chs2, Col/CHS2:chs2, and chs2-s1 plants. Bar = 100 μm.
The chs2 mutant is a dominant mutation, suggesting a gain-of-function substitution. To determine whether the chs2 phenotype was caused by the chs2 mutation, a 12-kb genomic fragment including the complete chs2 gene under the control of its own promoter (CHS2:chs2) was transformed into wild-type Col. Thirty-two out of 35 T1-independent transgenic lines showed all the chs2-conferred phenotypes under cold stress, including seedling lethality (Fig. 5C), high ion leakage (Fig. 2A), elevated PR1 expression (Fig. 5E), and extensive cell death (Fig. 5F). These data indicate that mutated chs2 recapitulates all the chs2-conferred phenotypes and therefore that CHS2 is RPP4. RPP4 encodes a TIR-NB-LRR-class R protein with high similarity to SNC1 (74% amino acid identity and 78% similarity). The Ser-389 residue in chs2 is very close to the putative GxP or GLPL motif in the ARC domain, which is conserved in many NB-LRR proteins (Rafiqi et al., 2009). This finding hence supports the importance of the ARC domain for the normal activity of R proteins.
The chs2-s1 Mutation Suppresses the Chilling Sensitivity of chs2
To further confirm that the mutation in RPP4 is responsible for the chs2 phenotype, we carried out a genetic suppressor screen in the chs2 background. M2 plants derived from EMS-mutagenized chs2 seeds were screened for mutants displaying wild-type morphology under cold stress. One such mutant, named chs2-s1 (for chs2 suppressor 1), was isolated (Fig. 5D). PR1 gene expression and the cell death phenotype were significantly suppressed in chs2-s1 (Fig. 5, E and F). This mutation was mapped to the original RPP4 locus. Sequencing analysis revealed a second point mutation of E to K at amino acid position 300 in chs2-s1, which resides close to the Walker B/Kinase 2 motif of the RPP4 NB domain (Fig. 5B). This motif was shown to be important for the function of NB-LRR proteins, and mutations in or close to this conserved motif might abrogate the activity of NB-LRR proteins (Bendahmane et al., 2002).
RPP4 Expression in chs2 at Different Conditions
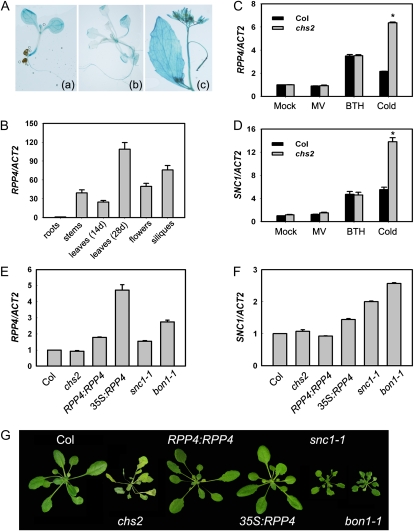
To elucidate the physiological function of RPP4, we examined its organ-specific expression in Arabidopsis. Transgenic plants harboring a GUS reporter gene driven by the RPP4 promoter were generated and analyzed. GUS staining revealed that RPP4 was expressed at low levels in leaves, stems, flowers, and siliques, and it was barely expressed in roots (Fig. 6A). This result is in agreement with public data from Genevestigator (https://www.genevestigator.com/gv/index.jsp) and was validated by quantitative reverse transcription (RT)-PCR analysis (Fig. 6B).
Figure 6.
Expression of RPP4 and SNC1 in chs2. A and B, Expression of RPP4 by GUS staining (A) and by real-time PCR (B). Total RNA was extracted from various tissues. The data represent means of three replicates ± sd. C and D, Expression of RPP4 (C) and SNC1 (D) under various treatments. Total RNA was extracted from plants treated with cold (4°C), methyl viologen (MV; 5 μm), or benzothiadiazole (BTH; 0.5 mm) for 24 h. E and F, Expression of RPP4 (E) and SNC1 (F) in 2-week-old 22°C-grown plants (Col, chs2, RPP4:RPP4, 35S:RPP4, snc1-1, and bon1-1) by real-time PCR. The data represent means of three replicates ± sd. * P < 0.01 (t test), significant difference from Col. All experiments were repeated three times with similar results. G, Phenotypes of the plants (Col, chs2, RPP4:RPP4, 35S:RPP4, snc1-1,and bon1-1) grown in soil at 22°C for 4 weeks and then cold treated at 4°C for 10 d.
RPP4 was expressed at relatively low levels in the plants, consistent with the low steady-state expression levels of R genes under normal conditions. However, R genes can be induced by certain stimuli such as pathogens and SA. Therefore, we investigated whether RPP4 expression was responsive to various stimuli. The expression of RPP4 was not induced by the oxidative inducer methyl viologen in either wild-type Col or chs2 plants (Fig. 6C). However, we found that RPP4 in the wild-type Col background was induced by benzothiadiazole (an SA analog) and cold. Strikingly, cold stress dramatically enhanced the induction of the mutated RPP4 in chs2 (Fig. 6C; Supplemental Fig. S4A). Cold-induced overexpression could be a consequence of feedback regulation upon R gene activation, which might account for the phenotypes of chs2 mutants under cold stress.
