The unique nature of the plant system as a self-sufficient, robust, and resilient organism requires the dynamic coordination of numerous signal transduction pathways in multiple organs and cell types with complementary functions to capture energy and nutrients, to orchestrate growth and development, to adjust or adapt to fluctuating environment, and to live with symbiotic or curb invasive microbes and animals. Plants “move” through their growth and developmental programs highly integrated with the complex environmental cues. Our understanding of plant signal transduction pathways has been greatly facilitated by the isolation and characterization of a wealth of mutant collections in Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), maize (Zea mays), wheat (Triticum aestivum), barley (Hordeum vulgare), pea (Pisum sativum), tomato (Solanum lycopersicum), Medicago truncatula, moss (Physcomitrella patens), Chlamydomonas reinhardtii, and many other plants. A quantum leap in the field was made possible when the molecular cloning of mutated genes and transgenic plant analysis became a reality in several reference plants in the past two decades. Although the mainstream research on signal transduction has focused on single signals and linear pathways, recent findings have revealed many previously unexpected signaling interconnections, manifesting both the complexity and reality (Fig. 1). The increasing availability of annotated plant genome sequences and large-scale genome-wide databases and computational tools have further empowered the dissection of signaling pathways based on traditional characterization in whole plants, organs, or tissues. Moving forward to discovering and connecting the cellular signaling networks, functional characterization of thousands of genes encoding large families of key regulatory components (Fig. 1) in single cell resolution with time kinetics will be the next challenge. Integrative approaches combing creative genetics, sensitive mass spectrometry, genome-wide screens, powerful bioinformatics tools and skills, versatile cell-based assays, and targeted mutagenesis will be critical for future discoveries (Fig. 2). Glc signaling networks and emerging research tools are briefly highlighted to help pave the way for future research in cellular signaling.
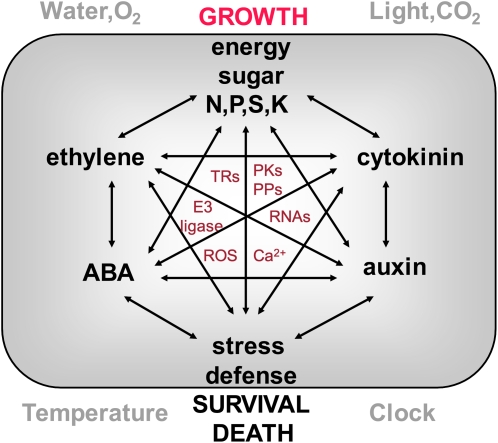
Figure 1.
Connect cellular signaling network in plants. Complex interactions of sugar, nutrient, energy, hormone, stress, and defense signaling pathways have emerged and will be further unraveled at the molecular and cellular level. Light, CO2, water, O2, temperature, and clock are global modulators of the signaling network, which is essential to the plant system output in growth, survival, senescence, or death. The functions of large families of key regulatory components, including more than 3,000 transcription regulators (TRs), 1,400 E3 ligases, more than 1,000 protein kinases (PKs) and protein phosphatases (PPs), and numerous regulatory RNAs, will be integrated in the various cellular signaling pathways. Calcium and reactive oxygen species (ROS) are versatile and universal cellular regulators that act through different sensors and signal relay partners to control diverse signal responses. Many more plant-signaling pathways and signaling mediators, e.g. hormones, peptides, chemicals, metabolites, lipids, receptors, channels, transporters, GTases, ATPases, chaperones, scaffolds, and diverse enzymes, are not presented here.
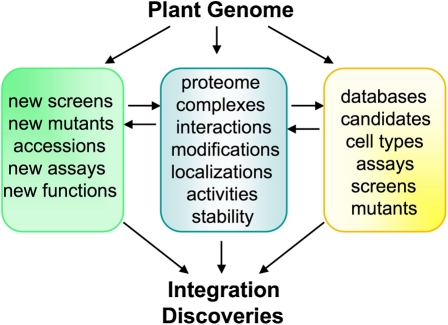
Figure 2.
