Abstract
Sulfate is required for the synthesis of sulfur-containing amino acids and numerous other compounds essential for the plant life cycle. The delivery of sulfate to seeds and its translocation between seed tissues is likely to require specific transporters. In Arabidopsis (Arabidopsis thaliana), the group 3 plasmalemma-predicted sulfate transporters (SULTR3) comprise five genes, all expressed in developing seeds, especially in the tissues surrounding the embryo. Here, we show that sulfur supply to seeds is unaffected by T-DNA insertions in the SULTR3 genes. However, remarkably, an increased accumulation of sulfate was found in mature seeds of four mutants out of five. In these mutant seeds, the ratio of sulfur in sulfate form versus total sulfur was significantly increased, accompanied by a reduction in free cysteine content, which varied depending on the gene inactivated. These results demonstrate a reduced capacity of the mutant seeds to metabolize sulfate and suggest that these transporters may be involved in sulfate translocation between seed compartments. This was further supported by sulfate measurements of the envelopes separated from the embryo of the sultr3;2 mutant seeds, which showed differences in sulfate partitioning compared with the wild type. A dissection of the seed proteome of the sultr3 mutants revealed protein changes characteristic of a sulfur-stress response, supporting a role for these transporters in providing sulfate to the embryo. The mutants were affected in 12S globulin accumulation, demonstrating the importance of intraseed sulfate transport for the synthesis and maturation of embryo proteins. Metabolic adjustments were also revealed, some of which could release sulfur from glucosinolates.
Sulfur is an essential macronutrient for all living organisms. In plants, sulfur is a constituent of the amino acids Met and Cys, the latter controlling protein folding and/or activity through reversible disulfide bond formation. As a constituent of Met, sulfur is important in the nutrition of animals and humans, which are unable to synthesize Met and rely on dietary sources. Furthermore, various compounds essential for plant growth and development are derived from sulfur-containing amino acids (e.g. glutathione, S-adenosylmethionine, ethylene, and biotin). In recent decades, reduced atmospheric pollution has led to increased sulfur deficiency in European agriculture (Schnug, 1991; McGrath et al., 1993; Zhao and McGrath, 1993). This is an agronomic issue, since many studies have reported that sulfur deficiency results in variations in crop yield and seed protein composition (Chandler et al., 1984; Higgins et al., 1986; Naito et al., 1994; Hirai et al., 1995; Higashi et al., 2006). Identifying the mechanisms involved in sulfur transport in plants, therefore, is of great interest for improving or maintaining seed yield and protein composition under such environmental fluctuations. Several studies have focused on sulfur acquisition by roots and transport to aerial parts of the plant (Smith et al., 1995, 1997; Takahashi et al., 2000; Shibagaki et al., 2002; Yoshimoto et al., 2002; Buchner et al., 2004a; Kataoka et al., 2004a; El Kassis et al., 2007), but to date, seed sulfur transport and metabolic adjustments of seeds to low-sulfur transport activity remain to be studied.
In plants, sulfur is usually taken up by the roots as sulfate and distributed within the tissues in this form. Tabe and Droux (2001) demonstrated that sulfate is the dominant form of sulfur found in the phloem supplying pods during lupin (Lupinus albus) seed development and that the seed is able to reduce and assimilate sulfate. If sulfate is not reduced, excess sulfate is stored in the vacuole (Kaiser et al., 1989; Martinoia et al., 2000, 2007). A recent transcriptome analysis of Medicago truncatula developing seeds suggested that seed tissues accommodate two distinct pathways of sulfur assimilation: most of the sulfate enters the embryo, being utilized for the synthesis of Cys and its incorporation into proteins, while sulfate in the seed coat and endosperm serves preferentially for the synthesis of defense-related sulfur compounds (Gallardo et al., 2007). This intertissue partitioning implies an active exchange of sulfate between the seed compartments.
Sulfate is delivered to plant tissues and cell compartments by specific sulfate transporters (SULTR), which are encoded by large gene families (12–14 members) in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa). Based upon sequence similarities, sulfate transporters can be subdivided into four groups (SULTR1 to -4), whose members possess 12 membrane-spanning domains and a STAS domain at the C terminus (Rouached et al., 2005), and a fifth group (SULTR5) lacking these features (Buchner et al., 2004b). In Arabidopsis, the SULTR5;2 gene has recently been demonstrated to encode a root high-affinity molybdate transporter (Tomatsu et al., 2007), which raises the question of the role of group 5 genes in sulfate transport. The differences in kinetics and expression patterns of groups 1 to 4 are suggestive of functional subtypes (Hawkesford, 2003). In recent years, sulfate transporters of groups 1 and 2, which are localized at the plasma membrane, have been the subject of several studies. Members of group 1 represent high-affinity transporters that facilitate the uptake of sulfate by the root (SULTR1;1 and SULTR1;2) or translocation of sulfate from source to sink (SULTR1;3; Smith et al., 1995, 1997; Takahashi et al., 2000; Shibagaki et al., 2002; Yoshimoto et al., 2002; El Kassis et al., 2007). Group 2 is composed of low-affinity sulfate transporter genes expressed in vascular tissues and reportedly involved in the translocation of sulfate within the plant (Takahashi et al., 2000; Buchner et al., 2004b; Awazuhara et al., 2005). Unlike groups 1 and 2, group 4 sulfate transporters have been localized in the tonoplast membrane, where they are proposed to mediate the efflux of sulfate from the vacuolar lumen into the cytoplasm (Kataoka et al., 2004b) and to play a role in redox homeostasis in seeds (Zuber et al., 2010).
While group 3 is the largest group of sulfate transporters in Arabidopsis, with five members (SULTR3;1 to -3;5), their roles are not yet established. These transporters are all predicted to be localized at the plasma membrane. This was confirmed for the SULTR3;5 transporter by particle bombardment of onion (Allium cepa) epidermal cells using a GFP-tagged SULTR3;5 fusion construct (Kataoka et al., 2004a). The data available in the literature for these genes indicate a differential expression in plant tissues, which is not stimulated by sulfur deficiency (Buchner et al., 2004a), and a role for SULTR3;5 in the root-to-shoot transport of sulfate in cooperation with SULTR2;1 in Arabidopsis under sulfur-deficient conditions (Kataoka et al., 2004a).
In this study, we observed that the Arabidopsis SULTR3 genes are expressed in developing seeds, especially in the tissues surrounding the embryo. To provide information about their possible contribution to seed sulfate transport and to the establishment of seed composition, we have characterized Arabidopsis T-DNA insertion lines for the five SULTR3 genes. While total sulfur content did not vary between wild-type and mutant seeds, the sulfate content was increased in mature seeds of four mutants out of five, which demonstrates reduced sulfate assimilation during seed development. A proteome analysis was performed on mature seeds from these mutant lines that revealed metabolic adjustments characteristic of a sulfur-deficiency response. Reduced maturation of embryo-specific proteins belonging to the 12S globulin family was also observed, consistent with a role for the SULTR3 transporters in sulfate translocation to the embryo.
RESULTS
Gene Expression of SULTR3 in Arabidopsis Plant Organs
We first studied by real-time quantitative reverse transcription (qRT)-PCR relative mRNA abundances of the group 3 Arabidopsis sulfate transporter genes (SULTR3;1; SULTR3;2, SULTR3;3, SULTR3;4, and SULTR3;5; Fig. 1A). RNAs from isolated seeds (7 and 10 d after flowering, corresponding to embryogenesis and seed-filling stages, respectively), isolated siliques (at 7–10 d after flowering), and other tissues (leaves, flowers, roots, and stems) were analyzed. All tissues were collected during the reproductive growth phase under unlimited sulfur availability. The qRT-PCR data, which were consistent with an available microarray-based transcriptomics data set (Toufighi et al., 2005; Supplemental Fig. S1), revealed a differential expression of the five group 3 sulfate transporters in plant organs (Fig. 1A). While SULTR3;1 was preferentially expressed in stems, SULTR3;2 and SULTR3;4 were more widely expressed in various organs. The SULTR3;3 gene was also expressed in several organs (mature seeds and leaves) but at relatively low levels compared with the other genes. Interestingly, SULTR3;5 was strongly expressed in mature seeds as compared with all other plant organs.
Figure 1.
Expression profiling of the five genes (SULTR3;1, SULTR3;2, SULTR3;3, SULTR3;4, and SULTR3;5) encoding sulfate transporters of group 3 in Arabidopsis plant and seed tissues. A, Relative mRNA quantity estimated by qRT-PCR in developing seeds separated from pods, in mature seeds, in siliques collected at 7 to 10 d after flowering (DAF), and in other tissues of wild-type plants (Col-0) grown in sulfur-sufficient conditions. 7 DAF, Embryogenesis; 10 DAF, early seed filling. Bars represent means ± sd of three independent measurements from two biological experiments. B, Expression data in seed tissues from the Harada-Goldberg Arabidopsis LCM GeneChip Data Set (Seedgenenetwork at http://estdb.biology.ucla.edu/ seed/, Gene Expression Omnibus accession series GSE12404). These data were normalized to the highest expression value set to 1 and represented as a color scale (Genesis software; Sturn et al., 2002) in which red and yellow represent the highest and lowest expression values, respectively. For each gene, the maximum expression level is indicated by a white dot.
