Abstract
Transgenic down-regulation of the Pt4CL1 gene family encoding 4-coumarate:coenzyme A ligase (4CL) has been reported as a means for reducing lignin content in cell walls and increasing overall growth rates, thereby improving feedstock quality for paper and bioethanol production. Using hybrid poplar (Populus tremula × Populus alba), we applied this strategy and examined field-grown transformants for both effects on wood biochemistry and tree productivity. The reductions in lignin contents obtained correlated well with 4CL RNA expression, with a sharp decrease in lignin amount being observed for RNA expression below approximately 50% of the nontransgenic control. Relatively small lignin reductions of approximately 10% were associated with reduced productivity, decreased wood syringyl/guaiacyl lignin monomer ratios, and a small increase in the level of incorporation of H-monomers (p-hydroxyphenyl) into cell walls. Transgenic events with less than approximately 50% 4CL RNA expression were characterized by patches of reddish-brown discolored wood that had approximately twice the extractive content of controls (largely complex polyphenolics). There was no evidence that substantially reduced lignin contents increased growth rates or saccharification potential. Our results suggest that the capacity for lignin reduction is limited; below a threshold, large changes in wood chemistry and plant metabolism were observed that adversely affected productivity and potential ethanol yield. They also underline the importance of field studies to obtain physiologically meaningful results and to support technology development with transgenic trees.
Composed of diverse layers of cellulose microfibrils and amorphous hemicelluloses within a matrix of pectins, proteins, and lignin, the secondary cell walls of plants are diverse in their morphology, chemistry, and physiological functions. Lignification is of particular interest, as it exhibits highly predictable temporal and spatial patterning and is the last major step in the structural reinforcement of cell walls before the protoplast is dissolved (Donaldson, 2001). To gain detailed insights into cell wall assembly, mutant or transgenic perturbations to lignin biosynthesis have been employed to alter native lignin content and monomer compositions (i.e. to shift ratios of syringyl [S], guaiacyl [G], and p-hydroxyphenyl [H] lignins; Porter et al., 1978; Miller et al., 1983; Baucher et al., 1996; Kajita et al., 1996; Lee et al., 1997; Anterola and Lewis, 2002; Davin et al., 2008a, 2008b; Patten et al., 2010a). In addition, such perturbations give needed insight into the role of lignin in providing resistance to mechanical (Mark, 1967; Niklas, 1992; Gindl and Teischinger, 2002) and biotic (Dixon and Paiva, 1995) stresses. Lignin affects xylem conductance and protects the vasculature from embolism by imparting a barrier between water under transpiration-induced tension in the xylem and the atmosphere (Raven, 1977; Boyce et al., 2004) and retards tissue digestion and decomposition by pathogens and herbivores. Economic incentives have also helped drive research on lignin reductions in wood because lignin is considered the principal cause of recalcitrance to chemical pulping and to simultaneous saccharification and fermentation to produce liquid biofuels (Huntley et al., 2003; Schubert, 2006; Jørgensen et al., 2007; Davin et al., 2008a, 2008b; Foust et al., 2008; Li et al., 2008; Yang and Wyman, 2008).
Because each of the major cell wall biopolymers has different functions, changes in one component should induce “compensatory” shifts in concentrations or compositions of the others. Indeed, altering lignin composition and content has been shown to have wide-ranging effects on cell wall morphology, including specification of cell identity and plant form (Davin et al., 2008a, 2008b). An early study of aspen (Populus tremuloides) down-regulated for 4-coumarate:coenzyme A ligase (4CL) reported that young trees had up to 45% less lignin, increased cellulose contents, and increased growth (Hu et al., 1999). These results led Hu and coworkers (1999) to hypothesize that enhanced growth and compensatory deposition of cell wall polysaccharides resulted from reduced carbon demand for lignin synthesis. However, these results were questioned on both analytical and biochemical grounds (Anterola and Lewis, 2002). Subsequent studies of greenhouse-grown aspen (Li et al., 2003; Hancock et al., 2007, 2008) and Chinese white poplar (Populus tomentosa; Jia et al., 2004) containing transgenes that suppress RNA expression of 4CL found no comparable growth enhancement.
4CL is generally considered to be the third step in the phenylpropanoid pathway. Consisting of a multigene family (Costa et al., 2005), 4CL is important for monolignol biosynthesis as well as for the generation of other secondary metabolites for plant defense in leaves and stem xylem tissues (Tsai et al., 2006). However, little is known about how down-regulation of 4CL can differentially affect the production of secondary metabolites and whether or not the types and amounts of the defense compounds produced may differ depending on the level of environmental stresses perceived by growing plants.
Because of the large differences in plant physiological behavior under field versus laboratory or greenhouse conditions, and the complex development of xylem in growing trees, field studies are essential to understand the level of lignin modification that might be economically useful yet also preserve tree health and productivity. Previous field studies with other forms of lignin modification have suggested that some kinds of perturbations might be tolerated (Pilate et al., 2002). However, comparable studies have not been reported on trees with lignin modifications induced by 4CL inhibition.
In this study, we report that 4CL down-regulation via antisense RNA was effective in reducing lignin contents of wood in field-grown trees. In agreement with more recent work (Li et al., 2003; Hancock et al., 2007) and in contrast to an early study (Hu et al., 1999), these changes did not promote increased growth rate. High levels of lignin reduction observed in approximately one-third of the transgenic events led to reduced growth and serious physiological abnormalities. In these low-lignin transgenic events, we identified and quantified significant nonlignin phenolic depositions and utilized a novel combination of cryofixation and confocal microscopy to visualize the in vivo distribution of these compounds within the wood. Finally, we determined that reductions in lignin content did not increase wood processability that would benefit fermentation to produce liquid biofuels.
RESULTS
4CL Transformants Had Reduced RNA Expression
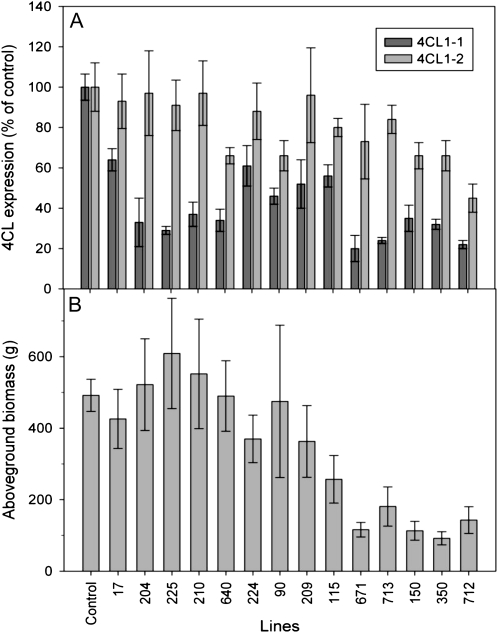
In this study, hybrid white poplar (Populus tremula × Populus alba) was transformed with Agrobacterium tumefaciens carrying an antisense aspen (P. tremuloides) Pt4CL1 gene construct with respect to the endogenous aspen Pt4CL1 (Li et al., 2003). To estimate levels of class 1 4CL down-regulation in the resulting transformants, primers specific for two genes, annotated herein as 4CL1-1 and 4CL1-2, were designed for quantitative reverse transcription (qRT)-PCR analyses. Both genes were initially identified based on BLAST searches against the black cottonwood (Populus trichocarpa) genome (version 1.1; Tuskan et al., 2006) using the aspen Pt4CL1 gene sequence. This yielded two homologs sharing 94% DNA sequence similarity and 89% amino acid identity (97% similarity), the so-called Ptr4CL3 and Ptr4CL5 (see “Materials and Methods”; Shi et al, (2010). Based on this in silico analysis, and after total RNA isolation from untransformed white poplar stem tissues, the 3′ untranslated regions of the homologs (4CL1-1 and 4CL1-2) to the two above P. trichocarpa 4CLs were sequenced. Expression of 4CL1-1 and 4CL1-2 in xylem tissues (harvested between internodes 5 and 6 in May 2007; for primer sequences, see Supplemental Table S1) using qRT-PCR found RNA expression down-regulated to 22% to 64% and 45% to 97% of controls, respectively (Fig. 1A).
Figure 1.
4CL1-1/4CL2-2 RNA transcript levels as measured by qRT-PCR (A), and biomass of 2-year-old control and transgenic white poplars (B). Events are arrayed by height from tallest (control) to shortest (event 712).
Tree Growth, Wood Chemistry, and Wood Color Were Altered in Many Transgenic Events
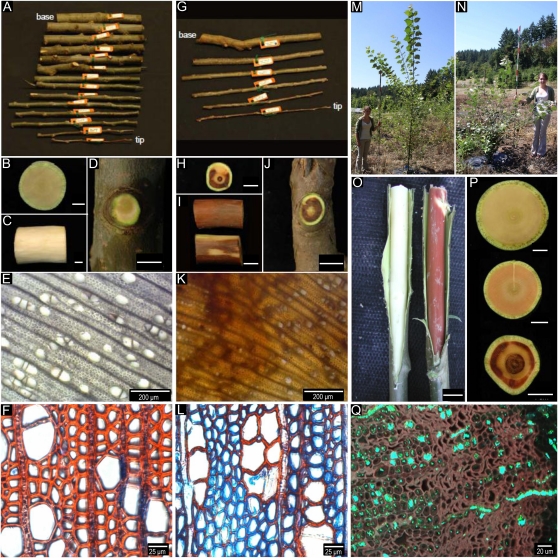
Reductions in 4CL expression were associated with reductions in aboveground biomass of the 2-year-old poplars (Fig. 1). Biomass reductions were greatest in five of the 14 transgenic events (150, 350, 671, 712, and 713) that were characterized by stem wood with patchy brown or reddish-brown color occupying about 24% to 60% on average of the cross-sectional area (hereafter called brown wood; Table I). Compared with the control (Fig. 2, A–E), the brown wood events often differed not only in wood color but in tree stature (Fig. 2, G–K). This difference in wood color often co-occurred with differences in cell shape and cell wall histochemistry, with reductions in phenolic content being most evident in the secondary walls of fibers compared with vessels (although vessels were often irregularly shaped or partially collapsed; Fig. 2, F and L). Most individuals from the brown wood events were largely stunted in growth and characterized by a branchy or shrubby appearance (Fig. 2, M and N). Brown or occasionally bright red wood was most abundant in distal branches (Fig. 2O). The altered coloration was associated with the deposition of phenolic “extractives” as well as radial bands of collapsed vessels and those fibers without a gelatinous G layer (Fig. 2Q; Supplemental Fig. S1). Although control wood (Fig. 2P, top) was distinctly different in color from brown wood (Fig. 2P, bottom), there were some transgenic events that had very little brown wood but did produce a slightly rose-colored wood (Fig. 2P, center).
