Abstract
Arabinogalactan-proteins (AGPs) are highly glycosylated hydroxyproline (Hyp)-rich glycoproteins that are frequently characterized by the presence of [Alanine-Hyp] ([AO]) repetitive units. AGP galactosyltransferase (GalT) activities in tobacco (Nicotiana tabacum) and Arabidopsis (Arabidopsis thaliana) microsomal membranes were studied here with an in vitro GalT reaction system, which used acceptor substrates composed of [AO] repetitive units, specifically, a chemically synthesized [AO]7 acceptor and a transgenically produced and deglycosylated d[AO]51 acceptor. Incorporation of [14C]Gal from UDP-[14C]Gal into the [AO]7 and d[AO]51 acceptors was observed following HPLC fractionation of the reaction products. Hyp-[14C]Gal monosaccharide and Hyp-[14C]Gal disaccharide were identified in the base hydrolysates of the GalT reaction products, indicating the presence of two distinct GalT activities for the addition of the first and second Gal residues to the [AO] peptide in both tobacco and Arabidopsis. Examination of the Arabidopsis Hyp:GalT activity using various acceptor substrates, including two extensin sequences containing SO4 modules and a [AP]7 peptide, indicated this activity was specific for peptidyl Hyp in AGP sequences. Mass spectrometry analysis demonstrated that only one Gal was added per peptide molecule to the C-terminal or penultimate Hyp residue of the [AO]7 peptide. In addition, [AO]7:GalT and d[AO]51:GalT activities were localized to the endomembrane system of Arabidopsis suspension-cultured cells following sucrose density gradient centrifugation. The in vitro assay reported here to detect GalT activities using AGP peptide and glycopeptide acceptor substrates provides a useful tool for the identification and verification of AGP-specific GalT proteins/genes and an entry point for elucidation of arabinogalactan biosynthesis for AGPs.
Arabinogalactan-proteins (AGPs) are highly glycosylated Hyp-rich glycoproteins (HRGPs) implicated in many physiological processes, including plant somatic embryogenesis, programmed cell death, wound responses, and hormone signaling pathways (Seifert and Roberts, 2007; Ellis et al., 2010). AGPs are defined by the presence of arabinogalactan (AG) polysaccharides and reside mainly at the plasma membrane-cell wall interface and in plant exudates (Langan and Nothnagel, 1997; Oxley and Bacic, 1999; Sherrier et al., 1999; Svetek et al., 1999; Lamport et al., 2006). These AG polysaccharides are added via O-glycosylation, which is widespread in plants and includes monogalactosylation of Ser residues and extensive modification of Hyp, which ranges from addition of oligoarabinosides to AG polysaccharide addition. The Hyp contiguity hypothesis predicts contiguous Hyp residues as sites of HRGP arabinosylation, while clustered noncontiguous Hyp residues are sites of galactosylation that give rise to AG polysaccharides on AGPs (Kieliszewski and Lamport, 1994; Kieliszewski et al., 1995; Tan et al., 2003). Tests of the hypothesis in gum arabic glycoprotein (Goodrum et al., 2000) and tobacco (Nicotiana tabacum; Shpak et al., 1999, 2001; Zhao et al., 2002; Tan et al., 2003; Held et al., 2004) using naturally occurring AGPs and synthetic genes encoding only clustered noncontiguous Hyp or contiguous Hyp confirmed that AG polysaccharide was added only to clustered, noncontiguous Hyp, while arabinosylation (oligosaccharides composed of approximately four Ara residues) occurred on contiguous Hyp blocks. The structure of a well-characterized Hyp-AG isolated from tobacco was recently elucidated (Tan et al., 2004; Fig. 1).
Figure 1.
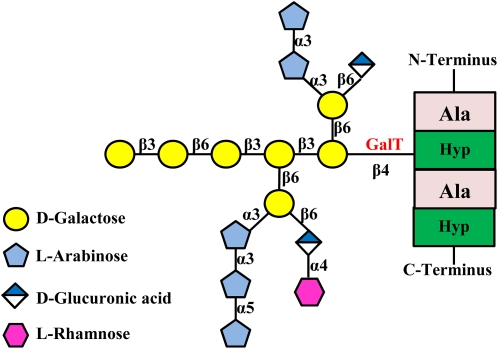
AG structure of an AGP molecule. The protein backbone containing a clustered noncontiguous Hyp motif is shown. Although the two Hyp residues in the protein backbone are both glycosylated with AG when expressed in tobacco cells, only one AG side chain is shown here for simplicity. The focus of this study is the GalT enzyme (shown in red) that adds the first Gal residue onto the AGP peptide backbone. Monosaccharide symbols used here are based on the Symbol and Text Nomenclature for Representation of Glycan Structure as proposed by the Consortium for Functional Glycomics (http://glycomics.scripps.edu/CFGnomenclature.pdf). Modified from Tan et al. (2004).
Based on the structure of this AG polysaccharide and given the specificity of glycosyltransferases (GTs), there may be as many as 15 transferase activities involved in the synthesis of this AG polysaccharide, namely, one peptidyl Hyp-β-galactosyltransferase, one α-(1,5)arabinosyltransferase, possibly four α-(1,3)arabinosyltransferases, three β-(1,3)galactosyltransferases, three β-(1,6)galactosyltransferases that add the three branch sites on the AG core, two β-(1,6)glucuronyltransferases, and one α-(1,4)rhamnosyltransferase. In other species, additional transferases are possible, as in Arabidopsis (Arabidopsis thaliana), where α-(1,2)fucosyltransferase is involved in AGP glycosylation (van Hengel and Roberts, 2002; Wu et al., 2010).
Despite the fact that plant cell walls contain substantial amounts of AGPs, there is an embarrassing lack of knowledge on the enzymology of AGP (and HRGP) biosynthesis (Ellis et al., 2010). Karr (1972) partially characterized a microsomal fraction that arabinosylated extensin peptides, and Bolwell (1986) suggested a lipid-linked intermediate might be involved in HRGP glycosylation. In addition, galactosyltransferase (GalT) activity associated with AGPs was identified in ryegrass (Lolium multiflorum; Mascara and Fincher, 1982) and pea (Pisum sativum; Hayashi and Maclachlan, 1984) membrane preparations; moreover, this activity in ryegrass was restricted to Golgi-derived membranes (Schibeci et al., 1984). Others identified an α-fucosyltransferase activity (Misawa et al., 1996) and a β-glucuronyltransferase activity (Endo et al., 2003) that might be involved in biosynthesis of radish (Raphanus sativus) root AGPs. The major reason for this lack of research progress is the difficulty in making appropriate acceptor substrates for these enzymes.
Very recently, however, some progress was made. A bioinformatics study looking for Arabidopsis homologs to mammalian β-(1,3)GalTs identified 20 putative Arabidopsis β-(1,3)GalTs (Qu et al., 2008), one of which was previously identified as a β-(1,3)GalT involved in the biosynthesis of protein-bound N-linked oligosaccharide (N-glycan; Strasser et al., 2007). Additionally, Oka et al. (2010) used an in vitro assay system to detect and localize Hyp:GalT activity in the endoplasmic reticulum (ER) of Arabidopsis using a chemically synthesized AGP peptide and variants thereof conjugated to fluorescein isothiocyanate via a γ-aminobutyric acid as acceptor substrates. Finally, Wu et al. (2010) identified and characterized two α-(1,2)fucosyltransferases encoded by AtFUT4 and AtFUT6 in Arabidopsis that are specific to AGPs.