Whereas group 3 genes were all expressed during seed development, their expression timing differed (Fig. 1A; Supplemental Fig. S1): SULTR3;1 and SULTR3;2 were expressed throughout seed development, while SULTR3;3 and SULTR3;5 were exclusively expressed at late stages, and SULTR3;4 transcripts specifically accumulated during embryogenesis and early seed filling. In addition to qRT-PCR profiles, we examined SULTR3 expression in seed tissues by exploring Laser Capture Microdissection/GeneChip data from Seedgenenetwork (Harada-Goldberg Arabidopsis LCM GeneChip Data Set [http://seedgenenetwork.net/], Gene Expression Omnibus accession series GSE12404; Le et al., 2010). All SULTR3 genes were strongly expressed in the tissues surrounding and/or attached to the embryo (Fig. 1B). SULTR3;1 and SULTR3;2 were preferentially expressed in the general seed coat and micropylar endosperm. SULTR3;3 and SULTR3;5 genes were strongly expressed in the three compartments of the endosperm (i.e. chalazal, peripheral, and micropylar) and in the chalazal seed coat. SULTR3;4 showed a distinct expression pattern, being strongly expressed in the suspensor and the micropylar endosperm. These expression profiles are consistent with a role in seed development for the SULTR3 genes, either in seed sulfate import or in sulfate translocation within the seed.
Genomic and Phenotypic Characterization of sultr3 Mutant Lines
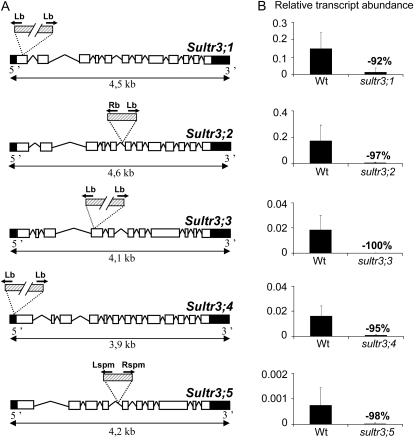
Mutant lines in the Columbia background (Col-0) for SULTR3;1, -3;2, -3;3, and -3;4 were from the SALK T-DNA insertions (Alonso et al., 2003), and the SULTR3;5 mutant line was from the SLAT collection (Tissier et al., 1999). All mutant lines, backcrossed once with Col-0, and the corresponding wild type were greenhouse grown under unlimited sulfur availability. For each plant analyzed, insertion presence and allele homozygosity were determined by PCR. Sequencing of the genomic regions flanking the T-DNA borders revealed possible multiple T-DNA insertions in the SULTR3;1, -3;3, and -3;4 genes: in the first exon of SULTR3;1 with a 36-bp deletion, in the fourth exon of SULTR3;3 with a 32-bp deletion, and in the promoter region of SULTR3;4 with a 27-bp deletion at position −180 relative to the ATG start codon (Fig. 2A). Single insertions were found in the SULTR3;2 and -3;5 genes: in the fifth intron of SULTR3;2 with a 35-bp deletion and in the fourth intron of SULTR3;5 (Fig. 2A). Detailed sequence and primer information are available in Supplemental Figure S2. The residual expression from the disrupted SULTR3 genes was measured by qRT-PCR using the gene-specific primers described in Supplemental Table S1. For the five mutant lines, mRNA abundance decreased by 92% to 100% as compared with the wild type (Fig. 2B). Finally, we have estimated the T-DNA copy number in the genome of each mutant using quantitative real-time PCR by comparison with three single-copy genes (see “Materials and Methods”). One to two T-DNA copies were identified (Supplemental Table S2), except for sultr3;2, which possesses five copies. The fact that two T-DNA left border sequences were identified in the SULTR3;1, -3;3, and -3;4 mutant lines suggests that these lines possess T-DNA insertions only in the SULTR3 genes (Fig. 2A; Supplemental Fig. S2).
Figure 2.
T-DNA insertion sites and residual expression from the disrupted SULTR3 genes. A, Structure of the SULTR3 genes with the insertion sites for the mutant lines SALK-023190 (sultr3;1), SALK-023980 (sultr3;2), SALK-031340 (sultr3;3), SALK-100362 (sultr3;4), and NASC-N112372 (sultr3;5). The exons are indicated by white boxes, untranslated regions by black boxes, and T-DNA by dashed boxes. PCR was performed by using specific primers for each gene binding upstream and downstream of the predicted T-DNA insertion and primers binding in the border regions of the corresponding T-DNA (black arrows). Detailed information about primers and sequences is available in Supplemental Figure S2. B, Residual expression level of the disrupted SULTR3 genes using the gene-specific primers listed in Supplemental Table S1 from RNA extracted from developing siliques (for the sultr3;1, sultr3;4, and sultr3;5 mutants versus the wild type [Wt]), flowers (for the sultr3;2 mutant versus the wild type), or leaves (for the sultr3;3 mutant versus the wild type). Bars represent means ± sd of three biological replicates. Lb and Lspm, Left border primers of the T-DNA from SALK and NASC lines, respectively; Rb and Rspm, right border primers of the T-DNA from SALK and NASC lines, respectively.
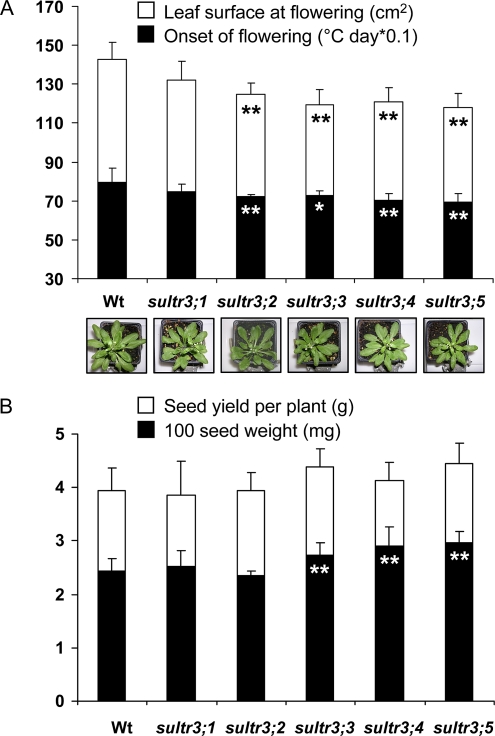
Phenotypic indicators of plant performance (onset of flowering, rosette leaf surface at flowering, and seed yield) were measured on the mutant and wild-type plants and submitted to statistical analyses to determine if there were significant differences between the wild-type and mutant plants (Fig. 3). Mutant plant growth was not arrested, but four group 3 mutants (sultr3;2, -3;3, -3;4, and -3;5) had an earlier onset of flowering and an average rosette leaf area approximately 20% smaller than the wild type (Fig. 3A). These phenotypic differences are similar to those observed under low-sulfate growth conditions (Nikiforova et al., 2004). In addition, sultr3;3, -3;4, and -3;5 had a weight of 100 seeds higher than the wild type (Fig. 3B). As no significant change in seed yield was observed, we assumed that this is due to there being fewer seeds per plant.
Figure 3.
Phenotypic characteristics of homozygous sultr3 mutant plants. A, Onset of flowering (black bars) and projected rosette leaf surface at flowering (white bars) for the wild-type (Wt) and mutant plants. B, Seed weight (black bars) and yield per plant (white bars) for the wild-type and mutant plants. Each bar represents the mean ± sd of four biological replicates (100 seed weight) or nine biological replicates (the other characters). The data were submitted to a Levene’s test to assess variance homogeneity and then to a Student’s t test to determine if there are statistically significant differences between wild-type and mutant plants. Asterisks represent P values: * P < 0.05, ** P < 0.01. [See online article for color version of this figure.]
Sulfur/Nitrogen/Carbon Contents and Anion and Thiol Levels in Mature Seeds of the sultr3 Mutant Lines
Total sulfur content, measured in mature seeds of each line, did not vary significantly between the mutants and the wild type (Fig. 4A). This finding and the absence of significant variation in seed yield indicate that sulfur compound allocation to the seed compartment (i.e. total seed mass) was not affected in all group 3 mutants. We further determined sulfate levels in mature seeds and leaves (Fig. 4B; Supplemental Fig. S3). Intriguingly, the sultr3;2, -3;3, -3;4, and -3;5 mutants possessed an average seed sulfate content from 1.7- to 2.3-fold higher than the wild type (Fig. 4B). In contrast, we did not find any significant variations for sulfate content in rosette leaves (Supplemental Fig. S3). By calculating the proportion of sulfur (S) accumulated in mature seeds in the form of sulfate [S (SO42−)], according to the formula S (SO42−) × 100/S (total), we observed that sulfur in sulfate form represents 6% of total sulfur content in the wild type, whereas it represents up to 16% in the sultr3 mutants. This increased S (SO42−) versus S (total) ratio is indicative of reduced sulfate assimilation in developing seeds of the mutant lines. To investigate whether the altered sulfate assimilation can be explained by defects in sulfate partitioning between the main seed tissues, the envelopes were separated from the mature embryo after a short period of imbibition for one selected mutant line and for the wild type, and then sulfate content was determined in both tissues and in the entire seed also subjected to a short period of imbibition. Knowing that at maturity the endosperm is only a thin layer and the seed coat is the major envelope surrounding the embryo, we have selected one mutant line for a SULTR3 gene strongly expressed in the seed coat. Among the two genes expressed in this tissue (SULTR3;1 and SULTR3;2; Fig. 1B), we have chosen to focus on SULTR3;2 because of the increased S (SO42−) versus S (total) ratio of the corresponding mutant seeds (Fig. 4). Interestingly, a statistically significant (P < 0.05) difference in sulfate partitioning between seed tissues was observed between the wild-type and sultr3;2 mutant seeds (Fig. 4B). The proportion of sulfate in the envelopes was higher (59.4%) than that observed for the wild-type seeds (45.7%). Also, despite not being statistically significant, a reduced proportion of sulfate was observed in the embryo for this mutant as compared with the wild type. This suggests a defect in sulfate translocation from the main envelopes to the embryo in the sultr3;2 developing seeds.
