Abstract
The microRNA159 (miR159) family represses the conserved GAMYB-like genes that encode R2R3 MYB domain transcription factors that have been implicated in gibberellin (GA) signaling in anthers and germinating seeds. In Arabidopsis (Arabidopsis thaliana), the two major miR159 family members, miR159a and miR159b, are functionally specific for two GAMYB-like genes, MYB33 and MYB65. These transcription factors have been shown to be involved in anther development, but there are differing reports about their role in the promotion of flowering and little is known about their function in seed germination. To understand the function of this pathway, we identified the genes and processes controlled by these GAMYB-like genes. First, we demonstrate that miR159 completely represses MYB33 and MYB65 in vegetative tissues. We show that GA does not release this repression and that these transcription factors are not required for flowering or growth. By contrast, in the absence of miR159, the deregulation of MYB33 and MYB65 in vegetative tissues up-regulates genes that are highly expressed in the aleurone and GA induced during seed germination. Confirming that these genes are GAMYB-like regulated, their expression was reduced in myb33.myb65.myb101 seeds. Aleurone vacuolation, a GA-mediated programmed cell death process required for germination, was impaired in these seeds. Finally, the deregulation of MYB33 and MYB65 in vegetative tissues inhibits growth by reducing cell proliferation. Therefore, we conclude that miR159 acts as a molecular switch, only permitting the expression of GAMYB-like genes in anthers and seeds. In seeds, these transcription factors participate in GA-induced pathways required for aleurone development and death.
The GAMYB or GAMYB-like genes encode a highly conserved family of R2R3 MYB domain transcription factors that have been implicated in GA signal transduction (Woodger et al., 2003). GAMYB was initially identified in the cereal aleurone, where its expression is induced by GA (Gubler et al., 1995). Here, it binds onto cis-acting GA-responsive elements, leading to the transcriptional activation of genes encoding hydrolases required for starch mobilization during seed germination (Gubler et al., 1999). GAMYB is also strongly expressed in cereal anthers, especially in the tapetum, where it is also induced by GA (Murray et al., 2003; Aya et al., 2009). Emphasizing its importance, microarray analysis found that GAMYB is responsible for the majority of GA-regulated gene expression in both rice (Oryza sativa) aleurone and anthers (Tsuji et al., 2006; Aya et al., 2009). In these tissues, GAMYB is involved in the programmed cell death (PCD) of both the aleurone and tapetum, and in both tissues this process is GA mediated (Guo and Ho, 2008; Aya et al., 2009). Conversely, GAMYB is negatively regulated by the microRNA (miRNA) family miR159 (Tsuji et al., 2006). In rice, mature miR159 is present throughout the plant but is absent in the seed (Tsuji et al., 2006). In the anther, miR159 is coexpressed with GAMYB and finely regulates the levels of this transcription factor (Tsuji et al., 2006).
In Arabidopsis (Arabidopsis thaliana), there is a clade of seven closely related GAMYB-like genes that are potential targets of the three different MIR159 genes (Rhoades et al., 2002). Deep sequencing has found that miR159a and miR159b are overwhelmingly the predominant forms (Fahlgren et al., 2007), and using T-DNA loss-of-function mutants, these two MIR159 genes were demonstrated to be functionally redundant, since a mir159ab double mutant displayed pleiotropic developmental defects (Allen et al., 2007). Although all seven GAMYB-like genes contain potential miR159-binding sites in Arabidopsis, only MYB33 and MYB65 appeared deregulated in mir159ab, a redundant gene pair with similar expression patterns and functions (Millar and Gubler, 2005; Allen et al., 2007). The significance of this deregulation was determined genetically, as all the developmental defects of mir159ab were suppressed in a mir159ab.myb33.myb65 quadruple mutant. This demonstrated that the pleiotropic phenotype seen in mir159ab is due solely to MYB33 and MYB65 activity. This was supported by the expression of a miR159-resistant mMYB33 transgene (carrying a synonymous mutation of the miR159-binding site; Palatnik et al., 2003) that could phenocopy mir159ab (Allen et al., 2007). Analysis of the reporter genes MYB33:GUS and mMYB33:GUS found that although MYB33 was transcribed broadly in the plant, miR159 appears to silence its expression everywhere but in seeds and anthers (Millar and Gubler, 2005). Therefore, similar to GAMYB in cereals, MYB33 protein is predominantly expressed in anthers and seeds. In anthers, rice GAMYB and MYB33/MYB65 are likely to play a similar role. The rice gamyb and myb33.myb65 mutants are male sterile due to the hypertrophy of the tapetum, which expands to occupy the entire locule, causing the microspores to degenerate (Kaneko et al., 2004; Millar and Gubler, 2005). In rice, this was demonstrated to be caused by a failure of the tapetum to undergo PCD (Aya et al., 2009). In seeds, the function of cereal GAMYB and MYB33 and MYB65 may also be conserved; however, no seed phenotype was apparent in myb33.myb65 (Millar and Gubler, 2005), although this may be due to further redundancy, as another close GAMYB-like family member, MYB101, is highly expressed in the seed (Penfield et al., 2006).
Unlike rice GAMYB, which is not involved in the transition to flowering (Kaneko et al., 2004), MYB33 and MYB65 have been implicated in the GA-signaling pathway regulating flowering under short-day conditions (Gocal et al., 2001; Achard et al., 2004). MYB33 mRNA levels were reported to increase in the shoot apex upon short-day to long-day shifts or GA application, treatments that also activate the expression of the flowering integrator gene LEAFY (LFY) and induce flowering (Gocal et al., 2001). As the LFY promoter has a potential MYB-binding site essential for its GA activation (Blazquez and Weigel, 2000) to which MYB33 can bind, it was predicted that MYB33 may be the transcription factor transducing the GA signal (Gocal et al., 2001). Supporting this hypothesis, the expression of a 35S:MIR159a transgene in the Arabidopsis ecotype Landsberg erecta decreased MYB33 and LFY steady-state mRNA levels and led to late flowering under short days (Achard et al., 2004). However, Schwab et al. (2005) also generated 35S:MIR159a transgenic Arabidopsis lines (ecotype Columbia) but could find no alteration to MYB33 and MYB65 transcript levels or flowering time. Although a possible difference in ecotypes could be the explanation, further analysis is required to clarify the roles of MYB33, MYB65, and miR159 in flowering.
Using physiological, microscopic, and molecular analyses on myb33.myb65 and mir159ab mutants, we found that miR159 acts as a molecular switch confining the expression of MYB33 and MYB65 to seeds and anthers. This repression is not released by GA, and consequently, MYB33 and MYB65 play no part in GA-mediated growth or flowering in vegetative tissues. We demonstrate that in seeds, these GAMYB-like genes together with MYB101 regulate the expression of genes that are induced by GA during germination and that they promote but are not essential for the progression of PCD in the aleurone.
RESULTS
MYB33 and MYB65 Are Not Essential for GA-Mediated Growth or Flowering
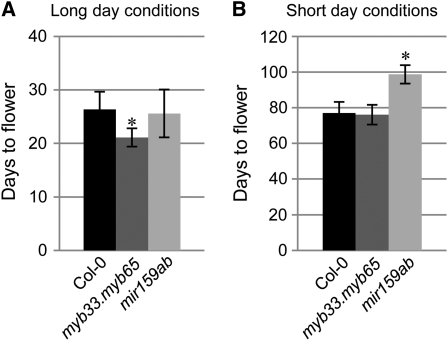
Previously, there have been conflicting reports on the involvement of MYB33 and miR159 in the promotion of GA-mediated flowering (Gocal et al., 2001; Achard et al., 2004; Schwab et al., 2005). In order to clarify the role of the miR159-MYB33/MYB65 pathway in GA-mediated floral induction, we examined the flowering time and GA response of the double mutants myb33.myb65 and mir159ab. Under long-day conditions, the flowering time of mir159ab was similar to the wild type (n = 34, P = 0.35), whereas myb33.myb65 flowered slightly earlier (n = 35, P = 4.71E-12; Fig. 1A). Under short-day conditions, in which GA promotes flowering (Wilson et al., 1992), myb33.myb65 had the same flowering time as the wild type (n = 30 and 31, respectively, P = 0.52; Fig. 1B). Conversely, mir159ab flowered later, and only 23 out of 32 plants flowered at 110 d after sowing. The flowering time of the 23 plants was delayed 20 d (P < 10−5; Fig. 1B). As mir159ab has a strong morphological phenotype, it is unsure whether this delay is due to the morphological defects of mir159ab or to MYB33 and MYB65 expression per se. However, it is clear that MYB33 and MYB65 deregulation in mir159ab does not promote flowering.
Figure 1.
Flowering time of myb33.myb65 and mir159ab. Flowering time under long-day (A) and short-day (B) conditions of wild-type Columbia-0 (Col-0), myb33.myb65, and mir159ab is shown. Error bars represent sd, and asterisks mark statistically significant changes.
