Branching habit is intrinsic to every higher plant species. Consider, for example, a pine tree (Pinus spp.) and an oak tree (Quercus spp.). Although the characteristic architecture of each species is mainly determined by genetics, the sessile nature of plants demands a high degree of plasticity. Environmental factors such as light, insects, crowding, and nutrient limitation can stimulate or limit branching and therefore change plant architecture. How do plants detect all these signals and channel them into one single decision: to grow or not to grow? A newly discovered class of hormones, the strigolactones, has a pivotal role in regulating branching habit. Studies on this group of carotenoid-derived molecules have provided exciting insights and guidance for future directions.
Control of branching was mainly associated with auxin and cytokinin until discoveries of mutants in Arabidopsis (Arabidopsis thaliana; more axillary growth [max]), rice (Oryza sativa; dwarf [d]), pea (Pisum sativum; ramosus [rms]), and petunia (Petunia hybrida; decreased apical dominance [dad]) indicated the existence of a factor with a strong effect on bud outgrowth independent of known phytohormones. Grafting experiment showed that mutant plants lacked a long-distance mobile signal that moves from the lower part of the plant to the shoots (Dun et al., 2009). A major breakthrough in understanding the nature of this unknown factor was the linkage of several branching mutants to Carotenoid Cleavage Dioxygenase7 (CCD7) and CCD8 (Sorefan et al., 2003; Booker et al., 2004). These enzymes can cleave carotenoids or carotenoid-derived molecules and are believed to be the key steps at the beginning of the pathway (Vogel et al., 2010; Walter et al., 2010). Knowing that the biologically active molecules are carotenoid derived led to the next major breakthrough, identification of an active molecule(s).
Strigolactones are terpenoid lactones that were initially characterized for their ability to trigger germination of parasitic plants when present in the roots exudates of their hosts. They also have a role in the interaction with arbuscular mycorrhizal fungi that improve nutrient uptake of a wide range of plants (Yoneyama et al., 2009). This class of compounds was postulated to be dependent upon the action of CCD7 and CCD8 and studies with mutants of those two genes confirmed that hypothesis (Gomez-Roldan et al., 2008; Umehara et al., 2008). Production of strigolactone in those mutants was impaired, establishing an essential role for CCD7 and CCD8 in their synthesis. Furthermore, the application of synthetic strigolactone restored the wild-type branching phenotype of those mutants, indicating that these molecules are either the active compounds or their precursors.
DISCOVERING GENES IN THE PATHWAY
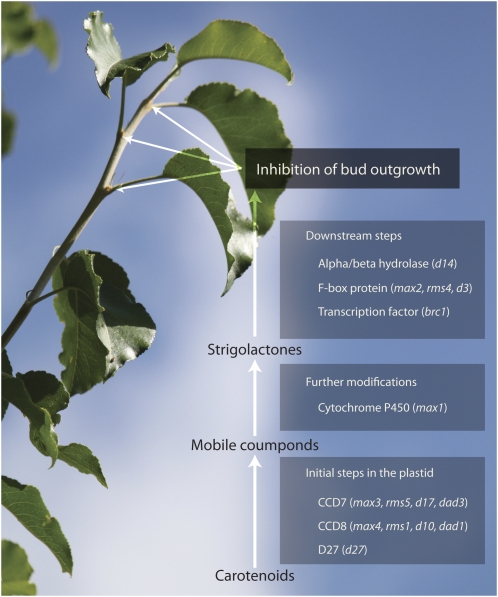
Combinations of strigolatone application and grafting experiments have permitted researchers to differentiate strigolactone synthesis mutants from subsequent modification and/or signal transduction mutants. In addition to CCD7 (MAX3, RMS5, D17, DAD3) and CCD8 (MAX4, RMS1, D10, DAD1), two other genes have been linked to synthesis (Fig. 1), a cytochrome P450 in Arabidopsis (MAX1) and an iron-containing protein in rice (D27; Booker et al., 2005; Lin et al., 2009). The phenotypes of each of these mutants can be restored to wild type by application of strigolactone in contrast with the downstream mutants (Beveridge and Kyozuka, 2010). Interestingly, D27, CCD7, and CCD8 are targeted to plastids while MAX1 seems to lack any plastid target peptide. Since carotenoids are highly hydrophobic and cannot move easily in the cell, they need to be substantially altered to become soluble and move out of the plastid. The plastid localization of CCD7, CCD8, and D27 suggests a multistep modification of the substrate by at least those three enzymes to produce a mobile compound that can be transported and further modified by downstream proteins such as MAX1. Given the substantial structural differences between carotenoids and strigolactones (Fig. 2), it seems unlikely that the four enzymes identified to date are the only enzymes in the biosynthetic pathway. Therefore, a major goal is to unequivocally identify all of the genes responsible for strigolactones synthesis. While mutant characterization has been the cornerstone of gene identification, we are now at a crossroad where most of the branching mutants in model species are characterized at a molecular level, forcing a reassessment of the stratagem. Elucidating the biosynthetic pathway will likely rely on either discovery of new mutants in untraditional species or an assemblage of alternative techniques, like protein-protein interaction and gene expression correlation. Although the use of mutants may seem like the most straightforward and simple option, it is not without a downside as mutants with gene redundancy tend to lack detectable phenotypes, preventing identification of important genes. Likewise, the low abundance of RNA and their correspondent proteins present major technical difficulties for this pathway.