Table I. Biomass accumulation and brown wood occurrence (±sd) measured for each of 14 transgenic events and controls.
Boldface values were significantly different (P < 0.05) from controls. The number of live trees sampled after 2 years of growth is represented by n.
| Event | n | Oven-Dry Aboveground Biomass | Brown Wood |
| g | % | ||
| Control | 31 | 492 ± 376 | 0 ± 1 |
| 17 | 12 | 426 ± 287 | 0 ± 1 |
| 204 | 11 | 522 ± 444 | 6 ± 17 |
| 225 | 12 | 609 ± 534 | 4 ± 10 |
| 210 | 11 | 552 ± 508 | 1 ± 2 |
| 640 | 10 | 490 ± 342 | 1 ± 2 |
| 224 | 11 | 370 ± 230 | 1 ± 1 |
| 90 | 10 | 475 ± 674 | 0 ± 1 |
| 209 | 9 | 363 ± 332 | 1 ± 2 |
| 115 | 11 | 257 ± 231 | 1 ± 2 |
| 671 | 10 | 116 ± 70 | 59 ± 34 |
| 713 | 10 | 181 ± 190 | 24 ± 31 |
| 150 | 9 | 113 ± 91 | 51 ± 37 |
| 350 | 10 | 92 ± 64 | 47 ± 30 |
| 712 | 7 | 143 ± 124 | 60 ± 33 |
Figure 2.
A to K, Phenotypic differences between stems of white poplar control event (A–E) and event 350 (G–K). A and G, After harvesting, 2-year-old tree stems were cut into approximately 30- to 40-cm sections. B to D and H to J, The stem base transverse and longitudinal sections of the control are yellow in color (B–D), whereas comparable locations in event 350 are largely red-brown (H–J). E and K, Light microscopy of transverse sections shows consistent wood color and large round vessels of the control versus the patchy brown color and, provisionally, a reduction in vessel size and frequencies in brown wood of event 350. F and L, Wood stained with safranin (red-colored wood indicates that the stain is bound to phenolics including lignin) and astra-blue (blue-colored wood indicates that the stain is bound to cellulose in the absence of lignin) from stem wood of the control (F) and poorly lignified wood from event 712 (L). M and N, Representative tree form of the control (M) and the “shrubby” brown wood-forming event 713 (N). O, The bright green color of a freshly harvested branch from the control line contrasts sharply with the red-colored wood from a similar branch from event 350. P, Wood color in transverse sections shows normal color (control; top), rose-colored wood (event 225; middle), and patchy brown wood (event 350; bottom). Q, Confocal microscopy of collapsed wood xylem from event 712 shows the distribution of phenolic extractives (blue-green fluorescence) in radial transit through ray parenchyma and after deposition, mostly into fiber cells. Most collapsed cells in this image were narrow vessels and normal fibers. The image represents a maximum projection of 36 optical sections at 1-μm intervals.
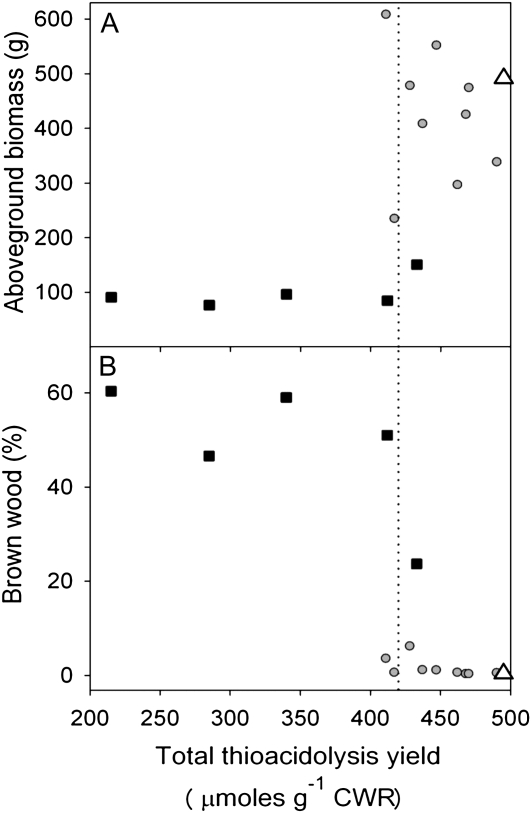
Compared with the control wood, total H/G/S thioacidolysis-releasable monomers were lower by up to approximately 40% (Fig. 3), suggesting that some events had substantial reductions in lignin content. There was no clear relationship between biomass and putative lignin content for transgenic events with modest decreases in lignin. However, there appeared to be a threshold of lignin reduction beyond which brown wood frequency increased dramatically and tree growth declined correspondingly (Fig. 3, dotted vertical line).
Figure 3.
Aboveground biomass (A) and stem brown wood percentage (B) plotted against total thioacidolysis yields. Each symbol is the mean value for the control or a transgenic event. White triangles, control; gray circles, normal transformants; black squares, brown wood transformants.
Brown Wood Was Enriched in Phenolics But Unchanged in Saccharification Rate
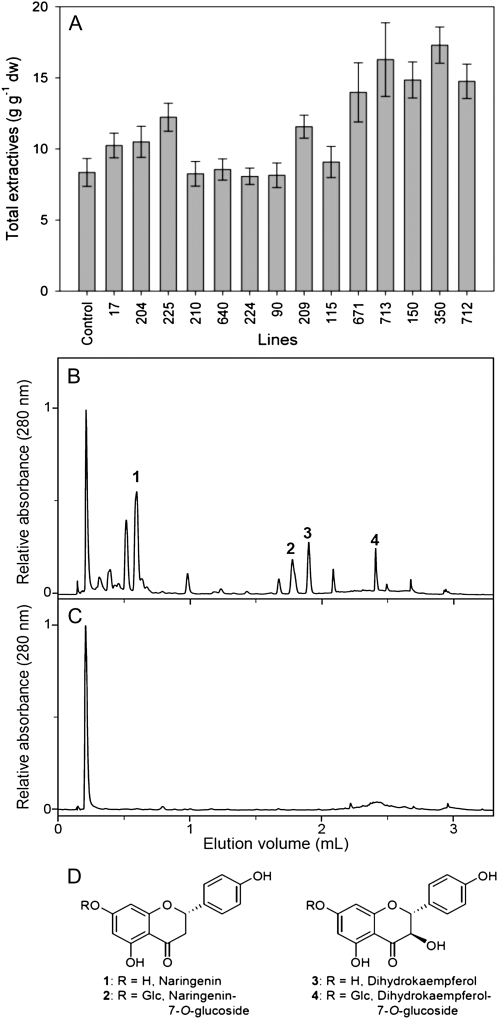
To investigate whether or not brown wood had greater amounts of extractives, stem wood from each event was extracted with toluene/ethanol, ethanol, or hot water. These data showed that extractive contents of seven of the events (17, 90, 115, 204, 210, 224, and 640) did not differ significantly from the controls (Fig. 4A). By contrast, the red-brown wood events had consistently higher extractive contents, averaging nearly twice the amount of the controls. Compared with the control line, each brown wood event had a significantly higher extractive content within the patches of brown wood, but normal colored wood outside of these patches were similar in extractive content to the controls (Table II). Because event 712 had the most different phenotype, this event was used to investigate the chemical nature of the extractives via ultra-performance liquid chromatography analysis and constituent identification (Fig. 4B). This procedure established that the major extractable phenolic constituents of brown wood were naringenin (1), dihydrokaempferol (3), and their corresponding glucosides (2 and 4; Fig. 4D). Each of these constituents was identified by comparison of its mass spectroscopic fragmentation pattern as well as with the corresponding authentic standard. In the control, these substances were present at nearly undetectable levels (Fig. 4C).
Figure 4.
A, Stem wood estimated extractive contents in control and transgenic white poplars. dw, Dry weight. B, Ultra-performance liquid chromatography elution profile of extractives isolated from event 712 showing the presence of naringenin (1), dihydrokaempferol (3), and their 7-O-glucosides 2 and 4. C, These extractives are absent in the control event under the conditions employed. D, Flavonoids known to be present in Populus species.
Table II. Cellulose and extractive contents (±sd) by brown wood presence or absence.