With this as a background, we report here on the identification and characterization of in vitro GalT activities that are likely involved in the initial steps of AGP glycosylation in both tobacco and Arabidopsis. This work was achieved by developing an in vitro AGP GalT assay using synthetic and transgenically produced AGP peptides as acceptor substrates, UDP-[14C]Gal as the sugar donor, and permeabilized microsomal membranes from tobacco BY2 and Arabidopsis suspension-cultured cells.
RESULTS
Development of a GalT Assay System with AGP-Like Peptides as Acceptor Substrates
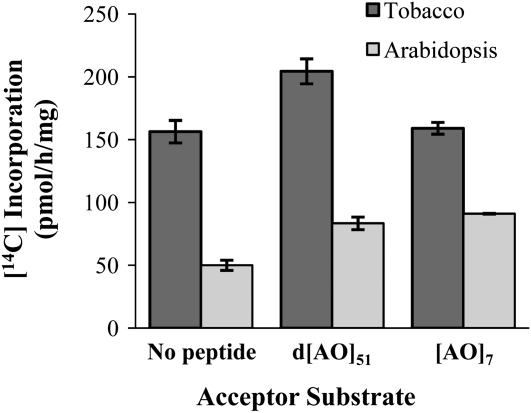
An in vitro GalT assay system was developed with UDP-[14C]Gal as the sugar donor and permeablized microsomal membranes from tobacco or Arabidopsis suspension-cultured cells as the enzyme resource. Two peptides with repetitive [AO] units, representing clustered noncontiguous Hyp motifs of AGPs, were used as acceptor substrates. One of the peptides, denoted as d[AO]51, contained an [AO] motif repeated 51 times. The d[AO]51 peptide was obtained from the [AO]51-EGFP (for enhanced GFP) fusion protein expressed in transgenic tobacco BY-2 cells; the EGFP tag was removed by trypsin digestion, and the AG side chains were removed by deglycosylation with hydrogen fluoride. The other peptide, namely [AO]7, was chemically synthesized and contained seven [AO] repeats. A control reaction was set up with the peptide acceptors excluded. After the reaction, unreacted UDP-[14C]Gal was removed with an ion-exchange resin column, and radioactivity in the reaction solution was counted. As shown in Figure 2, when tobacco permeabilized microsomal membranes were used as the enzyme resource, radioactivity detected in the d[AO]51:GalT reactions was higher than the control reaction as well as the [AO]7:GalT reactions, which displayed a similar level of radioactivity as the control reactions. Compared to the tobacco reactions, the Arabidopsis reactions showed much lower background levels and high, roughly equivalent levels of incorporation into both d[AO]51 and [AO]7.
Figure 2.
Total [14C]radiolabel incorporation into the GalT reaction product in the absence or presence of [AO]7 or d[AO]51 acceptor substrate. Permeabilized microsomal membranes from tobacco and Arabidopsis suspension-cultured cells served as the enzyme source. Reactions were done in triplicate, and mean values are presented.
Characterization of the GalT Assay Products by Reverse-Phase HPLC Analysis
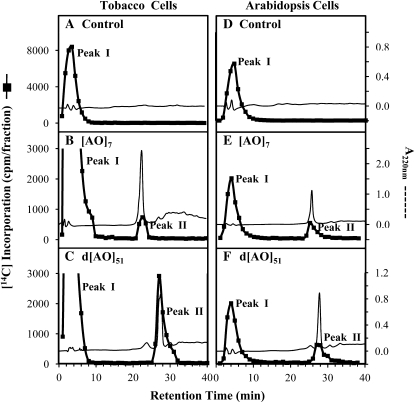
To confirm that [14C]radiolabel was transferred to the acceptors, reaction products were first fractionated by reverse-phase HPLC (RP-HPLC). RP-HPLC fractionation of the tobacco and Arabidopsis control reaction products resolved a single radioactive peak with a low retention time (Peak I in Fig. 3, A and D). Peak I was also detected in the experimental reactions with [AO]7 and d[AO]51 acceptor substrates (Fig. 3, B, C, E, and F). The identity of the components in this [14C]labeled fraction is unknown. The small A220 peaks appearing at the corresponding retention time were solvent peaks and did not represent protein components. It is possible that this fraction is composed of oligosaccharides with [14C]Gal incorporated into endogenous sugar acceptors during the GalT assay. Alternatively, the fraction may represent free [14C]Gal residues released during the assay by an endogenous galactosidase (Kato et al., 2003).
Figure 3.
RP-HPLC fractionation of the [AO]7:GalT and d[AO]51:GalT reaction products on a PRP-1 reverse-phase column. Radioactive Peak II coeluted with the [AO]7 or d[AO]51 acceptor substrate and was used for subsequent product analysis and in monitoring enzyme activity.
Another radioactive product peak coeluting with the [AO]7 and d[AO]51 acceptor substrates appeared only in the experimental reactions (Peak II in Fig. 3, B, C, E, and F). Control reactions, lacking the peptide acceptors, contained no such radioactive product peak. RP-HPLC fractionation provided evidence for incorporation of the [14C]label from UDP-[14C]Gal into the [AO]7 and d[AO]51 peptide acceptors in both the tobacco and Arabidopsis assay systems, including the [AO]7:GalT reaction in tobacco, which showed no substantial increase in radioactivity in the crude product analysis (Fig. 2).
To confirm that the [14C]radiolabel remained associated with Gal, RP-HPLC fractions containing the [14C]labeled GalT reaction product coeluting with the d[AO]51 peptide acceptor in the tobacco assay were pooled and subjected to total acid hydrolysis, which degraded the product into free amino acids and monosaccharides. The resulting [14C]labeled monosaccharide eluted as Gal following high-performance anion-exchange chromatography on a CarboPac PA-10 column (Dionex; Supplemental Fig. S1A). Similarly, [14C]labeled monosaccharide released from the Arabidopsis [AO]7:GalT reaction product eluted as Gal (Supplemental Fig. S1B). Such analysis confirmed that [14C]Gal was incorporated into the d[AO]51 and [AO]7 acceptors without any significant conversion into other monosaccharides.
Extent of AGP Peptide Galactosylation
To investigate how many Gal residues were incorporated into the reaction products, RP-HPLC fractions containing the [14C]labeled GalT reaction products coeluting with the peptide acceptors were pooled and subjected to base hydrolysis. Base hydrolysis degrades peptide bonds but keeps Hyp-glycosidic bonds intact (Kieliszewski and Shpak, 2001). The molecular sizes of the resulting [14C]labeled base hydrolysates were analyzed by gel filtration chromatography on a Bio-gel P2 column (Fig. 4). The rationale here was the extent of Gal addition would be reflected in the sizes of the resulting Hyp-glycosides. As shown in Supplemental Figure S2, A and B, Hyp generated from base hydrolysis of the [AO]7 peptide eluted as a degree of polymerization (DP) 3 size on the P2 column, which was identical to the elution profile of commercially available free Hyp amino acid.