Discover new cellular signaling pathways. All information for cellular signaling is embedded in the plant genome, which can be explored to discover and define signaling pathways and their interconnections through integrative approaches. The traditional genetic approach will be empowered with more specific and sensitive phenotypic screens to identify new mutants. Creative and thorough analysis of available natural variations/accessions and new assays for existing mutant collections will uncover new gene functions. The well-defined proteome with detailed information about protein complexes, interactions, modifications, localizations, activities, and stability will provide mechanistic and dynamic understanding of cellular signaling in plants. The growing power of comprehensive databases and computional analysis tools, from DNA, RNA, protein, to metabolite, will offer new ideas and predictions to select and characterize candidates in diverse signaling steps, and to integrate and model plant life. Finally, targeted mutagenesis and cell-type-specific and conditional mutants will be required for functional characterization of genes not currently accessible due to redundancy and lethality.
CONNECTING THE SUGAR SIGNALING NETWORK
Characterization of plant signaling pathways has been remarkably successful based on single signals (e.g. hormones, stresses, and microbial elicitors) and phenotypic or reporter-based observation of insensitive, constitutive, or hypersensitive response mutants, especially in Arabidopsis and rice. General frameworks, often with linear relationship of positive or negative regulators, for many plant signaling pathways have been established based on the initial signals applied in the mutant screens and molecular identification of the mutated genes. However, Arabidopsis mutant screens with an unconventional signal such as Glc (e.g. at high concentrations to trigger developmental arrest of seedlings) have revealed unexpected and extensive molecular connections between Glc and hormone signaling (Fig. 1). The effects of Suc are more complex and can act through Glc-, Fru-, or Suc-specific regulations (Gibson, 2005; Ramon et al., 2008; Hanson and Smeekens, 2009). The first compelling evidence for the interaction of Glc and hormone signaling is provided by the recognition that some Glc-insensitive (gin) mutants are ethylene overproduction (eto1) and constitutive ethylene signaling (ctr1) mutants. Consistently, all examined ethylene-insensitive mutants (etr1 and ein2, ein3, ein6) are Glc-oversensitive (glo) mutants. Molecular analysis supports the antagonistic relationship between Glc and ethylene via opposite control of master transcription factors (TFs), including EIN3 stability and ETHYLENE RESPONSE FACTOR1 transcripts at both high and low Glc levels. However, in a low nitrate condition, Glc repression of chlorophyll accumulation and photosynthesis gene expression mediated by the hexokinase1 (HXK1) Glc sensor is no longer interfered by constitutive ethylene signaling, indicating both HXK1-dependent and HXK1-independent signaling mechanisms in Glc and ethylene responses (Ramon et al., 2008; Cho et al., 2010). Thus, altering assay signals, growth conditions, and the use of informative marker genes for mutant analysis will reveal new signaling connections and uncover layers of complexity with seemingly simple phenotypes.
Similar mutant screens with Glc or Suc by many research groups have also uncovered a positive interaction between Glc and another hormone abscisic acid (ABA). Mutants in ABA biosynthesis (aba2, aba3) and signaling (ABA insensitive3 [abi3], abi4, abi5, abf2, abf3, abf4) are all Glc and Suc insensitive (Gibson, 2005; Ramon et al., 2008; Hanson and Smeekens, 2009). The transcripts for three different classes of TFs in ABA signaling, ABI3, ABI4, and ABI5, are elevated by Glc in an ABA-dependent manner. Glc activation of ABI4 and ABI5 can also be controlled in both HXK1-dependent and HXK1-independent manners and the state of ethylene signaling (Ramon et al., 2008; Cho et al., 2010). The tight link between Glc and ABA is further supported by the analysis of an ABA analog resistance mutant chotto1 (cho1) exhibiting Glc resistance. CHO1 encodes a putative TF with double AP2 domains and acts downstream of ABI4 to control genes involved in primary metabolism and stress response. Unexpectedly, the cho1 mutant displays insensitivity to high nitrate independent of abi4, suggesting multiple and independent functional roles of the same TF (Yamagishi et al., 2009). Although ABA signaling is mediated by complex interactions and expression patterns of distinct classes of TFs, Glc signaling likely activates ABA responses ubiquitously.