To test if overexpression of wild-type RPP4 would recapitulate the chs2 phenotype, we generated transgenic lines expressing wild-type RPP4 driven either by its native promoter (RPP4:RPP4) or by the cauliflower mosaic virus 35S promoter (35S:RPP4), and we analyzed their phenotypes under cold conditions. Interestingly, neither the RPP4:RPP4 nor 35S:RPP4 transgenic line, in which RPP4 was indeed overexpressed (Fig. 6E), exhibited chs2-like phenotypes at 4°C (Fig. 6G). Therefore, the chs2-conferred phenotypes are not simply caused by constitutive expression of RPP4 but rather by the amino acid substitution in chs2. All of these data indicate that chs2 is a gain-of-function mutant and that cold-induced overexpression of the mutated RPP4 gene is required for the chs2 phenotype.
chs2-Induced Chilling Sensitivity Is Independent of SNC1
Since the RPP5 locus R genes are coordinately regulated (Yi and Richards, 2007), we examined the expression of SNC1, a close homolog of RPP4, in the chs2 mutant. Similar expression patterns of SNC1 induction were found in wild-type Col and chs2 plants (Fig. 6D). SNC1 expression was induced by benzothiadiazole and cold stress in both genotypes. Moreover, chs2 plants accumulated higher levels of the SNC1 transcript than did cold-treated Col plants (Fig. 6D; Supplemental Fig. S4B).
To determine whether up-regulation of SNC1 also contributes to the chs2 phenotype, we tested the cold sensitivity of snc1-1 and bon1-1 plants, in which SNC1 is activated or derepressed (Yang and Hua, 2004; Li et al., 2007; Fig. 6F). Neither of them showed a chs2-like lethal phenotype at cold stress (Fig. 6G). In addition, we transformed the CHS2:chs2 clone into snc1-11 loss-of-function mutant plants. All 10 independent transgenic lines displayed a chs2-like chilling-sensitive phenotype (data not shown), indicating that the chs2 mutation confers a chs2 phenotype independent of SNC1.
chs2-Induced Chilling Sensitivity Is Independent of SA and NPR1
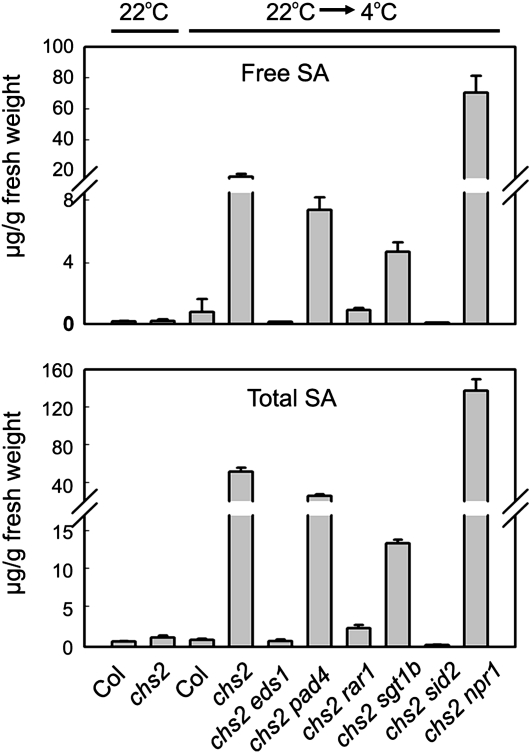
Because chs2 plants accumulated high levels of free SA and total SA after cold treatment (Fig. 4E), we then determined whether activation of the SA pathway is necessary for the chs2 phenotype by crossing chs2 with the SA-deficient mutant sid2-2 (Wildermuth et al., 2001). The chs2 sid2 double mutants exhibited chilling sensitivity and extensive cell death phenotypes similar to those of chs2 (Fig. 7, A and C). As expected, the levels of SA and total SA in the chs2 sid2 double mutants were reduced to a wild-type level under cold stress (Fig. 8). Therefore, the chs2-conferred chilling-sensitive phenotype does not require SA.
Figure 7.
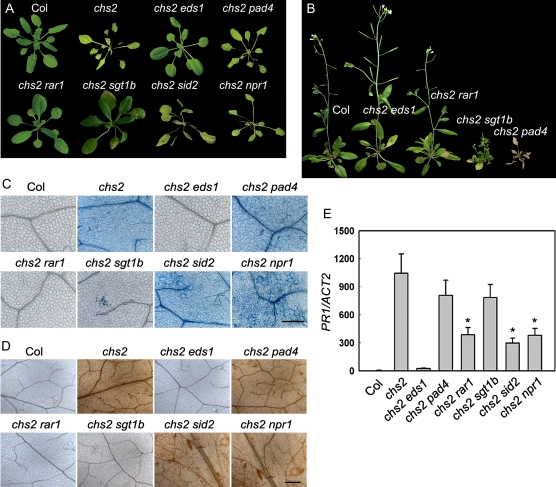
Phenotypes of the chs2 double mutants under cold conditions. Three-week-old 22°C-grown plants were treated at 4°C for 6 d (C–E), 14 d (A), or 5 weeks (B). A and B, Growth phenotypes of the double mutants under cold conditions. Representative plants are shown. C, Trypan blue staining of the leaves from the double mutants. Bar = 100 μm. Note that the photographs of 4°C-treated Col and chs2 plants stained with trypan blue are identical to those shown in Figure 2B. D, DAB staining of the leaves from the double mutants. Bar = 100 μm. E, PR1 gene expression in the double mutants by real-time PCR. The data represent means of three replicates ± sd. * P < 0.01 (t test), significant difference from chs2. All experiments were repeated three times with similar results.