Boldface values were significantly different (P < 0.05) from controls. CWR indicates oven-dry, extractive-free cell wall residue, and DW indicates oven-dry initial wood mass including extractives. For controls, n = 7; for normal wood, n = 4; and for brown wood, n = 3 (trees). Extractive and cellulose contents were determined gravimetrically. Toluene-ethanol extractives were estimated following soxhlet extraction, and hot water-soluble extractives were estimated following a series of water baths (see “Materials and Methods”). nd, Not determined.
| Event | Brown Wood Cellulose | Normal Wood Cellulose | Brown Wood Toluene-Ethanol Extractives | Normal Wood Toluene-Ethanol Extractives | Brown Wood Hot Water Extractives | Normal Wood Hot Water Extractives |
| % of CWR | % of DW | |||||
| Control | nd | 42.2 ± 0.9 | nd | 4.0 ± 0.6 | nd | 4.3 ± 1.1 |
| 17 | nd | 44.0 ± 1.1 | nd | 4.9 ± 0.5 | nd | 5.3 ± 1.0 |
| 204 | nd | 43.3 ± 2.3 | nd | 4.6 ± 1.1 | nd | 5.2 ± 0.8 |
| 225 | nd | 46.9 ± 1.9 | nd | 6.4 ± 1.4 | nd | 5.5 ± 0.3 |
| 210 | nd | 46.2 ± 1.5 | nd | 4.2 ± 0.6 | nd | 3.9 ± 0.9 |
| 640 | nd | 49.3 ± 1.0 | nd | 4.0 ± 0.4 | nd | 4.4 ± 0.9 |
| 224 | nd | 46.2 ± 3.0 | nd | 4.0 ± 0.5 | nd | 4.0 ± 0.5 |
| 90 | nd | 41.9 ± 3.0 | nd | 4.0 ± 0.9 | nd | 4.1 ± 0.6 |
| 209 | nd | 43.8 ± 14.1 | nd | 5.7 ± 0.6 | nd | 5.8 ± 0.8 |
| 115 | nd | 40.4 ± 2.6 | nd | 5.0 ± 0.9 | nd | 4.0 ± 1.0 |
| 671 | 47.2 ± 0.9 | 47.0 ± 1.6 | 9.5 ± 3.2 | 5.9 ± 1.5 | 6.2 ± 1.1 | 5.6 ± 2.1 |
| 713 | 38.6 ± 1.1 | 40.6 ± 1.2 | 19.5 ± 0.4 | 5.9 ± 2.8 | 8.0 ± 2.0 | 6.9 ± 1.7 |
| 150 | 45.9 ± 1.6 | 46.1 ± 2.0 | 11.2 ± 1.7 | 5.4 ± 1.4 | 8.5 ± 2.9 | 4.4 ± 0.8 |
| 350 | 42.0 ± 3.1 | 44.8 ± 2.5 | 16.2 ± 5.2 | 5.5 ± 1.5 | 7.4 ± 0.6 | 6.3 ± 0.7 |
| 712 | 51.0 ± 4.6 | 45.5 ± 0.9 | 9.5 ± 2.6 | 3.9 ± 0.5 | 8.7 ± 1.6 | 5.6 ± 1.6 |
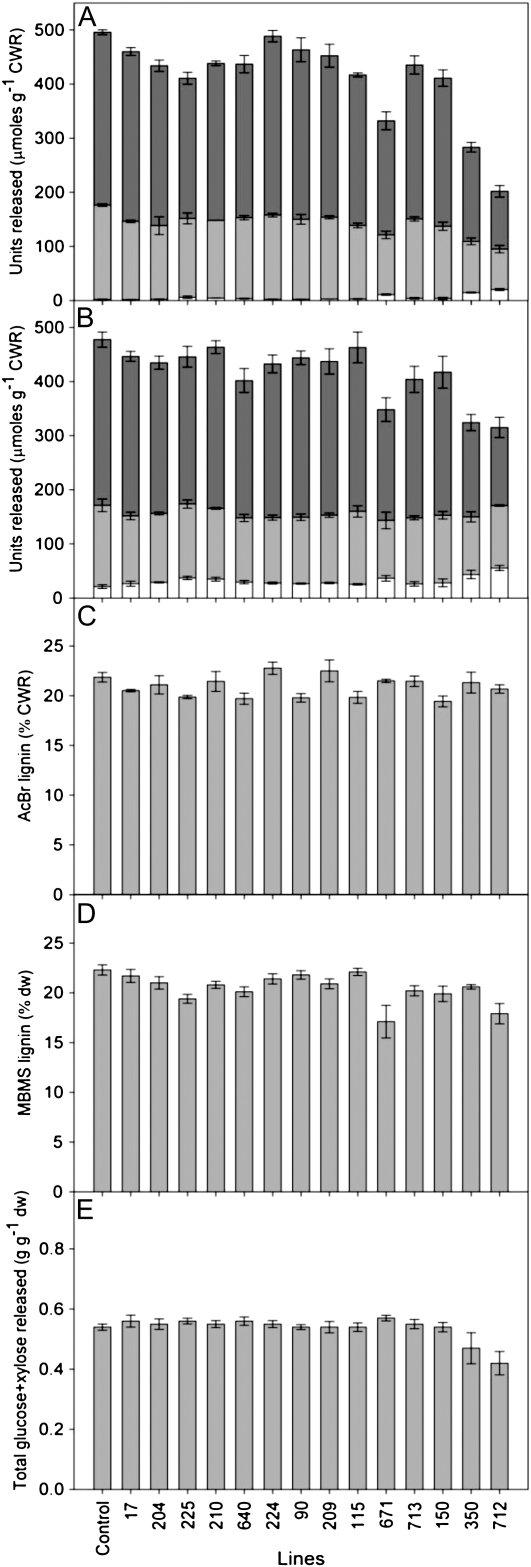
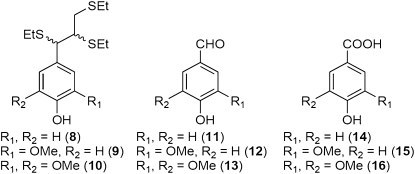
Among the transgenic events, solvent-extracted cell wall residues (CWRs) examined by thioacidolysis (Fig. 5A) and alkaline nitrobenzene oxidation (NBO; Fig. 5B) showed somewhat similar trends in total monomer release as well as in the proportions of S and G monomers. Thioacidolysis releases monomers 8 to 10 (Fig. 6), which are considered to originate from the cleavage of 8-O-4′ interunit linkages in lignin polymers, whereas NBO releases the corresponding benzaldehydes (11–13)/benzoic acids (14–16) via C7-C8 bond cleavage from a range of phenylpropanoids, including lignin (Fig. 6). In this study, the brown wood events gave lower amounts of released products for the highest levels of 4CL down-regulation. NBO analyses, however, released a higher proportion of H-derived monomers for each event. Despite the decreasing trends in total monomer release, putative acetyl bromide (AcBr) lignin contents showed little variation (Fig. 5C), as did molecular beam mass spectroscopy (MBMS)-based lignin estimates (Fig. 5D). To summarize, corresponding to reductions in 4CL expression (Fig. 1), the thioacidolysis/NBO methods for estimating monomeric compositions all indicated reductions in lignin contents in the brown wood samples (Fig. 5, A and B), these being accompanied, as expected, by reductions in S/G ratios (Table III).
Figure 5.
A and B, Releasable monomeric derivatives by thioacidolysis (A) and alkaline NBO (B). Dark gray bars represent S, light gray bars represent G, and white bars represent H monomers. C and D, Putative lignin contents as estimated by the AcBr lignin method (C) and MBMS (D). E, Sugar released during saccharification.
Figure 6.
Monomeric thioacidolysis (8–10) and alkaline NBO (11–16) products from lignins.
Table III. S/G ratios (±sd) as determined by thioacidolysis, NBO, and MBMS analyses.
Boldface values were significantly different (P < 0.05) from controls.
| Event | Thioacidolysis | NBO | MBMS |
| Control | 1.8 ± 0.08 | 2.0 ± 0.16 | 2.3 ± 0.2 |
| 17 | 2.2 ± 0.02 | 2.4 ± 0.03 | 2.5 ± 0.1 |
| 204 | 2.2 ± 0.14 | 2.2 ± 0.02 | 2.2 ± 0.4 |
| 225 | 1.8 ± 0.09 | 2.0 ± 0.04 | 2.0 ± 0.1 |
| 210 | 2.0 ± 0.03 | 2.3 ± 0.05 | 2.4 ± 0.1 |
| 640 | 1.9 ± 0.09 | 2.1 ± 0.09 | 2.2 ± 0.1 |
| 224 | 2.1 ± 0.10 | 2.4 ± 0.07 | 2.4 ± 0.2 |
| 90 | 2.1 ± 0.03 | 2.4 ± 0.14 | 2.6 ± 0.0 |
| 209 | 2.0 ± 0.19 | 2.3 ± 0.29 | 2.5 ± 0.1 |
| 115 | 2.1 ± 0.12 | 2.3 ± 0.09 | 2.5 ± 0.1 |
| 671 | 1.9 ± 0.19 | 1.9 ± 0.24 | 1.8 ± 0.6 |
| 713 | 1.9 ± 0.19 | 2.1 ± 0.13 | 2.2 ± 0.3 |
| 150 | 2.1 ± 0.22 | 2.1 ± 0.36 | 1.9 ± 0.5 |
| 350 | 1.8 ± 0.09 | 1.6 ± 0.08 | 1.7 ± 0.5 |
| 712 | 1.4 ± 0.11 | 1.3 ± 0.17 | 1.1 ± 0.4 |
The total Glc and Xyl release by enzymatic hydrolysis of nonextracted wood powder showed little variation among most of the transgenic events, with statistically significant differences only in events 350 and 712, which are two of the five brown wood events (Fig. 5E). The lack of a change in saccharification efficiency of pretreated poplar wood occurred despite reductions in lignin content (Fig. 5, A and B) and a trend toward higher cellulose contents of the brown wood events due to lignin levels being reduced (Table II).