To examine the GA response of myb33.myb65 and mir159ab, we applied a series of GA treatments and scored their flowering time under short-day conditions. The response of myb33.myb65 was similar to the wild type, since plants sprayed with GA flowered approximately 20 d before the control plants (Fig. 2A). However, the effect of GA on mir159ab plants was more subtle. Although the flowering time of GA-treated mir159ab was substantially later than GA-treated myb33.myb65 and the wild type, all GA-sprayed mir159ab plants flowered (n = 16), whereas only seven out of 12 ethanol-sprayed mir159ab plants flowered after 110 d. These data demonstrate that MYB33 and MYB65 are not the main effectors mediating the flowering response to GA.
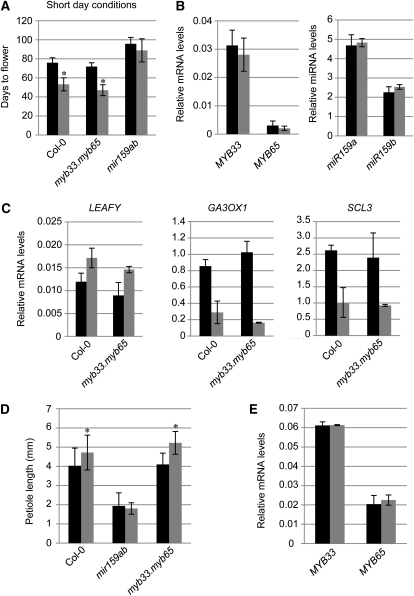
Figure 2.
GA response of the miR159-GAMYB pathway in vegetative tissues. A, Effect of GA and control (ethanol) treatments on the flowering time of wild-type Columbia-0 (Col-0) and mutant plants under short-day conditions. B, mRNA levels of MYB33 and MYB65 and the levels of mature miR159a and miR159b in wild-type SARs. C, LFY, GA3OX1, and SCL3 mRNA levels in wild-type Columbia-0 and myb33.myb65 SARs grown under short-day conditions. D, Petiole length of 23-d-old third leaves of plants grown under long-day conditions. E, mRNA levels of MYB33 and MYB65 in 32-d-old wild-type rosettes grown under short-day conditions. Black bars represent control (ethanol-treated) plants, and gray bars represent GA-treated plants. Error bars represent sd, and asterisks mark statistically significant changes.
We measured the effect of GA application on the steady-state transcript levels of MYB33 and MYB65 and the levels of mature miR159a and miR159b in the wild-type shoot apex regions (SARs). We also measured the transcript levels of the GA-responsive genes LFY, GIBBERELLIN3 β-HYDROXYLASE1 (GA3OX1; Cowling et al., 1998), and SCARECROW-LIKE3 (SCL3; Ogawa et al., 2003) in myb33.myb65 and wild-type SARs to confirm that the plants were responding to the GA treatment. The levels of MYB33, MYB65, miR159a, and miR159b remained unchanged after the application of GA (Fig. 2B). Conversely, the transcript levels of LFY were slightly up-regulated and the levels of GA3OX1 and SCL3 were down-regulated in GA-treated myb33.myb65 and wild-type plants (Fig. 2C). These controls confirmed that the plants had perceived and responded to the hormone.
We also analyzed the requirement of MYB33 and MYB65 for GA-mediated growth of long-day-grown rosettes. To quantify this, we measured the petiole length of third leaves, as this has been shown to be a good indicator of GA response in the ecotype Columbia (Gocal et al., 2001). Again, myb33.myb65 responded to GA similar to wild-type plants (n = 25; Fig. 2D) and MYB33 and MYB65 mRNA levels failed to change with GA application (Fig. 2E). In contrast, mir159ab did not display a growth response upon GA application (n = 25; Fig. 2D). Taken together, these data demonstrate that MYB33 and MYB65 are not required for flowering under short-day conditions or for GA-induced growth and that their mRNA levels are not up-regulated by GA in the Arabidopsis ecotype Columbia. Finally, no dramatic induction of MYB33 or MYB65 was detected in SARs in the Arabidopsis ecotype Landsberg erecta (Supplemental Fig. S1), implying that the mRNA levels of the GAMYB-like genes in SARs are independent of GA in both ecotypes.
miR159 Represses MYB33 Expression to Biologically Inconsequential Levels in Vegetative Tissues
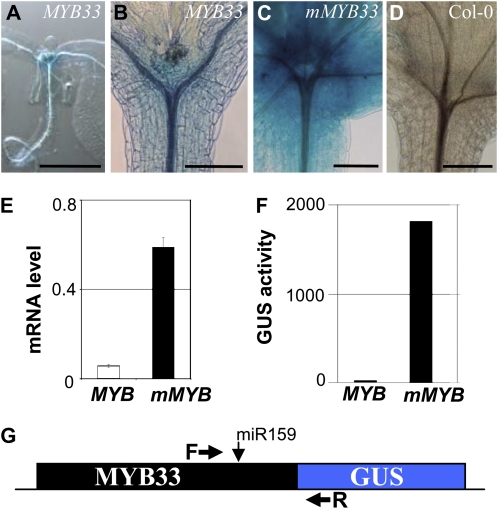
Since myb33.myb65 displayed a wild-type phenotype in all the physiological aspects tested, this raised the issue of whether MYB33 and MYB65 have any role in vegetative tissues. To determine this, we first analyzed the expression of a MYB33:GUS transgene in the SAR. We had previously shown that miR159 represses MYB33 in these tissues (Millar and Gubler, 2005). However, as MYB33 mRNA is still detectable at the SAR (Allen et al., 2007), we wanted to ascertain whether MYB33 protein is absent or whether it is restricted to a select group of cells by miR159. Of five independent MYB33:GUS lines that had previously shown strong staining in anthers (Millar and Gubler, 2005), very weak staining was only observable in three lines after extended staining periods (5 d) and tissue clearing (Fig. 3, A and B). In these three lines, GUS staining appeared present in all cells of the SAR (Fig. 3A). By contrast, the expression of the mMYB33:GUS transgene (miR159 resistant; Millar and Gubler, 2005) in five independent lines was extremely strong throughout the SAR, and staining appeared after only 16 h of incubation with the substrate (Fig. 3C). Taken together, these results suggest that MYB33 expression is strongly repressed by miR159 throughout the SAR. This applies to other vegetative tissues, since no GUS signal was detected in leaves or roots of MYB33:GUS plants even after prolonged staining.
Figure 3.
MYB33 is strongly repressed by miR159 in vegetative tissues. A to D, Histochemical staining for GUS activity in 14-d-old seedlings of MYB33:GUS after 5 d of staining (A and B), mMYB33:GUS after 16 h of staining (C), and wild-type Columbia-0 (Col-0) after 5 d of staining (D). Bars = 100 μm (A) and 200 μm (B–D). E, Levels of MYB33:GUS (MYB) and mMYB33:GUS (mMYB) mRNA in five independent lines detected by qRT-PCR. F, GUS activity in five independent MYB33:GUS and mMYB33:GUS averaged lines. Error bars represent sd. G, Illustration depicting the positions of the primers used to quantify the mRNA of the transgenes. The primers span the miRNA target site of the MYB33:GUS construct and therefore only detect uncleaved mRNA.
To further characterize the silencing of MYB33 by miR159, we measured both mRNA levels and GUS activity of the transgenes in five independent MYB33:GUS and mMYB33:GUS lines, where we assumed that GUS activity reflects MYB33:GUS protein levels. On average, mMYB33:GUS mRNA levels were approximately 10-fold higher than those of MYB33:GUS (Fig. 3E). In contrast, GUS activity levels were on average almost 400-fold higher in mMYB33:GUS compared with MYB33:GUS lines (Fig. 3F). As GUS activity in MYB33:GUS lines (4.64 ± 2.83) was only slightly higher than in nontransgenic lines (normalized to zero), such a high fold-level difference appears due to the almost complete absence of MYB33:GUS protein. Together, these data suggest that miR159 represses MYB33 expression not only by reducing mRNA levels but also by repressing the translation of any remaining MYB33 mRNA.
Finally, we carried out a microarray analysis on the SAR of 15-d-old myb33.myb65 plants. Very few genes were found to be differentially expressed between the wild type and myb33.myb65. Only two genes met the P ≤ 0.005, 2-fold change criteria (invertase [At1g62770], fold change = 2.19, P < 0.003; and expressed protein [At3g52060], fold change = 2.14, P < 0.003). Relaxation of the P value (P < 0.05) but with a 3-fold change cutoff again found only two genes to be differentially expressed (formaldehyde dehydrogenase [At5g14780], fold change = 3.2, P < 0.006; and nitrate reductase 1 [At1g77760], fold change = −3.84, P < 0.006). Thus, the transcriptomes of wild-type and myb33.myb65 SARs are almost identical, which supports the notion that miR159 completely represses these transcription factors under normal conditions in the SAR.