Figure 8.
SA accumulation in the double mutants under cold conditions. Three-week-old 22°C-grown plants were treated at 4°C for 6 d. Shown are mean values of free and total SA amount in different genotypes of three replicates ± sd. Similar results were observed in three independent experiments.
NPR1 is a master regulator of SA signaling and plant immunity (Cao et al., 1994). To examine the requirement for NPR1 in chs2-mediated signaling, a chs2 npr1 double mutant was generated and then characterized. The loss of NPR1 function, while significantly reducing PR1 expression, did not abrogate the chs2-mediated cold-sensitive morphology, cell death, or the accumulation of SA at low temperature (Figs. 7 and 8), indicating that NPR1 is dispensable for the chs2-conferred phenotype.
chs2-Induced Chilling Sensitivity Requires Multiple Signaling Components
To assess whether defense signaling components (including EDS1, PAD4, SGT1b, and RAR1) are involved in the chs2-mediated temperature signaling pathway, we first examined RPP4 expression in eds1-2 (Col; Bartsch et al., 2006), pad4-1 (Jirage et al., 1999), rar1-20 (Muskett et al., 2002), and sgt1b/eta3 (Gray et al., 2003) mutants. RPP4 expression was slightly down-regulated by eds1 and pad4 but not by rar1 or sgt1b (Supplemental Fig. S5). We also generated double mutants of chs2 with eds1-1 (Parker et al., 1996), pad4-1, rar1-20, and sgt1b/eta3 for further analyses.
Because eds1-1 is in the Wassilewskija accession, which does not contain the RPP4 gene, we compared the phenotypes of multiple chs2/EDS1and chs2 eds1 lines from the F2 population of chs2 crossed with eds1-1 to eliminate potential effects of mixed background. Among 211 F2 progeny, all 12 lines of chs2 eds1 and 25 lines of chs2/+ eds1 showed wild-type-like morphology at 4°C (Fig. 7A). Extensive cell death, elevated PR1 expression, and accumulation of H2O2 and SA under cold conditions were also totally suppressed in these chs2 eds1 and chs2/+ eds1 lines (Figs. 7 and 8). Moreover, all 14 chs2 EDS1 lines and 26 chs2/+EDS1 lines out of 211 F2 progeny we analyzed uniformly resembled chs2 phenotypes (data not shown). These results indicate that chs2 chilling sensitivity is dependent on EDS1.
The chs2 pad4 double mutant resembled the chs2 mutant in terms of morphology under cold, although the cold-induced lethal phenotype of chs2 pad4 was delayed slightly compared with the chs2 mutant (Fig. 7, A and B). Cell death, H2O2 accumulation, and PR1 gene expression in the chs2 pad4 double mutant were all comparable to those in chs2 under cold stress (Figs. 7 and 8). Therefore, the chs2-conferred phenotypes are largely independent of PAD4.
RAR1 and SGT1b were previously identified as regulators of various R genes (Austin et al., 2002; Muskett et al., 2002). rar1-20 completely suppressed chs2 cold-induced lethality at 4°C (Fig. 7, A and B). In accordance with the morphological phenotype, cell death and H2O2 accumulation were abolished in chs2 rar1-20 (Fig. 7, C and D). Cold-induced PR1 expression was partially suppressed in the chs2 rar1-20 double mutant (Fig. 7E). In addition, levels of SA in chs2 rar1-20 were restored to wild-type levels (Fig. 8). Therefore, the chs2-conferred phenotype requires RAR1.
chs2 sgt1b double mutant plants largely resembled wild-type plants 3 to 6 d after cold treatment, when chs2 started to exhibit a chilling defect. However, prolonged cold treatment (1–2 weeks) resulted in slightly yellow leaves in chs2 sgt1b (Fig. 7A). Moreover, chs2 sgt1b showed dwarfism with curly and chlorotic leaves after cold treatment for 5 weeks (Fig. 7B), which is characteristic of chs2 grown at 16°C to 18°C (Fig. 1E). The cell death phenotype and H2O2 accumulation were partially suppressed by the sgt1b mutation (Fig. 7, C and D). PR expression was partially compromised in chs2 sgt1b plants (Fig. 7E). In addition, SA accumulation in chs2 sgt1b was drastically reduced to one-fourth level compared with chs2 (Fig. 8). Taken together, these data indicate that the chs2 phenotype is partially dependent on SGT1b.
DISCUSSION
The Chilling Sensitivity of chs2 Is a Result of Activated Defense Responses
In this study, we characterized a previously reported chilling-sensitive mutant, chs2. This chs2 mutant exhibits yellowish leaves, increased ion leakage, damaged chloroplasts, ROS accumulation, extensive cell death, and consequent lethality at chilling temperatures (below 12°C). To our surprise, all the morphological and cell death phenotypes of chs2 under cold conditions are a result of the up-regulation of defense responses through the activated R gene RPP4. Chloroplast morphological change and ROS accumulation are observed in mutants showing cell death phenotypes (Tanaka et al., 2003; Dong et al., 2007; Hirashima et al., 2009). The accumulation of excess H2O2 in chs2 is likely due to programmed cell death induced by the activated RPP4 gene. This finding reveals a great impact of defense responses on cold sensitivity in plant growth and survival.