DISCUSSION
Tree Growth and Wood Color Varied Widely among Transgenic Events
The white poplar transgenics grown in the field for 2 years showed extensive variation in aboveground biomass, tree form, and wood color (Figs. 1–3; Table I). Nine of the 14 transgenic events had similar wood color to the controls, ranging from normally colored to rose or pink and rare or small patches of brown wood. These events grew reasonably well considering the variation inherent in field studies (approximately 52%–106% of control biomass; Figs. 2 and 3; Table I). The largest difference among transgenic events was associated with the proportion of brown wood (Figs. 2 and 3). These wood phenotypes, with 20% or more of their stem cross-sections as brown wood, were severely stunted (17%–31% of control biomass) and had a shrubby appearance (Figs. 2 and 3; Table I). Brown wood transformants also tended to exhibit shoot dieback late in the growing season and thus probably contributed to their shrubby form; these lines were also characterized by irregular or eccentric cambial activity (Voelker, 2009).
Similar to our results, previous transgenic poplar field trials found that lignin contents reduced by less than 10% did not appreciably change tree growth characteristics (Pilate et al., 2002), whereas reductions in lignin content by about 20% caused tree growth to be strongly reduced (Leplé et al., 2007). These reports contrast with that of increased growth in 4CL transgenic poplars grown in a greenhouse (Hu et al., 1999). In our study, 4CL down-regulation of poplars grown under field conditions resulted in considerable variation in productivity until a putative threshold was passed, at which point reductions in biomass, wood discoloration, and other pleiotropic effects became striking in the most strongly down-regulated events (Fig. 2). The lack of enhanced growth rate agrees with other studies of 4CL down-regulation, even ones that used a xylem-specific promoter, as in this study (Li et al., 2003; Hancock et al., 2007, 2008), rather than a constitutively expressed promoter (Hu et al., 1999). Moreover, studies conducted on other tree taxa (Jia et al., 2004; Wagner et al., 2009) and species lacking substantive secondary growth (Kajita et al., 1996; Lee et al., 1997) have reported either stunted phenotypes or no detectable growth enhancement from 4CL down-regulation. In agreement with these findings, Kirst and coworkers (2004) reported that while the expression of a number of expression quantitative trait loci markers associated with lignin biosynthesis was correlated with growth among interspecific backcross progeny of Eucalyptus, 4CL was not one of them. Taken together, these studies are consistent with the body of literature in showing a lack of increased growth rate for transgenic plants that have been modified in lignin biosynthesis (Anterola and Lewis, 2002; Davin et al., 2008a, 2008b; Li et al., 2010).
Wood Color Was Associated with Extractive Content and Deformed Stems
In five of the 14 transgenic events, brown wood exceeded 20% of the cross-sectional area near stem bases (Table I), was patchy in the transverse and longitudinal planes (Fig. 2, H–K and O–P [bottom]; Supplemental Fig. S2), and was also associated with modulated cambial activity, as indicated by stems with brown wood often having an irregular (i.e. noncircular) cross-sectional shape. Confocal microscopy indicated that brown wood appeared to be associated with copious deposition of putative phenolic extractives localized within the ray parenchyma and within fibers and vessels (Fig. 2Q). This pattern suggests that phenolics were synthesized in the parenchyma cells, which is analogous to the process that occurs during heartwood formation (Gang et al., 1998; Taylor et al., 2002; Patten et al., 2010b). Indeed, the metabolites 1 to 4 identified here in brown wood, as well as kaempferol (5), its 7-O-glucoside (6), and dihydroquercetin (7), have also been reported present in sapwood, heartwood, and knots of various aspen species (P. tremula, P. tremuloides, and Populus grandidentata) in amounts ranging from 11 to 82 mg g–1 dry weight (Fernandez et al., 2001; Pietarinen et al., 2006). Interestingly though, the amounts of dihydrokaempferol (3) were approximately 50- to 3,000-fold higher in “knotwood” of those species than in stem wood (Pietarinen et al., 2006). Often, trees that had traces of brown wood in lower stem sections had relatively greater amounts of brown or discolored wood at branch-to-stem junctions (Supplemental Fig. S3). Although the underlying causes of increased metabolite levels can only be speculated, our data and observations provisionally suggest that carbon reallocation away from lignification to other metabolic branches might have occurred in lines with the higher extractive levels. It should be emphasized, however, that from a biochemical perspective the underlying causes of increased metabolite deposition (such as phenolics) in both heartwood and knots remains poorly understood (Gang et al., 1998).
Putative shunt pathways and/or the accumulation of pathway metabolites resulting in abnormally pink, red, or brown xylem have been reported in other 4CL mutants (Kajita et al., 1996; Jia et al., 2004). Similarly, discolored wood has also been found when enzyme activity levels either upstream or downstream of 4CL have been altered (Porter et al., 1978; Miller et al., 1983; Baucher et al., 1996; Ralph et al., 1997; Tsai et al., 1998; Lapierre et al., 1999; Meyermans et al., 2000; Pilate et al., 2002; Jourdes et al., 2007; Leplé et al., 2007). Down-regulation of cinnamyl alcohol dehydrogenase, for example, produced red xylem that resulted from small amounts of sinapyl aldehyde entering the xylem cell wall region (Jourdes et al., 2007). This pigmentation, however, could readily be removed with methanol:1% HCl, a procedure generally utilized for anthocyanin floral pigment removal. Such solubilization behavior is indicative that this pigmentation was not part of the lignin macromolecule. In contrast, we found the brown or red color to remain after the same solvent extraction procedure, suggesting that some level of colored components other than these isolated flavonoids entered into the wood, as is generally observed to occur in heartwood. Although the mechanisms remain to be understood, our results suggest that the unintended production of xylem secondary metabolites is a likely a consequence of 4CL inhibition used to produce low-lignin trees.
Lignin Content Was Inversely Associated with Xylem Deformation
The highest levels of 4CL down-regulation gave rise to plants that were substantially reduced in lignin content, as indicated by thioacidolytic cleavage (Fig. 5A). These events were also reduced in size, developing a shrubbier appearance (Figs. 1–3). Furthermore, reductions in lignin contents led to lower wood strength and stiffness and an increase in the prevalence of tension wood (Voelker, 2009). It is known that tension wood formation is controlled by genes that are up-regulated by bending stress (Coutand et al., 2009), suggesting that greater brown wood occurrence at stem-to-branch junctions (i.e. knotwood) might have also been induced by a similar mechanism. These junctions are where mechanical stresses are concentrated and where the highest levels of so-called extractives are naturally deposited in wild trees.
When viewed with light microscopy, the vessels in brown wood tended to be distorted in shape (Fig. 2, F and L). Extensive anatomical investigations of low-lignin poplar wood (Kitin et al., 2010) suggest that this partial collapse can be caused by slide preparation or partial recovery of cells that were almost completely collapsed before preparation. In either case, the cell wall material was apparently weaker. Xylem collapse has also been noted in other low-lignin transgenic poplar (Coleman et al., 2008), and we observed similar cell morphology in transformants with the lowest lignin contents (Fig. 2Q; Supplemental Fig. S1). Interestingly, the uncollapsed cells were predominantly associated with tension wood fibers containing a G layer (Fig. 2Q; Supplemental Fig. S1). This observation is consistent with the lateral expansion of the G layer of tension wood fibers, which produces longitudinal contraction of the cell wall and resulting tensile stresses within those cells (Goswami et al., 2008). The mechanical stresses associated with wind-, rain-, and ice-induced bending, however, are greater in the field than in a greenhouse environment and should cause greater tension wood formation. In turn, the lateral expansion and shortening of tension wood fibers (Goswami et al., 2008) would locally exert compressive stresses on nearby cells. Because lignin is thought to be important in resisting compressive stresses (Mark, 1967; Niklas, 1992; Gindl and Teischinger, 2002), vasculature with lower lignin contents may more readily collapse (Fig. 2Q) due to compressive stresses surrounding tension wood fibers. The extensive variation in brown wood and xylem deformation seen in stems of these transgenic trees is likely to result from the well-known variation in degree of antisense down-regulation and its cell/tissue specificity during the development of transgenic plants (Anterola and Lewis, 2002). Because of this variation and the large natural variation in abiotic and biotic stresses that trigger changes in wood development, field trials are likely the most meaningful strategy to analyze the risks of such pleiotropy during tree growth.
Substantial Agreement among Studied Methods for the Analysis of Lignin Compositions
Thioacidolysis analyses showed that the nine “normal transformants” with infrequent patches of brown wood were characterized by similar monomer-releasable lignin-derived S or G moieties to the controls, with levels of approximately 400 to 500 versus 500 μmol g–1 CWR in the control (Fig. 5A). This overall reduction of about 20% compared with controls suggests that only small reductions in lignin content occurred in these events. The S/G ratios were also only modestly affected in these events based on thioacidolysis; S/G ratios ranged from 1.8 to 2.2 in these transgenics versus 1.8 in the controls, with the largest fluctuation due to variation in S content (Fig. 5A; Table III).
Comparable data were obtained using the NBO method (Fig. 5B), which gave similar trends, albeit with small levels of H units released in all events. Given the near absence of H units for thioacidolysis products from the same wood, this difference presumably results from non-lignin-derived moieties being detected by NBO analyses. Thus, taken together, these data suggest that the transgenics had reductions in thioacidolysis “lignin content” of up to approximately 20% compared with the control line. The AcBr “lignin” estimations suggested that reductions in lignin content were lesser, up to approximately 10% compared with the control line (Fig. 5C). However, this approximate reduction level could be underestimated due to the presence of remaining small amounts of UV light-absorbing extractives (flavonoids) that are not readily removed from woody tissue using existing solvent extraction procedures. This lack of complete solvent extractability has long been known to occur in heartwood/knot tissues (Gang et al., 1998). For these nine transformants, MBMS lignin contents determined on wood that had not undergone extraction were also somewhat similar to the AcBr results; these analyses estimated reductions of up to approximately 12%.