MYB33 and MYB65 Activate the Expression of Aleurone-Related Genes in mir159ab Vegetative Tissues
Next, we performed a transcriptomic analysis on the SAR of 15-d-old mir159ab plants in order to identify genes regulated by MYB33 and MYB65 and elucidate their biological role. Using a 2-fold change and a P < 0.005 cutoff, we found 121 up-regulated and 45 down-regulated genes in mir159ab when compared with the wild-type (for a complete list, see Supplemental Table S1). To validate that these genes were differentially expressed, quantitative real-time PCR (qRT-PCR) was performed on 36 of these genes that covered a broad range of fold-change levels. In all instances, gene expression was confirmed to be altered, and in many cases the fold changes determined by qRT-PCR were similar to those determined by the microarrays, implying that the microarray data were highly reliable (Table I; Supplemental Table S1). As expected, MYB33 and MYB65 were among the up-regulated genes in mir159ab, as they are no longer repressed by miR159 (Allen et al., 2007). Only one other predicted miR159 target was found to be up-regulated, OLIGOPEPTIDE TRANSPORTER1 (Supplemental Table S1), and no low-complementary targets were up-regulated (up to seven mismatches; http://bioinfo3.noble.org/miRNA/miRU.htm). This is consistent with the notion that plant miRNAs have highly specific effects on the transcriptome (Schwab et al., 2005). This also confirms that the majority of gene expression changes in mir159ab are due to MYB33 and MYB65 deregulation. In agreement with this, the analysis of gene expression in the rosettes of a mMYB33 transgenic line that had a phenotype indistinguishable from mir159ab plants (line 2; Allen et al., 2007) revealed similar gene expression changes to mir159ab (Table I).
Table I. Transcript profiling in mir159ab plants.
Fold change expression levels of select genes found to be misexpressed in mir159ab according to microarray analysis were measured by qRT-PCR in SARs, 28-d-old whole rosettes (WR), 3-d-old imbibed seeds (Seed), and flowers of mir159ab as well as in 28-d-old rosettes of transgenic lines carrying a mMYB33 construct (mMYB; Allen et al., 2007). The rice homologs of these genes that are regulated by GAMYB (Tsuji et al., 2006) in seeds (-S) or anthers (-A) are presented in the table with the percentage of similarity to the Arabidopsis counterparts and the alignment score calculated by the BLAST program in parentheses (National Center for Biotechnology Information; http://blast.ncbi.nlm.nih.gov/Blast.cgi).
| Arabidopsis Genome Initiative Identifier | Gene Title/Putative Function | Fold Change in mir159ab |
mMYBWR | Rice Homolog Up-Regulated by GAMYB |
|||||
| Array | SAR | WR | Seed | Flower | Gene Identifier | Similarity | |||
| Up-regulated genes in mir159ab | |||||||||
| At4g36880 | Cys proteinase (CP1) | 14.40 | 14.93 | 35.30 | −1.31 | 4.98 | 43.16 | AK071495-S | 67% (338) |
| At5g42650 | Allene oxide synthase (AOS) | 9.95 | 4.66 | 1.62 | 2.95 | 1.81 | 2.20 | ||
| At1g75750 | GA-regulated gene 1 (GASA1) | 6.49 | 5.92 | 5.48 | 1.84 | 3.64 | 2.00 | ||
| At4g28040 | Nodulin MtN21 transporter | 5.94 | 12.12 | 10.75 | 15.01 | 2.58 | 6.28 | AK106554-S | 53% (158) |
| AK070604-A | 47% (125) | ||||||||
| At1g44350 | Indole acetic acid-amino acid hydrolase 6 | 5.41 | 2.36 | 1.42 | 5.88 | 2.87 | – | AK110647-A | 64% (145) |
| At3g22880 | Meiotic recombination (DMC1) | 5.32 | 3.66 | 2.68 | 2.66 | 4.50 | 2.22 | ||
| At3g11440 | MYB65 | 4.10 | 3.81 | 4.93 | 2.85 | 5.13 | 2.09 | ||
| At5g49360 | Glycosyl hydrolase (BXL1) | 3.97 | 3.74 | 2.81 | −1.33 | 2.74 | 2.06 | AK072485-A | 43% (112) |
| At1g02640 | Glycosyl hydrolase (BXL2) | 3.96 | 4.07 | 4.50 | −1.12 | 2.93 | 4.34 | AK072485-A | 42% (126) |
| At3g48740 | Nodulin MtN3 transporter | 3.95 | 4.90 | 2.39 | −1.31 | 1.57 | 2.23 | AK103266-A | 75% (263) |
| At5g06100 | MYB33 | 3.69 | 3.75 | 2.85 | 2.96 | 3.45 | 10.72 | ||
| At3g45310 | Cys proteinase | 3.21 | 2.47 | 1.56 | −1.20 | 2.19 | – | AK071495-S | 55% (208) |
| At3g16380 | Poly(A)-binding protein (PAB6) | 3.17 | 5.90 | 1.21 | 1.56 | 1.31 | – | ||
| At3g47010 | Glycosyl hydrolase | 3.12 | 2.67 | 2.14 | 4.50 | 1.70 | 2.10 | AK072485-A | 69% (654) |
| At3g14067 | Subtilase protease | 2.58 | 1.87 | 1.43 | −1.23 | −1.41 | – | AK105112-A | 52% (437) |
| AK106823-A | 49% (280) | ||||||||
| At3g28220 | Metalloproteinase | 2.45 | 2.93 | 1.37 | 20.93 | 1.28 | – | ||
| At2g03710 | MADS box protein (AGL3) | 2.35 | 2.94 | 1.82 | 3.58 | 1.06 | 1.90 | ||
| At1g58270 | Metalloproteinase | 2.11 | 1.93 | 1.46 | −1.22 | 1.57 | – | ||
| At5g27360 | Sugar porter (SFP2) | 2.05 | 4.14 | 1.18 | 3.01 | −1.10 | – | AK069132-A | 42% (144) |
| At5g09220 | Amino acid permease (AAP2) | 2.05 | 2.35 | 1.38 | 1.43 | −1.30 | – | AK106814-A | 61% (346) |
| AK067118-S | 44% (117) | ||||||||
| Down-regulated genes in mir159ab | |||||||||
| At5g55450 | Protease inhibitor | −7.15 | −1.48 | −2.34 | −1.08 | −1.33 | −3.38 | ||
| At5g28640 | SSXT-related protein (AN3) | −2.36 | −1.45 | 1.02 | −1.51 | −1.10 | −1.35 | ||
| At2g19590 | 1-Aminocyclopropane-1-carboxylate oxidase 1 (ACO1) | −2.35 | −8.14 | −4.30 | −1.29 | −1.41 | −6.60 | AK058296-A | 61% (218) |
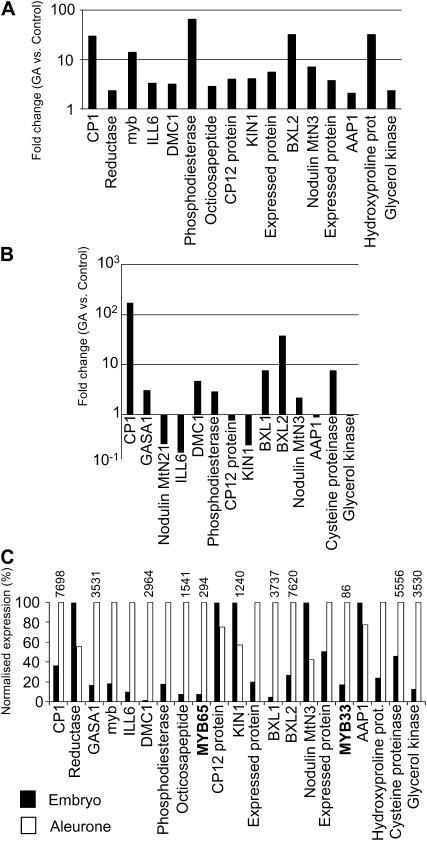
Interestingly, 16 out of the 39 genes that are more than 3-fold up-regulated in mir159ab are induced by GA in 3-, 6-, or 9-h-imbibed seeds of the GA-deficient mutant ga1-3, according to microarray data (Fig. 4A; Schmid et al., 2005). To confirm this, we carried out qRT-PCR analysis on 14 of the mir159ab up-regulated genes and found eight of them to be induced by GA in 30-h-imbibed ga1-3 seeds (Fig. 4B). This analysis identified three more genes that are up-regulated by GA in the seed: BETAXYLOSIDASE1 (BXL1; At5g49360), GA-REGULATED GENE1 (GASA1; At1g75750), and a putative Cys proteinase (CP; At3g45310). Therefore, approximately half of the genes identified on the arrays as being more than 3-fold up-regulated in mir159ab shoot apex tissues are induced by GA in the seed. Of these genes, CYSTEINE PROTEINASE1 (CP1; At4g36880), GASA1, and DISRUPTED MEIOTIC CONTROL1 (DMC1; At3g22880), three of the most up-regulated genes in our array, had been previously shown to be induced by GA in the seed (Bouquin et al., 2001; Ogawa et al., 2003).
Figure 4.
