Abscisic acid (ABA), a sesquiterpenoid phytohormone, was first discovered in the 1960s, and has since been found to be a key regulator of such diverse processes as dormancy, germination, seed development, and responses to biotic and abiotic stress (Cutler et al., 2010). Since the discovery of its structure, advances have been made in understanding ABA metabolism, synthesis, and hormone-responsive genes; however, the understanding of the early stage of ABA perception by the plant cell remained a mystery. During the past decade receptor proteins for the other traditional plant hormones (auxin, gibberellins, cytokinin, and ethylene) were discovered, while the identity of the ABA receptor remained elusive, creating controversy. Finally, in 2009, two research groups converged upon the PYR/PYL/RCAR family of soluble proteins in Arabidopsis (Arabidopsis thaliana), which evidence indicates are one type of ABA-binding receptor-like proteins (Ma et al., 2009; Park et al., 2009). This protein family has 14 members, almost all of which appear capable of forming an ABA-receptor complex that is able to activate the transcription of ABA-responsive genes. The use of multiple biochemical, chemical genetics, and proteomic approaches has provided the evidence to paint a remarkable picture of ABA binding, receptor complex formation, and initial downstream signaling. While the receptors and some important signal mediators have been elucidated, the molecular mechanisms by which they regulate the ABA signaling pathway continue to intrigue researchers and drive the field forward.
ABA RECEPTOR COMPLEX
After the discovery of the PYR/PYL proteins, a wave of crystallographic studies (Melcher et al., 2009; Miyazono et al., 2009; Nishimura et al., 2009; Santiago et al., 2009a; Yin et al., 2009) illuminated the structural basis for interaction between these soluble proteins, ABA, and type 2C protein phosphatases (PP2Cs). Nuclear magnetic resonance dynamic measurements of in-solution chemical shifts further corroborated the interaction of ABA with the PYR/PYL proteins. Collectively, these structural studies create the following model: ABA binds in a pocket of the PYR/PYL protein, which then undergoes a conformational shift to cover and isolate the hormone from the solvent. This binding exposes a hydrophobic surface on the PYR/PYL protein with which highly conserved residues of an individual PP2C can interact. The binding of the PP2C protein to the ABA-bound protein complex covers the active site of the phosphatase, thus acting as a competitive inhibitor of PP2C activity. While all members of the PYR/PYL family have highly conserved amino acid sequences in critical interaction domains, minor structural differences within the ABA-binding pocket, and on the hydrophobic PP2C interaction surface, are likely to play roles in the strength of interaction between receptors and ABA, and with different PP2C family members. While clade A PP2Cs have long been known for their role in negative regulation of ABA signaling (Rodriguez et al., 1998), these structural studies demonstrated their integral involvement in the ABA-receptor complex.
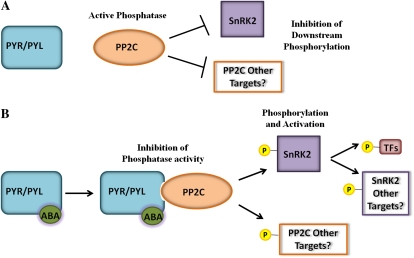
The discovery of the soluble ABA receptor proteins coincided with advancements in our understanding of the regulation of ABA-responsive genes by Ser-Thr kinases and PP2Cs. Specifically, a plant-specific kinase group known as the subfamily 2 SNF1-related kinases (SnRK2s), were found to play a vital role regulating ABA-induced gene expression (Fujii et al., 2009). In vitro assays were used to show that PP2Cs inhibit SnRK2-mediated phosphorylation of a family of basic Leu zipper transcription factors called ABFs/AREBs, which, when phosphorylated, act as positive regulators of ABA-responsive gene expression. Furthermore, these studies showed that in the presence of ABA these effects could be blocked by the addition of PYR/PYL proteins. Together, these results have led to a signaling model where ABA binding to PYR/PYL proteins inhibits PP2C activity and disrupts its interaction with target SnRK2s. This disruption permits the autophosphorylation and activation of the SnRK2 kinases, which then are free to phosphorylate ABF/AREB transcription factors, thereby activating ABA-responsive genes (Fig. 1). Furthermore, the SnRK2 kinase (OST1) has been shown to phosphorylate the slow-anion channel (SLAC1) in the presence of ABA, thus mediating ion flux from guard cells and inducing stomatal pore closure (Geiger et al., 2009; Lee et al., 2009).
Figure 1.
Current ABA receptor signaling model. A, In the absence of ABA, PP2C phosphatases interact with SnRK2 kinases to inhibit their autophosphorylation and activation. B, In the presence of ABA, inhibition of PP2C phosphatases by the ABA-receptor complex results in phosphorylation and activation of SnRK2 kinases, which in turn phosphorylate transcription factors that promote transcription of ABA-responsive genes. Other targets of PP2C phosphatases and SnRK kinases are not currently fully defined, but may play important roles in downstream ABA signaling.