A recent study shows surprisingly that overexpression of the SnRK1.1 protein kinase (Arabidopsis KIN10; Snf1-related PK), which is implicated in the global regulation of transcription and metabolism (Baena-González et al., 2007; Baena-González and Sheen, 2008), causes synergistic hypersensitive response to Glc and ABA (Jossier et al., 2009). Although the expression of more than 1,000 KIN10-regulated genes are negatively correlated with Glc, Suc, and CO2 levels in plant cells, a supply of Glc can nevertheless activate overexpressed SnRK1.1 and lead to the altered expression of other genes, PATHOGENESIS RELATED1(PR1), PR2, and PR5, markers of salicylic acid and stress or immune responses (Ramon et al., 2008; Jossier et al., 2009). As SnRK1.1 is included in a heterotrimeric complex, it is proposed that various noncatalytic subunits may allow SnRK1.1 to respond to various stimuli depending on the conditions, the tissues, cell types, or the cellular compartment. The study brings a new interaction between Glc and ABA signaling during transition from the heterotrophic to the autotrophic stage (Jossier et al., 2009). How long-term SnRK1.1 overexpression enhances Glc and ABA sensitivity requires further elucidation. In suspension culture protoplast assays, Suc deprivation and auxin or ABA stimulate SnRK1 promoter activity after 3 d of cultivation in the dark (Radchuk et al., 2009), adding layers of complexity in sugar and hormone interactions.
Recent studies in the SnRK1-repressed pea embryos provide new evidence for its role in coordinated cotyledon emergence and growth via cytokinin-mediated auxin transport and/or distribution. SnRK1 is also required for ABA synthesis and/or signal transduction at an early stage, promoting TCA cycle and amino acid synthesis, essential for later embryo maturation and nutrient storage (Radchuk et al., 2009). The molecular basis underlying SnRK1 regulation of multiple processes is partially revealed by a mesophyll protoplast-based screen, which identified specific basic Leu zippers TFs (bZIP1, 2, 11, 53, 63) as G-box binding factors that act synergistically and redundantly to control KIN10 early target genes. The cell-based assay in combination with cell-type-specific gene expression profile illustrates a powerful approach to functionally screen the 75-member bZIP TFs family with diverse roles (Baena-González et al., 2007; Baena-González and Sheen, 2008; Ramon et al., 2008). A comprehensive analysis of bZIP53 and its interacting partners has uncovered the ternary complex formation between the bZIP heterodimers (bZIP53 and bZIP10/25) and ABI3 important for the expression of seed maturation genes in Arabidopsis (Alonso et al., 2009). It is likely that SnRK1 plays critical roles throughout the embryogenesis process from proliferation to seed maturation and desiccation. Extensive bZIP heterodimerizations (bZIP1, 11, 44, 53, and bZIP10, 25, 63) have also been revealed in maize and Arabidopsis mesophyll protoplasts (Kang et al., 2010). Arabidopsis plant studies uncover the critical functions of bZIP1 and bZIP11 in controlling seedling development and amino acid metabolism, respectively (Hanson et al., 2008; Kang et al., 2010). A creative systems approach has identified bZIP1 in an organic nitrogen-responsive gene network that is also regulated by the master clock control TF CCA1 (Gutiérrez et al., 2008). Interestingly, besides embryogenesis and seed development, SnRK1 and HXKs also play essential roles during seed germination and seedling growth in rice (Cho et al., 2009; Lee et al., 2009). Using the regulation of the α-amylase gene promoter as a model system, various MYB TFs (MYBS1 and MYBGA) have been shown to act downstream of rice SnRK1A to integrate Glc repression, gibberellic acid and ABA signaling, and hypoxia response (Lu et al., 2007; Lee et al., 2009).