chs2 mutants contain a gain-of-function mutation (S389F) in the TIR-NB-LRR-type R gene RPP4. The S389F mutation is located in the NB-ARC1 domain of RPP4. The plant NB-ARC domain has been shown to be responsible for ATP binding and hydrolysis (Tameling et al., 2002; Ueda et al., 2006). The NB-ARC domain serves as a molecular switch for R protein activity, and its action is dependent on its nucleotide-binding state (ATP/ADP). Some R protein mutations affecting the ATP-binding domain will inactivate the protein (Dinesh-Kumar et al., 2000; Tao et al., 2000; Howles et al., 2005; Ueda et al., 2006; van Ooijen et al., 2008); in contrast, reduced ATP hydrolysis with normal ATP binding can result in constitutive activation of some R proteins (Takken et al., 2006; Ade et al., 2007; van Ooijen et al., 2008). We hypothesize that the chs2 mutation might interfere with ATP hydrolysis, thus causing a gain-of-function activity. It is possible that low temperature induces a conformational change within chs2, resulting in an active signaling state (on state) under cold conditions. In accordance with this study, a number of mutants with deregulated R-like proteins have been shown to have temperature-dependent autoimmune responses (Xiao et al., 2003; Yang and Hua, 2004; Zhou et al., 2008; Alcazar et al., 2009).
Temperature Sensitivity and R Genes
Many gain-of-function mutations of R genes confer temperature sensitivity. However, their temperature-sensitive ranges can be different. RPP4 and SNC1 are highly homologous in their predicted amino acid sequences; in addition, their gene structures are very similar, including their position at the RPP5 locus and their numbers of exons and introns (van der Biezen et al., 2002). A gain-of-function snc1-1 mutant shows a growth-defective phenotype and activated defense responses at 22°C but not at 28°C (Yang and Hua, 2004). Nevertheless, snc1-1 can survive and set seeds even at temperatures of 4°C to 22°C. In contrast, chs2 shows obvious defense activation at 16°C to 18°C and is lethal at temperatures below 12°C. As these R or R-like genes share downstream signaling components such as EDS1 (Li et al., 2001), the temperature sensitivity likely comes from R genes, as different mutants have different ranges of temperature sensitivity. This was demonstrated recently by altering the temperature sensitivity of defense responses through manipulating R genes. Specific missense mutations in SNC1 and N genes could retain defense responses normally inhibited at elevated temperatures, and additional missense mutations in the SNC1 protein reverse the temperature sensitivity of defense responses (Zhu et al., 2010). Thus, differences in temperature sensitivity and sensitivity range are most likely due to varying temperature sensitivity in R protein, and different forms of NB-LRR proteins mediate temperature sensitivity in plant immune responses by conformationally transitioning between off and on states (Zhu et al., 2010).
RPP4 Regulates Cold Response and Defense Responses via Both Common and Distinct Signaling Mediators
Previous studies show that RPP4 confers resistance to H. parasitica, which requires the action of multiple signaling components including DTH9, EDS1, PAD4, NPR1, NDR1, PAL, PBS2, PBS3, SGT1b, RAR1, RPS5, SID1, SID2, and SA. In this study, we found that chs2 is dependent on EDS1, SGT1, and RAR1 but is independent of PAD4, NPR1, and SA. This result indicates that the signaling components required for the temperature sensitivity of chs2 mutants show similarities and differences with those required for RPP4 function in pathogen resistance. The different genetic requirement of chs2 and RPP4 might be due to the nature of the mutation in the CHS2 protein. The molecular mechanism by which chs2 regulates temperature-dependent cell death is still unknown, and the subcellular localization of RPP4 or CHS2 remains unclear. Further study on the protein localization, protein activities, and suppressors of chs2 will shed more light on the function of RPP4 in the regulation of temperature-dependent cell death and the interconnected mechanisms of cold stress and defense signaling.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants of the accessions Col and Wassilewskija were used in this study. The chs2-1 and chs2-2 (Schneider et al., 1995) mutants were obtained from the Arabidopsis Biological Resource Center (ABRC; stock nos. CS6298 and CS6299). Plants were grown at 22°C or 4°C under a long-day (16 h of light/8 h of dark) photoperiod at 100 μmol m−2 s−1 with 50% to 70% relative humidity in soil or on MS medium (Sigma) containing 2% Suc and 0.8% agar.
Genetic Mapping and Cloning of the CHS2 Gene
The chs2-2 seeds were treated with 0.3% EMS for 8 h. Approximately 20,000 M2 plants (derived from 5,000 M1 seeds) were screened at 4°C for chs2-s mutants with a wild-type phenotype.
To map the chs2-2 mutation, a homozygous chs2-2 mutant (Col background) was crossed with Ler. The F1 plants from the cross were self-fertilized, and the resulting F2 seeds were collected. The segregating F2 population seedlings with a wild-type phenotype were used for mapping. A total of 3,000 F2 plants were selected. Genomic DNA from these F2 plants was extracted and used for PCR-based mapping with simple sequence length polymorphism and derived cleaved-amplified polymorphic sequence (dCAPS) markers. Additional mapping markers were developed based on insertions/deletions identified from the Cereon Arabidopsis polymorphism and Ler sequence collection (www.arabidopsis.org). Genomic DNA corresponding to candidate genes was PCR amplified from the mutant and sequenced to identify the mutation.
To map chs2-s1 mutations, the F2 populations were derived from genetic crossing between the mutants (in Col) and Ler. Bulked segregation analysis was performed with simple sequence length polymorphism, cleaved-amplified polymorphic sequence, and dCAPS markers.