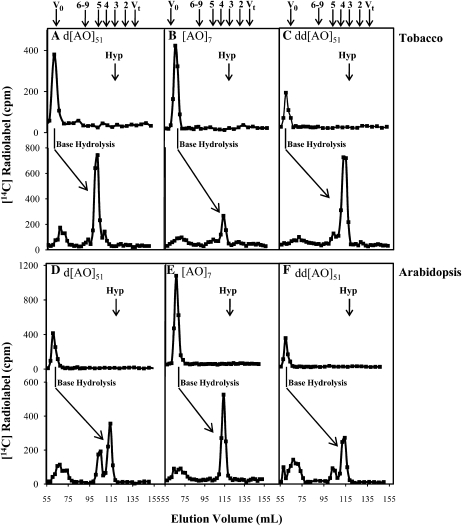
Figure 4.
Bio-gel P2 fractionation of the RP-HPLC purified GalT reaction product. Elution profiles of the d[AO]51:GalT reaction product before and after base hydrolysis are shown for tobacco reactions (A) and Arabidopsis reactions (D); elution profiles of the [AO]7:GalT reaction product before and after base hydrolysis are shown for tobacco reactions (B) and Arabidopsis reactions (E); elution profiles of the dd[AO]51:GalT reaction product before and after base hydrolysis are shown for tobacco reactions (C) and Arabidopsis reactions (F); the elution position of free Hyp amino acid (corresponding to DP3) is shown with an arrow in each panel. The column was calibrated with high-Mr dextran (V0), Gal (Vt), xylo-oligosaccharides with DP2 to 5, and xyloglucan-oligosaccharides with DP6 to 9; their elution positions are indicated with arrows at the top of the figure.
Before base hydrolysis treatment, intact [14C]labeled d[AO]51 and [AO]7 peptides eluted at the void volume (V0) of the P2 column, as shown in the top portion of each panel in Figure 4. When tobacco permeabilized microsomal membranes were used as the enzyme source, the base hydrolysate of the d[AO]51 reaction product mainly eluted as DP5 (Fig. 4A), while the major base hydrolysate released from [AO]7 reaction product eluted as DP4 (Fig. 4B). Given that Hyp eluted as DP3 (Supplemental Fig. S2) under the same conditions, the results indicated that Hyp residues in [AO]7 were glycosylated with one Gal residue, while Hyp residues in d[AO]51 had glycosyl side chains corresponding to two Gal residues. Note that Hyp, but not Ala residues, are known sites of glycosylation for AGPs.
In the case of the d[AO]51 acceptor, it was possible that two monosaccharide sugars were added to one Hyp residue in the GalT assay. However, it could not be excluded that one Gal residue might preexist on the Hyp residues of d[AO]51 because of incomplete deglycosylation during the preparation of this acceptor substrate. To test this possibility, double deglycosylated d[AO]51 (dd[AO]51) was produced by subjecting d[AO]51 to a second hydrogen fluoride deglycosylation reaction and used as an acceptor in the GalT assay. The base hydrolysate generated from the tobacco dd[AO]51 reaction products eluted as DP4 on the Bio-gel P2 column (Fig. 4C), indicating not only that d[AO]51 was incompletely deglycoslyated, but also providing evidence for the presence of two GalT activities in tobacco microsomal membranes. One activity, a Hyp:GalT activity, transfers the initial Gal onto Hyp residues in dd[AO]51 and [AO]7, while the second activity, a likely Gal:GalT activity, transfers the next Gal residue onto galactosylated Hyp in the incompletely deglycosylated d[AO]51.
When Arabidopsis permeablized microsomal membranes were used as the enzyme source, a similar picture emerged. Here, the base hydrolysate from the [AO]7:GalT reaction product eluted as DP4 on the Bio-gel P2 column (Fig. 4E). Moreover, the base hydrolysate of the dd[AO]51:GalT product eluted mainly as DP4 with a small amount of DP5, whereas the base hydrolysate of the d[AO]51:GalT product eluted with considerably more DP5 relative to DP4 (Fig. 4, D and F). The result indicated that Arabidopsis, like tobacco, contained two GalT activities; however, the relative activity of the putative Gal:GalT responsible for the DP5 peak was considerably greater in tobacco than in Arabidopsis.
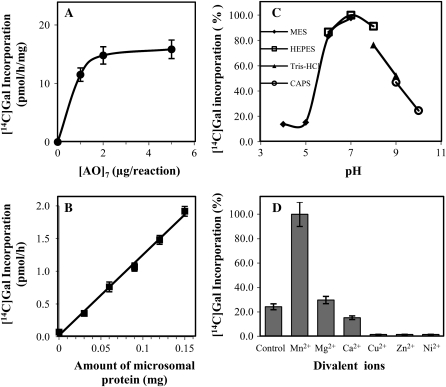
Enzymatic Characteristics of the [AO]7:GalT Activity in Arabidopsis Suspension-Cultured Cells
Since Arabidopsis material will be a better source for identification of the AGP GalT enzyme(s) using proteomic strategies given its complete genome sequence, Arabidopsis [AO]7:GalT activity was further characterized as described below. With a total of 150 μg of microsomal proteins in the assay system, [AO]7:GalT activity approached saturation when 2 μg of [AO]7 was included in the reaction mix (Fig. 5A). With 2 μg [AO]7 in the GalT assay system, incorporation of [14C]Gal increased proportionally with respect to the amount of microsomal protein up to 150 μg using an incubation time of 2 h (Fig. 5B). The [AO]7:GalT activity had a pH optimum of 7 (Fig. 5C) and a temperature optimum of 40°C (data not shown). The [AO]7:GalT activity increased approximately 4-fold in the presence of Mn2+ but was unchanged with Mg2+, in comparison to the control (Fig. 5D). In addition, the presence of Ca2+, Cu2+, Zn2+, and Ni2+ divalent ions inhibited activity to different extents (Fig. 5D).
Figure 5.
Biochemical characteristics of the Arabidopsis [AO]7:GalT activity. Data are the average of triplicate assays. A, Relationship between [AO]7 concentration and incorporation of [14C]Gal into [AO]7. B, Relationship between microsomal protein concentration and incorporation of [14C]Gal into [AO]7. C, Effect of pH on enzyme activity. D, Effect of different divalent ions (5 mm) on enzyme activity.
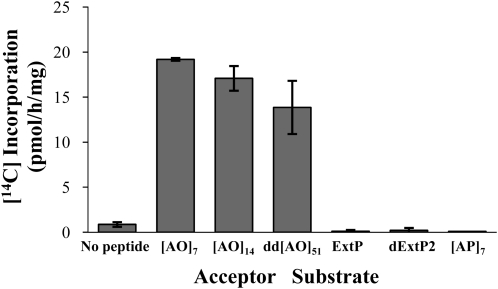
The specificity of the Arabidopsis Hyp:GalT activity was investigated using various acceptor substrates (Fig. 6). Namely, [AO]7, [AO]14, and dd[AO]51 were used as AGP peptides with repetitive [AO] units of various lengths. Incorporation of [14C]radiolabel decreased with increasing lengths of these [AO] acceptor substrates. In contrast, [AP]7 containing seven repetitive [Ala-Pro] units did not serve as an acceptor substrate, indicating that peptidyl Hyp was required for galactosylation. In addition, two extensin sequences were used as potential acceptor substrates, a chemically synthesized extensin peptide (ExtP) designed based on a repetitive sequence (i.e. SOOOOYVYSSOOOOY) found in several Arabidopsis extensins (e.g. At4g08410) and a deglycosylated tomato (Solanum lycopersicum) P2 extensin (dExtP2) containing numerous SOOOOVYK and YK repeat units (Smith et al., 1986). Both of these extensin sequences, which contained contiguous peptidyl Hyp as well as other amino acid sequences, failed to act as acceptors, indicating this Hyp:GalT activity was specific for AGP sequences containing noncontiguous peptidyl Hyp.