Many genes up-regulated in mir159ab are GA regulated in seeds and preferentially expressed in the aleurone. A, Early GA induction in ga1-3 seeds of 16 genes up-regulated in mir159ab. B, Late GA response in ga1-3 seeds of 14 up-regulated genes in mir159ab as determined by qRT-PCR. C, Normalized expression levels for MYB33, MYB65, and the GA-induced genes in the aleurone and embryo. Numbers on top of the bars are absolute expression values in the aleurone. Data for A were obtained from AtGenExpress (Schmid et al., 2005), and data for C were obtained from the Arabidopsis eFP browser (Winter et al., 2007).
According to microarray data, many of these seed GA-responsive genes are predominantly expressed in the aleurone, where MYB33 and MYB65 are also preferentially transcribed (Fig. 4C; Penfield et al., 2006, Winter et al., 2007). Moreover, nine of these genes had an aleurone microarray expression value of over 1,000, indicating that they are very strongly expressed in this tissue (Fig. 4C; Penfield et al., 2006, Winter et al., 2007). In addition, many of the up-regulated genes encoded transporters and hydrolases (Supplemental Table S2). Most of the transporters up-regulated in the array are predicted to transport nutrients like sugars, amino acids, and proteins. Among the up-regulated hydrolases, seven were predicted to encode proteases and five were predicted or have been confirmed to be involved in cell wall degradation (Supplemental Table S2). Functions such as this would be consistent with the secretory role of the aleurone and its final progression to PCD through autophagy (Fath et al., 2000). All these data strongly support the notion that the GAMYB class of transcription factors are positive regulators of GA signaling during seed germination (Gubler et al., 1995; Woodger et al., 2003). Finally, the function of these GAMYB transcription factors is highly conserved, as many common homologs are up-regulated by GAMYB in mir159ab vegetative tissues and in anthers and embryoless half seeds of rice (Table I; Tsuji et al., 2006).
As MYB33 and MYB65 are globally deregulated in mir159ab (Allen et al., 2007), we examined whether these aleurone-related genes have also been up-regulated throughout mir159ab. qRT-PCR found that CP1, GASA1, DMC1, BLX1, and BXL2 genes were all up-regulated in whole rosette and floral tissues of mir159ab (Table I). The fact that CP1, BLX1, and BLX2 were not induced in mir159ab seeds probably reflects the fact that they are already highly expressed in this tissue. However, in addition to these aleurone-related genes, many other genes were found to be globally affected (Table I; Supplemental Table S1). This includes ALLENE OXIDASE SYNTHASE1 and ACC OXIDASE1, which code for key enzymes in jasmonic acid and ethylene biosynthesis, respectively. This suggests that the overall gene expression changes observed in mir159ab likely result from the alteration of numerous pathways.
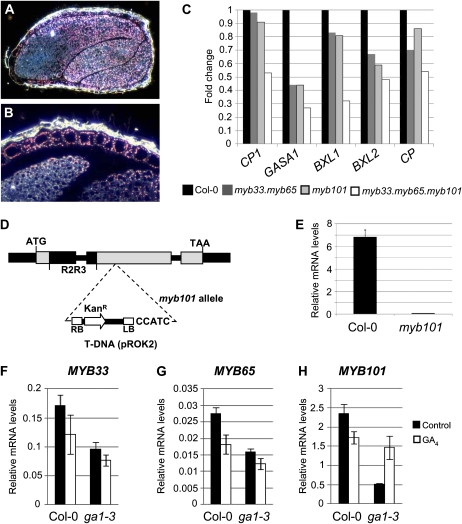
The Up-Regulated mir159ab Genes Are Down-Regulated in myb33.myb65.myb101 Seeds
The up-regulation in mir159ab of many genes highly expressed in the aleurone prompted us to examine whether MYB33 protein is in fact present in this tissue. To examine this, we stained and sectioned 30-h-imbibed MYB33:GUS seeds. MYB33 expression was detected in the embryo (Fig. 5A) and was especially strong throughout the aleurone layer (Fig. 5B), confirming that the presence of GAMYB activity in the aleurone is conserved in both monocotyledonous and dicotyledonous plants. If the genes up-regulated in mir159ab SAR are activated by MYB33 and MYB65 during germination, then their expression should be down-regulated in myb33.myb65 seeds. We measured the mRNA abundance of CP1, CP, GASA1, BXL1, and BXL2 in 30-h-imbibed myb33.myb65 seeds. We chose those genes because they were up-regulated by GA in 30-h-imbibed ga1-3 seeds (Fig. 4B). All these genes were slightly down-regulated in myb33.myb65 with the exception of CP1 (Fig. 5C). However, further redundancy of GAMYB activity in seeds is likely, as MYB101, another GAMYB-like gene, is highly transcribed in the aleurone (Penfield et al., 2006). We obtained a myb101 mutant from the SALK collection (SALK_061355) in which the T-DNA had inserted within the second exon of the gene (Fig. 5D). qRT-PCR analysis determined that the expression of the myb101 allele was 100-fold lower compared with the MYB101 allele (Fig. 5E). Despite this, the myb101 mutant displayed a wild-type phenotype. In myb101 seeds, the levels of GASA1, BXL1, and BXL2 were reduced to the same extent as in myb33.myb65 seeds (Fig. 5C). Using this myb101 allele, we obtained the myb33.myb65.myb101 triple mutant that displayed a wild-type phenotype except for male sterility, similar to myb33.myb65. The mRNA levels of CP1, CP, GASA1, BXL1, and BXL2 were reduced 2-fold or more in myb33.myb65.myb101 seeds (Fig. 5C). These data demonstrate that these three transcription factors are regulating similar genes in seeds and that there exists a tight regulatory relationship between GAMYB-like activity and the mRNA abundance of these five genes, whether it is in the aleurone of wild-type plants or in vegetative tissues of mir159ab.
Figure 5.
Identification of GAMYB-like regulated genes in the seed. A and B, Visualization of MYB33:GUS expression (pink crystals) in the seed (A) and aleurone (B) with dark-field optics. C, Fold changes in mRNA levels of the GA-responsive genes CP1, GASA1, BXL1, BXL2, and CP in myb33.myb65, myb101, and myb33.myb65.myb101 mutants as determined by qRT-PCR. D, Genomic structure of the myb101 allele (SALK_061355). The conserved R2R3 MYB domain (R2R3) is represented in the gene. E, MYB101 mRNA levels in myb101 mutant seeds. F to H, mRNA levels of MYB33, MYB65, and MYB101 in wild-type Columbia-0 (Col-0) and ga1-3 seeds treated with GA. Error bars represent sd.
As CP1, CP, GASA1, BXL1, and BXL2 are GA regulated in the seed, we investigated whether any of the three GAMYB-like genes could be candidates for transducing this GA signal in seeds by measuring their mRNA levels in GA-treated ga1-3 seeds. Only MYB101 mRNA levels were increased upon GA treatment (Fig. 5, F–H). Therefore, although MYB33 and MYB65 are controlling the expression of seed GA-regulated genes, their mRNA abundance does not appear GA regulated in the seed.
MYB33, MYB65, and MYB101 Promote PCD in the Aleurone
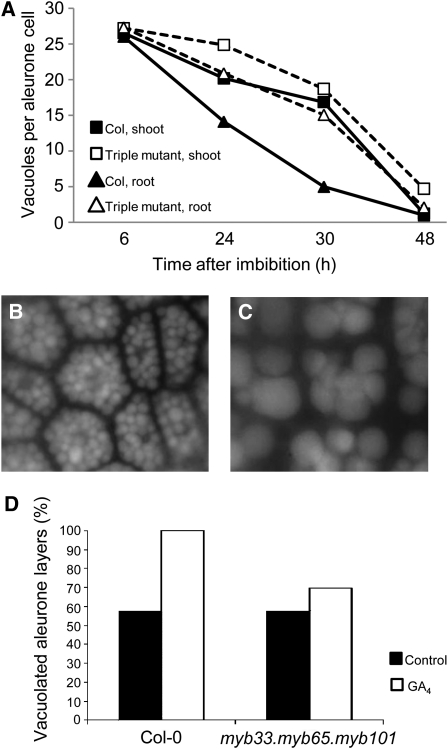
As GAMYB has been implicated in the PCD of the aleurone in barley (Hordeum vulgare; Guo and Ho, 2008), we examined whether this process is compromised in myb33.myb65.myb101. We visualized the aleurone layers of myb33.myb65.myb101 under UV light, which causes protein storage vacuoles (PSVs) to fluoresce (Bethke et al., 2007). Shortly after imbibition, aleurone cells contain many PSVs, but during germination they coalesce, resulting in a decrease in their numbers (Bethke et al., 2007). At late stages of germination, only one big lytic vacuole occupies the aleurone cell. This process is called vacuolation. We determined the vacuolation rate of the myb33.myb65.myb101 aleurone layers. We distinguished between the area of the aleurone that is in contact with the embryo shoot and the area of the aleurone in contact with the radicle, as they have different vacuolation rates (Bethke et al., 2007). The vacuolation rate of the area of the aleurone in contact with the radicle was slower in myb33.myb65.myb101 compared with the wild type, as 24 and 30 h after imbibition the mutant aleurone cells contained more PSVs (P < 0.0001; Fig. 6A). However, the triple mutant was able to vacuolate completely after 48 h. No difference was seen in the aleurone in contact with the embryo shoot.