Plasmid Construction and Plant Transformation
A 12-kb PstI genomic fragment containing the RPP4 promoter and coding region was cloned from bacterial artificial chromosome clone F5D3 (ABRC) into the binary vector pCAMBIA1300 (CAMBIA) to generate the RPP4:RPP4 construct. A 1.0-kb EcoRV-EcoRI genomic fragment containing the chs2 mutation was amplified by PCR using CHS2-1F and CHS2-1R from the genomic DNA of chs2 plants and used to replace the wild-type fragment in RPP4:RPP4 to generate the CHS2:chs2 construct.
An 8.3-kb genomic fragment containing the RPP4 coding region and 3′ untranslated region from RPP4:RPP4 was cloned into the binary vector pGreen-0229 (Hellens et al., 2000) to generate the 35S:RPP4 construct.
For the CHS2:GUS fusion, a 1.46-kb genomic fragment upstream of the CHS2 ATG start codon was amplified by PCR using the CHS2-p1F and CHS2-p1R primers (Supplemental Table S1) and fused with the GUS reporter gene in the binary vector pZPGUS2 (Diener et al., 2000).
Agrobacterium tumefaciens strain GV3101 carrying different constructs was used to transform wild-type (Col) plants via floral dip transformation (Clough and Bent, 1998).
Genetic Analysis
To generate double mutants, chs2-2 was crossed to eds1-1 (Parker et al., 1996), pad4-1 (Jirage et al., 1999), rar1-20 (Muskett et al., 2002), sgt1b/eta3 (Gray et al., 2003), sid2-2 (Wildermuth et al., 2001), and npr1 (Durrant and Dong, 2004). The F2 progeny were specifically genotyped. Homozygosity of the chs2 mutation was identified using dCAPS markers and the CHS2-2F and CHS2-2R primers (Supplemental Table S1).
Ion Leakage and Pro Content Assays
The electrolyte leakage test was performed as described previously (Lee et al., 2002). Three-week-old plants grown in soil under normal conditions were treated at 4°C for different periods of time. The percentage of electrolyte leakage was calculated as the percentage of conductivity before versus after autoclaving. Pro content was measured as described by Bates et al. (1972).
SA Measurement
Free SA and total SA were extracted and measured from 3-week-old plants grown at 22°C or treated at 4°C for 6 d as described with some modifications (Li et al., 1999). The last extracted residue was dissolved in acetonitrile and analyzed by HPLC using 5% acetate (pH 3.2) as the mobile phase.
Analysis of Chlorophyll, and Electron Microscopy
Total chlorophylls were determined as described previously (Huang et al., 2009). Sections of leaf tissue were prepared for electron microscopic analysis as described (Huang et al., 2009).
Histochemical Staining Assay
Trypan blue staining and DAB staining were performed as described previously (Bowling et al., 1997; Thordal-Christensen et al., 1997). Histochemical detection of GUS activity was performed as described previously (Yang et al., 2006).
Quantitative RT-PCR
Total RNA was isolated from 10-d-old seedlings on MS plates or 21-d-old seedlings in soil using TRIzol (Invitrogen) followed by treatment with RNase-free DNase I (Takara). Two micrograms of RNA was subjected to first-strand cDNA synthesis using Moloney murine leukemia virus reverse transcriptase (Promega) and an oligo(dT)18 primer. The primers used for real-time PCR are listed in Supplemental Table S1. Real-time PCR was performed using SYBR Green PCR Master Mix (Takara). Analysis was performed using the Applied Biosystems PRISM 7500 real-time PCR system. The primer efficiencies were measured and the relative expression levels were calculated as described previously (Miura et al., 2007).
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: RPP4/CHS2, At4g16860; PAD4, At3g52430; EDS1, At3g48090; NPR1, At1g64280; SID2, At1g74710; NDR1, At3g20600; RAR1, At5g51700; SGT1b, At4g11260; SNC1, At4g16890; PR1, At2g14610; PR2, At3g57260; CBF1, At4g25490; CBF2, At4g25470; CBF3, At4g25480; RD29A, At5g52310; COR47, At1g20440; ZAT12, At5g59820; APX1, At1g07890; CAT1, At1g20630; FER1, At5g01600; CSD1, At1g08830; ACT2, At3g18780.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The effect of light on the chs2 phenotype under cold stress.
Supplemental Figure S2. Expression of ROS-associated genes in chs2 plants under cold stress.
Supplemental Figure S3. Relative mRNA levels of cold-responsive genes in chs2.
Supplemental Figure S4. Expression of RPP4 and SNC1 in chs2 under cold stress.
Supplemental Figure S5. Expression of RPP4 in eds1, pad4, rar1, and sgt1b mutants.
Supplemental Table S1. Gene-specific primers used in this study.
Supplementary Material
Acknowledgments
We thank Jian Hua for her helpful discussion of the manuscript and providing plasmids. We thank Jeffery L. Dangl, Julia Dewdney, Xinnian Dong, Xin Li, Jane E. Parker, Brain J. Staskawicz, and the ABRC for mutant seeds.