Emerging findings have brought numerous exciting new molecular links to support the central roles of SnRK1 (Arabidopsis KIN10/11) in sugar and stress signaling (Baena-González and Sheen, 2008; Ramon et al., 2008; Wingler and Roitsch, 2008; Hanson and Smeekens, 2009). For example, SnRK1 is implicated in resistance to geminivirus infection (Shen et al., 2009), hypersensitive response (Szczesny et al., 2010), and sugar reallocation to roots to tolerate herbivory (Schwachtje et al., 2006). Trehalose-6-P inhibits SnRK1 activity and promotes biosynthesis processes in growth tissues (Zhang et al., 2009). Comprehensive analysis of global transcript levels illustrates the close link between sugar, light, and circadian responses (Usadel et al., 2008) and KIN10-regulated genes (Baena-González and Sheen, 2008). An Arabidopsis WD40 domain-containing myoinnositol polyphosphate 5-phosphase (5PTase13) directly interacts and modulates SnRK1 activity, and plays a regulatory role linking inositol, sugar, and stress signaling (Ananieva et al., 2008). The 5ptase13 mutant is Glc and ABA resistant. However, it remains to be determined whether the 5PTase activity is required for this new function. KIN10/11 also interact with a NAC domain TF, ATAF1/ANAC002. Silencing of the ATAF1 subfamily TFs supports its positive roles in plant development and a potential function in abiotic and biotic stress responses (Kleinow et al., 2009). It will be essential to dissect and define the activities of different TFs acting downstream of KIN10/11, which activate genes involved in catabolism but repress genes important for anabolism (Baena-González et al., 2007; Baena-González and Sheen, 2008; Ramon et al., 2008).
As a plant’s life relies on and revolves around its sugar-generating and sugar-utilization activity, Glc has emerged as a central signaling molecule that interacts with multiple growth hormone signaling pathways via diverse mechanisms and regulators. Studies with mutants and transgenic plants support HXK1-mediated positive relationship between auxin and Glc, while cytokinin reduces Glc responses (Fig. 1; Ramon et al., 2008). On the other hand, KIN10 represses genes involved in auxin, brassinosteroid, and jasmonate metabolism (Baena-González et al., 2007). For Suc-induced expression of anthocyanin biosynthetic genes in Arabidopsis, ABA and jasmonate enhance sugar responses, whereas gibberellic acid blocks sugar activation (Loreti et al., 2008). Numerous studies have suggested that plants possess multiple sugar signaling pathways and the regulation of anthocyanin genes appears to be independent of the HXK1 sensor (Ramon et al., 2008). The regulation of inorganic phosphate starvation genes is dramatically enhanced by sugars via unknown mechanisms, likely through HXK1-dependent and KIN10/11 signaling (Karthikeyan et al., 2007; Fragoso et al., 2009). The differential localization of KIN10 and KIN11 in the chloroplasts in response to inorganic phosphate status is an interesting observation but requires more vigorous examination (Fragoso et al., 2009). As SnRK1 does not bind to Glc directly, the identification of its metabolite regulators (besides ATP and AMP) remains an important future challenge.
The Arabidopsis regulator of G protein signaling-1 (AtRGS1) protein contains seven transmembrane-spanning domains and a C-terminal RGS domain. Genetic evidence supports a novel role of AtRGS1 in sensing high concentration of d-Glc at the plasma membrane. Using fluorescence resonance energy transfer, it is shown that 6% d-Glc but not l-Glc transiently alters the interaction between AtGPA1 and AtRGS1 in vivo. Biochemical analysis suggests that AtGPA1 is a unique heterotrimeric G-protein α-subunit that binds constitutively to GTP. It is suggested that high Glc modulates the GTPase-activating protein activity of AtRGS1 to inhibit AtGPA1 (Johnston et al., 2007). This Glc-sensing mechanism is distinct from the HXK1 pathway. As G-protein subunits have been implicated in functioning in multiple plant hormone and stress signaling pathways, further studies may uncover more Glc signaling interactions (Ramon et al., 2008; Hanson and Smeekens, 2009). Recent characterization of a new sugar-insensitive (sis3) mutant has identified a RING E3 ligase involved in protein degradation, which may bring a new sugar link to other signaling pathways (Vierstra, 2009; Huang et al., 2010). Molecular characterization of high sugar response8 and oversensitive to sugar1 mutants has identified new players involved in Ara synthesis and putative methyltransferase activity, respectively (Li et al., 2007; Gao et al., 2008). Apparently, actin cytoskeleton is required for some HXK1-mediated Glc responses (Balasubramanian et al., 2007). It will be interesting to examine the hormone responses of these new sugar mutants and determine their molecular and cellular links in the sugar and hormonal signaling network (Fig. 1; Ramon et al., 2008).