Only in brown wood events did significant reductions in both cleavable monomer amounts and H/S/G ratios occur. In the case of events 350, 671, and 712, the thioacidolysis yields were between about 30% and 55% lower than the control, indicative of a substantial reduction in lignin content (Fig. 5A). Depending on the method, S/G ratios declined to as little as 1.1 to 1.4 in event 712 as compared with 1.8 to 2.3 in the control line (Table III). This trend would be expected when lignin reductions of this magnitude are encountered and affect the secondary cell walls of fibers (Fig. 2L) that normally have a greater proportion of S monomers compared with the middle lamellae, cell corners, and vessel cell walls (Saka and Goring, 1985; Donaldson, 2001; Nakashima et al., 2008).
Small but significant amounts of H-derived thioacidolysis monomeric units were observed in the brown wood transformants, in agreement with other data on mutant plants with significant reductions in lignin contents (Coleman et al., 2008; Patten et al., 2010a). Additionally, while the NBO data showed a similar trend in H units released (Fig. 5B), this method resulted in greater amounts of H units released across all events, presumably reflecting the presence of nonextractable flavonoids and other nonlignin components released by NBO. Interestingly, in the brown wood events, the putative “AcBr lignin” contents were very similar to that of the control event (95%–98%; Fig. 5C), thus highlighting the limitations of the “AcBr lignin” method for tissues harboring nonlignin phenolics, as it measures releasable UV light-absorbing substances (Anterola and Lewis, 2002). The MBMS lignin estimates were also comparable to those of the AcBr lignin method (Fig. 5, C and D), suggesting limitations in this analytical technique as well. Interestingly, the S/G ratio was 2.3 in the control event, whereas it ranged from 2.6 to 2.0 in most of the transgenics, except for those containing brown-colored wood, where the range was approximately 1.1 to 2.2 (Table III). For “normal transformants,” the MBMS method generally gave higher S/G ratios than thioacidolysis results, whereas for brown wood events, the S/G ratios were similar among methods (Table III).
Wood Chemical Constituents and Saccharification
By necessity, a decrease in lignin proportion will result in greater proportions of cellulose and/or hemicelluloses within cell walls. Therefore, greater saccharification efficiency (sugar release per unit of biomass) for low-lignin xylem is expected and has been observed in alfalfa (Medicago sativa; Chen and Dixon, 2007). After taking into account brown wood abundance (Table I) and the cellulose contents of brown versus normal colored wood (Table II), we found that cellulose contents increased as expected as lignin contents declined among events (r2 = 0.26, P = 0.05), with linear regression-predicted values ranging from 432 to 486 mg g–1 CWR. Yet, saccharification yields of pretreated poplar wood did not increase despite the decreased lignin. Rather, they differed very little across control and transgenic events, except for the two most severely affected brown wood events, 712 and 350 (Fig. 5E). Both of these events had significantly lower total sugar release as compared with control wood (Fig. 5E). It also should be noted that a constant extent of saccharification (g g–1 biomass) suggests that hydrolysis decreased on a per unit cellulose basis in events with lowered lignin contents. Although somewhat counterintuitive, these data argue that normal wood with lignin removed during pretreatment may provide better structural access for enzymatic degradation of cellulose than transgenic wood with inherently less lignin. Additionally, in the most severely affected brown wood events, extractives may have inhibited enzymatic hydrolysis if they were not fully removed by the steam pretreatment conditions employed for the assays. Both of these potential mechanisms support the view that cellulose accessibility strongly governs cellulase digestibility of lignocellulosic tissue (Jeoh et al., 2007).
CONCLUSION
Very little is known about the ecophysiological effects of the diverse transgenic perturbations to lignin metabolism that have been proposed as means to accelerate the domestication of biofuel crops. We found that 4CL down-regulation of poplars grown in a field environment did not have increased growth rates and displayed important physiological vulnerabilities when lignin contents were strongly reduced. Moreover, the most strongly lignin-reduced events did not yield increases in fermentation efficiency that could benefit biofuel production. These results suggest a need for more extensive field trials, early in scientific development, to guide the efficient and ecologically sound development of transgenic tree varieties for biofuel production.
MATERIALS AND METHODS
Plant Genotypes and Transformation
Hybrid white poplar (Populus tremula × Populus alba ′INRA-France 717-1B4'!) was used for all transformations essentially as described by Filichkin et al. (2006). Three constructs were cotransformed using three Agrobacterium tumefaciens C58 strains. These included an antisense aspen (Populus tremuloides) 4CL1 construct in an antisense orientation and a sense sweetgum (Liquidambar styraciflua) LsCAld5H construct, both driven by the aspen 4CL1 promoter (Li et al., 2003). The third construct was a putative sterility gene, Att35S, that included a barnase gene driven by the poplar LEAFY promoter (Wei et al., 2007). Only transformants with the antisense 4CL1 construct alone, based on PCR analyses of regenerated transgenic plants, were propagated and used further. To ensure that transformation events were independent, a single clone per individual explant was selected for further propagation after confirmation of transgene presence.
Genomic DNA was isolated from young white poplar leaves using a Plant DNAeasy Kit (Qiagen), with approximately 25 to 50 ng of DNA used as a template for PCR. Transgene presence was confirmed using P. tremuloides 4CL1-specific primers (5′-CAGGAATGCTCTGCACTCTG-3′ and 5′-ATGAATCCACAAGAATTCAT-3′) to amplify a 1.6-kb product. The PCR conditions used for 30 cycles were as follows: 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min; the resulting PCR products were separated on a 1% agarose gel and stained with ethidium bromide.
Plant Preparation and Field Trial Establishment
PCR-positive events were propagated in vitro (Filichkin et al., 2006), with 50- to 60-d-old plantlets transferred to soil in small pots (5.7 × 8.3 cm) in a greenhouse. These were grown for 2 months under a 16-h/8-h photoperiod with supplemental lighting (April to May 2005) and then transferred to tubular pots (6.7 × 24.8 cm) for another 2 months (June to July 2005). A total of 14 transgenic events (i.e. independent gene insertions) with 10 to 17 ramets plus 108 nontransformed controls were produced. Plants were then moved to an outdoor, covered shadehouse for 3 months of acclimatization at ambient temperature and photoperiod in Corvallis, Oregon (August to October 2005). Transgenic controls were not employed with this white poplar clone, since the transformation protocol used in our laboratory, greenhouses, and field sites suggests very small and usually undetectable somaclonal variation (Strauss et al., 2004).
The field trial, planted in November 2005 with dormant plants, was conducted just outside Corvallis, Oregon (44.65° N, 123.3° W, 140-m elevation). Mean annual precipitation at the site is 130 cm, with June through September usually being very dry. The frost-free period ranges from 160 to 210 d, with mean maximum and minimum temperatures over the period of 23.2°C and 8.6°C. Soil at the site is a well-drained, silty, clay loam in the top approximately 15 cm that transitions to clay at a depth of approximately 40 cm. All trees were regularly hand watered during the 2006 growing season, and permanent drip irrigation was installed for the 2007 growing season. The planting arrangement was a randomized complete block with 10 to 15 ramets from each of the 14 transgenic events and a control event planted at a square spacing with 3 m between trees. To minimize the influence of competing vegetation, the bare soil surrounding each tree was covered with nursery ground cloth, and a glyphosate herbicide was applied to rows between trees at the beginning of the 2007 growing season.
Estimation of 4CL Expression Levels in Transgenic Events
In May 2007, bark tissues from four or five ramets per event and six control trees were excised with the developing xylem and then individually sampled between internodes 5 and 6. A modified Qiagen RNA extraction protocol was used (Busov et al., 2003), with the resulting RNA samples treated with DNaseI (TURBO DNA-free kit; Ambion, Applied Biosystems). RNA from four ramets for each event was pooled prior to RT-PCR analyses. First-strand synthesis of cDNA from 1 μg of total RNA for each sample using qRT-PCR was next carried out according to SuperScript III First-Strand Synthesis System general guidelines (Invitrogen). Each RT reaction was divided into aliquots and diluted 10 times, with 1 μL used as template for the PCRs.
The expression of the two endogenous 4CL1-1 and 4CL1-2 genes, homologous to those in Populus trichocarpa, was assessed by real-time PCR. BLAST searches against the P. trichocarpa genome (version 1.1; Tuskan et al., 2006) using the 4CL1 gene sequence from P. tremuloides (GenBank accession no. AF041049) yielded two homologs that share 94% DNA sequence similarity and 89% amino acid identity (97% similarity). The P. trichocarpa genome gene model for 4CL1-1 was grail3.0100002702 (Joint Genome Institute annotation Ptr4CL3) and the gene model for 4CL1-2 was fgenesh4_pg.C_LG_III001773 (Joint Genome Institute annotation Ptr4CL5; Shi et al., 2010). The 3′ untranslated regions were verified using total RNA from stem tissues of untransformed control trees (GeneRacer kit; Invitrogen). Based on the sequences obtained, primers specific for each gene were designed for qRT-PCR (for primer sequences, see Supplemental Table S1). The polyubiquitin gene (UBQ14; P. trichocarpa version 1.1. gene model estExt_fgenesh4_pm.C_LG_XI0348) was used as an internal control gene, because it was previously found to have the lowest developmental variance of 10 “housekeeping” genes studied in poplar (Brunner et al., 2004; for primer sequences, see Supplemental Table S1).
Final concentration of the primers was 0.5 μm. Conditions for all PCRs were as follows: 50°C for 2 min, 95°C for 2 min, followed by 40 cycles of 95°C for 30 s, 61°C for 30 s, and 72°C for 30 s. Transcript levels of 4CL1-1 and 4CL1-2 and the housekeeping gene were determined from standard curves of the control sample sequentially diluted five times. The amounts of 4CL1-1 and 4CL1-2 were then divided by the housekeeping reference amounts to obtain normalized expression levels of 4CL1-1 and 4CL1-2 genes.