Figure 4.
Nitrogen/carbon/sulfur, thiol, and anion contents in mature seeds of wild-type (Wt) and sultr3 mutant plants. A, Nitrogen/carbon/sulfur seed contents (%) measured by high-performance ionic chromatography. B, Chloride, nitrate, sulfate, phosphate, malate, and citrate seed contents (μmol g−1) measured by high-performance ionic chromatography. C, Cys and glutathione (GSH) contents (nmol g−1) measured by HPLC. Sulfur in the form of sulfate represents 6.3%, 8.9%, 15.7%, 14.2%, 16.3%, and 11.8% of total sulfur in mature seeds of the wild type, sultr3;1, -3;2, -3;3, -3;4, and -3;5, respectively. Bars represent means ± sd of at least four biological replicates. The table in B shows mean ± sd of sulfate percentage in the entire mature seed that is partitioned between the embryo and the surrounding envelopes in the wild type and sultr3;2 after 90 h of seed imbibition (three biological replicates and three technical replicates from each biological replicate). * P < 0.05, ** P < 0.01 (Student’s t test).
We further measured the levels of free Cys and glutathione in mature seeds and in leaves of each mutant (Fig. 4C; Supplemental Fig. S3). Interestingly, Cys level was decreased by 2.6-, 1.8-, and 1.6-fold in mature seeds of sultr3;1, sultr3;2, and sultr3;3 (Fig. 4C), respectively, whereas it was not affected in leaves (Supplemental Fig. S3). This could be indicative of a reduced activity of the sulfur assimilation pathways leading to Cys synthesis in the developing mutant seeds. This is consistent with the reduced sulfate use efficiency observed for the sultr3;2 and -3;3 mutant seeds. This also suggests that an alteration of sulfur metabolism, such as a reduced release and/or conversion of Cys from other precursors than sulfate, may occur in the sultr3;1 mutant seeds not significantly affected for seed sulfate content.
Finally, other mineral and organic major anion contents (nitrate, phosphate, chloride, malate, and citrate) were measured in seeds of each mutant, and the overall nitrogen and carbon contents were determined (Fig. 4B). Mature seed nitrate contents of the sultr3;3 and -3;4 mutants were decreased by 1.7- and 2.3-fold, respectively, compared with the wild type. In mature seeds of the sultr3;4 mutant, a significant decrease of chloride content was also observed reaching a much lower value (1.4-fold) than in the other lines. No variation was detected for the other anions and elements measured.
Proteome Analysis of SULTR3 Mutant Seeds
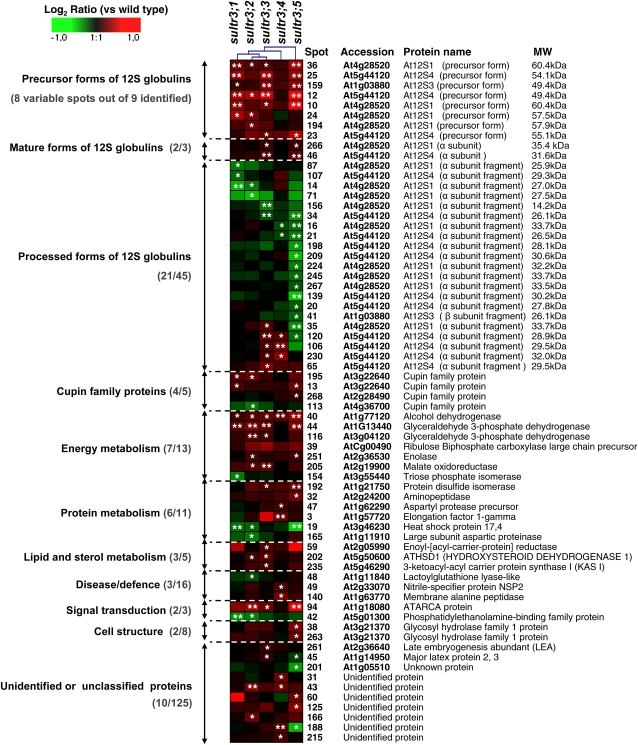
To further explore whether changes in sulfate transport capacity may have modified seed protein composition, a proteome analysis of mature seeds from the group 3 Arabidopsis mutants and the wild type was performed. For each line, proteins were extracted from four biological replicates and separated in duplicate by two-dimensional (2D) electrophoresis. Image analysis of Coomassie Brilliant Blue gels using the Samespots software allowed the quantification of protein abundance for 273 well-resolved spots (Supplemental Fig. S4; Supplemental Tables S3 and S4). Statistical analyses (Levene’s and Student’s t tests) identified 68 spots for which the relative abundance in mature seeds varied between the wild type and at least one mutant line (Fig. 5). Of these, 41 and 23 spots showed an increased and a decreased accumulation level, respectively, in seeds of the mutant lines, whereas four spots (spots 35, 45, 120, and 188) displayed distinct abundance variations according to the mutant (Fig. 5). Interestingly, the seed proteome changes observed for sultr3;1, -3;2, and -3;3 mutants affected for free Cys content (Fig. 4C) were relatively similar, whereas several proteins vary specifically in the sultr3;4 and -3;5 mutants (Fig. 5).
Figure 5.
Clustered expression ratios of the 68 seed proteins differentially accumulated in the five sultr3 mutant lines as compared with the wild type. Each row represents a single protein, and each column represents a mutant line. Log2 expression ratios were calculated for the different mutants relative to the wild type and visualized using the Genesis software (Sturn et al., 2002). The color scale is indicated on the top. Red indicates an overaccumulation of the protein in mutant seeds, green indicates a decreased abundance of the protein in mutant seeds, and black indicates no change in protein abundance between the mutant and wild-type seeds. * P < 0.05, ** P < 0.01 (variance analysis followed by a t test). Accession number, protein name, and ontological classification (Bevan et al., 1998) are indicated for each spot. Within each class, the number of proteins identified whose abundance varied versus the total number of proteins identified is indicated in parentheses. Detailed information on the proteins is listed in Supplemental Tables S3, S4, and S5 (see also the proteome reference map in Supplemental Fig. S4).
To study the putative functions of these proteins, we further annotated 152 spots detected on 2D gels by using proteome reference maps previously established for Arabidopsis mature seeds (Gallardo et al., 2001, 2002; Rajjou et al., 2004; Higashi et al., 2006; Supplemental Fig. S4; Supplemental Tables S3 and S4). We have checked the identity of 53 spots by tryptic peptide mass fingerprinting or liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS). In all cases, their identity was confirmed (Supplemental Table S5).
Variation in 12S Globulin Abundances
Polypeptides corresponding to the four 12S globulin isoforms (At12S1 [At4g28520], At12S2 [At1G03890], At12S3 [At1g03880], and At12S4 [At5g44120]) were identified on 2D gels from mature seeds (57 spots; Supplemental Table S3). Of these 57 spots, the abundance of 32 varied significantly in at least one group 3 mutant as compared with the wild type. 12S globulins are synthesized as precursors, which are posttranslationally cleaved to produce α- and β-subunits. These subunits are further processed by limited C-terminal proteolysis during maturation (Higashi et al., 2006). Based on the C-terminal sequences of the spots identified on 2D gels, and on their experimental molecular mass and pI, we were able to distinguish the precursor, mature subunits, and proteolyzed forms for the different isoforms. Nine spots corresponded to residual precursor forms, three spots to intact α-subunits from At12S1 (spot 261), At12S3 (spot 259), and At12S4 (spot 46), and 36 spots to proteolyzed α-subunits (Fig. 5; Supplemental Table S4).
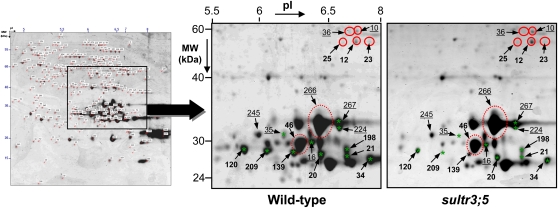
These detailed annotations revealed that all 12S globulin spots whose abundance decreased in sultr3 seeds were proteolyzed α- and/or β-subunits (18 spots; Fig. 5; Supplemental Table S4). In contrast, the 12S globulin spots that increased in sultr3 seeds corresponded to residual precursors (eight spots) and intact At12S1 and At12S4 subunits (two spots; Figs. 5 and 6). In the sultr3;3 and -3;4 mutant seeds, we also found an increased abundance of some of the less proteolyzed At12S1 and At12S4 subunits (i.e. higher molecular mass; spots 35, 65, 106, 120, and 230 in Fig. 4). The mutant displaying the most marked changes in 12S globulin accumulation was sultr3;5 (Figs. 5 and 6). In contrast, very few 12S globulins vary in sultr3;4 mutant seeds as compared with the wild type. In summary, all sultr3 mutants are affected in storage protein accumulation and/or processing, the most affected being the sultr3;5 mutant and the least affected being sultr3;4.