Figure 6.
Incorporation of [14C]radiolabel into acceptor substrates containing various Hyp motifs. [AO]7, [AO]14, and dd[AO]51 contain 7, 14, and 51 [AO] units, respectively. A chemically synthesized ExtP and tomato dExtP2 contain repetitive SO4 units. [AP]7 contains seven [AP] units. Enzyme reactions were done in triplicate, and mean values are presented.
The requirement of a specific nucleotide sugar donor for [AO]7 glycosylation was also tested. Arabidopsis microsomal membranes specifically incorporated [14C]Gal, but not [14C]Glc, [14C]Xyl, or [14C]Fuc from their corresponding nucleotide sugars, into the [AO]7 acceptor substrate (Table I).
Table I. Incorporation of [14C]radiolabel into the [AO]7 acceptor substrate when various nucleotide sugars were used as the sugar donor.
| Sugar Donor | [14C]Incorporation |
| pmol h−1 mg−1 | |
| UDP-[14C]Gal | 11.50 ± 0.19 |
| UDP-[14C]Glc | 0.15 ± 0.03 |
| UDP-[14C]Xyl | 0.01 ± 0.02 |
| GDP-[14C]Fuc | 0.03 ± 0.00 |
Identification of the Arabidopsis [AO]7:GalT Reaction Product by Electrospray Ionization-Tandem Mass Spectrometry Analysis
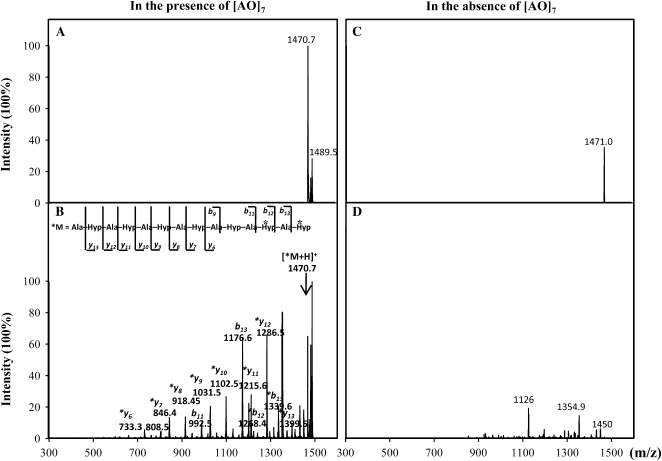
When nonradioactive (cold) UDP-Gal was used in the GalT reactions as the sugar donor, the resulting RP-HPLC purified [AO]7:GalT reaction product was analyzed by electrospray ionization-tandem mass spectrometry (ESI-MS/MS) analysis. No product peak was visible in either the [AO]7:GalT product or the control product by ESI-MS under full scan mode (data not shown) due to a low signal-to-noise ratio. In MS/MS analysis, however, a mass-to-charge ratio (m/z) 1470.7 ion corresponding to the putative galactosylated [AO]7 with one Gal attached was isolated in the [AO]7 assay product (Fig. 7A). The identity of the 1470.7 ion was confirmed by ion fragmentation using the collision-induced dissociation technique, which generated a series of peaks with their m/z matching the theoretical fragments from [AO]7 with either a single Gal attached or not (Fig. 7B). The fragmentation pattern of the m/z 1470.7 ion also indicated that the attachment site of the Gal residue was at Hyp-12 or Hyp-14. Although an ion with a m/z of 1471.0 was isolated from the control assay product (Fig. 7C), the fragmentation pattern of the m/z 1471.0 ion was not related to the [AO]7 substrate or to galactosylated [AO]7 (Fig. 7D). Moreover, we were unable to detect or isolate ions corresponding to [AO]7 peptide with more than one Gal attached from either the [AO]7 assay product or control product.
Figure 7.
ESI-MS/MS analysis of the Arabidopsis [AO]7:GalT reaction product. The ion with m/z of 1470.7 corresponding to glycosylated [AO]7 with one Gal attached was isolated from the [AO]7:GalT reaction product (A). The fragments generated from ion 1470.7 following collision-induced dissociation (B) were matched with the [AO]7 fragments or galactosylated [AO]7 fragments (b and y ions with asterisks) based on their calculated m/z. In ion 1470.7, the Gal attachment site was deduced to be the Hyp residue at position 12 or position 14 (i.e. Hyp residues with asterisks in the peptide). A peak with m/z of 1471.0 was isolated from the control reaction (C). However, fragmentation of ion 1471.0 did not show peaks relevant to [AO]7 fragments or the galactosylated [AO]7 product (D).
Subcellular Localization of [AO]7:GalT and d[AO]51:GalT Activities in Arabidopsis Suspension-Cultured Cells
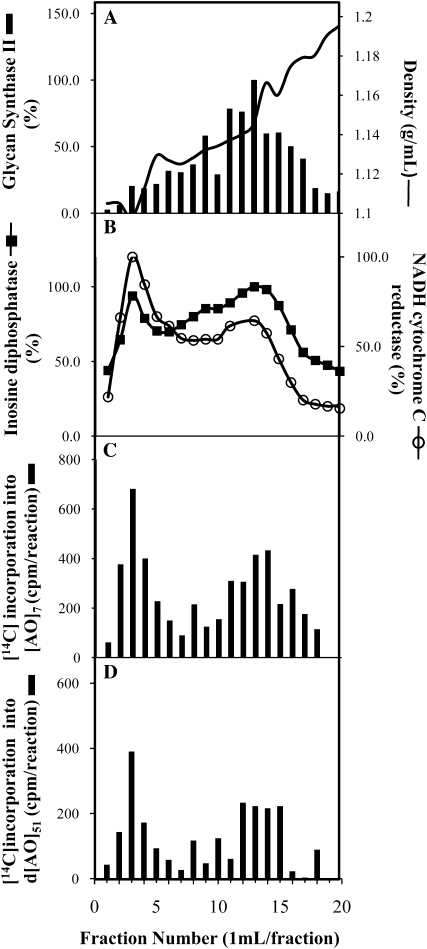
A continuous Suc density gradient was employed to fractionate microsomal membranes, followed by marker enzyme assays to identify subcellular membrane systems. Although various combinations of gradient conditions were used, including different Suc gradients, with or without the addition of EDTA (1 or 5 mm) or Mg2+ (1 or 5 mm), it was not possible to completely resolve ER membranes from Golgi membranes. The best separation of ER and Golgi membranes was achieved with a 30% to 45% (w/v) Suc gradient in the presence of 1 mm EDTA (Fig. 8A), in which both NADH cytochrome C reductase (an ER marker) and inosine diphosphatase (a Golgi marker) showed two activity peaks (Fig. 8B). However, NADH cytochrome C reductase showed higher activity in the peak of lower density (fractions 2–5), while inosine diphosphatase had greater activity in the higher density peak (fractions 7–16). The plasma membrane exhibited a broad distribution with a single peak around fraction 13, as indicated by using glucan synthase II as a marker (Fig. 8A). The gradient fractions were used for GalT assays. With both [AO]7 and d[AO]51 as acceptor substrates, two GalT activity peaks corresponding to the ER and Golgi marker enzyme activities were detected (Fig. 8, C and D).