Figure 1.
Schematic representation of the strigolactone pathway leading to inhibition of bud outgrowth.
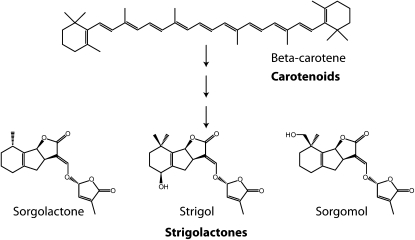
Figure 2.
Structures of some strigolactones found in sorghum root exudates and the predicted initial substrate beta-carotene.
Until now, only a few genes have been associated with processes downstream of strigolactones synthesis (Fig. 1). One of these genes encodes an α/β hydrolase that has been characterized in rice (D14). Exogenous application of the synthetic strigolactone GR24 does not rescue the phenotype of d14. Moreover, this mutant accumulates a higher level of an endogenous strigolactone than the wild type, demonstrating a post-strigolactone synthesis function for this protein (Arite et al., 2009). Another gene that has been characterized in numerous species and is essential for the response to the active compound encodes a nuclear localized F-box protein (MAX2, RMS4, D3). That group of proteins is often implicated in protein degradation by mediating targeted ubiquitination (Stirnberg et al., 2007). One compelling idea is that this F-box protein controls the fate of growth-related transcription factors. In Arabidopsis, one transcription factor, BRANCHED1 (BRC1), has been linked to the strigolactone pathway. BRC1 is down-regulated in max mutants buds and up-regulated in response to application of strigolactone (Aguilar-Martínez et al., 2007; Mashiguchi et al., 2009). As with the genes in the upstream part of the pathway, the challenge lies in identification of the functions of all these genes.
FROM GENES TO MOLECULES
Despite the identification of a role for strigolactones in branching, our knowledge of the biosynthetic pathway remains limited. Activity of CCD7 and CCD8 in vitro and in carotenoid-expressing bacteria points to beta-carotene as the most likely initial substrate (Schwartz et al., 2004). However, other carotenoids cannot be ruled out as the starting point for the pathway. The characterization of theses enzymes presents unique problems because of carotenoid hydrophobicity and the difficulty to replicate the plastid organization ex planta. Nonetheless, identification of the primary carotenoid substrate or substrates will be essential to define the other steps of strigolactones synthesis.
Multiple strigolactones have been identified both within and between species (Fig. 2; Yoneyama et al., 2009). These differences could be attributed to either multiple initial substrates or, more likely, to subsequent modifications. The structural diversity also raises questions about possible differential activities of those molecules on bud outgrow inhibition, as the ability of the downstream proteins to recognize, modify, and/or degrade them could substantially vary. Diverse activities between strigolactones have already been observed for parasitic weed germination and hyphal branching (Yoneyama et al., 2009; Akiyama et al., 2010). Correlation of plant branching with the various strigolactones would provide insights into why plants produce a range of these molecules. However, characterizing the diversity of strigolactones in plant tissue will be particularly challenging, as the accurate detection of the molecules is limited at this point to root exudates. Today, very few laboratories have the expertise to quantify strigolactones, limiting our ability to evaluate strigolactone biology.
HORMONE INTERACTIONS
While identification of the components of the pathway is essential for our understanding of branching control, it is important to remember that strigolactone is part of an intricate web connecting environmental cues and hormonal response. Altering one compound impacts the balance of all the others, triggering a complex and fine-tuned response. Consider for example the interactions of strigolactones with auxin. Transcript levels of CCD7 and CCD8 are positively correlated with auxin in several species. In Arabidopsis, auxin signaling and auxin transcriptional repressor mutants (auxin resistant1 and bodenlos) exhibit reduced expression of the two CCD genes. Moreover, application of a strigolactone to these mutants reduces their increased branching phenotypes (Brewer et al., 2009; Hayward et al., 2009). These results suggest a feedback loop between auxin and the strigolactone pathway in which auxin promotes strigolactone synthesis which, in turn, inhibits bud outgrowth. While this interaction seems important, auxin is not the only factor that regulates strigolactone production. The pea RMS2, which has yet to be characterized at the molecular level, seems to control an auxin-independent feedback signal that influences the expression of CCD8 (Beveridge et al., 2009). Likewise, abscisic acid synthesis mutants in tomato (Solanum lycopersicum; notabilis, sitiens, and flacca) have reduced strigolactone content compared to the wild type, suggesting an impact of that hormone on strigolactone biosynthesis through an unknown mechanism (López-Ráez et al., 2010). Besides those factors, phosphorous deficiency has been shown to cause a significant increase of strigolactones in root exudates (López-Ráez et al., 2008). Together, these experiments demonstrate the diversity of elements regulating the pathway and illustrate the challenge for mapping out the circuitry controlling the branching response.