Figure 6.
Accumulation profiles of 12S globulins in mature seeds of wild-type and sultr3;5 homozygous mutant lines. Sections of 2D gels from total seed proteins of wild-type (on the left) and sultr3;5 mutant (on the right) lines are enlarged. Red and green represent the polypeptides whose levels increased and decreased, respectively, in mutant seeds compared with the wild type. Precursor and mature forms of 12S globulins are shown in solid circles and dashed circles, respectively, and fragmented forms are indicated by stars. Underlined and boldface numbers represent At12S1 and At12S4 isoforms, respectively. The seed proteome map shown on the left is enlarged and appears in Supplemental Figure S4.
Variations in Nonstorage Protein Abundances
The group 3 mutants also displayed variations in the levels of 26 nonstorage proteins (out of 68) compared with the wild type. The nonstorage proteins identified in this study were classified in functional categories according to the gene ontology of Bevan et al. (1998; Fig. 5; Supplemental Table S3). These functional annotations revealed that the spots whose abundance varied in the group 3 mutant seeds are related to energy (seven spots out of 13 identified), protein metabolism (six spots out of 11), lipid and sterol metabolism (three spots out of five), defense (three spots out of 16), cell structure (two spots out of eight), and signal transduction (two spots out of three).
Several variations observed at the proteome level in the sultr3;1, -3;2, -3;3, and -3;5 mutants are consistent with downstream regulation observed at the metabolome level in Arabidopsis seedlings under sulfur-stress conditions. These are depicted in Supplemental Figure S5. For example, a decreased level of an enzyme implicated in the synthesis of lactoylglutathione from glutathione (lactoylglutathione lyase; spot 48 in Fig. 5; Supplemental Fig. S5) was observed, which is consistent with a decreased level of glutathione in sulfate-starved seedlings (Hirai et al., 2003; Nikiforova et al., 2003, 2005). As a second example, a sharp decrease in the amount of a phosphatidylethanolamine-binding protein (spot 42 in Fig. 5; Supplemental Fig. S5) was observed in seeds of several group 3 mutants, which is consistent with an overall decrease in lipid content in sulfur-starved plants, including a decrease of phosphatidylethanolamine content (Nikiforova et al., 2005). Finally, we observed an increased accumulation of a signal transduction-related ATARCA protein (spot 94 in Fig. 5; Supplemental Fig. S5), whose gene expression is induced in response to auxin (Vahlkamp and Palme, 1997). This corroborates previous data reporting a surplus metabolic flux via auxin and activation of auxin-induced genes during sulfur deficiency (Kutz et al., 2002; Nikiforova et al., 2003).
Modulation of several other metabolic pathways that have not previously been related to sulfur stress appeared to occur in all group 3 mutants, except sultr3;4. Notably, we observed an overaccumulation of several enzymes associated with glycolysis, namely glyceraldehyde 3-phosphate dehydrogenase (spots 44, 116, and 187), enolase (spot 251), and malate oxidoreductase (spot 205), in seeds of these mutants (Fig. 5; Supplemental Fig. S5). The shift to glycolysis might be a way to maintain carbon flow and energy production for reserve synthesis during seed development. Interestingly, we observed an overaccumulation of an alcohol dehydrogenase in the sultr3 mutant seeds (spot 40 in Fig. 5; Supplemental Fig. S5). The increased level of this enzyme, which is involved in fermentation, may help to boost glycolysis, since anaerobic respiration has been considered as the key catalytic process for regenerating NAD+ essential for glycolysis (Tadege et al., 1999; Fernie et al., 2004). Moreover, one key enzyme of the Calvin cycle, ribulose-1,5-biphosphate carboxylase (spot 39 in Fig. 5; Supplemental Fig. S5), was overaccumulated in sultr3 mutants that may be important in recycling the carbon released.
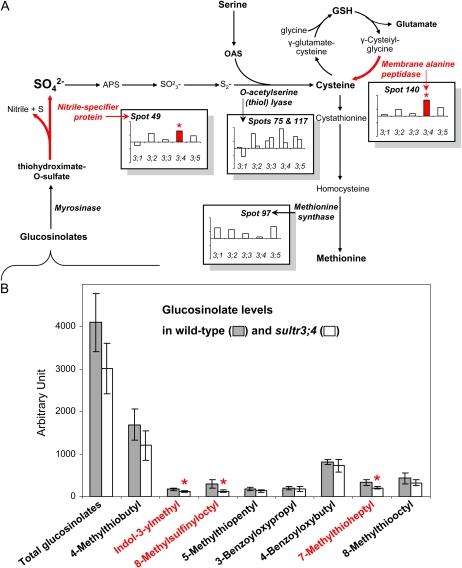
Interestingly, among the proteins overaccumulated in the sultr3;4 mutant seeds were two proteins releasing sulfur compounds. These corresponded to a defense-related enzyme (spot 140 in Figs. 5 and 7A) whose homolog in animals has been implicated in the conversion of glutathione to Cys (Lieberman et al., 1995; Habib et al., 2003) and a nitrile-specifier protein NSP2 (spot 49 in Figs. 5 and 7A) involved in glucosinolate catabolism.
Figure 7.
Suggested compensation mechanisms in sultr3;4 mutant seeds. A, Sulfate assimilation pathways and the regulations in mutant seeds reflected by proteomic analyses. The bars indicate protein abundance variation in mature seeds (log2 volume ratios in mutant seeds versus wild-type seeds). APS, Adenosine-5′-phosphosulfate; GSH, glutathione; OAS, O-acetylserine. B, Glucosinolate levels determined in wild-type and sultr3;4 mutant seeds. The bars represent means ± sd of four biological replicates. Asterisks and red color indicate significant variations (Student’s t test, P < 0.05). See Figure 4C for Cys and GSH levels in mature seeds of the sultr3 mutants. [See online article for color version of this figure.]
Glucosinolate Levels in the sultr3;4 Mutant Seeds
Because the proteomics data obtained for the sultr3;4 mutant suggested compensation mechanisms involving glucosinolate degradation (Fig. 7A), we have determined the levels of glucosinolates by ultra-performance (UP)LC-MS/MS in the wild-type and sultr3;4 mutant seeds. It is worth noting that this study provides, to our knowledge, the most comprehensive list available of glucosinolates in Arabidopsis seeds, with eight forms identified (Fig. 7B): 4-methylthiobutyl (mass-to-charge ratio [m/z] 420), indol-3-ylmethyl (m/z 447), 8-methylsulfinyloctyl (m/z 492), 5-methylthiopentyl (m/z 434), 3-benzoyloxypropyl (m/z 480), 4-benzoyloxybutyl (m/z 494), 7-methylthioheptyl (m/z 462), and 8-methylthiooctyl (m/z 476). Although the decreasing trend of total glucosinolate level in sultr3;4 mutant seeds was not statistically different compared with the wild type, three individual glucosinolates display significantly decreased accumulation (Fig. 7B). These are indol-3-ylmethyl glucosinolate, 8-methylsulfinyloctyl glucosinolate, and 7-methylthioheptyl glucosinolate.
DISCUSSION
At the beginning of this study, there was no information available about the possible role of the group 3 plasma membrane-localized sulfate transporters in seeds. By comparing seed composition of Arabidopsis T-DNA mutants for the five genes of this group, we observed a striking increase in sulfate contents of sultr3;2, -3;3, -3;4, and -3;5 mutant seeds compared with the wild type and an unchanged sulfate content in the sultr3;1 mutant seeds (Fig. 4B). These findings contrast with the reduced sulfate level in mutant seeds for the SULTR2;1 transporter known to control the import of sulfate into developing seeds (Awazuhara et al., 2005). Therefore, none of the SULTR3 genes, taken individually, may be essential for sulfate uptake in the developing Arabidopsis seed. However, the remarkable increased S (SO42−) versus S (total) ratio (i.e. the quantity of sulfur in the form of sulfate versus total sulfur) in seeds of four of the five mutants compared with the wild type indicates a reduction in the capacity of the developing mutant seeds to assimilate sulfate, thus implying a function for the corresponding transporters within the developing seed. Since all SULTR3 genes were preferentially expressed in embryo-surrounding tissues, it may be that SULTR3 transporters control the translocation of sulfate to the maturing embryo and, when defective, result in an elevated level of sulfate in the envelopes. By studying the partitioning of sulfate in seed tissues from the sultr3;2 mutant, we indeed observed an increased proportion of sulfate in the envelopes compared with the wild type (Fig. 4B). The reduced exchange of sulfate between the seed compartments may have influenced sulfur metabolism, previously shown to operate in the different seed tissues (Gallardo et al., 2007). Indeed, free Cys level was reduced up to 50% in seeds of this mutant but also in the sultr3;1 and sultr3;3 mutants compared with the wild type (Fig. 4C). Moreover, several proteome changes observed for sultr3 mutant seeds parallel those seen by transcriptomics and/or metabolomics in Arabidopsis seedlings, leaves, or roots under sulfur-deficient conditions (Hirai et al., 2003; Maruyama-Nakashita et al., 2003; Nikiforova et al., 2003, 2005; Supplemental Fig. S5). These results point to a role for SULTR3 transporters in sulfate translocation between seed tissues, presumably to provide sulfate to the developing embryo. Nevertheless, one should note that the use of single loss-of-function alleles for each gene somewhat limits the conclusions that can be drawn with regard to the function of each group 3 sulfate transporter.