Figure 8.
Fractionation of mixed membranes of Arabidopsis on a Suc gradient in the presence of 1 mm EDTA. Each fraction was assayed for: A, density and glucan synthase II (a plasma membrane marker); B, activities of antimycin A-insensitive NADH cytochrome C reductase (an ER marker) and IDPase (a Golgi apparatus marker); C, [AO]7:GalT activity; and D, d[AO]51:GalT activity.
DISCUSSION
Considering GTs are linkage specific (Keegstra and Raikhel, 2001), the complex AG structure implies the involvement of multiple GalTs in AG biosynthesis. Using chemically synthesized and transgenically produced AGP peptides as acceptor substrates and UDP-[14C]Gal as the sugar donor, an in vitro GalT assay was developed to detect GalT activities involved in AGP glycosylation in tobacco and Arabidopsis. Product analysis by RP-HPLC and monosaccharide analysis showed that [14C]Gal was incorporated into AGP peptides in the GalT assay (Fig. 3; Supplemental Fig. S1). In examining the extent of AGP peptide galactosylation in the [AO]7, d[AO]51, and dd[AO]51 reaction products (Fig. 4), evidence for two distinct GalT activities was detected. The first activity, a Hyp:GalT activity, catalyzes the addition of Gal onto peptidyl Hyp residues, as demonstrated in reactions with [AO]7 and dd[AO]51 acceptor substrates. The second activity, a probable Gal:GalT activity, extends the sugar side chain with a second Gal as demonstrated in reactions with the d[AO]51 acceptor substrate and was more pronounced in tobacco compared to Arabidopsis.
The Arabidopsis Hyp:GalT activity observed here is similar to that reported by Oka et al. (2010) with respect to its properties (pH optimum, temperature optimum, requirement for Mn2+, and specificity for UDP-Gal; Fig. 5, Table I) and localization to the endomembrane system (Fig. 8). However, further analysis of the Hyp:GalT activity here revealed that it was specific for AGP sequences and not for other related protein sequences, including extensins with their characteristic SO4 repeat units. This finding is consistent with the Hyp contiguity hypothesis, which states that noncontiguous Hyp residues are sites of arabinogalactan polysaccharide addition, whereas contiguous Hyp residues are sites for the addition of Ara oligosaccharides (Kieliszewski and Lamport, 1994; Kieliszewski et al., 1995; Tan et al., 2003). In addition, ESI-MS/MS analysis of the [AO]7 product demonstrated that only one Gal was added per peptide molecule to the C-terminal or penultimate Hyp residue of this peptide. Whether this observation reflects the enzymatic mechanism associated with this Hyp:GalT activity or the in vitro nature of this work awaits further investigation. It should also be noted that Oka et al. used a different in vitro assay system involving a chemically synthesized AGP14 peptide and variants thereof conjugated to fluorescein isothiocyanate via a γ-aminobutyric acid linker as acceptor substrates. Regardless, such chemically synthesized peptides, whether it be [AO]7 or AGP14, were only useful in demonstrating the addition of the first Gal residue to Hyp and provide no evidence for a second Gal:GalT activity, which could only be detected when deglycosylated AGP sequences were used as acceptor substrates. These observations also highlight the need to develop appropriate acceptor substrates to detect specific enzyme activities in the AG biosynthetic pathway as the small amount of product formed in these assays is apparently insufficient to serve as an effective substrate for the next glycosylation reaction.
In ryegrass, AGP GalT activity was reported to incorporate [14C]Gal into endogenous acceptor substrates, forming 66% ethanol insoluble products with (1,6)-linked galactosyl residues (Mascara and Fincher, 1982). AGP GalT activity was also detected in radish roots and catalyzed addition of Gal residues onto exogenous β-(1,3)-galactotriose acceptors through β-(1,6)-linkages (Kato et al., 2003). In these two studies, AGP GalT activities were involved in elongation of AG side chains and so are likely to be different from the Hyp:GalT activity identified here. Indeed, the Hyp:GalT activity exhibited distinct features from the ryegrass GalT and radish GalT activity. For example, Mg2+ did not activate the Arabidopsis Hyp:GalT activity as it did for ryegrass GalT activities. The optimal temperature of Arabidopsis Hyp:GalT (40°C) is higher than that of radish GalT (30°C) and of ryegrass GalT (between 10°C and 25°C).
N-glycosylation and O-glycosylation are the two major types of protein glycosylation. The biosynthesis of N-glycans involves assembling an oligosaccharide precursor structure linked to lipid carriers. The oligosaccharide precursor is then transferred en block from lipid carriers to the protein N-glycosylation sites (Varki et al., 1999). In contrast, O-glycosylation is often viewed as the sequential addition of single sugar residues to the polypeptide, as exemplified by two well-defined processes, O-mannosylation (Goto, 2007) and mucin type O-glycosylation (Hanisch, 2001). The identification of Hyp:GalT activity and likely Gal:GalT activity for the formation of a disaccharide galactosyl chain on Hyp as well as the lack of detection of en block transfer in this study is consistent with the notion that Gal residues are transferred sequentially to the protein backbone in AGP glycosylation. In addition, the recent identification and characterization of two AGP fucosyltransferases that have the ability to fucosylate AGPs lacking terminal Fuc residues is also consistent with this notion of sequential sugar addition (Wu et al., 2010).
Protein N-glycosylation is initiated in ER but completed in Golgi apparatus. Oka et al. (2010) proposed a hypothetical model for AG biosynthesis in which the first Gal residue is added to peptidyl Hyp in an AGP backbone by GalT in the ER with further AG side chain elongation occurring in the Golgi apparatus. The subcellular localization of the Hyp:GalT and Gal:GalT activities here is consistent with this model in that these enzyme activities were localized in the endomembrane system (Fig. 8). Cloning and tagging of the Hyp:GalT and Gal:GalT enzymes in the future should allow for more precise subcellular localizations. Moreover, the subcellular localization of AGP GalT activities in ryegrass (Mascara and Fincher, 1982; Schibeci et al., 1984) and radish root (Kato et al., 2003), AGP fucosyltransferase activity in radish root (Misawa et al., 1996), an AGP fucosyltransferase enzyme in tobacco leaves (Wu et al., 2010), and AGP arabinosyltransferase activity in tobacco cultured cells (Kawasaki, 1987) likewise are consistent with involvement of the endomembrane system, particularly the Golgi apparatus, in AG biosynthesis.
Although a large number of GTs are predicted to be involved with the biosynthesis of plant cell wall components, only a few GTs are unambiguously identified and functionally characterized (Keegstra and Raikhel, 2001). One of the main obstacles for GT identification is the lack of specific enzyme activity assays. Here, an in vitro assay was developed that allows for the detection of two distinct AGP GalT activities in tobacco and Arabidopsis microsomal membranes. Clearly, this radioactive, in vitro assay to detect GalT activity using AGP peptide and glycopeptide acceptor substrates provides a valuable tool for the identification of AGP GalT proteins/genes and serves as an entry point for the elucidation of AG biosynthesis for AGPs. Finally, this assay can be readily modified using different AG acceptor substrates along with appropriate radiolabeled sugar nucleotides to detect other GT activities involved with AG biosynthesis.