INTEGRATIVE APPROACHES FOR FUTURE DISCOVERY
The emerging view of the sugar-signaling network is complex and involved in a plethora of cellular processes from embryogenesis to senescence. The sugar signals are sensed directly by multiple sugar sensors or indirectly by sugar-derived metabolites. Specific signaling pathways sensing and relaying Suc and sugar signals other than Glc, as well as metabolic signaling molecules remain to be discovered. New sugar and metabolite sensors, besides the universal HXK1 and the novel AtRGS1 Glc sensors and the energy sensor KIN10/11, likely exist to accommodate the diverse sugar responses discovered in the past two decades. Further studies will provide more surprising regulatory mechanisms underlying sugar and hormone connections in different organs and cell types at various developmental stages (Gibson, 2005; Baena-González and Sheen, 2008; Ramon et al., 2008; Hanson and Smeekens, 2009). More complex interactions between sugar and hormonal signaling in stress responses and organ-specific senescence will likely emerge in future research (Wingler and Roitsch, 2008). To deconvolute the signaling complexity and reconcile controversial observations, it will be very important to precisely define specific functions of HXK1 and KIN10/11 in different subcellular compartments with distinct partners and downstream signaling components in different physiological context and developmental stages of specific cell types (Gibson, 2005; Cho et al., 2006; Baena-González and Sheen, 2008; Ramon et al., 2008; Hanson and Smeekens, 2009). Valuable cell type transcriptome atlas generated by laser microdissection and GFP-based cell sorting will facilitate detailed genetic, molecular, and biochemical characterization of cellular signaling in plants (Brady and Provart, 2009; Jiao et al., 2009; Moreno-Risueno et al., 2010).
The recognition of functional overlaps and lethality hurdles in genetic manipulations, direct and indirect phenotypes, and dynamic feedback responses will assist in designing more sensitive and specific mutant screens and analyses. Genetic and molecular characterization of natural accessions and the large pool of mutant collections will reveal new gene functions and establish previously unexpected molecular links in the cellular signaling network. Ambitious mutant screens and molecular identification of mutations will be greatly accelerated by the powerful cloning by deep sequencing method (Fig. 2; Cuperus et al., 2010).
The biochemical and molecular understanding of cellular signaling mechanisms mediated by protein components will be empowered by the available large-scale proteome data, including protein expression and quantity in different cellular compartments and organelles, protein complex analysis, protein-protein interaction databases, posttranslational modifications, and characterization of protein activities and stability (Fig. 2; Brady and Provart, 2009; Ding et al., 2009; De Bodt et al., 2010). Systematic and hypothesis-driven experiments based on the wealth of accumulated knowledge and databases from DNA, RNA, protein, to metabolite are now a reality. The large-scale high-quality and quantitative data can be explored to discover regulatory candidates in biologically relevant context by integrating databases and computational tools (Fig. 2; Brady and Provart, 2009; De Bodt et al., 2010; Ruffel et al., 2010). It is also important to be able to obtain targeted mutations and to design new mutants using zinc finger nucleases and novel means in plants for functional characterization (Fig. 2; Zhang et al., 2010).
Acknowledgments
Former and current lab members are greatly appreciated for sharing their excitement, knowledge, and discoveries.