Interestingly, among the common changes observed at the level of the proteome in the sultr3 mutant seeds were changes in the accumulation of embryo-specific storage proteins of the 12S globulin family, reflecting reduced processing (Fig. 5). Similar observations have been made for mature seeds of Arabidopsis grown under sulfate-deficient conditions (Higashi et al., 2006). The underlying cause of the alteration of globulin processing, proposed by Higashi et al. (2006) to be due to the repression of protease synthesis under conditions of low sulfate availability, remains unclear and merits further investigation. Interestingly, the different mutants were differentially affected in 12S globulin accumulation and processing. The most affected mutant, sultr3;5, is strongly expressed in late seed maturation compared with other developmental stages and organs (Figs. 1 and 6). This suggests that a major role of SULTR3;5 in seeds is to provide sulfur for maturation of embryo proteins. While SULTR3;5 is the most studied sulfate transporter of group 3, its biological activity still remains unclear. Indeed, although a Lotus japonicus SULTR3;5 homolog, SST1, has been shown to complement a yeast sulfate transporter-deficient mutant (Krusell et al., 2005), the Arabidopsis SULTR3;5 transporter has been reported to exhibit no sulfate uptake activity in the yeast expression system but to act in a synergic way by enhancing the sulfate transport activity of SULTR2;1 when the genes encoding both proteins are coexpressed in a yeast mutant defective in sulfate transport (Kataoka et al., 2004a). Complementary qRT-PCR experiments performed in our study show that SULTR3;5 and SULTR2;1 are coexpressed in Arabidopsis roots (Supplemental Fig. S6), but in developing seeds, the expression patterns of the two genes were highly contrasted: SULTR3;5 is strongly expressed in maturing seeds, whereas SULTR2;1 is expressed to a lesser extent during early stages of seed development (Supplemental Fig. S6). From these data and the contrasting seed sulfate contents between the sultr3;5 (this study) and sultr2;1 (Awazuhara et al., 2005) mutants, it seems unlikely that the two transporters cooperate in developing seeds under our culture conditions (sulfur sufficient).
While the capacity of the sultr3;4 mutant seeds to assimilate sulfate was significantly reduced, this mutant is the least affected for 12S globulin accumulation and for other proteins related to sulfur stress (Fig. 5; Supplemental Fig. S5). Therefore, it seems that this mutant is able to compensate for the limited availability of sulfate. Gene expression profiling during seed development (Fig. 1A; Supplemental Fig. S1) suggests that SULTR3;4 plays a role at early stages, when sulfur compounds (e.g. glucosinolates, glutathione) can still be transported from other plant parts to developing seeds in response to sulfur stress and thus provide alternative sources of sulfur. Moreover, SULTR3;4 is the only gene of group 3 strongly expressed in the suspensor (Fig. 1B), an organ that has been reported to be the major route of nutrient uptake for the embryo (Yeung and Meinke, 1993). Interestingly, one of the few differences specifically observed in the sultr3;4 mutant seed proteome was an overaccumulation of two proteins that may release sulfur from alternative sulfur sources (Fig. 7A). The first protein is a membrane Ala peptidase (spot 140 in Figs. 5 and 7A) described in animals as leading to Cys formation via the degradation of glutathione (Lieberman et al., 1995). The overaccumulation of this protein could reflect adaptive mechanisms to provide Cys through the hydrolysis of glutathione, which is one of the major forms of storage and transport of sulfur (Foyer et al., 2001). The second is the nitrile-specifier protein NSP2 (spot 49 in Figs. 5 and 7A) involved in the hydrolysis of glucosinolates to nitrile, sulfur, and sulfate (Wittstock and Halkier, 2002; Grubb and Abel, 2006; Kissen and Bones, 2009), which suggests an increased catabolism of these compounds in sultr3;4 developing seeds. Glucosinolates are transported from all parts of the plant into developing seeds, and most of the glucosinolates contained in oilseeds are located in the embryo (Josefsson, 1970; Gijzen et al., 1989). Their catabolism may release sulfate in the seed embryo for assimilatory pathways. Sulfate deficiency causes an up-regulation of genes encoding proteins related to the degradation of glucosinolates (two putative thioglucosidases [Maruyama-Nakashita et al., 2003; Hirai et al., 2005], a myrosinase-binding protein, and a myrosinase precursor [Nikiforova et al., 2003]), lending further support to a role for glucosinolates as a source of sulfur under stress conditions. Interestingly, among the three glucosinolates whose abundance decreased in the sultr3;4 mutant seeds was glucobrassicin (i.e. indol-3-ylmethyl glucosinolate; Fig. 7B), reported to be degraded preferentially among all other glucosinolates during sulfate deprivation (Kutz et al., 2002). These data reinforce the hypothesis, based on proteomics data, that glucosinolate catabolism has occurred during sultr3;4 mutant seed development to counteract sulfur stress and maintain embryo metabolism. This also suggests new directions toward maintaining seed sulfur assimilation under conditions of low sulfate availability.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Genotyping
We studied Arabidopsis (Arabidopsis thaliana) T-DNA insertion lines (Col-0) from the Nottingham Arabidopsis Stock Centre for the genes encoding group 3 sulfate transporters: SULTR3;1 (At3g51895), SULTR3;2 (At4g02700), SULTR3;3 (At1g23090), SULTR3;4 (At3g15990), and SULTR3;5 (At5g19600; Tissier et al., 1999; Alonso et al., 2003). These T-DNA lines, previously used by El Kassis et al. (2007), have the following accession numbers: SALK-023190 (SULTR3;1), SALK-023980 (SULTR3;2), SALK-031340 (SULTR3;3), SALK-100362 (SULTR3;4), and NASC-N112372 (SULTR3;5). The T-DNA borders of these mutant lines were sequenced in this study after PCR by using specific primers for each gene binding upstream and downstream of the predicted T-DNA insertion and primers binding in the border regions of the corresponding T-DNA. Detailed information about the primers and sequences are available in Supplemental Figure S2. The five T-DNA lines were all backcrossed one time to the Col-0 wild type before analysis. Col-0-derived homozygous insertion lines and the corresponding Col-0 wild type were grown under unlimited sulfur conditions (peat-perlite mixture) and fertilized three times a week by subirrigation (nitrogen:phosphorus:potassium, 20:20:20) in a greenhouse with supplemental light to 16 h per day. For genotyping, DNA was extracted from a leaf disc by using the cetyl-trimethyl-ammonium bromide method from Doyle and Doyle (1990). A PCR screen was performed by using specific primers for each gene binding upstream and downstream of the predicted T-DNA insertion as well as one primer binding in the left border region of the corresponding T-DNA (for information about primer sequences for genotyping, see Supplemental Table S1).
Estimation of T-DNA Copy Number by Quantitative Real-Time PCR
T-DNA copy number in the genome was estimated for each mutant by quantitative real-time PCR using the delta cycle threshold method, as described previously by Mason et al. (2002) and Skulj et al. (2008). In this method, T-DNA copy number was determined by comparing the quantitative data obtained for the T-DNA insert to three endogenous genes (At12S4 [At5g44120], SULTR1;1 [At4G08620], and SULTR4;1 [AT5G13550]) present in single copy in the Arabidopsis genome. Information about primers is deposited in Supplemental Table S1. Real-time PCR was performed on a LightCycler 480 Real-Time PCR System (Roche) using the MESA GREEN qPCR MasterMix Plus for SYBR Assay (Eurogentec). Real-Time PCR was performed in 10-μL mixtures containing 1× MESA GREEN qPCR MasterMix Plus for SYBR, 100 nmol μL−1 forward and reverse primers, and 2.5 μL of DNA. A melting curve analysis was applied to check PCR specificity by heating PCR products from 59°C to 96°C. Standard curves were obtained for each primer pair, and correlation coefficients of the standard curves all exceeded 0.99. Each reaction was performed in triplicate from four biological replicates and repeated two times (Supplemental Table S2).
Measurements of Phenotypic Characters
Rosette leaf area and seed yield were measured from nine individual plants. Leaf area was quantified from the rosette scan at flowering using the Visilog 5.4 software (Noesis). The onset of flowering was scored from nine individual plants and expressed as °C per day. Seed weight was determined from three seed samples of 10 mg (around 400 seeds) collected on four individual plants. All phenotypic data were subjected to a Levene’s test to assess variance homogeneity, and then a Student’s t test was performed to compare each mutant with the wild type using the Statistica 7.0 software (StatSoft).