References
- Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE. (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci USA 95: 10306–10311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ade J, DeYoung BJ, Golstein C, Innes RW. (2007) Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci USA 104: 2531–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcazar R, Garcia AV, Parker JE, Reymond M. (2009) Incremental steps toward incompatibility revealed by Arabidopsis epistatic interactions modulating salicylic acid pathway activation. Proc Natl Acad Sci USA 106: 334–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin MJ, Muskett P, Kahn K, Feys BJ, Jones JD, Parker JE. (2002) Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295: 2077–2080 [DOI] [PubMed] [Google Scholar]
- Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE. (2006) Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18: 1038–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates LS, Waldren RP, Teare ID. (1972) Rapid determination of free proline for water-stress studies. Plant Soil 39: 205–207 [Google Scholar]
- Bendahmane A, Farnham G, Moffett P, Baulcombe DC. (2002) Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J 32: 195–204 [DOI] [PubMed] [Google Scholar]
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X. (1997) The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9: 1573–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X. (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6: 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 [DOI] [PubMed] [Google Scholar]
- Century KS, Holub EB, Staskawicz BJ. (1995) NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc Natl Acad Sci USA 92: 6597–6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SM, Mur LA, Wood JE, Scott IM. (2004) Salicylic acid dependent signaling promotes basal thermotolerance but is not essential for acquired thermotolerance in Arabidopsis thaliana. Plant J 38: 432–447 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Davletova S, Schlauch K, Coutu J, Mittler R. (2005) The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol 139: 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener AC, Li H, Zhou W, Whoriskey WJ, Nes WD, Fink GR. (2000) Sterol methyltransferase 1 controls the level of cholesterol in plants. Plant Cell 12: 853–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh-Kumar SP, Tham WH, Baker BJ. (2000) Structure-function analysis of the tobacco mosaic virus resistance gene N. Proc Natl Acad Sci USA 97: 14789–14794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF. (2009) Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21: 972–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong CH, Hu X, Tang W, Zheng X, Kim YS, Lee BH, Zhu JK. (2006) A putative Arabidopsis nucleoporin, AtNUP160, is critical for RNA export and required for plant tolerance to cold stress. Mol Cell Biol 26: 9533–9543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Deng Y, Mu J, Lu Q, Wang Y, Xu Y, Chu C, Chong K, Lu C, Zuo J. (2007) The Arabidopsis Spontaneous Cell Death1 gene, encoding a zeta-carotene desaturase essential for carotenoid biosynthesis, is involved in chloroplast development, photoprotection and retrograde signalling. Cell Res 17: 458–470 [DOI] [PubMed] [Google Scholar]
- Du L, Ali GS, Simons KA, Hou J, Yang T, Reddy AS, Poovaiah BW. (2009) Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457: 1154–1158 [DOI] [PubMed] [Google Scholar]
- Durrant WE, Dong X. (2004) Systemic acquired resistance. Annu Rev Phytopathol 42: 185–209 [DOI] [PubMed] [Google Scholar]
- Fryer MJ, Oxborough K, Mullineaux PM, Baker NR. (2002) Imaging of photo-oxidative stress responses in leaves. J Exp Bot 53: 1249–1254 [PubMed] [Google Scholar]
- Gilmour SJ, Artus NN, Thomashow MF. (1992) cDNA sequence analysis and expression of two cold-regulated genes of Arabidopsis thaliana. Plant Mol Biol 18: 13–21 [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Fowler SG, Thomashow MF. (2004) Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol Biol 54: 767–781 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Rogers EE, Ausubel FM. (1996) Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143: 973–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Muskett PR, Chuang HW, Parker JE. (2003) Arabidopsis SGT1b is required for SCF(TIR1)-mediated auxin response. Plant Cell 15: 1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy CL. (1990) Cold acclimation and freezing stress tolerance: role of protein metabolism. Annu Rev Plant Physiol Plant Mol Biol 41: 187–223 [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hideg E, Barta C, Kalai T, Vass I, Hideg K, Asada K. (2002) Detection of singlet oxygen and superoxide with fluorescent sensors in leaves under stress by photoinhibition or UV radiation. Plant Cell Physiol 43: 1154–1164 [DOI] [PubMed] [Google Scholar]
- Hirashima M, Tanaka R, Tanaka A. (2009) Light-independent cell death induced by accumulation of pheophorbide a in Arabidopsis thaliana. Plant Cell Physiol 50: 719–729 [DOI] [PubMed] [Google Scholar]
- Howles P, Lawrence G, Finnegan J, McFadden H, Ayliffe M, Dodds P, Ellis J. (2005) Autoactive alleles of the flax L6 rust resistance gene induce non-race-specific rust resistance associated with the hypersensitive response. Mol Plant Microbe Interact 18: 570–582 [DOI] [PubMed] [Google Scholar]
- Huang X, Zhang X, Yang S. (2009) A novel chloroplast-localized protein EMB1303 is required for chloroplast development in Arabidopsis. Cell Res 19: 1205–1216 [DOI] [PubMed] [Google Scholar]