Figure 6.
The Arabidopsis GAMYB-like genes promote aleurone PCD. A, Number of PSVs per aleurone cell in wild-type Columbia-0 (Col) and myb33.myb65.myb101 (triple mutant) seeds at different time points during germination. n = 50 cells, and sd ranges from 0 to 7.71. Error bars have been omitted for clarity. B and C, Typical images of aleurone cells incubated at 30°C for 5 d without (B) and with (C) GA. D, Percentage of vacuolated aleurone layers after 5 d of incubation at 30°C with or without GA (n = 20).
It has been previously shown that isolated aleurone layers that are incubated at 30°C do not vacuolate (Fig. 6B) unless they are supplemented with GA (Fig. 6C; Bethke et al., 2007). To determine if the Arabidopsis GAMYB-like genes are involved in this GA process, we incubated myb33.myb65.myb101 and wild-type aleurone layers at 30°C with and without GA and counted the number of aleurone layers that had vacuolated after 5 d. When incubated in the control medium, 50% of the wild-type and mutant aleurone layers had vacuolated after 5 d (Fig. 6D). However, only 70% of the mutant aleurone layers incubated with GA vacuolated compared with 100% of the wild-type aleurone layers (Fig. 6D). These data demonstrate that MYB33, MYB65, and MYB101 promote, but are not essential for, the vacuolation of the aleurone. Accordingly, the triple mutant seeds germinate as efficiently as wild-type seeds under normal conditions (data not shown).
MYB33 and MYB65 Antagonize Growth in Vegetative Tissues through Inhibition of Cell Proliferation
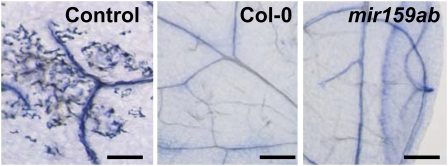
As the up-regulated genes in mir159ab might be involved in the progression of PCD in the aleurone, we wanted to determine if cells in the aerial organs of mir159ab were undergoing PCD as a result. However, staining of 13-d-old rosettes with the dye trypan blue, which stains dead and dying cells, failed to find any evidence of enhanced PCD in mir159ab rosettes (Fig. 7). Therefore, we carried out a microscopic analysis of mir159ab to determine the consequences of the up-regulation of MYB33 and MYB65 in mir159ab.
Figure 7.
Trypan blue staining of third leaves of 13-d-old wild-type Columbia-0 (Col-0) and mir159ab plants. Necrotic rosette leaves from 6-week-old wild-type plants were used as a positive control. Bars = 2 mm. [See online article for color version of this figure.]
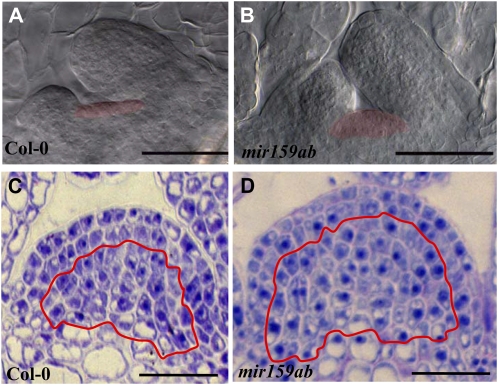
Anatomical differences appeared very early in mir159ab development. The shoot apical meristem (SAM) of 4-d-old mir159ab plants appeared dome shaped and enlarged, compared with the flat SAM of wild-type plants (n = 5; Fig. 8, A and B). An enlarged SAM was also present in 14-d-old mir159ab plants (n = 2; Fig. 8, C and D). The organization of the tissue layers of the mir159ab SAM appeared normal, as the L1, L2, and L3 layers were clearly distinguishable, although the mir159ab SAM had more cells in L3 and subtending meristematic (nonvacuolated) tissue (Fig. 8D). Newly initiated mir159ab rosette leaves are flat but gradually curl upward with time (Allen et al., 2007). We studied the structure of 24-d-old fifth leaves of mir159ab that were curled at that age. They were either transversely sectioned (Fig. 9, A and B) or cryofractured and analyzed with scanning electron microscopy (cryo-SEM; Fig. 9, C and D). The most notable difference in mir159ab leaves was that mesophyll cells were considerably larger than their wild-type counterparts (Fig. 9, A and B). Cell size measurements determined that mir159ab palisade and spongy mesophyll cells were approximately 217% and 274% larger, respectively (Fig. 9E). mir159ab leaves also had approximately 58% fewer mesophyll cells per mm2 than wild-type leaves (Fig. 9F). Since mir159ab leaves are also smaller, this implies that they contain far fewer mesophyll cells.
Figure 8.
mir159ab displays a hypertrophic SAM. A and B, Differential interference contrast microscopy of cleared tissue from 4-d-old wild-type Columbia-0 (Col-0; A) and mir159ab (B) seedlings. SAM regions have been shaded. C and D, Median longitudinal cross-sections of 14-d-old wild-type (C) and mir159ab (D) SAMs. The red line is delimiting the L3 and subtending meristematic region. Bars = 50 μm.
Figure 9.
Alterations to leaf development in mir159ab plants. A and B, Transverse sections of 24-d-old wild-type Columbia-0 (Col-0; A) and mir159ab (B) fifth leaves. C and D, Cryo-fracture of 24-d-old wild-type (C) and mir159ab (D) fifth leaves. E and F, Differences between the wild type and mir159ab concerning mesophyll cell size (E) and cell density (F). G to J, SEM analysis of wild-type adaxial (G) and abaxial (H) as well as mir159ab adaxial (I) and abaxial (J) surfaces. K and L, Epidermal cell number (K) and cell size (L). M and N, Midvein cross-sections in the wild type (M) and mir159ab (N). P, Phloem; X, xylem. O to R, Venation pattern of 14-d-old wild-type (O) and mir159ab (Q) cotyledons as well as wild-type (P) and mir159ab (R) first leaves. Error bars represent sd, and bars = 50 μm (A–D, G–J, M, and N) and 200 μm (O–R). [See online article for color version of this figure.]
Further analysis was performed using SEM on the epidermal surfaces of mir159ab and wild-type leaves (Fig. 9, G–J). Similar to the mir159ab mesophyll layers, both abaxial and adaxial surfaces of mir159ab had fewer cells when compared with the wild type (Fig. 9K). However, the mir159ab epidermal cells were smaller (Fig. 9L). This reduction in cell number and size was more pronounced on the adaxial surface, causing the ratio between the lengths of the adaxial and abaxial surfaces to be reduced from 0.98 ± 0.02 (n = 2) in the wild type to 0.79 ± 0.02 (n = 2) in mir159ab leaves. This may have resulted in the upward curling of the leaves. Furthermore, the number of cells composing the vascular bundles was considerably reduced in both xylem and phloem (Fig. 9, M and N), and the venation pattern of cotyledons and leaves was also simpler (Fig. 9, O–R). This, together with the fact that all organs of mir159ab plants are smaller (Allen et al., 2007), suggests that the major consequence of the deregulation of MYB33 and MYB65 in mir159ab is a reduction of cell proliferation.
We reasoned that this disruption of cell proliferation could be due to the up-regulation of cell cycle inhibitor genes. We measured the expression levels of the seven members of the KIP-RELATED PROTEIN (KRP; De Veylder et al., 2001) family of cell cycle inhibitors and found that KRP7 was up-regulated throughout mir159ab (Supplemental Fig. S2). However, an RNA interference construct generated to silence KRP7 in the mir159ab background failed to suppress the mir159ab phenotype (Supplemental Fig. S2). This suggested that the increased KRP7 levels in mir159ab are only indicative of the decrease in cell proliferation. However, it is clear that the expression of MYB33 and MYB65 in vegetative tissues antagonizes growth through the disruption of cell proliferation, an outcome completely counterintuitive to the notion that these GAMYB-like genes promote GA-mediated growth.
DISCUSSION
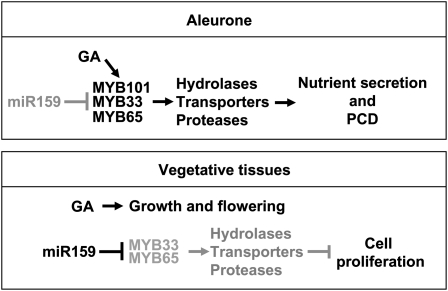
Although much is known about plant miRNAs and their regulatory relationship with their target genes, what downstream genes or processes these regulatory modules control is in most instances unknown. Here, we have identified a set of genes activated by MYB33 and MYB65 that are usually strongly expressed in the aleurone but are globally up-regulated in mir159ab. GAMYB-like activity in the aleurone promotes PCD, whereas in rosette tissues it antagonizes cell proliferation (Fig. 10). Although these activated genes are GA up-regulated, this is restricted to seeds, as MYB33/MYB65 activity appears fully suppressed in vegetative tissues by miR159, where they are not involved in GA-mediated growth or flowering. Based on these findings, miR159 could be regarded as a molecular switch that confines MYB33 and MYB65 to seeds and anthers, where we have shown here that they promote PCD in the aleurone, and it is likely that they also do so in the tapetum.