RNA Extraction and Transcript Level Determination
Silique and seed samples at three stages of development (7 and 10 d after flowering and mature seeds) and tissue samples (flowers, roots, leaves, and stems) were collected on two independent batches of plants. Frozen tissues were ground in liquid nitrogen, and total RNA was extracted according to Chang et al. (1993). RNAs (10 μg) were incubated with 10 units of RNase-free RQ1 DNase (Promega). Non-reverse-transcribed RNA samples were checked for the absence of contaminating genomic DNA by PCR using primers for the constitutively expressed elongation factor α-chain gene (At5g60390). DNA-free RNA was converted into first-strand cDNA. Samples were reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad). The qRT-PCR was performed for each SULTR3 gene and for the SULTR2;1 gene using the real-time SYBR Green method on a Bio-Rad iQ5 thermal cycler using iQ SYBR Green Supermix (Bio-Rad) and gene-specific primers (Supplemental Table S1). To establish the presence of a single PCR product and the absence of primer-dimers, melting analysis (i.e. heat dissociation of oligonucleotides) was applied immediately after PCR by heating PCR products from 59°C to 96°C. Normalization for cDNA quantity was performed for each template using the elongation factor gene (At5g60390) as a control according to the relative standard curve method (delta cycle threshold; see Bio-Rad manual instructions). The expression stability of the control gene in the different test samples was verified by comparison with two other constitutively expressed genes encoding a ubiquitin (At4g27960) and a protein phosphatase (At1g13320; data not shown). This qRT-PCR method was also used to estimate the expression level of the disrupted SULTR3 genes using the gene-specific primers listed in Supplemental Table S1 from RNA extracted from developing siliques (for the sultr3;1, sultr3;4, and sultr3;5 mutants), flowers (sultr3;2 mutant), or leaves (sultr3;3 mutant).
Determination of Anion, Carbon/Nitrogen/Sulfur, Glutathione, and Cys Contents
For each line, rosette leaves and mature seeds harvested from four individual plants were ground in liquid nitrogen. Seed tissues were manually dissected from three biological replicates of 50 mature seeds for the sultr3;2 and wild-type lines (repeated three times) using the microscope after a 90-min imbibition in Milli-Q water at 4°C. Anions were extracted in Milli-Q water heated at 70°C for 20 min from 50 mg of frozen powder or from the isolated seed tissues. The extract was centrifuged at least three times at 20,000g for 10 min at 4°C. Anion content of the final clear supernatant was determined by high-performance ionic chromatography (LC20 Dionex) using a IonPaq AS11 column and a sodium hydroxide linear gradient (1–22 mm), as described (El Kassis et al., 2007). Contents of total sulfur, nitrogen, and carbon were measured from 10 mg of dried powder of seed samples using an elemental analyzer (Vario EL elemental analyzer system; Elementar). Glutathione and Cys contents were quantified by HPLC after derivatization. The derivatization procedure and separation of thiol derivatives were performed as described by Wirtz et al. (2004) from 50 mg of seeds and leaves. All data were subjected to a Levene’s test to assess variance homogeneity, and then a Student’s t test was performed to compare each mutant with the wild type using the Statistica 7.0 software (StatSoft). Only differences with P < 0.05 were considered significant.
Total Seed Protein Extraction and 2D Electrophoresis
The seed samples subjected to proteomics were immediately frozen in liquid nitrogen after harvest to avoid changes in protein abundance that can be induced by storage. Total proteins were extracted as described by Gallardo et al. (2003) from 20 mg of mature seeds collected on four individual plants. Protein concentration was measured according to Bradford (1976). A constant volume (57 μL) of the protein extracts (around 200 μg of proteins) was used for isoelectrofocusing that corresponded to a constant seed weight (2 mg). Proteins were separated in duplicate from the four biological seed samples using gel strips forming an immobilized nonlinear pH 3 to 10 gradient (Immobiline DryStrip, 24 cm; GE Healthcare/Amersham Biosciences). Strips were rehydrated in the IPGphor system (GE Healthcare/Amersham Biosciences) for 7 h at 20°C with the thiourea/urea lysis buffer containing 2% (v/v) Triton X-100, 20 mm dithiothreitol, and the protein extracts. Isoelectrofocusing was performed at 20°C in the IPGphor system for 7 h at 50 V, 1 h at 300 V, 2 h at 3.5 kV, and 7 h at 8 kV. Prior to the second dimension, each gel strip was incubated at room temperature for 2 × 15 min in 2 × 15 mL of equilibration buffer as described by Gallardo et al. (2002). Proteins were separated on vertical polyacrylamide gels according to Gallardo et al. (2002).
Protein Staining and Quantification
Gels were stained with Coomassie Brilliant Blue G-250 (Bio-Rad) according to Mathesius et al. (2001). Image acquisition was done using the Odyssey Infrared Imaging System (LI-COR Biosciences) at 700 nm with a resolution of 169 μm. Image analyses and spot volume quantification were realized using the Progenesis Samespots version 2.0 software (Nonlinear Dynamics) according to the instruction manual. For each gel, normalized spot volumes were calculated as the ratio of each spot volume to total spot volume in the gel (arbitrary unit). Eight gels (four biological replicates and two technical replicates from each biological replicate) were used and analyzed for each wild-type and mutant line. Molecular mass and pI were calculated according to the migration of standard proteins (Bio-Rad 2D SDS-PAGE standards). The protein spots were hierarchically clustered using the Genesis software (version 1.7.2; Sturn et al., 2002) following the average linkage clustering method. All data were submitted to statistical analyses (Levene’s test followed by a Student’s t test) using the Statistica 7.0 software (StatSoft). Only differences with P < 0.05 were considered significant.
Protein Identification
Spots were annotated using proteome reference maps previously established for Arabidopsis mature seeds (Gallardo et al., 2001, 2002; Rajjou et al., 2004; Higashi et al., 2006; http://www.seed-proteome.com). The identity of 53 spots was confirmed by nano-LC-MS/MS (Q-TOF-Ultima Global equipped with a nano-ESI source coupled with a Cap LC nano-HPLC system; Waters Micromass) as described by Gallardo et al. (2007) and used in a parallel study of seed composition of a mutant for a sulfate transporter belonging to group 4 (Zuber et al., 2010). Detailed information about protein digestion and MS data acquisition is listed in Supplemental Table S5. Peak lists of precursor and fragment ions were matched to proteins in the National Center for Biotechnology Information nonredundant database (March 2008; 7,387,702 sequences, 2,551,671,261 residues, taxonomy of Arabidopsis) using the MASCOT version 2.2 program (Matrix Science). The MASCOT search parameters are described in Supplemental Table S4. Only matches with individual ion scores above 20 were considered.
Measurements of Glucosinolates
For the extraction of polar compounds, 50 mg of mature Arabidopsis seeds from four biological replicates was ground for 1.5 min in liquid nitrogen at 1,500 rpm with a Mikro-Dismembrator S ball mill (Sartorius Stedim Biotech). A total of 700 μL of methanol containing 500 μm sinigrin as an internal standard (2-propenyl glucosinolate; Sigma-Aldrich) was immediately added, the tubes were sealed and sonicated at 50°C for 20 min, and then 400 μL of chloroform was added before another 5 min of sonication at 50°C. Then, 600 μL of ultrapure water (Millipore) was added, and samples were shaken with a vortex mixer and centrifuged for 2 min at 4,000g to collect the methanol/water supernatant, which was stored at −20°C for precipitation. Samples were centrifuged for 15 min at 16,000g, and 200 μL of the extract was dried in a vacuum concentrator and solubilized in 150 μL of ultrapure water. The identification and characterization of glucosinolates from the seed samples was performed using a Waters Quattro Premier XE (Waters) equipped with an electrospray ionization source and coupled to an Acquity UPLC system (Waters). Chromatographic separation was achieved using an Acquity UPLC BEH C18 column (100 × 2.1 mm, 1.7 μm; Waters) coupled to an Acquity UPLC BEH C18 precolumn (2.1 × 5 mm, 1.7 μm; Waters). The mobile phase consisted of water (A) and acetonitrile (B), both containing 0.1% formic acid. The run was started by 2 min of 98% A, then a linear gradient was applied to reach 85% A at 5.5 min, then 0% A at 10 min, followed by an isocratic run using B during 2.8 min. Finally, the return to initial conditions (98% A) was achieved in 2.2 min. The total run time was 15 min. The column was operated at 28°C with a flow rate of 0.4 mL min−1 (sample injection volume of 3 μL). Nitrogen generated from pressurized air in an N2G nitrogen generator (Mistral) was used as drying and nebulizing gas. The nebulizer gas flow was set to approximately 50 L h−1 and the desolvation gas flow to 900 L h−1. The interface temperature was set at 400°C and the source temperature at 135°C. The capillary voltage was set at 3.4 kV and the cone voltage at 25 V, and the ionization was in negative mode. Low-mass and high-mass resolution were 15 for both mass analyzers, ion energies 1 and 2 were 0.5 V, entrance and exit potential were 2 and 1 V, and detector (multiplier) gain was 650 V. Data acquisition and analysis were performed with the MassLynx software (version 4.1) running under Windows XP professional on a Pentium personal computer. The seed samples were first analyzed in full-scan mode (m/z 90–600). The selected ion recording MS mode was used to determine parent mass transition of the internal standard sinigrin (m/z 358) and of eight glucosinolates of m/z 420, 434, 447, 462, 476, 480, 492, and 494. Glucosinolate fragmentation was performed by collision-induced dissociation with argon at 1.0 × 10−4 mbar using daughter scan. The collision energy was set to 20 V, and the following transitions were used for glucosinolate identification: [M-H]− > 275, 259, 195, 97, and 96, as described by Mellon et al. (2002) and Cataldi et al. (2007). The parent mass and characteristic fragment ions were used to identify glucosinolates without ambiguity. Peak areas of 4-methylthiobutyl glucosinolate (m/z 420) and 4-benzoyloxybutyl glucosinolate (m/z 494) were the highest (Fig. 7B), in good agreement with glucosinolate contents previously reported in Arabidopsis seeds (Petersen et al., 2002; Brown et al., 2003).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At3g51895, At4g02700, At1g23090, At3g15990, and At5g19600.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Comparison of the qRT-PCR data obtained in this study for the Arabidopsis genes belonging to group 3 sulfate transporters with the corresponding data from a publicly available expression atlas (Toufighi et al., 2005).