Tree Growth
Tree heights and basal diameters were measured in November 2005 (planting), 2006, and 2007 for all of the 10 to 15 ramets of each of the 14 transgenic events plus the 32 controls that constituted the initial planting. At the end of 2007, six control trees and three or four trees that spanned the range of tree size for each transgenic event were harvested to determine allometric estimates of oven-dried aboveground biomass. These relationships were used with diameter and height measurements to estimate biomass for each individual at the end of the 2007 growing season. These estimates, after 2 years of growth, were used to compare mean biomass among events.
Brown Wood
Cross-sectional areas of brown wood were estimated near the base of each tree by overlaying a grid of dots on a transparent plastic sheet over three cross-sections from each tree at three heights (stem base and 20 and 40 cm from ground level) and then recording the relative frequency of brown wood as compared with the entire cross-sectional wood area.
Estimated Lignin Contents and Monomeric Compositions
Control and transgenic trees were harvested in November 2007, with the stem of each cut into sections (approximately 30–35 cm each) except for two trees from event 712 (712-7 and 712-14) that had only basal sections 6 and 15 cm long, respectively, with a number of branches growing from it. For each stem, the most basal 4- to 6-cm section (or the entire base for 712-7 and 712-14) was sampled and the bark was removed. Each section was then reduced to small pieces with a chisel, freeze dried, ground in a Waring blender in the presence of liquid N2, and finally ball milled (Fritsch planetary mill) for 2 to 3 h until a homogenous powder was obtained. Extractive-free CWRs were next obtained as described by Patten et al. (2005) with successive extraction with ethanol:toluene (1:1, v/v), ethanol, and water of each powdered sample (approximately 1 g). Lignin contents were estimated by the AcBr method (Iiyama and Wallis, 1988) as modified (Jourdes et al., 2007), whereas lignin monomeric compositions were estimated using thioacidolysis (Rolando et al., 1992; Blee et al., 2001) and NBO (Iiyama and Lam, 1990; Patten et al., 2005)
Lignin Estimations Using MBMS
MBMS is a high-throughput method that uses nonextracted wood flour samples. It is based on pyrolysis mass spectrometry (described below) and after calibration gives both lignin and S/G estimates. Estimated lignin values were corrected to approximate Klason lignin values by using an internal standard developed at the National Renewable Energy Laboratory, where multiple MBMS spectra of National Institute of Standards and Technology standard 8492 (Populus deltoides) were averaged and lignin was estimated by summing the peak corresponding to lignin degradation products (Evans and Milne, 1987). A correction factor was then determined by dividing the Klason lignin value for the National Institute of Standards and Technology 8492 standard by the lignin value determined by MBMS. This correction factor was then applied to the remaining samples.
A custom-built MBMS device using an Extrel model TQMS C50 mass spectrometer was used for pyrolysis vapor analysis (Evans and Milne, 1987; Tuskan et al., 1999). Minor modifications were made to incorporate a commercially available autosampler inlet pyrolysis system (Sykes et al., 2009). The autosampler furnace was electronically maintained at 500°C, and the interface was set to 350°C. The 3.2-mm transfer line was wrapped in heat tape and heated to approximately 350°C measured with thermocouples. Helium gas (2 L min−1) was used to carry the pyrolysis vapors from the pyrolyzer to the mass spectrometer. The residence time of the pyrolysis vapors in the reactor pyrolysis zone has been estimated to be less than 10 ms and is short enough that secondary cracking reactions are minimal.
Stem samples of four to eight trees per event/control line were air dried (not extracted) and milled to 20 mesh using a Wiley minimill. For each sample, approximately 4 mg of biomass was introduced into the quartz pyrolysis reactor via 80-μL deactivated stainless steel Eco-Cups (Frontier Lab). Samples were randomized throughout the experimental run to eliminate bias due to possible spectrometer drift. Discs of glass fiber filter paper (type A/D) cover the top of the sample to prevent sample from coming out of the cup during injection. Mass spectral data from mass-to-charge ratio 30 to 450 were acquired on a Merlin Automation data system (version 2.0) using 22.5-eV electron impact ionization.
Extractive Contents
Extractive contents of oven-dry wood were determined gravimetrically using basal stem wood from four transformants per event and seven control trees. For these analyses, brown and nonbrown woody tissues were separated and ground at the same time using a Dremel tool (Robert Bosch Tool Corp.). Each sample (approximately 1 g, air-dried wood) was then weighed and heat sealed in a polyester filter bag (mesh size of 25 μm; ANKOM Technology). Extractive contents were estimated in two steps. First, the amounts of toluene-ethanol solubles were determined by reweighing oven-dried bags, following soxhlet extraction with toluene:ethanol (3:1, v/v) and ethanol (24 h each). The hot water-soluble extractive amounts were also determined by reweighing oven-dried bags following their immersion in a distilled water bath at 90°C for 2 h, with this procedure repeated twice using fresh distilled water. (Changes in bag mass were calculated by including three blank bags at each step, and this accounted for less than 1% of the bag mass for all steps.)
Characterization of Naringenin, Dihydrokaempferol, and Their Glucosides
For wood extractive component identification, powdered wood samples (approximately 5 mg) from a control tree and event 712 were individually extracted with methanol:water (8:2, v/v; 10 mL) by sonication for 10 min at room temperature. Crude extracts were centrifuged (3,000g) for 10 min, with each supernatant (approximately 8 mL) individually dried under a stream of nitrogen. Each residue was then redissolved in methanol:water (8:2, v/v; 1 mL) and passed through a syringe filter (0.2 μm pore size; Nalgene, Thermo Scientific), with aliquots (1 μL) subjected to HPLC/MS analyses as described below.
Chromatographic analyses were carried out using a Waters Acquity ultra-performance liquid chromatography system, coupled with diode array and mass spectrometric (Thermo Finnigan; atmospheric pressure chemical ionization mode) detection. Separations employed a reverse-phase Acquity BEH column (C18; 50 × 2.1 mm, 1.7-μm particle size; Waters) with a Vanguard precolumn (5 mm × 2.1, 1.7-μm particle size; Waters) at a flow rate of 300 μL min–1 and a solvent system as follows: A (water:acetic acid, 97:3, v/v) and B (CH3CN) in an A:B ratio of 95:5 for 6 min, with linear gradients of A:B 60:40 in 6.5 min, A:B 55:45 in 6 min, and finally A:B 0:100 in 2 min, with the latter held for 2 min. MS data were in agreement with previously published data (Le Gall et al., 2003) and those obtained from authentic standards.
Cellulose Contents
Cellulose contents of the extracted wood samples described above were estimated by reweighing oven-dried bags after lignins and hemicelluloses were removed. The noncellulosic constituents were removed using the same heat-sealed polyester filter bags described above with the sodium chlorite method outlined by Green (1963). Correction for blank bags was again less than 1%.
Saccharification
A portion of basal stem sections (from about 0 to 20 cm height) of four to seven trees per transgenic event and eight control trees were debarked, and the wood was ground into powder. Samples from 10 trees were selected at random to be run twice to test for analytical precision, for a total of 90 trees and 100 samples. Triplicate samples were subjected to high-throughput pretreatment and enzyme hydrolysis (Decker et al., 2009; Selig et al., 2010). Briefly, 5.0 ± 0.3 mg of 20- to 80-mesh wood powder was loaded into a 96-well format pretreatment reactor using a Symyx Powdernium MTM solids-dispensing robot (Symyx Technologies). Weights for each sample were recorded, and sugar release was adjusted to a mass basis. Wells for blanks, enzyme-only, sugar standards, and biomass-only controls were included on each reactor plate. Total volume capacity for each well was 417 μL. Each well was loaded with 300 μL of water as a catalyst, sealed with high-temperature aluminum foil seals, gasketed and clamped into stacks of five to 10 reactors, and heated with steam to 180°C for 40 min in a modified Parr reactor. After rapid cooling with cold water, the reactor plates were centrifuged to remove condensation from the underside of the seals. The seals were pierced, and 40 μL of enzyme in citrate buffer (1.0 m, pH 5.0) was added. The enzyme cocktail was loaded on a mass basis and consisted of Spezyme CP cellulase (Genencor-Danisco) loaded at 70 mg protein g–1 initial biomass and Novo188 β-glucosidase (Novozymes) loaded at 2.5 mg protein g–1 initial biomass. After resealing, the reactor plates were incubated static at 40°C for 72 h. The extent of enzymatic hydrolysis was evaluated by quantitation of Glc and Xyl released during the digestion using enzyme-linked sugar assay kits (Megazyme International). After adding Glc and Xyl standards to standard wells in each reactor, a 1:20 dilution of the digestion mixture was made in water on a separate assay plate. Aliquots from the assay plate were analyzed via the enzyme-linked assays below on a Molecular Devices Spectromax 190 plate reader at the wavelengths indicated.
Glc was measured via a Glc oxidase/peroxidase enzyme-linked assay.
Xyl was quantified using a Xyl dehydrogenase enzyme-linked assay.
Microscopy
For light microscopy, thin hand sections were stained with safranin and astra-blue following Jourez et al. (2001). Images were captured with a digital CCD camera (Q Imaging; Micropublisher 5.0 RTV) interfaced with a bright-field light microscope (Nikon E400).
To visualize phenolics, a branch characterized by extensive brown wood formation was selected and flash frozen in liquid nitrogen. The cryofixed branch was cut into 2- to 3-cm-long segments at –12°C in a walk-in freezer. Then, the segments were planed in a frozen state (–10°C to –30°C) on a sliding microtome and freeze dried. Transverse and longitudinal planed surfaces were observed with a confocal microscope (LSM 510; Carl Zeiss) using single-track, triple-channel imaging with 405-, 488-, and 543-nm laser lines and emission filters (BP 420–480, BP 530–600, and BP 604–625). Phenolic depositions had strong blue and green autofluorescence, while the autofluorescence of unstained cell walls in the blue and green spectrum was not visualized in the confocal images because of its considerably lower intensity in comparison with the phenolics. The autofluorescence of cell walls was imaged in the red spectrum (604–625).