- Hwang CF, Bhakta AV, Truesdell GM, Pudlo WM, Williamson VM. (2000) Evidence for a role of the N terminus and leucine-rich repeat region of the Mi gene product in regulation of localized cell death. Plant Cell 12: 1319–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iba K. (2002) Acclimative response to temperature stress in higher plants: approaches of gene engineering for temperature tolerance. Annu Rev Plant Biol 53: 225–245 [DOI] [PubMed] [Google Scholar]
- Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J. (1999) Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci USA 96: 13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoth C, Ringler J, Dangl JL, Eulgem T. (2007) Arabidopsis WRKY70 is required for full RPP4-mediated disease resistance and basal defense against Hyaloperonospora parasitica. Mol Plant Microbe Interact 20: 120–128 [DOI] [PubMed] [Google Scholar]
- Lee BH, Kapoor A, Zhu J, Zhu JK. (2006) STABILIZED1, a stress-upregulated nuclear protein, is required for pre-mRNA splicing, mRNA turnover, and stress tolerance in Arabidopsis. Plant Cell 18: 1736–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Guo Y, Ohta M, Xiong L, Stevenson B, Zhu JK. (2002) LOS2, a genetic locus required for cold-responsive gene transcription encodes a bi-functional enolase. EMBO J 21: 2692–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Nam J, Park HC, Na G, Miura K, Jin JB, Yoo CY, Baek D, Kim DH, Jeong JC, et al. (2007) Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J 49: 79–90 [DOI] [PubMed] [Google Scholar]
- Li X, Clarke JD, Zhang Y, Dong X. (2001) Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol Plant Microbe Interact 14: 1131–1139 [DOI] [PubMed] [Google Scholar]
- Li X, Zhang Y, Clarke JD, Li Y, Dong X. (1999) Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98: 329–339 [DOI] [PubMed] [Google Scholar]
- Li Y, Yang S, Yang H, Hua J. (2007) The TIR-NB-LRR gene SNC1 is regulated at the transcript level by multiple factors. Mol Plant Microbe Interact 20: 1449–1456 [DOI] [PubMed] [Google Scholar]
- Los DA, Murata N. (2004) Membrane fluidity and its roles in the perception of environmental signals. Biochim Biophys Acta 1666: 142–157 [DOI] [PubMed] [Google Scholar]
- Lyons JM. (1973) Chilling injury in plants. Annu Rev Plant Physiol 24: 445–466 [Google Scholar]
- Mauch-Mani B, Slusarenko AJ. (1996) Production of salicylic acid precursors is a major function of phenylalanine ammonia-lyase in the resistance of Arabidopsis to Peronospora parasitica. Plant Cell 8: 203–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayda E, Mauch-Mani B, Vera P. (2000) Arabidopsis dth9 mutation identifies a gene involved in regulating disease susceptibility without affecting salicylic acid-dependent responses. Plant Cell 12: 2119–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell JM, Cuzick A, Can C, Beynon J, Dangl JL, Holub EB. (2000) Downy mildew (Peronospora parasitica) resistance genes in Arabidopsis vary in functional requirements for NDR1, EDS1, NPR1 and salicylic acid accumulation. Plant J 22: 523–529 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9: 490–498 [DOI] [PubMed] [Google Scholar]
- Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM. (2007) SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19: 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muskett PR, Kahn K, Austin MJ, Moisan LJ, Sadanandom A, Shirasu K, Jones JD, Parker JE. (2002) Arabidopsis RAR1 exerts rate-limiting control of R gene-mediated defenses against multiple pathogens. Plant Cell 14: 979–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanjo T, Kobayashi M, Yoshiba Y, Sanada Y, Wada K, Tsukaya H, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. (1999) Biological functions of proline in morphogenesis and osmotolerance revealed in antisense transgenic Arabidopsis thaliana. Plant J 18: 185–193 [DOI] [PubMed] [Google Scholar]
- Noel L, Moores TL, van Der Biezen EA, Parniske M, Daniels MJ, Parker JE, Jones JD. (1999) Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11: 2099–2112 [PMC free article] [PubMed] [Google Scholar]
- O’Kane D, Gill V, Boyd P, Burdon R. (1996) Chilling, oxidative stress and antioxidant responses in Arabidopsis thaliana callus. Planta 198: 371–377 [DOI] [PubMed] [Google Scholar]
- Parker JE, Holub EB, Frost LN, Falk A, Gunn ND, Daniels MJ. (1996) Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8: 2033–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T, Schutze K, Fey V, Sherameti I, Oelmuller R. (2003) Chloroplast redox control of nuclear gene expression: a new class of plastid signals in interorganellar communication. Antioxid Redox Signal 5: 95–101 [DOI] [PubMed] [Google Scholar]
- Rafiqi M, Bernoux M, Ellis JG, Dodds PN. (2009) In the trenches of plant pathogen recognition: role of NB-LRR proteins. Semin Cell Dev Biol 20: 1017–1024 [DOI] [PubMed] [Google Scholar]
- Ravet K, Touraine B, Boucherez J, Briat JF, Gaymard F, Cellier F. (2009) Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis. Plant J 57: 400–412 [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Davletova S, Liang H, Mittler R. (2004) The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J Biol Chem 279: 11736–11743 [DOI] [PubMed] [Google Scholar]
- Scheel D. (1998) Resistance response physiology and signal transduction. Curr Opin Plant Biol 1: 305–310 [DOI] [PubMed] [Google Scholar]