Supplemental Figure S2. Sequences of T-DNA borders for the five sultr3 mutants, with the primer sets used.
Supplemental Figure S3. Sulfate and thiol contents in wild-type and mutant (homozygous) rosette leaves.
Supplemental Figure S4. Proteome map of mature Arabidopsis seeds (Col-0 wild type).
Supplemental Figure S5. Main metabolic pathways affected in the sultr3 mutant seeds, adapted from Catusse et al. (2008).
Supplemental Figure S6. Expression profiling of genes encoding sulfate transporters SULTR3;5 and SULTR2;1.
Supplemental Table S1. Primer sets for quantitative PCR and genotyping.
Supplemental Table S2. Real-time PCR estimation of T-DNA copy number in the sultr3 mutants.
Supplemental Table S3. Proteomics data sets from mature seeds in wild-type and mutant lines.
Supplemental Table S4. Detailed information on seed storage proteins in the wild-type and mutant lines.
Supplemental Table S5. Detailed information on LC-MS/MS protein identification.
Supplementary Material
Acknowledgments
We acknowledge the Nottingham Arabidopsis Stock Centre for providing us with the Arabidopsis T-DNA insertion lines. We sincerely thank John Harada (Section of Plant Biology, Division of Biological Sciences, University of California, Davis) and Bob Goldberg (Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles) for permission to refer to gene expression data from the Seedgenenetwork Web site. We also thank Myriam Sanchez, Jean Potier, and Mario Rega (Legume Ecophysiology and Genetics Research Unit, INRA Center at Dijon) for their very valuable technical support and Judith Burstin (Legume Ecophysiology and Genetics Research Unit, INRA Center at Dijon) for critical reading of the manuscript and helpful discussions.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Awazuhara M, Fujiwara T, Hayashi H, Watanabe-Takahashi A, Takahashi H, Saito K. (2005) The function of SULTR2;1 sulfate transporter during seed development in Arabidopsis thaliana. Plant Physiol 125: 95–105 [Google Scholar]
- Bevan M, Bancroft I, Bent E, Love K, Goodman H, Dean C, Bergkamp R, Dirkse W, Van Staveren M, Stiekema W, et al. (1998) Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 391: 485–488 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brown PD, Tokuhisa JG, Reichelt M, Gershenzon J. (2003) Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62: 471–481 [DOI] [PubMed] [Google Scholar]
- Buchner P, Stuiver E, Westerman S, Wirtz M, Hell R, Hawkesford M, De Kok L. (2004a) Regulation of sulfate uptake and expression of sulfate transporter genes in Brassica oleracea as affected by atmospheric H2S and pedospheric sulfate nutrition. Plant Physiol 136: 3396–3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner P, Takahashi H, Hawkesford MJ. (2004b) Plant sulphate transporters: co-ordination of uptake, intracellular and long-distance transport. J Exp Bot 55: 1765–1773 [DOI] [PubMed] [Google Scholar]
- Cataldi T, Rubino A, Lelario F, Bufo SA. (2007) Naturally occurring glucosinolates in plant extracts of rocket salad (Eruca sativa L.) identified by liquid chromatography coupled with negative ion electrospray ionization and quadrupole ion-trap mass spectrometry. Rapid Commun Mass Spectrom 21: 2374–2388 [DOI] [PubMed] [Google Scholar]
- Catusse J, Strub J-M, Job C, Van Dorsselaer A, Job D. (2008) Proteome-wide characterization of sugarbeet seed vigor and its tissue specific expression. Proc Natl Acad Sci USA 105: 10262–10267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler PM, Spencer D, Randall PJ, Higgins T. (1984) Influence of sulphur nutrition on developmental patterns of some major pea seed proteins and their mRNAs. Plant Physiol 75: 651–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113–116 [Google Scholar]
- Doyle JL, Doyle JJ. (1990) Isolation of plant DNA from fresh tissue. Focus 12: 13–15 [Google Scholar]
- El Kassis E, Cathala N, Rouached H, Fourcroy P, Berthomieu P, Terry N, Davidian JC. (2007) Characterization of a selenate-resistant Arabidopsis mutant: root growth as a potential target for selenate toxicity. Plant Physiol 143: 1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Carrari F, Sweetlove LJ. (2004) Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport. Curr Opin Plant Biol 7: 254–261 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Theodoulou FL, Delrot S. (2001) The functions of inter- and intracellular glutathione transport systems in plants. Trends Plant Sci 6: 486–492 [DOI] [PubMed] [Google Scholar]
- Gallardo K, Firnhaber C, Zuber H, Héricher D, Belghazi M, Henry C, Küster H, Thompson R. (2007) A combined proteome and transcriptome analysis of developing Medicago truncatula seeds: evidence for metabolic specialization of maternal and filial tissues. Mol Cell Proteomics 6: 2165–2179 [DOI] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SPC, Puype M, Demol H, Vandekerckhove J, Job D. (2001) Proteomic analysis of Arabidopsis seed germination and priming. Plant Physiol 126: 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SPC, Puype M, Demol H, Vandekerckhove J, Job D. (2002) Proteomics of Arabidopsis seed germination: a comparative study of wild-type and gibberellin-deficient seeds. Plant Physiol 129: 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Le Signor C, Vandekerckhove J, Thompson RD, Burstin J. (2003) Proteomics of Medicago truncatula seed development establishes the time frame of diverse metabolic processes related to reserve accumulation. Plant Physiol 133: 664–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijzen M, McGregor I, Séguin-Swartz G. (1989) Glucosinolate uptake by developing rapeseed embryos. Plant Physiol 89: 260–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb CD, Abel S. (2006) Glucosinolate metabolism and its control. Trends Plant Sci 11: 89–100 [DOI] [PubMed] [Google Scholar]
- Habib GM, Shi ZZ, Cuevas AA, Lieberman MW. (2003) Identification of two additional members of the membrane-bound dipeptidase family. FASEB J 17: 1313–1315 [DOI] [PubMed] [Google Scholar]
- Hawkesford MJ. (2003) Transporter gene families in plants. The sulphate transporter gene family: redundancy or specialization? Physiol Plant 117: 115–163 [Google Scholar]
- Higashi Y, Hirai MY, Fujiwara T, Naito S, Noji M, Saito K. (2006) Proteomic and transcriptomic analysis of Arabidopsis seeds: molecular evidence for successive processing of seed proteins and its implication in the stress response to sulphur nutrition. Plant J 48: 557–571 [DOI] [PubMed] [Google Scholar]
- Higgins TJ, Chandler PM, Randall PJ, Spencer D, Beach LR, Blagrove RJ, Kortt AA, Inglis AS. (1986) Gene structure, protein structure, and regulation of the synthesis of a sulphur-rich protein in pea seeds. J Biol Chem 261: 11124–11130 [PubMed] [Google Scholar]
- Hirai MY, Fujiwara T, Awazuhara M, Kimura T, Noji M, Saito K. (2003) Global expression profiling of sulphur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-l-serine as a general regulator of gene expression in response to sulphur nutrition. Plant J 33: 651–663 [DOI] [PubMed] [Google Scholar]
- Hirai MY, Fujiwara T, Chino M, Naito S. (1995) Effects of sulfate concentrations on the expression of a soybean seed storage protein gene and its reversibility in transgenic Arabidopsis thaliana. Plant Cell Physiol 36: 1331–1339 [PubMed] [Google Scholar]
- Hirai MY, Klein M, Fujikawa Y, Yano M, Goodenowe DB, Yamazaki Y, Kanaya S, Nakamura Y, Kitayama M, Suzuki H. (2005) Elucidation of gene-to-gene and metabolite-to-gene networks in Arabidopsis by integration of metabolomics and transcriptomics. J Biol Chem 280: 25590–25595 [DOI] [PubMed] [Google Scholar]
- Josefsson E. (1970) Glucosinolate content and amino acid composition of rapeseed (Brassica napus) meal as affected by sulphur and nitrogen nutrition. PhD thesis University of Lund, Lund, Sweden [Google Scholar]
- Kaiser G, Martinoia E, Schroppelmeier G, Heber U. (1989) Active-transport of sulfate into the vacuole of plant cells provides halotolerance and can detoxify SO2. J Plant Physiol 133: 756–763 [Google Scholar]