MATERIALS AND METHODS
Suspension Culture of Arabidopsis Cells
The tobacco (Nicotiana tabacum, BY2) and Arabidopsis suspension-cultured cells (Arabidopsis thaliana, ecotype Columbia) were maintained in liquid NT-1 media (Arabidopsis Biological Resource Center; http://abrc.osu.edu/cell_culture_handling.html) on a rotary shaker (120 rpm) at 24°C. NT-1 media (pH 5.8) contains 4.3 g/L Murashige and Skoog salts (Caisson), 30 g/L Suc, 180 mg/L KH2PO4, 100 mg/L myoinositol, 1 mg/L thiamine HCl, and 0.44 mg/L 2,4-dichlorophenoxacetic acid. Cells were subcultured weekly (1:10, v/v) into fresh culture media. The Arabidopsis cells were obtained from and established by Axelos et al. (1992).
Preparation of Microsomal Membranes from Tobacco and Arabidopsis Suspension-Cultured Cells
Microsomal membranes were prepared as described earlier (Zeng et al., 2008). The method was modified for the use of suspension-cultured cells as the starting material. Briefly, 140 g of mid-log phase cells (7 d cultures) were harvested and thoroughly washed with double distilled water at 4°C and resuspended in 70 mL extraction buffer (0.1 m HEPES-KOH, pH 7, 0.4 m Suc, 0.1% BSA, 1 mm dithiothreitol, 5 mm MgCl2, 5 mm MnCl2, 1 mm phenylmethylsulfonyl fluoride, and one tablet of Roche complete protease inhibitor cocktail). Cells were disrupted in a Brinkmann Homogenizer (model CPU11; Brinkmann Instruments) at speed 9 for 3 min (stopping for 30 s every 30 s during disruption). The homogenate was filtered through two layers of miracloth, and the filtrate was centrifuged at 3,000g for 20 min. The resulting supernatant was layered over a 1.8 m Suc cushion buffer and centrifuged at 100,000g for 60 min. The uppermost layer was discarded without disturbing the membrane containing interphase layer. A discontinuous Suc gradient was implemented by sequentially layering 1.1 and 0.25 m Suc solutions onto the interphase layer and centrifuging at 100,000g for 60 min. The microsomal membranes enriched at the 0.25/1.1 m Suc interphase were collected and pelleted by another centrifugation at 100,000g for 30 min. The pellet was resuspended in 700 μL extraction buffer and stored at −80°C until use.
Standard Assay for GalT Activity
The standard GalT reaction mixture (75 μL) consisted of permeabilized microsomal membranes (approximately 150 μg total protein), acceptor substrate peptide (50 μg), and UDP-[14C]Gal (approximately 3 μm, 90,000 cpm, 465 cpm/pmol; PerkinElmer Life Sciences). To achieve permeabilization, 50 μL of the microsome preparations were treated with 1% Triton X-100 (15 min, 4°C), followed by ultracentrifugation at 100,000g for 45 min. The pellet was resuspended in 50 μL extraction buffer (BSA eliminated). The [AO]7 acceptor substrate was a chemically synthesized peptide (GenScript). The sequence of [AO]7 was [Ala-Hyp-Ala-Hyp-Ala-Hyp-Ala-Hyp-Ala-Hyp-Ala-Hyp-Ala-Hyp]. The d[AO]51 peptide was obtained from the [AO]51-EGFP fusion protein expressed in tobacco BY-2 cells as described previously (Tan et al., 2003). In the control reaction, no acceptor substrate was included. Reactions were incubated at room temperature for 2 h and terminated by mixing with 400 μL anion-exchange resin (DOWEX 1X8-100 resin; Sigma-Aldrich; 1:1 v/v in double distilled water). The resin mixture was loaded on a Zeba spin column (Pierce) and centrifuged at 15,000g for 1 min. While the unreacted UDP-[14C]Gal was retained by the ion-exchange resin beads, the flow-through contained the incorporated [14C]radiolabeled product and was analyzed with an LS6500 multipurpose scintillation counter (Beckman).
Analysis of the GalT Assay Products by RP-HPLC
The GalT assay product purified by the ion-exchange resin was loaded onto a polymeric reverse-phase column (PRP-1, 5 μm, 4.1 × 150 mm; Hamilton) equilibrated with buffer A (2% acetonitrile and 0.1% trifluoroacetic acid). Reaction products were eluted with isotonic buffer A for 10 min followed by a linear gradient of buffer B (100% acetonitrile with 0.1% trifluoroacetic acid) from 2% to 100% in 90 min at a flow rate of 0.5 mL/min. Chromatography was monitored by absorption at 220 nm. The eluate was collected in 0.5 mL fractions and counted for radioactivity in cpm.
Monosaccharide Composition Analysis of the GalT Assay Product
The RP-HPLC fractions containing [14C]radiolabeled peptides were collected and freeze dried. The dried powder was dissolved in double distilled water and subjected to total acid hydrolysis (2 m trifluoroacetic acid, 121°C, 60 min). Excess trifluoroacetic acid was removed by a freeze drying-dissolving (with double distilled water) cycle repeated three times. The product from total acid hydrolysis was dissolved in deionized water and analyzed on a CarboPac PA10 column (4 × 250 mm; Dionex) in a BioLC system using pulsed amperometric detection (ED50 electrochemical detector; Dionex). The column was equilibrated at a flow rate of 0.5 mL/min with 200 mm NaOH for 10 min, double distilled water for 10 min, and 2.5 m NaOH for 30 min. The sample was eluted with 2.5 m NaOH at a flow rate of 0.5 mL/min. Each fraction (0.5 mL) was counted for radioactivity in cpm. Fuc, Glc, Gal, Ara, and Xyl monosaccharide (Acros Organics) were used as standards.
Hyp [14C]Galactoside Profile Analysis
The RP-HPLC fractions containing [14C]radiolabeled peptides were pooled and freeze dried. The dried powder was dissolved in double distilled water and analyzed by size exclusion chromatography before or after base hydrolysis (0.44 m NaOH, 18 h, 105°C). Size exclusion chromatography was performed with a Bio-gel P2 column (90 × 1.5 cm) under gravity with degassed double distilled water. Each fraction (2.3 mL) was counted for radioactivity in cpm. The base hydrolysate of [AO]7 as well as pure Hyp amino acid (Sigma-Aldrich) was eluted on Bio-gel P2 under the same elution conditions as described above for comparison. Hyp fractions were analyzed using a colorimetric method described previously (Lamport and Miller, 1971). The column system was calibrated with high-Mr dextran (V0), Gal (Vt), xylo-oligosaccharides (DP2–5), and xyloglucan-oligosaccharides (DP6–9; Megazyme).
Characterization of the Arabidopsis [AO]7:GalT Activity
The standard GalT assay was modified for the enzymatic characterization tests using [AO]7 peptide as the acceptor substrate. The assay product from each reaction was fractionated on RP-HPLC to specifically measure incorporated [14C]radiolabel into acceptor substrates.
To test the effect of pH, permeabilized microsomal membranes were dissolved in test buffers at a final concentration of 100 mm and used for the GalT assay. Test buffers included MES-KOH buffer at pH 4, 5, 6, and 7; HEPES-KOH buffer at pH 6, 7, and 8; Tris-HCl buffer at pH 8 and 9; and CAPS-KOH buffer at pH 10.
To test the effect of divalent ions, microsomal membranes were extracted with divalent ions eliminated from the extraction buffer. Permeabilized microsomal membranes were dissolved in extraction buffer with divalent ions eliminated and distributed into aliquots. MnCl2, MgCl2, CaCl2, CuCl2, NiCl2, and ZnSO4 at a final concentration of 5 mm were added when the GalT assays were performed. Deionized distilled water instead of divalent ions was added in the control reaction.