Statistical Analyses
Least-squares regression methods were used to assess relationships between tree form and size. To compare trait values among the control event and transgenic events, we conducted ANOVA tests. Traits were first compared with a global ANOVA (PROC GLM, SAS version 9.2; SAS Institute). Further analyses compared means among the control event and transgenic events with Tukey’s honestly significant difference tests to control for type 1 experiment-wise error.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AF041049.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expanded confocal image from Figure 2Q of collapsed vasculature, extractives, and tension wood.
Supplemental Figure S2. Axial and cross-sectional distribution patterns of brown wood.
Supplemental Figure S3. Examples of brown wood occurrence in stem and branch-stem junctions in event 210.
Supplemental Table S1. Primer sequences used for qRT-PCR.
Supplementary Material
Acknowledgments
We thank Cathleen Ma and Elizabeth Etherington for their roles in propagating the trees and managing the field trial. We thank Dr. Joe Chappell and two anonymous reviewers for their helpful comments on the manuscript. We are also indebted to the laboratory of Dr. Vincent Chiang for the gene construct and to Val Cleland and Kristen Falk for their help in data collection.
References
- Anterola AM, Lewis NG. (2002) Trends in lignin modification: a comprehensive analysis of the effects of genetic manipulations/mutations on lignification and vascular integrity. Phytochemistry 61: 221–294 [DOI] [PubMed] [Google Scholar]
- Baucher M, Chabbert B, Pilate G, Van Doorsselaere J, Tollier MT, Petit-Conil M, Cornu D, Monties B, Van Montagu M, Inzé D, et al. (1996) Red xylem and higher lignin extractability by down-regulating a cinnamyl alcohol dehydrogenase in poplar. Plant Physiol 112: 1479–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blee K, Choi JW, O’Connell AP, Jupe SC, Schuch W, Lewis NG, Bolwell GP. (2001) Antisense and sense expression of cDNA coding for CYP73A15, a class II cinnamate-4-hydroxylase, leads to a delayed and reduced production of lignin in tobacco. Phytochemistry 57: 1159–1166 [DOI] [PubMed] [Google Scholar]
- Boyce CK, Zwieniecki MA, Cody GD, Jacobsen C, Wirick S, Knoll AH, Holbrook NM. (2004) Evolution of xylem lignification and hydrogel transport regulation. Proc Natl Acad Sci USA 101: 17555–17558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner AM, Yakovlev IA, Strauss SH. (2004) Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol 18: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busov VB, Meilan R, Pearce DW, Ma C, Rood SB, Strauss SH. (2003) Activation tagging of a dominant gibberellin catabolism gene (GA 2-oxidase) from poplar that regulates tree stature. Plant Physiol 132: 1283–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Dixon RA. (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol 25: 759–761 [DOI] [PubMed] [Google Scholar]
- Coleman HD, Samuels AL, Guy RD, Mansfield SD. (2008) Perturbed lignification impacts tree growth in hybrid poplar: a function of sink strength, vascular integrity, and photosynthetic assimilation. Plant Physiol 148: 1229–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa MA, Bedgar DL, Moinuddin SGA, Kim KW, Cardenas CL, Cochrane FC, Shockey JM, Helms GL, Amakura Y, Takahashi H, et al. (2005) Characterization in vitro and in vivo of the putative multigene 4-coumarate CoA ligase network in Arabidopsis: syringyl lignin and sinapate/sinapyl alcohol derivative formation. Phytochemistry 66: 2072–2091 [DOI] [PubMed] [Google Scholar]
- Coutand C, Martin L, Leblanc-Fournier N, Decourteix M, Julien JL, Moulia B. (2009) Strain mechanosensing quantitatively controls diameter growth and PtaZFP2 gene expression in poplar. Plant Physiol 151: 223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davin LB, Jourdes M, Patten AM, Kim KW, Vassão DG, Lewis NG. (2008a) Dissection of lignin macromolecular configuration and assembly: comparison to related biochemical processes in allyl/propenyl phenol and lignan biosynthesis. Nat Prod Rep 25: 1015–1090 [DOI] [PubMed] [Google Scholar]
- Davin LB, Patten AM, Jourdes M, Lewis NG. (2008b) Lignins: a twenty first century challenge. Himmel ME, , Biomass Recalcitrance: Deconstructing the Plant Cell Wall for Bioenergy. Blackwell Publishing, Oxford, pp 213–305 [Google Scholar]
- Decker SR, Brunecky R, Tucker MP, Himmel ME, Selig MJ. (2009) High-throughput screening techniques for biomass conversion. Bioenerg Res 2: 179–192 [Google Scholar]
- Dixon RA, Paiva NL. (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7: 1085–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson LA. (2001) Lignification and lignin topochemistry: an ultrastructural view. Phytochemistry 57: 859–873 [DOI] [PubMed] [Google Scholar]
- Evans RJ, Milne TA. (1987) Molecular characterization of the pyrolysis of biomass. Energy Fuels 1: 123–137 [Google Scholar]
- Fernandez MP, Watson PA, Breuil C. (2001) Gas chromatography-mass spectrometry method for the simultaneous determination of wood extractive compounds in quaking aspen. J Chromatogr A 922: 225–233 [DOI] [PubMed] [Google Scholar]
- Filichkin SA, Meilan R, Busov VB, Ma C, Brunner AM, Strauss SH. (2006) Alcohol-inducible gene expression in transgenic Populus. Plant Cell Rep 25: 660–667 [DOI] [PubMed] [Google Scholar]
- Foust TD, Ibsen KN, Dayton DC, Hess RJ, Kenney KE. (2008) The biorefinery. Himmel ME, , Biomass Recalcitrance: Deconstructing the Plant Cell Wall for Bioenergy. Blackwell Publishing, Oxford, pp 7–37 [Google Scholar]
- Gang DR, Fujita M, Davin LB, Lewis NG. (1998) The “abnormal lignins”: mapping heartwood formation through the lignan biosynthetic pathway. Lewis NG, Sarkanen S, , Lignin and Lignan Biosynthesis. ACS Symposium Series, Vol 697 American Chemical Society, Washington, DC, pp 389–421 [Google Scholar]
- Gindl W, Teischinger A. (2002) Axial compression strength of Norway spruce related to structural variability and lignin content. Composites Part A 33: 1623–1628 [Google Scholar]
- Goswami L, Dunlop JW, Jungnikl K, Eder M, Gierlinger N, Coutand C, Jeronimidis G, Fratzl P, Burgert I. (2008) Stress generation in the tension wood of poplar is based on the lateral swelling power of the G-layer. Plant J 56: 531–538 [DOI] [PubMed] [Google Scholar]
- Green JW. (1963) Wood cellulose. Methods Carbohydr Chem 3: 9–21 [Google Scholar]
- Hancock JE, Bradley KL, Giardina CP, Pregitzer KS. (2008) The influence of soil type and altered lignin biosynthesis on the growth and above and belowground biomass allocation of Populus tremuloides. Plant Soil 308: 239–253 [Google Scholar]
- Hancock JE, Loya WM, Giardina CP, Li L, Chiang VL, Pregitzer KS. (2007) Plant growth, biomass partitioning and soil carbon formation in response to altered lignin biosynthesis in Populus tremuloides. New Phytol 173: 732–742 [DOI] [PubMed] [Google Scholar]
- Hu WJ, Harding SA, Lung J, Popko JL, Ralph J, Stokke DD, Tsai CJ, Chiang VL. (1999) Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat Biotechnol 17: 808–812 [DOI] [PubMed] [Google Scholar]
- Huntley SK, Ellis D, Gilbert M, Chapple C, Mansfield SD. (2003) Significant increases in pulping efficiency in C4H-F5H-transformed poplars: improved chemical savings and reduced environmental toxins. J Agric Food Chem 51: 6178–6183 [DOI] [PubMed] [Google Scholar]
- Iiyama K, Lam TBT. (1990) Lignin in wheat internodes. Part 1. The reactivities of lignin units during alkaline nitrobenzene oxidation. J Sci Food Agric 51: 481–491 [Google Scholar]
- Iiyama K, Wallis AFA. (1988) An improved acetyl bromide procedure for determining lignin in woods and wood pulps. Wood Sci Technol 22: 271–280 [Google Scholar]
- Jeoh T, Ishizawa CI, Davis MF, Himmel ME, Adney WS, Johnson DK. (2007) Cellulase digestibility or pretreated biomass is limited by cellulose accessibility. Biotechnol Bioeng 98: 112–122 [DOI] [PubMed] [Google Scholar]
- Jia C, Zhao H, Wang H, Xing Z, Du K, Song Y, Wei J. (2004) Obtaining the transgenic poplars with low lignin content through down-regulation of 4CL. Chin Sci Bull 49: 905–909 [Google Scholar]
- Jørgensen H, Kristensen JB, Felby C. (2007) Enzymatic conversion of lignocellulose into fermentable sugars: challenges and opportunities. Biofuel Bioprod Bioref 1: 119–134 [Google Scholar]
- Jourdes M, Cardenas CL, Laskar DD, Moinuddin SGA, Davin LB, Lewis NG. (2007) Plant cell walls are enfeebled when attempting to preserve native lignin configuration with poly-p-hydroxycinnamaldehydes: evolutionary implications. Phytochemistry 68: 1932–1956 [DOI] [PubMed] [Google Scholar]
- Jourez B, Riboux A, Leclercq A. (2001) Anatomical characteristics of tension wood and opposite wood in young inclined stems of poplar (Populus euramericana cv. ‘Ghoy’). IAWA J 22: 133–157 [Google Scholar]