- Kataoka T, Hayashi N, Yamaya T, Takahashi H. (2004a) Root-to-shoot transport of sulfate in Arabidopsis: evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol 136: 4198–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T, Watanabe-Takahashi A, Hayashi N, Ohnishi M, Mimura T, Buchner P, Hawkesford MJ, Yamaya T, Takahashi H. (2004b) Vacuolar sulfate transporters are essential determinants controlling internal distribution of sulfate in Arabidopsis. Plant Cell 16: 2693–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissen R, Bones AM. (2009) Nitrile-specifier proteins involved in glucosinolate hydrolysis in Arabidopsis thaliana. J Biol Chem 284: 12057–12070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusell L, Krause K, Ott T, Desbrosses G, Krämer U, Sato S, Nakamura Y, Tabata S, James EK, Sandal N, et al. (2005) The sulfate transporter SST1 is crucial for symbiotic nitrogen fixation in Lotus japonicus root nodules. Plant Cell 17: 1625–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutz A, Müller A, Hennig P, Kaiser WM, Piotrowski M, Weiler EW. (2002) A role for nitrilase 3 in the regulation of root morphology in sulphur-starving Arabidopsis thaliana. Plant J 30: 95–106 [DOI] [PubMed] [Google Scholar]
- Le BH, Cheng C, Bui AQ, Wagmaister JA, Henry KF, Pelletier J, Kwong L, Belmonte M, Kirkbride R, Horvath S, et al. (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA 107: 8063–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman MW, Barrios R, Carter BZ, Habib GM, Lebovitz RM, Rajagopalan S, Sepulveda AR, Shi ZZ, Wan DF. (1995) γ-Glutamyl transpeptidase: what does the organization and expression of a multipromoter gene tell us about its functions? Am J Pathol 147: 1175–1185 [PMC free article] [PubMed] [Google Scholar]
- Martinoia E, Maeshima M, Neuhaus HE. (2007) Vacuolar transporters and their essential role in plant metabolism. J Exp Bot 58: 83–102 [DOI] [PubMed] [Google Scholar]
- Martinoia E, Massonneau A, Frangne N. (2000) Transport processes of solutes across the vacuolar membrane of higher plants. Plant Cell Physiol 41: 1175–1186 [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Inoue E, Watanabe-Takahashi A, Yamaya T, Takahashi H. (2003) Transcriptome profiling of sulphur-responsive genes in Arabidopsis reveals global effects of sulphur nutrition on multiple metabolic pathways. Plant Physiol 132: 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason G, Provero P, Vaira AM, Accotto GP. (2002) Estimating the number of integrations in transformed plants by quantitative real-time PCR. BMC Biotechnol 2: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathesius U, Keijzers G, Natera SH, Weinman JJ, Djordjevic MA, Rolfe BG. (2001) Establishment of a root proteome reference map for the model legume Medicago truncatula using the expressed sequence tag database for peptide mass fingerprinting. Proteomics 1: 1424–1440 [DOI] [PubMed] [Google Scholar]
- McGrath SP, Zhao F, Crosland AR, Salmon SE. (1993) Sulphur status in British wheat grain and its relationship with quality parameters. Asp Appl Biol 36: 317–326 [Google Scholar]
- Mellon FA, Bennett RN, Holst B, Williamson G. (2002) Intact glucosinolate analysis in plant extracts by programmed cone voltage electrospray LC/MS: performance and comparison with LC/MS/MS methods. Anal Biochem 306: 83–91 [DOI] [PubMed] [Google Scholar]
- Naito S, Hirai MY, Chino M, Komeda Y. (1994) Expression of a soybean (Glycine max [L.] Merr.) seed storage protein gene in transgenic Arabidopsis thaliana and its response to nutritional stress and to abscisic acid mutations. Plant Physiol 104: 497–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforova V, Freitag J, Kempa S, Adamikn M, Hesse H, Hoefgen R. (2003) Transcriptome analysis of sulphur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. Plant J 33: 633–650 [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Gakière B, Kempa S, Adamik M, Willmitzer L, Hesse H, Hoefgen R. (2004) Towards dissecting nutrient metabolism in plants: a systems biology case study on sulphur metabolism. J Exp Bot 55: 1861–1870 [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Kopka J, Tolstikov V, Fiehn O, Hopkins L, Hawkesford MJ, Hesse H, Hoefgen R. (2005) Systems rebalancing of metabolism in response to sulphur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant Physiol 138: 304–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen BL, Chen S, Hansen CH, Olsen CE, Halkier BA. (2002) Composition and content of glucosinolates in developing Arabidopsis thaliana. Planta 214: 562–571 [DOI] [PubMed] [Google Scholar]
- Rajjou L, Gallardo K, Debeaujon I, Vandekerckhove J, Job C, Job D. (2004) The effect of α-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol 134: 1598–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouached H, Berthomieu P, El Kassis E, Cathala N, Catherinot V, Labesse G, Davidian JC, Fourcroy P. (2005) Structural and functional analysis of the C-terminal STAS (Sulfate Transporter and Anti-sigma Antagonist) domain of the Arabidopsis thaliana sulfate transporter SULTR1.2. J Biol Chem 280: 15976–15983 [DOI] [PubMed] [Google Scholar]
- Schnug E. (1991) Sulphur nutritional status of European crops and consequences for agriculture. Sulphur Agric 15: 7–12 [Google Scholar]
- Shibagaki N, Rose A, McDermott JP, Fujiwara T, Hayashi H, Yoneyama T, Davies JP. (2002) Selenate-resistant mutants of Arabidopsis thaliana identify Sultr1;2, a sulfate transporter required for efficient transport of sulfate into roots. Plant J 29: 475–486 [DOI] [PubMed] [Google Scholar]
- Skulj M, Okrslar V, Jalen S, Jevsevar S, Slanc P, Strukelj B, Menart V. (2008) Improved determination of plasmid copy number using quantitative real-time PCR for monitoring fermentation processes. Microb Cell Fact 7: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FW, Ealing PM, Hawkesford MJ, Clarkson T. (1995) Plant members of a family of sulfate transporters reveal functional subtypes. Proc Natl Acad Sci USA 92: 9373–9377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FW, Hawkesford MJ, Ealing PM, Clarkson DT, Vanden Berg PJ, Belcher AR, Warrilow AG. (1997) Regulation of expression of a cDNA from barley roots encoding a high affinity sulphate transporter. Plant J 12: 875–884 [DOI] [PubMed] [Google Scholar]
- Sturn A, Quackenbush J, Trajanoski Z. (2002) Genesis: cluster analysis of microarray data. Bioinformatics 18: 207–208 [DOI] [PubMed] [Google Scholar]
- Tabe LM, Droux M. (2001) Sulphur assimilation in developing lupin cotyledons could contribute significantly to the accumulation of organic sulphur reserves in the seed. Plant Physiol 126: 176–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M, Dupuis I, Kuhlemeier C. (1999) Ethanolic fermentation: new functions for an old pathway. Trends Plant Sci 4: 320–325 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Watanabe-Takahashi A, Smith FW, Blake-Kalff M, Hawkesford MJ, Saito K. (2000) The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant J 23: 171–182 [DOI] [PubMed] [Google Scholar]
- Tissier AF, Marillonnet S, Klimyuk V, Patel K, Angel Torres M, Murphy G, Jones JDG. (1999) Multiple independent defective suppressor-mutator transposon insertions in Arabidopsis: a tool for functional genomics. Plant Cell 11: 1841–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomatsu H, Takano J, Takahashi H, Watanabe-Takahashi A, Shibagaki N, Fujiwara T. (2007) An Arabidopsis thaliana high-affinity molybdate transporter required for efficient uptake of molybdate from soil. Proc Natl Acad Sci USA 104: 18807–18812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. (2005) The Botany Array Resource: e-northerns, expression angling, and promoter analyses. Plant J 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Vahlkamp L, Palme K. (1997) AtArcA. Accession No. U77381, the Arabidopsis thaliana homolog of the tobacco ArcA gene (PGR97-145). Plant Physiol 115: 8639411456 [Google Scholar]
- Wirtz M, Droux M, Hell R. (2004) O-Acetylserine(thiol)lyase: an enigmatic enzyme of plant cysteine biosynthesis revisited in Arabidopsis thaliana. J Exp Bot 55: 1785–1798 [DOI] [PubMed] [Google Scholar]
- Wittstock U, Halkier BA. (2002) Glucosinolate research in the Arabidopsis era. Trends Plant Sci 7: 263–270 [DOI] [PubMed] [Google Scholar]
- Yeung EC, Meinke DW. (1993) Embryogenesis in angiosperms: development of the suspensor. Plant Cell 5: 1371–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto N, Takahashi H, Smith FW, Yamaya T, Saito K. (2002) Two distinct high-affinity sulphate transporters with different inducibilities mediate uptake of sulphate in Arabidopsis roots. Plant J 29: 465–473 [DOI] [PubMed] [Google Scholar]
- Zhao F, McGrath SP. (1993) Assessing the risk of sulphur deficiency in cereals. J Sci Food Agric 63: 119 [Google Scholar]
- Zuber H, Davidian JC, Wirtz M, Hell R, Belghazi M, Thompson R, Gallardo K. (2010) Sultr4;1 mutant seeds of Arabidopsis have an enhanced sulphate content and modified proteome suggesting metabolic adaptations to altered sulphate compartmentalization. BMC Plant Biol 10: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.