The importance of the discovery of the soluble ABA receptor protein family was underscored by a wave of ABA-related reports that have flooded the top journals this past year, and with SCIENCE magazine naming it a runner-up for breakthrough of the year, 2009 (The News Staff, 2009), a very rare occurrence for plant-related discoveries. The recent pace of progress in the ABA field has both begun to answer old questions as well as prompt new, focused investigations of ABA synthesis, transport, receptor recognition, and regulation of downstream signaling.
ALTERNATE PP2C AND SNRK2 TARGETS
In the case of other plant hormone systems (e.g. gibberellin, auxin, and jasmonic acid), hormone interaction with a receptor leads to rapid degradation of target proteins that in turn release transcription factors directly affecting activation of hormone-responsive genes (Dharmasiri et al., 2005; Griffiths et al., 2006; Chini et al., 2007). Signal transduction by protein degradation is an irreversible mechanism requiring de novo protein synthesis to accomplish feedback desensitization to the hormone signal. In the case of ABA, it seems that kinases/phosphatases are the main effector proteins that drive the cellular response to the hormone in a highly reversible manner. The balance of phosphorylation and dephosphorylation can be quickly altered by the effector proteins, and may explain the broad effects of ABA. Thus, activation, as well as desensitization, can be achieved on a short time scale, allowing for distinct responses to a variety of physiological conditions in which ABA may be involved.
It has been postulated that the downstream targets of the ABA receptor complex may be much larger than currently understood. As SnRK2s are known targets of the PP2Cs, and downstream basic Leu zipper transcription factors targets for activated SnRK2s, many more proteins may be PP2C and SnRK2 targets and act as important signaling intermediates in the ABA pathway. Identifying downstream target proteins whose phosphorylation status is altered by the ABA receptor complex, or downstream activated kinases, is of critical importance to defining the broader signaling mechanisms of ABA. This information may provide the link necessary to generate hypotheses on how ABA regulates cation homeostasis, as well as salt and drought tolerance.
DIFFERENTIAL BINDING OF ABA AND PP2C MEMBERS TO PYR/PLY RECEPTORS
With 14 members of the PYR/PYL receptor family and nine members of the PP2C family, the possibility is raised that the ABA signal-receptor complex functions under combinatorial control. The first level of control may come from the interaction between ABA and the receptor proteins. The diversity of the PYR/PYL members may allow for biological sensing of gradients of ABA concentration, allowing unique reactions to a range of ABA states, as may be indicated by the receptors showing differential selectivity to ABA isomers (Park et al., 2009). The PYR/PYL proteins have been classified into subfamilies I, II, and III based on amino acid sequence identity, but whether subfamily members bind to distinct downstream effectors has not been shown. In fact, recent quantitative data have shown that some family members have 50- to 90-fold differences in Kd dissociation constants (Ma et al., 2009; Yin et al., 2009). Currently PYR/PYL members from each subfamily have been purified and crystallized, but full structural elucidation has only been reported for ABA receptors belonging to subclass I (PYR1, PYL1, and PYL2; Melcher et al., 2009; Miyazono et al., 2009; Nishimura et al., 2009; Santiago et al., 2009a; Yin et al., 2009). Solving the crystal structures for other subfamily members may reveal a structural basis for differences in ABA affinity, and provide a new understanding of the molecular mechanisms unique to each of the variety of responses that may be elicited by different levels of ABA.
A second level of signal regulation may occur between receptors and PP2Cs, as specificity in PYR/PYL binding with individual PP2C family members has previously been shown (Santiago et al., 2009b). The differential interaction of ABA with PYR/PYL proteins and their selective interaction with PP2C family members may prove to reflect their different physiological roles in the ABA response in vivo. It remains to be determined if a structural basis can be identified for differences in properties among receptor family members, and if these properties explain the broad action of ABA. A detailed examination of PP2C binding affinities, as well as their cell type specificity and subcellular localization will answer these questions.
POTENTIAL FOR AGONIST AND ANTAGONIST DEVELOPMENT AND APPLICATION
While the natural redundancy in receptor proteins previously provided serious challenges to geneticists interested in understanding ABA, the number of receptor family members may provide useful chemical targets and opportunities for unique regulation by exogenously applied synthetic molecules. Differential selectivity for the naturally occurring S-(+) and the unnatural R-(−) ABA observed with distinct members of the receptor family suggests that the receptors may contain variable residues within their ligand-binding pockets, providing the basis for selective receptor activation. The isolation of the selective ABA agonist, pyrabactin, is a notable step toward using synthetic small molecules to control the ABA signaling pathway (Park et al., 2009). Of particular interest would be the identification and use of small molecule antagonists and agonists for particular PYR/PYL family members. The selective manipulation of receptor family members would allow for the elucidation of PYR/PYL functions, and may help clarify the connection between ABA and plant response to biotic and abiotic stresses. Furthermore, the receptor family of proteins is highly conserved in crop species, making it likely that progress in the elucidation of the structure-function relationships of individual receptor proteins and use of synthetic small molecules, will translate into the enhancement of crop tolerance to stressors in the agricultural setting. Genetic methods of influencing ABA perception in crops may also provide advancements in plant stress tolerance.