To test the enzyme specificity for different acceptor substrates, the standard GalT assay was performed with 2 μg of various acceptor substrates. The [AO]14, [AO]7, and ExtP acceptor substrates were chemically synthesized peptides (GenScript). The sequence of [AO]14 is [Ala-Hyp-Ala-Hyp-Ala-Hyp-Ala-Hyp-Ala-Hyp-Ala-Hyp-Ala-Hyp-Ala-Hyp-Ala-Hyp-Ala-Hyp-Ala-Hyp-Ala-Hyp-Ala-Hyp-Ala-Hyp]. The sequence of [AP]7 is [Ala-Pro-Ala-Pro-Ala-Pro-Ala-Pro-Ala-Pro-Ala-Pro-Ala-Pro]. The sequence of ExtP is [Ser-Hyp-Hyp-Hyp-Hyp-Tyr-Val-Tyr-Ser-Ser-Hyp-Hyp-Hyp-Hyp-Tyr]. The dExtP2 was isolated from tomato (Solanum lycopersicum) suspension-cultured cells and deglycosylated as described previously (Smith et al., 1986). dExtP2 has a highly periodic structure that is composed of di-and octapeptides, with sequences of [Tyr-Lys] and [Ser-Hyp-Hyp-Hyp-Hyp-Val-Tyr-Lys], respectively.
To analyze the enzyme specificity for sugar donors, the standard activity assay was performed with [AO]7 as the acceptor substrate and various [14C]labeled nucleotide sugars (90,000 cpm) as the sugar donor. The nucleotide sugars tested included UDP-[14C]Glc (MP Biomedicals), UDP-[14C]Xyl (PerkinElmer Life Sciences), and GDP-[14C]Fuc (PerkinElmer Life Sciences).
ESI-MS/MS of the [AO]7:GalT Assay Product
The GalT assay was conducted under standard assay conditions except that UDP-[14C]Gal was replaced by nonradioactive (cold) UDP-Gal (12 μm). The cold GalT assay products were fractionated by RP-HPLC, and fractions were collected according to the retention time of the glycopeptide peak as determined by a preceding run with radiolabeled products. RP-HPLC purified products were lyophilized and dissolved in 50% methanol containing 0.1% acetic acid. The sample was introduced into the ESI source at 3 μL/min using a Cole-Parmer 74900 syringe pump. ESI-MS/MS spectra were acquired on an Esquire 6000 Ion Trap analyzer (Bruker Daltonics) operated in positive ion mode with a capillary voltage of 4,000 V, drying gas temperature of 300°C, drying gas flow rate of 5 L/min, and a nebulizer pressure of 10 p.s.i. Nitrogen was used as both the nebulizing gas and drying gas. Samples from the control or experimental assay product were subjected to a full scan from m/z 600 to 3000. In MS/MS analysis, precursor ions with the target m/z values were isolated from the full scan spectrum and fragmented by collision-induced dissociation with helium as the collision gas and a collision voltage of 1 V. Target m/z values for precursor ions were set as 1470, 1632, and 1794, corresponding to the theoretical m/z values of protonated [AO]7 with one, two, and three Gal residues attached. Instrument control and data acquisition were performed with Esquire 5.0 software.
Continuous Sucrose Gradient Centrifugation
Linear 30% to 45% (w/v) Suc gradients (20 mL) were poured over 4-mL cushions of 62% Suc (w/v) in a centrifugation tube by Auto Densi-Flow (Labconco). All solutions are made up in 0.1 m HEPES-KOH (pH 7) in the presence of 1 mm EDTA. Microsomal membranes from Arabidopsis suspension-cultured cells (1 mL) were layered on top of the Suc gradient. After centrifugation (100,000g, 16 h, 4°C; Beckman Coulter SW32 rotor), 20 equal fractions were collected from the top of the gradient downward with the electric gradient fractionator Auto Densi-Flow (Labconco).
The cytochrome C reductase (an ER marker), inosine diphosphatase (a Golgi marker), and glycan synthase II (a plasma membrane marker) in each gradient fraction were measured according to procedures described previously (Shore and Maclachlan, 1975; Gibeaut and Carpita, 1990; Dhugga and Ray, 1994).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Monosaccharide analysis of the RP-HPLC purified tobacco d[AO]51:GalT reaction product and the Arabidopsis [AO]7:GalT reaction product.
Supplemental Figure S2. Bio-gel P2 fractionation of free Hyp amino acid or Hyp amino acid produced following base hydrolysis of the [AO]7 acceptor substrate.
Supplementary Material
Acknowledgments
We thank Glen Jackson of the Department of Chemistry and Biochemistry at Ohio University for assistance with analysis of ESI-MS/MS data.
References
- Axelos M, Curie C, Mazzolini L, Bardet C, Lescure B. (1992) A protocol for transient gene expression in Arabidopsis thaliana protoplasts isolated from cell suspension cultures. Plant Physiol Biochem 30: 123–128 [Google Scholar]
- Bolwell GP. (1986) Microsomal arabinosylation of polysaccharide and elicitor-induced carbohydrate-binding glycoprotein in french bean. Phytochemistry 25: 1807–1813 [Google Scholar]
- Dhugga KS, Ray PM. (1994) Purification of 1,3-beta-D-glucan synthase activity from pea tissue. Two polypeptides of 55 kDa and 70 kDa copurify with enzyme-activity. Eur J Biochem 220: 943–953 [DOI] [PubMed] [Google Scholar]
- Ellis M, Egelund J, Schultz C, Bacic A. (2010) Arabinogalactan-proteins (AGPs): key regulators at the cell surface? Plant Physiol 153: 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Maruyama Y, Kotake T, Tsumuraya Y. (2003) Beta-glucuronosyltransferase from radish primary roots involving in the synthesis of arabinogalactan-proteins. Plant Cell Physiol 44: S99–S99 [Google Scholar]
- Gibeaut DM, Carpita NC. (1990) Separation of membranes by flotation centrifugation for in vitro synthesis of plant-cell wall polysaccharides. Protoplasma 156: 82–93 [Google Scholar]
- Goodrum LJ, Patel A, Leykam JF, Kieliszewski MJ. (2000) Gum arabic glycoprotein contains glycomodules of both extensin and arabinogalactan-glycoproteins. Phytochemistry 54: 99–106 [DOI] [PubMed] [Google Scholar]
- Goto M. (2007) Protein O-glycosylation in fungi: diverse structures and multiple functions. Biosci Biotechnol Biochem 71: 1415–1427 [DOI] [PubMed] [Google Scholar]
- Hanisch FA. (2001) O-glycosylation of the mucin type. Biol Chem 382: 143–149 [DOI] [PubMed] [Google Scholar]
- Hayashi T, Maclachlan G. (1984) Glycolipids and glycoproteins formed from UDP-galactose by pea membranes. Phytochemistry 23: 487–492 [Google Scholar]
- Held MA, Tan L, Kamyab A, Hare M, Shpak E, Kieliszewski MJ. (2004) Di-isodityrosine is the intermolecular cross-link of isodityrosine-rich extensin analogs cross-linked in vitro. J Biol Chem 279: 55474–55482 [DOI] [PubMed] [Google Scholar]
- Karr AL. (1972) Isolation of an enzyme system which will catalyze the glycosylation of extensin. Plant Physiol 50: 275–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi Y, Tsumuraya Y, Hashimoto Y, Nakano H, Kovac P. (2003) In vitro biosynthesis of galactans by membrane-bound galactosyltransferase from radish (Raphanus sativus L.) seedlings. Planta 217: 271–282 [DOI] [PubMed] [Google Scholar]