- Schneider JC, Hugly S, Somerville CR. (1995) Chilling-sensitive mutants of Arabidopsis. Plant Mol Biol Rep 13: 11–17 [Google Scholar]
- Seo PJ, Kim MJ, Park JY, Kim SY, Jeon J, Lee YH, Kim J, Park CM. (2010) Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis Plant J 61: 661–671 [DOI] [PubMed] [Google Scholar]
- Seo PJ, Lee AK, Xiang F, Park CM. (2008) Molecular and functional profiling of Arabidopsis pathogenesis-related genes: insights into their roles in salt response of seed germination. Plant Cell Physiol 49: 334–344 [DOI] [PubMed] [Google Scholar]
- Snider CS, Hsiang T, Zhao G, Griffith M. (2000) Role of ice nucleation and antifreeze activities in pathogenesis and growth of snow molds. Phytopathology 90: 354–361 [DOI] [PubMed] [Google Scholar]
- Someya N, Niinuma K, Kimura M, Yamaguchi I, Hamamoto H. (2004) Pattern of N gene-mediated systemic hypersensitive response and turnover of viral replicase protein in tobacco. Arch Virol 149: 2105–2113 [DOI] [PubMed] [Google Scholar]
- Stokes TL, Richards EJ. (2002) Induced instability of two Arabidopsis constitutive pathogen-response alleles. Proc Natl Acad Sci USA 99: 7792–7796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken FL, Albrecht M, Tameling WI. (2006) Resistance proteins: molecular switches of plant defence. Curr Opin Plant Biol 9: 383–390 [DOI] [PubMed] [Google Scholar]
- Tameling WI, Elzinga SD, Darmin PS, Vossen JH, Takken FL, Haring MA, Cornelissen BJ. (2002) The tomato R gene products I-2 and MI-1 are functional ATP binding proteins with ATPase activity. Plant Cell 14: 2929–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R, Hirashima M, Satoh S, Tanaka A. (2003) The Arabidopsis-accelerated cell death gene ACD1 is involved in oxygenation of pheophorbide a: inhibition of the pheophorbide a oxygenase activity does not lead to the “stay-green” phenotype in Arabidopsis. Plant Cell Physiol 44: 1266–1274 [DOI] [PubMed] [Google Scholar]
- Tao Y, Yuan F, Leister RT, Ausubel FM, Katagiri F. (2000) Mutational analysis of the Arabidopsis nucleotide binding site-leucine-rich repeat resistance gene RPS2. Plant Cell 12: 2541–2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB. (1997) Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11: 1187–1194 [Google Scholar]
- Tsutsui T, Kato W, Asada Y, Sako K, Sato T, Sonoda Y, Kidokoro S, Yamaguchi-Shinozaki K, Tamaoki M, Arakawa K, et al. (2009) DEAR1, a transcriptional repressor of DREB protein that mediates plant defense and freezing stress responses in Arabidopsis. J Plant Res 122: 633–643 [DOI] [PubMed] [Google Scholar]
- Ueda H, Yamaguchi Y, Sano H. (2006) Direct interaction between the tobacco mosaic virus helicase domain and the ATP-bound resistance protein, N factor during the hypersensitive response in tobacco plants. Plant Mol Biol 61: 31–45 [DOI] [PubMed] [Google Scholar]
- van der Biezen EA, Freddie CT, Kahn K, Parker JE, Jones JD. (2002) Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J 29: 439–451 [DOI] [PubMed] [Google Scholar]
- van Ooijen G, Mayr G, Kasiem MM, Albrecht M, Cornelissen BJ, Takken FL. (2008) Structure-function analysis of the NB-ARC domain of plant disease resistance proteins. J Exp Bot 59: 1383–1397 [DOI] [PubMed] [Google Scholar]
- Warren RF, Henk A, Mowery P, Holub E, Innes RW. (1998) A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell 10: 1439–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren RF, Merritt PM, Holub E, Innes RW. (1999) Identification of three putative signal transduction genes involved in R gene-specified disease resistance in Arabidopsis. Genetics 152: 401–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Xiao S, Charoenwattana P, Holcombe L, Turner JG. (2003) The Arabidopsis genes RPW8.1 and RPW8.2 confer induced resistance to powdery mildew diseases in tobacco. Mol Plant Microbe Interact 16: 289–294 [DOI] [PubMed] [Google Scholar]
- Xin Z, Browse J. (1998) Eskimo1 mutants of Arabidopsis are constitutively freezing-tolerant. Proc Natl Acad Sci USA 95: 7799–7804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Z, Mandaokar A, Chen J, Last RL, Browse J. (2007) Arabidopsis ESK1 encodes a novel regulator of freezing tolerance. Plant J 49: 786–799 [DOI] [PubMed] [Google Scholar]
- Yang S, Hua J. (2004) A haplotype-specific resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell 16: 1060–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Yang H, Grisafi P, Sanchatjate S, Fink GR, Sun Q, Hua J. (2006) The BON/CPN gene family represses cell death and promotes cell growth in Arabidopsis. Plant J 45: 166–179 [DOI] [PubMed] [Google Scholar]
- Yi H, Richards EJ. (2007) A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing. Plant Cell 19: 2929–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Goritschnig S, Dong X, Li X. (2003) A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15: 2636–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Mosher S, Tian M, Sassi G, Parker J, Klessig DF. (2008) The Arabidopsis gain-of-function mutant ssi4 requires RAR1 and SGT1b differentially for defense activation and morphological alterations. Mol Plant Microbe Interact 21: 40–49 [DOI] [PubMed] [Google Scholar]
- Zhu J, Jeong JC, Zhu Y, Sokolchik I, Miyazaki S, Zhu JK, Hasegawa PM, Bohnert HJ, Shi H, Yun DJ, et al. (2008) Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proc Natl Acad Sci USA 105: 4945–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Qian W, Hua J. (2010) Temperature modulates plant defense responses through NB-LRR proteins. PLoS Pathog 6: e1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.