References
- Alonso R, Oñate-Sánchez L, Weltmeier F, Ehlert A, Diaz I, Dietrich K, Vicente-Carbajosa J, Dröge-Laser W. (2009) A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 21: 1747–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananieva EA, Gillaspy GE, Ely A, Burnette RN, Erickson FL. (2008) Interaction of the WD40 domain of a myoinositol polyphosphate 5-phosphatase with SnRK1 links inositol, sugar, and stress signaling. Plant Physiol 148: 1868–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein J, Sheen J. (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Sheen J. (2008) Convergent energy and stress signaling. Trends Plant Sci 13: 474–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian R, Karve A, Kandasamy M, Meagher RB, Moore B. (2007) A role of F-actin in hexokinase-mediated glucose signaling. Plant Physiol 145: 1423–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Provart NJ. (2009) Web-queryable large-scale data sets for hypothesis generation in plant biology. Plant Cell 21: 1034–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JI, Ryoo N, Eom JS, Lee DW, Kim HB, Jeong SW, Lee YH, Kwon YK, Cho MH, Bhoo SH, et al. (2009) Role of the rice hexokinases OsHXK5 and OsHXK6 as glucose sensors. Plant Physiol 149: 745–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, Sheen J, Yoo SD. (2010) Low glucose uncouples HXK1-dependent sugar signaling from stress and defense hormone ABA and C2H4 responses in Arabidopsis. Plant Physiol 152: 1180–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, Yoo SD, Sheen J. (2006) Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127: 579–589 [DOI] [PubMed] [Google Scholar]
- Cuperus JT, Montgomery TA, Fahlgren N, Burke RT, Townsend T, Sullivan CM, Carrington JC. (2010) Identification of MIR390a precursor processing defective mutants in Arabidopsis by direct genome sequencing. Proc Natl Acad Sci USA 107: 466–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S, Carvajal D, Hollunder J, Van den Cruyce J, Movahedi S, Inze D. (2010) CORNET: a user-friendly tool for data mining and integration. Plant Physiol 152: 1167–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Richter T, Chen M, Fujii H, Seo YS, Xie M, Zheng X, Kanrar S, Stevenson RA, Dardick C, et al. (2009) A rice kinase-protein interaction map. Plant Physiol 149: 1478–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragoso S, Espındola L, Paez-Valencia J, Gamboa A, Camacho Y, Martınez-Barajas E, Coello P. (2009) SnRK1 isoforms AKIN10 and AKIN11 are differentially regulated in Arabidopsis plants under phosphate starvation. Plant Physiol 149: 1906–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Xin Z, Zheng ZL. (2008) The OSU1/QUA2/TSD2-encoded putative methyltransferase is a critical modulator of carbon and nitrogen nutrient balance response in Arabidopsis. PLoS ONE 3: e1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson SI. (2005) Control of plant development and gene expression by sugar signaling. Curr Opin Plant Biol 8: 93–102 [DOI] [PubMed] [Google Scholar]
- Gutiérrez RA, Stokes TL, Thum K, Xu X, Obertello M, Katari MS, Tanurdzic M, Dean A, Nero DC, McClung CR, et al. (2008) Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci USA 105: 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson J, Hanssen M, Wiese A, Hendrik MMWB, Smeekens S. (2008) The sucrose regulated transcription factor bZIP11 affects amino acid metabolism by regulating the expression of ASPARAGINE SYNTHETASE1 and PROLINE DEHYDROGENASE2. Plant J 53: 935–949 [DOI] [PubMed] [Google Scholar]
- Hanson J, Smeekens S. (2009) Sugar perception and signaling—an update. Curr Opin Plant Biol 12: 1–6 [DOI] [PubMed] [Google Scholar]
- Huang Y, Li CY, Pattison DL, Gray WM, Park S, Gibson SI. (2010) SUGAR-INSENSITIVE3, a RING E3 ligase, is a new player in plant sugar response. Plant Physiol 152: 1889–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Tausta SL, Gandotra N, Sun N, Liu T, Clay NK, Ceserani T, Chen M, Ma L, Holford M, et al. (2009) A transcriptome atlas of rice cell types uncovers cellular, functional and developmental hierarchies. Nat Genet 41: 258–263 [DOI] [PubMed] [Google Scholar]
- Johnston CA, Taylor JP, Gao Y, Kimple AJ, Grigston JC, Chen JG, Siderovski DP, Jones AM, Willard FS. (2007) GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. Proc Natl Acad Sci USA 104: 17317–17322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossier M, Bouly JP, Meimoun P, Arjmand A, Lessard P, Hawley S, Hardie DG, Martine Thomas M. (2009) SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signaling in Arabidopsis thaliana. Plant J 59: 316–328 [DOI] [PubMed] [Google Scholar]