Figure 10.
Proposed model of the miR159-GAMYB regulatory pathway in Arabidopsis. GAMYB-like proteins are present in the aleurone, indicating low activity of miR159. In this tissue, we hypothesize that they transduce the GA signal for the activation of GA-induced genes, which leads to nutrient secretion and the progression of PCD. Conversely, strong miR159 activity fully represses MYB33 and MYB65 in vegetative tissues, ensuring that aleurone-related genes remain inactive to allow the progression of growth. GA appears not associated with the miR159-GAMYB regulatory module in these tissues.
miR159a and miR159b Act as “Switch miRNAs” in Vegetative Tissues
We have three lines of evidence supporting the notion that despite MYB33 and MYB65 being consistently transcribed throughout Arabidopsis, their protein levels are suppressed to biologically insignificant levels except in seeds and anthers. First, the very weak expression of MYB33:GUS contrasts to the intense expression of the mMYB33:GUS transgene in vegetative tissues, implying that although MYB33 is being transcribed, very little MYB33 protein accumulates. This silencing appears not only due to the reduction of MYB33 mRNA levels but also to the inhibition of the translation of the remaining MYB33 transcript, as highlighted by the discrepancies between mRNA and GUS activity levels of the MYB33:GUS transgene (Fig. 3, E and F). miRNA-mediated translational repression mechanisms appear common in plants (Brodersen et al., 2008), which in this case may fully ensure the complete silencing of MYB33. This complete silencing is supported by the wild-type phenotype of myb33.myb65 at the vegetative stage and the negligible differences between the SAR transcriptomes of the wild type and myb33.myb65. In this respect, miR159 could be regarded as a switch miRNA (Bartel and Chen, 2004), fully repressing MYB33 and MYB65. As miR159a and miR159b are redundant, they must be made in large excess to carry out this repression, and this is especially so considering that miR159a is 10-fold more abundant than miR159b (Rajagopalan et al., 2006; Fahlgren et al., 2007), but a mir159a mutant still appears phenotypically indistinguishable from the wild type (Allen et al., 2007). Thus, increasing miR159 levels even further would be predicted to have little effect, and this is what Schwab et al. (2005) found when they overexpressed miR159a, as 35S:MIR159a transgenic plants did not exhibit any aberrant phenotype apart from male sterility and had unaltered mRNA levels of MYB33 and MYB65.
MYB33 and MYB65 Are Not Essential in the GA Response for Vegetative Growth or Flowering
Previous reports found MYB33 mRNA levels up-regulated by GA and associated with the induction of LFY and flowering under short days (Achard et al., 2004). However, our work shows that MYB33 and MYB65 are not essential for flowering under short-day conditions, as the myb33.myb65 double mutant has a wild-type flowering time and responds to GA treatments to the same extent as wild-type plants (Figs. 1 and 2). It could be argued that in the absence of these transcription factors, the other members of the GAMYB-like family take over their role in the flowering pathway. However, this seems highly improbable, as transcription of these members is restricted to anthers and seeds (Zimmermann et al., 2004) and they were not up-regulated in the SAR of myb33.myb65 (data not shown). Furthermore, no flowering time-related genes were up-regulated in mir159ab according to the microarray analysis, consistent with the fact that mir159ab exhibits a delayed flowering time, going against the view that MYB33 and MYB65 are activators of flowering. Finally, we failed to detect any GA induction of MYB33 or MYB65 mRNA in vegetative tissues upon GA treatment, despite the plants displaying clear physiological GA responses. Furthermore, as the levels of miR159 did not change with GA application, it is unlikely that MYB33 and MYB65 protein levels increase due to the release of the translational repression by miR159. Therefore, our data suggest that MYB33 and MYB65 do not transduce the GA signal in vegetative tissues of Arabidopsis.
The GAMYB-like Genes Promote GA-Mediated PCD in the Aleurone
However, we have linked MYB33, MYB65, and MYB101 to a GA-mediated response in the aleurone. This process appears conserved between monocotyledonous and dicotyledonous plants but has been best characterized in cereals. Here, the embryo produces GA during germination that stimulates the aleurone and transforms it into a secretory tissue that synthesizes a spectrum of hydrolases for the mobilization of nutrients in the endosperm. These hydrolases are synthesized de novo from amino acids that arise from the breakdown of the proteins stored in the PSVs that are numerous in mature aleurone cells. As a consequence of the hydrolysis, these organelles swell and coalesce to form a big lytic vacuole that at the end of germination collapses and results in cell death (for review, see Fath et al., 2000). GAMYB in cereals activates the expression of hydrolytic enzymes in response to GA (Gubler et al., 1999) and is also involved in the progression of PCD (Guo and Ho, 2008). Although Arabidopsis aleurone has been less characterized, it is known that it also undergoes GA-mediated vacuolation (Bethke et al., 2007), as the cellular sources of nutrients are catabolized and exported to the growing seedling. Our work shows that MYB33, MYB65, and MYB101 are involved in these GA-regulated aleurone processes. We have demonstrated that the vacuolation of the myb33.myb65.myb101 aleurone is impaired when compared with the wild type (Fig. 6). Interestingly, there is a differential response between the aleurone cells surrounding the zone at which the root penetrates the endosperm and aleurone cells on the shoot side. As the aleurone layer and the penetration of the radicle through the endosperm are factors controlling seed dormancy (Bethke et al., 2007), the GAMYB-like genes could be involved in controlling this important seed trait. In addition to this slower rate of vacuolation, there is a reduction in the expression of genes that would be predicted to be involved in this process. First, the expression of two highly expressed Cys proteinases was reduced in myb33.myb65.myb101 germinating seeds. Proteases are associated with the mobilization of storage proteins early during germination, but at later stages they are correlated with autolysis and cell death (for review, see Fath et al., 2000). We speculate that CP1 and CP may be important Cys proteinases carrying out such functions. Supporting this, the rice ortholog of CP1 has been related to PCD in the anther. The oscp1 loss-of-function mutant is male sterile (Lee et al., 2004), and Li et al. (2006) showed that the expression of CP1 is up-regulated by the transcription factor TAPETUM DEGENERATION RETARDATION (TDR) that is required for the PCD of the tapetum to occur. CP1 is most likely a direct target of TDR, as this transcription factor is able to bind to the CP1 promoter (Li et al., 2006).
Second, the closely related genes BXL1 and BXL2 are similarly reduced in myb33.myb65.myb101. They encode β-xylosidases that are predicted to be targeted to the extracellular matrix (Goujon et al., 2003). BXL1 has been characterized further and has been found to have α-l-arabinofuranosidase and β-d-xylosidase activity (Minic et al., 2004), so it can hydrolyze xylans, the major component of the hemicelluloses of the cell wall. It has been shown to be induced by sugar starvation, possibly to release sugars from the cell wall and provide a source of carbon (Lee et al., 2007). Moreover, there is evidence that BXL1 is required for weakening the outer primary cell of the seed coat to allow the release of the mucilage upon seed imbibition (Arsovski et al., 2009). Therefore, we propose that BXL1 and BXL2 may participate in an analogous role in the aleurone, degrading and weakening the aleurone cell wall, as during imbibition aleurone cells that are located near the radicle become spherical, indicating that the cell walls are thinning and weakening (Bethke et al., 2007), a process that may precede the degradation of the aleurone cell wall to allow radicle penetrance. All four of these genes (CP1, CP, BXL1, and BXL2) plus many other hydrolases and transporters identified as being regulated by MYB33 and MYB65 are GA regulated in the seed, giving further credence that these genes are part of the aleurone response to GA. Moreover, the mRNA levels of GASA1, BXL1, BXL2, and CP correlate tightly with GAMYB-like activity, with intermediate reductions in myb33.myb65 and myb101 but even further reductions in myb33.myb65.myb101. This demonstrated that these three GAMYB-like genes are rate limiting in terms of the expression of these genes; however, they are redundant with regard to the PCD of the aleurone, as no changes in vacuolation rates were seen in myb33.myb65 or myb101 mutants (data not shown). Interestingly, only MYB101 mRNA levels were found to be up-regulated by GA in the seeds (Fig. 5). MYB65 and MYB33 might be regulated by GA posttranscriptionally or, alternatively, they might not be regulated by GA at all. However, the fact that only the triple mutant displayed a slower vacuolation rate together with an altered response to GA (Fig. 6) suggests that all three transcription factors are involved in the transduction of the GA signal. In conclusion, we propose that we have not only identified the GAMYB-like transcription factors involved in these GA-mediated aleurone pathways but also likely many genes that encode hydrolases and transporters that may be the effectors of the processes of secretion and PCD in the aleurone (Fig. 10). However, to prove the latter, it is necessary to determine whether the GA induction of these aleurone-related genes is attenuated in a myb33.myb65.myb101.ga1-3 quadruple mutant and to demonstrate that vacuolation is impaired in mutants of these aleurone-related genes.