A BRANCHING FUTURE
As we unravel all the components of this newly disclosed pathway, we can sense the potential importance of these discoveries for plant science. Besides helping us understand how plants regulate their architecture and interact with arbuscular mycorrhizal fungi, understanding this pathway has the potential to significantly impact agriculture, as it has for many other phytohormones. Examples could include ways to manage germination of parasitic weeds responsible for massive crop losses in developing world as well as control of lateral branching in forestry to produce higher quality woods with fewer knots. Regardless of what will emerge from that field, we can predict that this branching pathway will remain a topic of interest for the next decades.
References
- Aguilar-Martínez JA, Poza-Carrión C, Cubas P. (2007) Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K, Ogasawara S, Ito S, Hayashi H. (2010) Structural requirements of strigolactones for hyphal branching in AM fungi. Plant Cell Physiol 51: 1104–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J. (2009) d14, a strigolactone insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol 50: 1416–1424 [DOI] [PubMed] [Google Scholar]
- Beveridge CA, Dun EA, Rameau C. (2009) Pea has its tendrils in branching discoveries spanning a century from auxin to strigolactones. Plant Physiol 151: 985–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge CA, Kyozuka J. (2010) New genes in the strigolactone-related shoot branching pathway. Curr Opin Plant Biol 13: 34–39 [DOI] [PubMed] [Google Scholar]
- Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O. (2004) MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signalling molecule. Curr Biol 14: 1232–1238 [DOI] [PubMed] [Google Scholar]
- Booker J, Sieberer T, Wright W, Williamson L, Willett B, Stirnberg P, Turnbull C, Srinivasan M, Goddard P, Leyser O. (2005) MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev Cell 8: 443–449 [DOI] [PubMed] [Google Scholar]
- Brewer PB, Dun EA, Ferguson BJ, Rameau C, Beveridge CA. (2009) Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol 150: 482–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun EA, Brewer PB, Beveridge CA. (2009) Strigolactones: discovery of the elusive shoot branching hormone. Trends Plant Sci 14: 364–372 [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC, et al. (2008) Strigolactone inhibition of shoot branching. Nature 455: 180–194 [DOI] [PubMed] [Google Scholar]
- Hayward A, Stirnberg P, Beveridge C, Leyser O. (2009) Interactions between auxin and strigolactone in shoot branching control. Plant Physiol 151: 400–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Wang R, Qian Q, Yan M, Meng X, Fu Z, Yan C, Jiang B, Su Z, Li J, et al. (2009) DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21: 1512–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Ráez JA, Charnikhova T, Gómez-Roldán V, Matusova R, Kohlen W, De Vos R, Verstappen F, Puech-Pages V, Bécard G, Mulder P, et al. (2008) Tomato strigolactones are derived from carotenoids and their biosynthesis is promoted by phosphate starvation. New Phytol 178: 863–874 [DOI] [PubMed] [Google Scholar]
- López-Ráez JA, Kohlen W, Charnikhova T, Mulder P, Undas AK, Sergeant MJ, Verstappen F, Bugg TDH, Thompson AJ, Ruyter-Spira C, et al. (2010) Does abscisic acid affect strigolactone biosynthesis? New Phytol 187: 343–354 [DOI] [PubMed] [Google Scholar]
- Mashiguchi K, Sasaki E, Shimada Y, Nagae M, Ueno K, Nakano T, Yoneyama K, Suzuki Y, Asami T. (2009) Feedback-regulation of strigolactone biosynthetic genes and strigolactone-regulated genes in Arabidopsis. Biosci Biotechnol Biochem 73: 2460–2465 [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Qin X, Loewen M. (2004) The biochemical characterization of two carotenoid cleavage enzymes from Arabidopsis indicates that a carotenoid-derived compound inhibits lateral branching. J Biol Chem 279: 46940–46945 [DOI] [PubMed] [Google Scholar]
- Stirnberg P, Furner IJ, Leyser HMO. (2007) MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J 50: 80–94 [DOI] [PubMed] [Google Scholar]
- Sorefan K, Booker J, Haurogné K, Goussot M, Bainbridge K, Foo E, Chatfield S, Ward S, Beveridge C, Rameau C, et al. (2003) MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev 17: 1469–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, et al. (2008) Inhibition of shoot branching by new terpenoid plant hormone. Nature 455: 195–200 [DOI] [PubMed] [Google Scholar]
- Vogel JT, Walter MH, Giavalisco P, Lytovchenko A, Kohlen W, Charnikhova T, Simkin AJ, Goulet C, Strack D, Bouwmeester HJ, et al. (2010) SlCCD7 controls strigolactone biosynthesis, shoot branching and mycorrhiza-induced apocarotenoid formation in tomato. Plant J 61: 300–311 [DOI] [PubMed] [Google Scholar]
- Walter MH, Floss DS, Strack D. (2010) Apocarotenoids: hormones, mycorrhizal metabolites and aroma volatiles. Planta 232: 1–17 [DOI] [PubMed] [Google Scholar]
- Yoneyama K, Xie X, Yoneyama K, Takeuchi Y. (2009) Strigolactones: structures and biological activities. Pest Manag Sci 65: 467–470 [DOI] [PubMed] [Google Scholar]