Finally, another interesting area of ABA research has been the endogenous synthesis of this plant hormone in humans. It has been suggested that ABA is an endogenous proinflammatory cytokine in human granulocytes and can stimulate the secretion of insulin in pancreatic beta cells (Bruzzone et al., 2008). This finding, along with others that indicate that ABA can up-regulate the peroxisome proliferator-activated receptor (Bassaganya-Riera et al., 2010), has put ABA on the map as a potential pharmacological treatment for human diabetes. In fact, ABA has structural similarities to conventional thiazolidinedione diabetes drugs, and dietary supplementation of ABA in mice was found to decrease fasting Glc concentrations (Guri et al., 2007). If this work is substantiated and ABA’s role in mammalian physiology becomes compelling, a role for ABA in nonplant systems may prove useful not only as a health tool, but also as a basic research tool, in many other nonplant systems.
The discovery of the soluble ABA receptor protein family is a spring board to launch new avenues for ABA research. Advances in the last year alone included the identification of multiple plasma membrane ABA transporters (Kang et al., 2010; Kuromori et al., 2010), insights into ABA’s connection with the circadian clock signaling pathway (Castells et al., 2010), and an examination of ABA’s role in localizing reactive oxygen species in guard cells for stomatal closure (Leshem et al., 2010; Zhao et al., 2010). The pace shows no sign of slowing.
References
- Bassaganya-Riera J, Skoneczka J, Kingston DG, Krishnan A, Misyak SA, Guri AJ, Pereira A, Carter AB, Minorsky P, Tumarkin R, et al. (2010) Mechanisms of action and medicinal applications of abscisic acid. Curr Med Chem 17: 467–478 [DOI] [PubMed] [Google Scholar]
- Bruzzone S, Bodrato N, Usai C, Guida L, Moreschi I, Nano R, Antonioli B, Fruscione F, Magnone M, Scarfi S, et al. (2008) Abscisic acid is an endogenous stimulator of insulin release from human pancreatic islets with cyclic ADP ribose as second messenger. J Biol Chem 283: 32188–32197 [DOI] [PubMed] [Google Scholar]
- Castells E, Portoles S, Huang W, Mas P. (2010) A functional connection between the clock component TOC1 and abscisic acid signaling pathways. Plant Signal Behav 5: 409–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, Garcia-Casado G, Lopez-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. (2005) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al-Rasheid KA, et al. (2009) Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci USA 106: 21425–21430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun TP, et al. (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guri AJ, Hontecillas R, Si H, Liu D, Bassaganya-Riera J. (2007) Dietary abscisic acid ameliorates glucose tolerance and obesity-related inflammation in db/db mice fed high-fat diets. Clin Nutr 26: 107–116 [DOI] [PubMed] [Google Scholar]
- Kang J, Hwang JU, Lee M, Kim YY, Assmann SM, Martinoia E, Lee Y. (2010) PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc Natl Acad Sci USA 107: 2355–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T, Miyaji T, Yabuuchi H, Shimizu H, Sugimoto E, Kamiya A, Moriyama Y, Shinozaki K. (2010) ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc Natl Acad Sci USA 107: 2361–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Lan W, Buchanan BB, Luan S. (2009) A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci USA 106: 21419–21424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshem Y, Golani Y, Kaye Y, Levine A. (2010) Reduced expression of the v-SNAREs AtVAMP71/AtVAMP7C gene family in Arabidopsis reduces drought tolerance by suppression of abscisic acid-dependent stomatal closure. J Exp Bot 61: 2615–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Melcher K, Ng LM, Zhou XE, Soon FF, Xu Y, Suino-Powell KM, Park SY, Weiner JJ, Fujii H, Chinnusamy V, et al. (2009) A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462: 602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K, Miyakawa T, Sawano Y, Kubota K, Kang HJ, Asano A, Miyauchi Y, Takahashi M, Zhi Y, Fujita Y, et al. (2009) Structural basis of abscisic acid signalling. Nature 462: 609–614 [DOI] [PubMed] [Google Scholar]
- Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, Schroeder JI, Getzoff ED. (2009) Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science 326: 1373–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez PL, Benning G, Grill E. (1998) ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett 421: 185–190 [DOI] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Marquez JA. (2009a) The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462: 665–668 [DOI] [PubMed] [Google Scholar]
- Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park SY, Marquez JA, Cutler SR, Rodriguez PL. (2009b) Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J 60: 575–588 [DOI] [PubMed] [Google Scholar]
- The News Staff (2009) Breakthrough of the year: the runners-up. Science 326: 1600–1607 [DOI] [PubMed] [Google Scholar]
- Yin P, Fan H, Hao Q, Yuan X, Wu D, Pang Y, Yan C, Li W, Wang J, Yan N. (2009) Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat Struct Mol Biol 16: 1230–1236 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Stanley BA, Zhang W, Assmann SM. (2010) ABA-regulated G protein signaling in Arabidopsis guard cells: a proteomic perspective. J Proteome Res 9: 1637–1647 [DOI] [PubMed] [Google Scholar]