Analogous Gene Functions in the Aleurone and Tapetal Cell Layers?
As in cereals, Arabidopsis GAMYB-like expression is strong in the aleurone and tapetum (Fig. 5; Millar and Gubler, 2005). These tissues share analogous biological functions, providing rapidly dividing organs (the embryo and pollen grains) with nutrients and undergoing PCD in the process (for review, see Rogers, 2005). In myb33.myb65, the tapetum fails to degenerate (Millar and Gubler, 2005), and also aleurone PCD is compromised in myb33.myb65.myb101. Thus, it is likely that a subset of the genes identified in this study are activated by MYB33 and MYB65 in the anther or even in both aleurone and anther. For instance, DMC1 and CP1 were found to be induced by GA in the seed, but DMC1 is also involved in the progression of meiosis (Couteau et al., 1999) and CP1 is necessary for male fertility in rice (Lee et al., 2004). Interestingly, microarray analysis in rice found that GAMYB activates different sets of genes in anthers and seeds (Tsuji et al., 2006), which appears at odds with the scenario in Arabidopsis, as many of the aleurone-related genes were globally up-regulated in mir159ab, including flowers (Table I). However, it is unknown whether these aleurone-related genes are indeed the most MYB33/MYB65 up-regulated set of genes in Arabidopsis anthers, especially considering that the most up-regulated gene in mir159ab flowers we found was only 5-fold higher. Furthermore, although PCD is common to both the aleurone and tapetum, it is likely that MYB33 and MYB65 are performing many specialized roles in the anther that do not occur in the aleurone. Therefore, microarray analysis of wild-type versus myb33.myb65 anthers would be needed to resolve this question. Moreover, the anther is a tissue where MYB101 is strongly transcribed (Allen et al., 2007), adding further complexity. Unlike the seed, myb33.myb65 anthers display a mutant phenotype, suggesting subfunctionalization of MYB101 with regard to MYB33 and MYB65. As a myb101 mutant is not male sterile, a detailed comparison of myb33.myb65 with myb33.myb65.myb101 may be needed to uncover any role MYB101 has in the anther.
MYB33 and MYB65 Inhibit Cell Proliferation in Vegetative Tissues
It is clear from our transcript profiling analysis that many genes that are normally transcribed strongly in the aleurone have been globally up-regulated in mir159ab. Despite this occurring, we can find no evidence for PCD taking place in mir159ab rosette tissues. Instead, reduced cell proliferation occurs in mir159ab, as indicated by our anatomical analysis of mir159ab leaves that exhibit fewer cells in all the tissues and also larger cells in the mesophyll. This larger cell volume in association with reduced cell numbers is indicative of the phenomenon called “compensation,” which consists of the increase in the volume of each cell triggered by a reduction in cell proliferation in order to maintain a wild-type leaf size (Tsukaya, 2008). Reduced cell proliferation could be a secondary effect to the activation of PCD processes that usually occur in the aleurone. In animal systems, the coordination of PCD and cell proliferation has been comprehensively demonstrated, where arrest or disruption of the cell cycle is a common feature of cells that eventually undergo PCD (Gilchrist, 1998). Furthermore, it has been shown that many PCD factors such as p53 are able to trigger cell cycle arrest and apoptosis (Aylon and Oren, 2007). According to our hypothesis, the activation of MYB33/MYB65 in mir159ab vegetative tissues is not sufficient to lead to death, as we showed by staining rosette leaves with trypan blue (Fig. 7), but it does lead to a reduction in cell numbers.
MATERIALS AND METHODS
Plant Material, Growing Conditions, and Physiological Analysis
All Arabidopsis (Arabidopsis thaliana) seeds were sterilized and stratified at 4°C in the dark and then grown in 22°C growth cabinets under fluorescent illumination of 130 to 150 μmol m−2 s−1 on long-day (16 h of light) or short-day (10 h of light) photoperiods in Metro-Mix soil. The double mutants myb33.myb65 and mir159ab have been described previously (Millar and Gubler, 2005; Allen et al., 2007). myb33.myb65 was in a mixed Columbia-6 (myb33) and Columbia-0 (myb65-2) background, and ga1-3 was in a Columbia-0 background.
For determination of flowering time, we scored days to flowering when flowers were visible in the shoot apex by naked eye. For the GA induction of flowering under short-day conditions, 22-d-old plants were sprayed twice weekly for 3 weeks with 20% ethanol or 100 μm GA4 dissolved in 20% ethanol, and their flowering time was recorded as described above. For gene expression analysis, plants were harvested 2 h after the fourth treatment, and SARs (containing hypocotyl, SAM, and leaf primordia smaller than 0.5 cm) or rosettes were isolated for analysis. For gene expression analysis on SARs of Landsberg erecta, we proceeded as mentioned above but plants were sprayed at 13 d after sowing, as this ecotype flowers earlier. For measuring the effect of GA on petiole elongation, long-day-grown plants were GA treated as above on days 11, 13, 15, and 19 after sowing. Plants were harvested 4 d later and photographed, and petioles were measured with ImageJ (National Institutes of Health). All experiments were repeated twice. Student’s t tests were used to compare mean values.
Microscopy
Cryo-SEM was performed by a modification of the method of Huang et al. (1994). Leaves were inserted into blocks, immediately frozen in liquid nitrogen, and then loaded into a cryo-transfer unit. The leaves were either fractured for examination of internal tissues or left intact for analysis of epidermal cells and then gold coated prior to cryo-SEM imaging. All samples were examined with a Cambridge S360 SEM apparatus.
For analysis of meristem and leaf structures, samples were fixed, dehydrated in a graded ethanol series and infiltrated, and embedded with LR White resin (London Resin Co.). Semithin (2 μm) sections were stained with toluidine blue. Vein patterns were visualized by clearing cotyledons and third leaves with 70% ethanol. For histochemical localization of GUS activity, we proceeded as described by Millar and Gubler (2005) and then cleared the tissue with a saturated chloral hydrate solution. Trypan blue staining was performed as described by Van Wees (2002).
For the visualization of the aleurone layers, seeds were sown on 0.6% agarose plates, stratified overnight, and then incubated in the growth chamber for 6, 24, 30, and 48 h. Then, the aleurone layers were isolated and visualized as described by Bethke et al. (2007) with a Leica DMLB microscope. For each time point, we scored five aleurone cells per seed in a total of five seeds per genotype (n = 25), and we repeated the experiment twice. For determining the response of the aleurone to GA at 30°C, aleurone layers were isolated from seeds imbibed for 1 to 3 h and then mounted on a slide with water or 30 μm GA4. Slides were placed in a humid chamber and incubated at 30°C for 5 d prior to visualization of the aleurones. We scored 20 seeds per genotype and treatment, and we analyzed three biological replicates.
Morphometrical Analysis of mir159ab Leaves
To estimate the size of the epidermal cells, we collected four to seven cryo-SEM images throughout the adaxial and abaxial surfaces of four leaves per genotype. We then counted the number of cells per SEM image and divided the area of the picture by the number of cells to obtain average cell sizes.
We analyzed a single cross-section of two Columbia-0 and two mir159ab fifth leaves with the program ImageJ. These cross-sections were perpendicular to the midvein and taken from the middle of the leaf. We measured the lengths of the adaxial and abaxial surfaces on the whole leaf section, determined the adaxial-abaxial ratio of each leaf, and then calculated the mean. To determine the number of epidermal cells, we counted the number of adaxial and abaxial epidermal cells in the sections. To calculate the density of cells in the mesophyll, we counted the number of mesophyll cells in the sections and divided it by the area of the sectioned leaf. Finally, we measured the area of 20 palisade and 20 spongy mesophyll cells per leaf and then calculated the mean to obtain mesophyll cell size.
Microarray Analysis
Transcriptomic analysis was performed using Affymetrix GeneChip Arabidopsis Genome ATH1 microarrays. Three biological replicates were analyzed for each genotype (Columbia-0, myb33.myb65, mir159ab), with each array representing a single biological replicate. We isolated total RNA as described (Chang et al., 1993) from the SAR (shoot apices that included hypocotyl, meristem, and leaf primordia shorter than 0.5 cm) of 15-d-old plants grown under long-day conditions. The quality of each total RNA sample was verified with an Agilent Bioanalyzer 2100 and the Agilent Eukaryotic Total RNA Nano assay kit (Agilent Technologies). For each sample, biotinylated copy RNA was prepared according to the standard Affymetrix single-amplification protocol from 5 μg of total RNA (Expression Analysis Technical Manual; Affymetrix). Following hybridizations, array quality was assessed using quality-control metrics implemented in GCOS 1.4 (Affymetrix) and software procedures available in R/Bioconductor (Bioconductor version 2.4.0; Gentleman et al., 2004) and at the Web site of the Centre for Bioinformation Science (http://cbis.anu.edu.au/software.html). Based on these metrics, the quality of all array hybridizations was assessed as satisfactory. CEL files were next imported into Partek Genomics Suite version 6.3 (Partek) and normalized by quantile normalization following Robust Multi-array Analysis background correction with adjustment of probe cell intensities to correct for probe sequence effects. Probe set summarization was done with the median polish option. One-factor ANOVA was carried out on the log2-transformed expression values of each of the 22,810 probe sets in Partek. Probe sets with an uncorrected ANOVA P < 0.005 and with a fold change of 2 or greater between any two experimental groups were selected for further investigation.