- Kajita S, Katayama Y, Omori S. (1996) Alterations in the biosynthesis of lignin in transgenic plants with chimeric genes for 4-coumarate:coenzyme A ligase. Plant Cell Physiol 37: 957–965 [DOI] [PubMed] [Google Scholar]
- Kirst M, Myburg AA, De León JPG, Kirst ME, Scott J, Sederoff R. (2004) Coordinated genetic regulation of growth and lignin revealed by quantitative trait locus analysis of cDNA microarray data in an interspecific backcross of Eucalyptus. Plant Physiol 135: 2368–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitin P, Voelker SL, Meinzer FC, Beeckman H, Strauss SH, Lachenbruch B. (2010) Tyloses and phenolic deposits in xylem vessels impede water transport in low-lignin transgenic poplars: a study by cryo-fluorescence microscopy. Plant Physiol 154: 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre C, Pollet B, Petit-Conil M, Toval G, Romero J, Pilate G, Leplé JC, Boerjan W, Ferret V, De Nadai V, et al. (1999) Structural alterations of lignins in transgenic poplars with depressed cinnamyl alcohol dehydrogenase or caffeic acid O-methyltransferase activity have an opposite impact on the efficiency of industrial kraft pulping. Plant Physiol 119: 153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Meyer K, Chapple C, Douglas CJ. (1997) Antisense suppression of 4-coumarate:coenzyme A ligase activity in Arabidopsis leads to altered lignin subunit composition. Plant Cell 9: 1985–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gall G, DuPont MS, Mellon FA, Davis AL, Collins GJ, Verhoeyen ME, Colquhoun IJ. (2003) Characterization and content of flavonoid glycosides in genetically modified tomato (Lycopersicon esculentum) fruits. J Agric Food Chem 51: 2438–2446 [DOI] [PubMed] [Google Scholar]
- Leplé JC, Dauwe R, Morreel K, Storme V, Lapierre C, Pollet B, Naumann A, Kang KY, Kim H, Ruel K, et al. (2007) Downregulation of cinnamoyl-coenzyme A reductase in poplar: multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure. Plant Cell 19: 3669–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhou Y, Cheng X, Sun J, Marita JM, Ralph J, Chiang VL. (2003) Combinatorial modification of multiple lignin traits in trees through multigene cotransformation. Proc Natl Acad Sci USA 100: 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Bonawitz D, Weng JK, Chapple C. (2010) The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. Plant Cell 22: 1620–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Weng JK, Chapple C. (2008) Improvement of biomass through lignin modification. Plant J 54: 569–581 [DOI] [PubMed] [Google Scholar]
- Mark R. (1967) Cell Wall Mechanics of Tracheids. Yale University Press, New Haven, CT [Google Scholar]
- Meyermans H, Morreel K, Lapierre C, Pollet B, De Bruyn A, Busson R, Herdewijn P, Devreese B, Van Beeumen J, Marita JM, et al. (2000) Modifications in lignin and accumulation of phenolic glucosides in poplar xylem upon down-regulation of caffeoyl-coenzyme A O-methyltransferase, an enzyme involved in lignin biosynthesis. J Biol Chem 275: 36899–36909 [DOI] [PubMed] [Google Scholar]
- Miller JE, Geadleman JL, Marten GC. (1983) Effect of the brown midrib-allele on maize silage quality and yield. Crop Sci 23: 493–496 [Google Scholar]
- Nakashima J, Chen F, Jackson L, Shadle G, Dixon RA. (2008) Multi-site genetic modification of monolignol biosynthesis in alfalfa (Medicago sativa): effects on lignin composition in specific cell types. New Phytol 179: 738–750 [DOI] [PubMed] [Google Scholar]
- Niklas KJ. (1992) Plant Biomechanics. University of Chicago Press, Chicago [Google Scholar]
- Patten AM, Cardenas CL, Cochrane FC, Laskar DD, Bedgar DL, Davin LB, Lewis NG. (2005) Reassessment of effects on lignification and vascular development in the irx4 Arabidopsis mutant. Phytochemistry 66: 2092–2107 [DOI] [PubMed] [Google Scholar]
- Patten AM, Jourdes M, Cardenas CL, Laskar DD, Nakazawa Y, Chung BY, Franceschi VR, Davin LB, Lewis NG. (2010a) Probing native lignin macromolecular configuration in A. thaliana in specific cell wall types: further insights into limited substrate degeneracy and assembly of the lignins of ref8, fah 1-2 and C4H:F5H lines. Mol Biosyst 6: 499–515 [DOI] [PubMed] [Google Scholar]
- Patten AM, Wolcott MP, Davin LB, Lewis NG. (2010b) Trees: a remarkable bounty. Verpoorte R, , Comprehensive Natural Products Chemistry II, Vol 3. Discovery, Development and Modification of Bioactivity. Elsevier, Oxford, pp 1173–1296 [Google Scholar]
- Pietarinen SP, Willför SM, Vikström FA, Holmbom BR. (2006) Aspen knots, a rich source of flavonoids. J Wood Chem Technol 26: 245–258 [Google Scholar]
- Pilate G, Guiney E, Holt K, Petit-Conil M, Lapierre C, Leplé JC, Pollet B, Mila I, Webster EA, Marstorp HG, et al. (2002) Field and pulping performances of transgenic trees with altered lignification. Nat Biotechnol 20: 607–612 [DOI] [PubMed] [Google Scholar]
- Porter KS, Axtell JD, Lechtenberg VL, Colenbrander VF. (1978) Phenotype, fiber composition, and in vitro dry matter disappearance of chemically induced brown midrib (bmr) mutants of sorghum. Crop Sci 18: 205–208 [Google Scholar]
- Ralph J, MacKay JJ, Hatfield RD, O’Malley DM, Whetten RW, Sederoff RR. (1997) Abnormal lignin in a loblolly pine mutant. Science 277: 235–239 [DOI] [PubMed] [Google Scholar]
- Raven JA. (1977) The evolution of vascular land plants in relation to supracellular transport processes. Adv Bot Res 5: 153–219 [Google Scholar]
- Rolando C, Monties B, Lapierre C. (1992) Thioacidolysis. Lin SY, Dence CW, , Methods in Lignin Chemistry. Springer-Verlag, Berlin, pp 334–349 [Google Scholar]
- Saka S, Goring DAI. (1985) Localization of lignins in wood cell walls. Higuchi T, , Biosynthesis and Biodegradation of Wood Components. Academic Press, Orlando, FL, pp 51–62 [Google Scholar]
- Schubert C. (2006) Can biofuels finally take center stage? Nat Biotechnol 24: 777–784 [DOI] [PubMed] [Google Scholar]
- Selig MJ, Tucker MP, Sykes RW, Reichel KL, Brunecky R, Himmel ME, Davis MF, Decker SR. (2010) Lignocellulose recalcitrance screening by integrated high-throughput hydrothermal pretreatment and enzymatic saccharification. Ind Biotechnol 6: 104–111 [Google Scholar]
- Shi R, Sun YH, Li Q, Heber S, Sederoff R, Chiang VL. (2010) Towards a systems approach for lignin biosynthesis in Populus trichocarpa: transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol 51: 144–163 [DOI] [PubMed] [Google Scholar]
- Strauss SH, Brunner AM, Busov VB, Ma C, Meilan R. (2004) Ten lessons from 15 years of transgenic Populus research. Forestry 77: 455–465 [Google Scholar]
- Sykes R, Yung M, Novaes E, Kirst M, Peter G, Davis M. (2009) High-throughput screening of plant cell-wall composition using pyrolysis molecular beam mass spectroscopy. Mielenz JR, , Biofuels: Methods and Protocols. Humana Press, New York, pp 169–183 [DOI] [PubMed] [Google Scholar]
- Taylor AM, Gartner BL, Morrell JJ. (2002) Heartwood formation and natural durability: a review. Wood Fiber Sci 34: 587–611 [Google Scholar]
- Tsai CJ, Harding SA, Tschaplinski TJ, Lindroth RL, Yuan Y. (2006) Genome-wide analysis of the structural genes regulating defense phenylpropanoid metabolism in Populus. New Phytol 172: 47–62 [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Popko JL, Mielke MR, Hu WJ, Podila GK, Chiang VL. (1998) Suppression of O-methyltransferase gene by homologous sense transgene in quaking aspen causes red-brown wood phenotypes. Plant Physiol 117: 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan G, West D, Bradshaw HD, Neale D, Sewell M, Wheeler N, Megraw B, Jech K, Wiselogel A, Evans R, et al. (1999) Two high-throughput techniques for determining wood properties as part of a molecular genetics analysis of hybrid poplar and loblolly pine. Appl Biochem Biotechnol 77–79: 55–65 [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Voelker SL. (2009) Functional decreases in hydraulic and mechanical properties of field-grown transgenic poplar trees caused by modification of the lignin synthesis pathway through downregulation of the 4-coumarate:coenzyme A ligase gene. PhD thesis Oregon State University, Corvallis [Google Scholar]
- Wagner A, Donaldson L, Kim H, Phillips L, Flint H, Steward D, Torr K, Koch G, Schmitt U, Ralph J. (2009) Suppression of 4-coumarate-CoA ligase in the coniferous gymnosperm Pinus radiata. Plant Physiol 149: 370–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Meilan R, Brunner AM, Skinner JS, Ma C, Gandhi HT, Strauss SH. (2007) Field trial detects incomplete barstar attenuation of vegetative cytotoxicity in Populus trees containing a poplar LEAFY promoter:barnase sterility transgene. Mol Breed 19: 69–85 [Google Scholar]
- Yang B, Wyman CE. (2008) Pretreatment: the key to unlocking low-cost cellulosic ethanol. Biofuel Bioprod Biorefin 2: 26–40 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.