- Kawasaki S. (1987) Synthesis of arabinose-containing cell-wall precursors in suspension-cultured tobacco cells. 3. Purification and some properties of the major component. Plant Cell Physiol 28: 187–197 [Google Scholar]
- Keegstra K, Raikhel N. (2001) Plant glycosyltransferases. Curr Opin Plant Biol 4: 219–224 [DOI] [PubMed] [Google Scholar]
- Kieliszewski MJ, Lamport DTA. (1994) Extensin: repetitive motifs, functional sites, posttranslational codes, and phylogeny. Plant J 5: 157–172 [DOI] [PubMed] [Google Scholar]
- Kieliszewski MJ, Oneill M, Leykam J, Orlando R. (1995) Tandem mass-spectrometry and structural elucidation of glycopeptides from a hydroxyproline-rich plant-cell wall glycoprotein indicate that contiguous hydroxyproline residues are the major sites of hydroxyproline O-arabinosylation. J Biol Chem 270: 2541–2549 [DOI] [PubMed] [Google Scholar]
- Kieliszewski MJ, Shpak E. (2001) Synthetic genes for the elucidation of glycosylation codes for arabinogalactan-proteins and other hydroxyproline-rich glycoproteins. Cell Mol Life Sci 58: 1386–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamport DT, Kieliszewski MJ, Showalter AM. (2006) Salt stress upregulates periplasmic arabinogalactan proteins: using salt stress to analyse AGP function. New Phytol 169: 479–492 [DOI] [PubMed] [Google Scholar]
- Lamport DTA, Miller DH. (1971) Hydroxyproline arabinosides in the plant kingdom. Plant Physiol 48: 454–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langan KJ, Nothnagel EA. (1997) Cell surface arabinogalactan-proteins and their relation to cell proliferation and viability. Protoplasma 196: 87–98 [Google Scholar]
- Mascara T, Fincher GB. (1982) Biosynthesis of arabinogalactan-protein in Lolium multiflorum (ryegrass) endosperm cells. 2. In vitro incorporation of galactosyl residues from UDP-galactose into polymeric products. Aust J Plant Physiol 9: 31–45 [Google Scholar]
- Misawa H, Tsumuraya Y, Kaneko Y, Hashimoto Y. (1996) Alpha-L-fucosyltransferases from radish primary roots. Plant Physiol 110: 665–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T, Saito F, Shimma YI, Yoko-o T, , Nomura Y, , Matsuoka K, , Jigami Y. (2010) Characterization of endoplasmic reticulum-localized UDP-d-galactose: hydroxyproline O-galactosyltransferase using synthetic peptide substrates in Arabidopsis. Plant Physiol 152: 332–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxley D, Bacic A. (1999) Structure of the glycosylphosphatidylinositol anchor of an arabinogalactan protein from Pyrus communis suspension-cultured cells. Proc Natl Acad Sci USA 96: 14246–14251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu YM, Egelund J, Gilson PR, Houghton F, Gleeson PA, Schultz CJ, Bacic A. (2008) Identification of a novel group of putative Arabidopsis thaliana beta-(1,3)-galactosyltransferases. Plant Mol Biol 68: 43–59 [DOI] [PubMed] [Google Scholar]
- Schibeci A, Pnjak A, Fincher GB. (1984) Biosynthesis of arabinogalactan-protein in Lolium multiflorum (Italian ryegrass) endosperm cells. Subcellular distribution of galactosyltransferases. Biochem J 218: 633–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert GJ, Roberts K. (2007) The biology of arabinogalactan proteins. Annu Rev Plant Biol 58: 137–161 [DOI] [PubMed] [Google Scholar]
- Sherrier DJ, Prime TA, Dupree P. (1999) Glycosylphosphatidylinositol-anchored cell-surface proteins from Arabidopsis. Electrophoresis 20: 2027–2035 [DOI] [PubMed] [Google Scholar]
- Shore G, Maclachlan GA. (1975) The site of cellulose synthesis. Hormone treatment alters the intracellular location of alkali-insoluble beta-1,4-glucan (cellulose) synthetase activities. J Cell Biol 64: 557–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak E, Barbar E, Leykam JF, Kieliszewski MJ. (2001) Contiguous hydroxyproline residues direct hydroxyproline arabinosylation in Nicotiana tabacum. J Biol Chem 276: 11272–11278 [DOI] [PubMed] [Google Scholar]
- Shpak E, Leykam JF, Kieliszewski MJ. (1999) Synthetic genes for glycoprotein design and the elucidation of hydroxyproline-O-glycosylation codes. Proc Natl Acad Sci USA 96: 14736–14741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Muldoon EP, Willard JJ, Lamport DTA. (1986) Tomato extensin precursors P1 and P2 are highly periodic structures. Phytochemistry 25: 1021–1030 [Google Scholar]
- Strasser R, Bondili JS, Vavra U, Schoberer J, Svoboda B, Glossl J, Leonard R, Stadlmann J, Altmann F, Steinkellner H, et al. (2007) A unique beta 1,3-galactosyltransferase is indispensable for the biosynthesis of N-glycans containing lewis a structures in Arabidopsis thaliana. Plant Cell 19: 2278–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetek J, Yadav MP, Nothnagel EA. (1999) Presence of a glycosylphosphatidylinositol lipid anchor on rose arabinogalactan proteins. J Biol Chem 274: 14724–14733 [DOI] [PubMed] [Google Scholar]
- Tan L, Leykam JF, Kieliszewski MJ. (2003) Glycosylation motifs that direct arabinogalactan addition to arabinogalactan-proteins. Plant Physiol 132: 1362–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Qiu F, Lamport DT, Kieliszewski MJ. (2004) Structure of a hydroxyproline (Hyp)-arabinogalactan polysaccharide from repetitive Ala-Hyp expressed in transgenic Nicotiana tabacum. J Biol Chem 279: 13156–13165 [DOI] [PubMed] [Google Scholar]
- van Hengel AJ, Roberts K. (2002) Fucosylated arabinogalactan-proteins are required for full root cell elongation in Arabidopsis. Plant J 32: 105–113 [DOI] [PubMed] [Google Scholar]
- Varki A, Cummings R, Esko J, Freeze H, Hart G, Marth J. (1999) O-glycans, Ed 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Wu YY, Williams M, Bernard S, Driouich A, Showalter AM, Faik A. (2010) Functional identification of two nonredundant Arabidopsis α(1,2)fucosyltransferases specific to arabinogalactan proteins. J Biol Chem 285: 13638–13645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Chatterjee M, Faik A. (2008) UDP-xylose-stimulated glucuronyltransferase activity in wheat microsomal membranes: characterization and role in glucurono(arabino)xylan biosynthesis. Plant Physiol 147: 78–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZD, Tan L, Showalter AM, Lamport DT, Kieliszewski MJ. (2002) Tomato LeAGP-1 arabinogalactan-protein purified from transgenic tobacco corroborates the Hyp contiguity hypothesis. Plant J 31: 431–444 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.