Gene Expression Analysis
For determination of mRNA levels, RNA isolation and qRT-PCR were performed as described by Allen et al. (2007) with the primers listed in Supplemental Table S3. Mature miR159 levels were quantified with the TaqMan MicroRNA Assays (Applied Biosystems) following the manufacturer’s instructions. For these assays, RNA from shoot apices of short-day-grown plants was extracted using TRIZOL (Invitrogen), and a 10-ng sample was retrotranscribed with the TaqMan MicroRNA RT kit (Applied Biosystems) following the kit protocol. In each reaction, we included the stem-loop RT primers for either miR159a or miR159b and also the normalization gene sno101. A total of 1.33 μL of reverse transcription-PCR product was used in 20-μL qRT-PCR. Three technical replicates were done per sample, and we analyzed two different biological replicates.
GUS activity in transgenic plants was determined using the fluorogenic substrate 4-methylumbelliferyl-β-d-glucuronide as described by Jefferson et al. (1987) with 100-mg leaf discs. The fluorescence was measured using the FLUOstar OPTIMA multidetection plate reader.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. MYB33 and MYB65 mRNA levels are not induced upon GA treatment in Landsberg erecta.
Supplemental Figure S2. KRP7 mRNA levels are elevated in mir159ab.
Supplemental Table S1. Microarray analysis of mir159ab SARs.
Supplemental Table S2. Molecular function classification of mir159ab up-regulated genes.
Supplemental Table S3. Primers used in this work.
Supplementary Material
Acknowledgments
We thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutant SALK_061355. Thanks are due to Cheng Huang for his assistance with SEM. Rod King and Jayne Griffiths kindly provided GA4 and ga1-3 seeds, respectively, and much advice. We also thank Jose M. Barrero for his comments on the manuscript.
References
- Achard P, Herr A, Baulcombe DC, Harberd NP. (2004) Modulation of floral development by a gibberellin-regulated microRNA. Development 131: 3357–3365 [DOI] [PubMed] [Google Scholar]
- Allen RS, Li JY, Stahle MI, Dubroue A, Gubler F, Millar AA. (2007) Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc Natl Acad Sci USA 104: 16371–16376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsovski AA, Popma TM, Haughn GW, Carpita NC, McCann MC, Western TL. (2009) AtBXL1 encodes a bifunctional β-d-xylosidase/α-l-arabinofuranosidase required for pectic arabinan modification in Arabidopsis mucilage secretory cells. Plant Physiol 150: 1219–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aya K, Ueguchi-Tanaka M, Kondo M, Hamada K, Yano K, Nishimura M, Matsuoka M. (2009) Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell 21: 1453–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Oren M. (2007) Living with p53, dying of p53. Cell 130: 597–600 [DOI] [PubMed] [Google Scholar]
- Bartel DP, Chen CZ. (2004) Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet 5: 396–400 [DOI] [PubMed] [Google Scholar]
- Bethke PC, Libourel IGL, Aoyama N, Chung YY, Still DW, Jones RL. (2007) The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiol 143: 1173–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez MA, Weigel D. (2000) Integration of floral inductive signals in Arabidopsis. Nature 404: 889–892 [DOI] [PubMed] [Google Scholar]
- Bouquin T, Meier C, Foster R, Nielsen ME, Mundy J. (2001) Control of specific gene expression by gibberellin and brassinosteroid. Plant Physiol 127: 450–458 [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. (1993) A simple method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113–116 [Google Scholar]
- Couteau F, Belzile F, Horlow C, Grandjean O, Vezon D, Doutriaux MP. (1999) Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell 11: 1623–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling RJ, Kamiya Y, Seto H, Harberd NP. (1998) Gibberellin dose-response regulation of GA4 gene transcript levels in Arabidopsis. Plant Physiol 117: 1195–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GTS, Krols L, Terras P, Landrieu I, Van der Schueren E, Maes S, Naudts M, Inze D. (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13: 1653–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, et al. (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fath A, Bethke P, Lonsdale J, Meza-Romero R, Jones R. (2000) Programmed cell death in cereal aleurone. Plant Mol Biol 44: 255–266 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DG. (1998) Programmed cell death in plant disease: the purpose and promise of cellular suicide. Annu Rev Phytopathol 36: 393–414 [DOI] [PubMed] [Google Scholar]
- Gocal GF, Sheldon CC, Gubler F, Moritz T, Bagnall DJ, MacMillan CP, Li SF, Parish RW, Dennis ES, Weigel D, et al. (2001) GAMYB-like genes, flowering, and gibberellin signaling in Arabidopsis. Plant Physiol 127: 1682–1693 [PMC free article] [PubMed] [Google Scholar]
- Goujon T, Minic Z, El Amrani A, Lerouxel O, Aletti E, Lapierre C, Joseleau JP, Jouanin L. (2003) AtBXL1, a novel higher plant (Arabidopsis thaliana) putative beta-xylosidase gene, is involved in secondary cell wall metabolism and plant development. Plant J 33: 677–690 [DOI] [PubMed] [Google Scholar]
- Gubler F, Kalla R, Roberts JK, Jacobsen JV. (1995) Gibberellin-regulated expression of a myb gene in barley aleurone cells: evidence for Myb transactivation of a high-Pl α-amylase gene promoter. Plant Cell 7: 1879–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Raventos N, Keys M, Watts R, Mundy J, Jacobsen JV. (1999) Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant J 17: 1–9 [DOI] [PubMed] [Google Scholar]
- Guo WJ, Ho THD. (2008) An abscisic acid-induced protein, HVA22, inhibits gibberellin-mediated programmed cell death in cereal aleurone cells. Plant Physiol 147: 1710–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CX, Canny MJ, Oates K, McCully ME. (1994) Planing frozen hydrated plant specimens for SEM observation and Edx microanalysis. Microsc Res Tech 28: 67–74 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Inukai Y, Ueguchi-Tanaka M, Itoh H, Izawa T, Kobayashi Y, Hattori T, Miyao A, Hirochika H, Ashikari M, et al. (2004) Loss-of-function mutations of the rice GAMYB gene impair α-amylase expression in aleurone and flower development. Plant Cell 16: 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Matsumura Y, Soga K, Hoson T, Koizumi N. (2007) Glycosyl hydrolases of cell wall are induced by sugar starvation in Arabidopsis. Plant Cell Physiol 48: 405–413 [DOI] [PubMed] [Google Scholar]
- Lee S, Jung KH, An GH, Chung YY. (2004) Isolation and characterization of a rice cysteine protease gene, OSCP1, using T-DNA gene-trap system. Plant Mol Biol 54: 755–765 [DOI] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, et al. (2006) The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AA, Gubler F. (2005) The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17: 705–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minic Z, Rihouey C, Do CT, Lerouge P, Jouanin L. (2004) Purification and characterization of enzymes exhibiting β-d-xylosidase activities in stem tissues of Arabidopsis. Plant Physiol 135: 867–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray F, Kalla R, Jacobsen J, Gubler F. (2003) A role for HvGAMYB in anther development. Plant J 33: 481–491 [DOI] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwalhara A, Kamiya Y, Yamaguchi S. (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15: 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu XL, Schommer C, Schwab R, Carrington JC, Weigel D. (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Penfield S, Li Y, Gilday AD, Graham S, Graham IA. (2006) Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 18: 1887–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20: 3407–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. (2002) Prediction of plant microRNA targets. Cell 110: 513–520 [DOI] [PubMed] [Google Scholar]
- Rogers HJ. (2005) Cell death and organ development in plants. Curr Top Dev Biol 71: 225–261 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8: 517–527 [DOI] [PubMed] [Google Scholar]
- Tsuji H, Aya K, Ueguchi-Tanaka M, Shimada Y, Nakazono M, Watanabe R, Nishizawa NK, Gomi K, Shimada A, Kitano H, et al. (2006) GAMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers. Plant J 47: 427–444 [DOI] [PubMed] [Google Scholar]
- Tsukaya H. (2008) Controlling size in multicellular organs: focus on the leaf. PLoS Biol 6: 1373–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wees S. (2002) Trypan blue stain for fungi, oomycetes, and dead plant cells. Weigel D, Glazebrook J, , Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 86–87 [Google Scholar]
- Wilson RN, Heckman JW, Somerville CR. (1992) Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol 100: 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodger FJ, Millar A, Murray F, Jacobsen JV, Gubler F. (2003) The role of GAMYB transcription factors in GA-regulated gene expression. J Plant Growth Regul 22: 176–184 [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.