- Kang SG, Price J, Lin PC, Hong JC, Jang JC. (2010) The Arabidopsis bZIP1 transcription factor is involved in sugar signaling, protein networking, and DNA binding. Mol Plant 3: 361–373 [DOI] [PubMed] [Google Scholar]
- Karthikeyan AS, Varadarajan DK, Jain A, Held MA, Carpita NC, Raghothama KG. (2007) Phosphate starvation responses are mediated by sugar signaling in Arabidopsis. Planta 225: 907–918 [DOI] [PubMed] [Google Scholar]
- Kleinow T, Himbert S, Krenz B, Jeske H, Koncz C. (2009) NAC domain transcription factor ATAF1 interacts with SNF1-related kinases and silencing of its subfamily causes severe developmental defects in Arabidopsis. Plant Sci 177: 360–370 [Google Scholar]
- Lee KW, Chen PW, Lu CA, Chen S, Ho TH, Yu SM. (2009) Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Sci Signal 2: ra61. [DOI] [PubMed] [Google Scholar]
- Li Y, Smith C, Corke F, Zheng L, Merali Z, Ryden P, Derbyshire P, Waldron K, Bevan MW. (2007) Signaling from an altered cell wall to the nucleus mediates sugar-responsive growth and development in Arabidopsis thaliana. Plant Cell 19: 2500–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E, Povero G, Novi G, Solfanelli C, Alpi A, Perat P. (2008) Gibberellins, jasmonate and abscisic acid modulate the sucrose-induced expression of anthocyanin biosynthetic genes in Arabidopsis. New Phytol 179: 1004–1016 [DOI] [PubMed] [Google Scholar]
- Lu CA, Lin CC, Lee KW, Chen JL, Huang LF, Ho SL, Liu HJ, Hsing YI, Yu SM. (2007) The SnRK1A protein kinase plays a key role in sugar signaling during germination and seedling growth of rice. Plant Cell 19: 2484–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Busch W, Benfey PN. (2010) Omics meet networks—using systems approaches to infer regulatory networks in plants. Curr Opin Plant Biol 13: 126–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radchuk R, Emery RJN, Weier D, Vigeolas H, Geigenberger P, Lunn JE, Feil R, Weschk W, Weber H. (2009) Sucrose non-fermenting kinase 1 (SnRK1) coordinates metabolic and hormonal signals during pea cotyledon growth and differentiation. Plant J 61: 324–338 [DOI] [PubMed] [Google Scholar]
- Ramon M, Rolland F, Sheen J. (2008) Sugar sensing and signaling in Arabidopsis. The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0117, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffel S, Krouk G, Coruzzi GM. (2010) A systems view of responses to nutritional cues in Arabidopsis: toward a paradigm shift for predictive network modeling. Plant Physiol 152: 445–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwachtje J, Minchin PEH, Jahnke S, van Dongen JT, Schittko U, Baldwin IT. (2006) SNF1-related kinases allow plants to tolerate herbivory by allocating carbon to roots. Proc Natl Acad Sci USA 103: 12935–12940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Reyes MI, Hanley-Bowdoin L. (2009) Arabidopsis protein kinases GRIK1 and GRIK2 specifically activate SnRK1 by phosphorylating its activation loop. Plant Physiol 150: 996–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesny R, Büttner D, Escolar L, Schulze S, Seiferth A, Bonas U. (2010) Suppression of the AvrBs1-specific hypersensitive response by the YopJ effector homolog AvrBsT from Xanthomonas depends on a SNF1-related kinase. New Phytol 187: 1058–1074 [DOI] [PubMed] [Google Scholar]
- Usadel B, Blasing OE, Gibon Y, Retzlaff K, Hohne M, Gunther M, Stitt M. (2008) Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol 146: 1834–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra R. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Wingler AT, Roitsch T. (2008) Metabolic regulation of leaf senescence: interactions of sugar signalling with biotic and abiotic stress responses. Plant Biol 10: 50–62 [DOI] [PubMed] [Google Scholar]
- Yamagishi K, Tatematsu K, Yano R, Preston J, Kitamura S, Takahashi H, McCourt P, Kamiya Y, Nambara E. (2009) CHOTT1, a double AP2 domain protein of Arabidopsis thaliana, regulates germination and seedling growth under excess supply of glucose and nitrate. Plant Cell Physiol 50: 330–340 [DOI] [PubMed] [Google Scholar]
- Zhang F, Maeder ML, Unger-Wallace E, Hoshaw JP, Reyond D, Christian M, Li X, Pierick C, Dobbs D, Peterson T, et al. (2010) High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc Natl Acad Sci USA 107: 12028–12033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Primavesi LF, Jhurreea D, Andraloj PJ, Mitchell RAC, Powers SJ, Schluepmann H, Delatte T, Wingler A, Paul MJ. (2009) Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol 149: 1860–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]