THE IMPORTANCE OF TRANSCRIPTIONAL REGULATION IN PHENOTYPIC VARIATION
Ever since the work by Jacob and Monod on the Lac operon, scientists have appreciated that the control of gene expression is one of the most important points of regulation in biology. Although many other layers of regulation exist, the possibility of control at the start point of production of RNA and therefore also of protein makes transcriptional initiation the primary site for regulating gene expression. These early experiments in Escherichia coli provided the first example of mechanisms by which the abundance of proteins encoded by DNA could be altered by external factors. Today, understanding how the genotype of an organism is translated into its phenotype is central to many questions in biology, including those that deal with both the origins of phenotypic variation and evolutionary change and the design of strategies to modify form and function through plant biotechnology. It is generally believed that variation in gene expression is an important source of phenotypic diversity, although recently it has been suggested that much of the variation in transcriptomes between Arabidopsis (Arabidopsis thaliana) accessions is not manifest as phenotypic variation, with only a few influential “hot spots” causing major phenotypic variation, a situation referred to as phenotypic buffering (Fu et al., 2009).
Relatively recent technical advances in the availability of whole-genome sequences, transcriptome array, and sequence data have underlined further the complex challenges in relating genotype to phenotype, particularly in higher eukaryotes such as plants. The level of transcription conferred by the basal transcriptional machinery is effectively zero, and the transcription of individual genes is controlled by the combined activity of multiple transcription factors (TFs) binding to cis-elements, which operate on a modular basis, in their promoters. This combinatorial control, involving both transcriptional activators and repressors, integrates signals, and results in diverse outcomes on target gene expression. Typically only a portion of the genes in a genome are expressed at any given time and place, for instance in response to developmental or environmental signals mediated by TFs. TFs are classified into families primarily on the basis of their conserved DNA-binding domains, and both the number of families as well as the number of members in each family have increased during the evolution of ever more complex organisms. Gene and genome duplication is a recurring theme in eukaryotic evolution, notably in higher plants, and the duplication of TF genes with associated subfunctionalization is essentially a symmetry breaking process and therefore associated with increasing complexity.
THE EXPANSION OF TF GENE FAMILIES WITH INCREASING COMPLEXITY OF ORGANISMS
A higher plant like Arabidopsis is estimated to have more than 2,000 genes encoding TFs representing about 60 protein families defined by the conserved folds of their DNA-binding domains (http://plntfdb.bio.uni-potsdam.de/v3.0/; Pérez-Rodríguez et al., 2010). This represents about 7% of the total number of genes in Arabidopsis; similar proportions are found in other angiosperm species. One example of evolution working through the expansion of membership of a TF gene family can be observed for the MADS-box genes, one class of which primarily controls aspects of flower development. More than 100 MADS-box genes are recognized in flowering plants such as Arabidopsis (Parenicova et al., 2003), but only about 20 in the moss Physcomitrella patens (Rensing et al., 2008).
At a wider taxonomic level, the MYB family of TFs has three members in vertebrates, and just one member in invertebrates such as Drosophila. In plants, the family has been significantly expanded to include five 3R MYB proteins similar to c-Myb in animals, and two large plant-specific families, the R2R3 MYB family with 126 members in Arabidopsis and the 1R family with 83 members in Arabidopsis (Bailey et al., 2008). In both R2R3 and 1R MYB families, members are associated with plant-specific functions (Martin and Paz-Ares, 1997; Dubos et al., 2010). In P. patens, the R2R3 MYB family is estimated to have 48 members, suggesting that this family has also been expanded in the angiosperm lineage since it diverged from its common ancestor with mosses, 400 million years ago (Rensing et al., 2008).
While substantial differences in the number of TFs could explain differences among organisms of very different complexity, it is much less clear why Arabidopsis, poplar (Populus spp.), and maize (Zea mays) should be phenotypically so different while containing a comparable set of genes (Britten and Davidson, 1969; Wilson, 1975). In animal evolutionary developmental biology this problem is known as the Hox paradox, the observation that a set of homologous genes, the homeobox-containing Hox TF genes, have given rise to very divergent body plans in different species. These differences in body plan are thought to arise from differences in the expression of the conserved Hox genes rather than differences in gene function or, indeed, changes in their target genes through variation in cis-acting regulatory motifs. Many scientists, consequently, have been persuaded that evolution of form is primarily dependent on selection of variation in cis-acting regulation of transcription.
ARE REGULATORY CHANGES IN GENES ENCODING TFs THE PREDOMINANT MECHANISMS FOR GENERATING NOVEL PHENOTYPES?
In 1998 Doebley and Lukens proposed that “...change in the cis-regulatory elements of transcriptional regulators provides a predominant mechanism for the generation of novel phenotypes” (Doebley and Lukens, 1998, p. 1075). The strongest evidence for this comes from study of the gene encoding the teosinte branched TCP TF whose activity has influenced (negatively) the degree of branching in the transition during domestication from teosinte to maize, largely through changes in its expression pattern (Doebley et al., 1997; Wang et al., 1999; Lukens and Doebley, 2001; Doebley, 2004). Some studies, such as those of Weber et al. (2007), have subsequently shown strong association of major traits distinguishing species to be associated with regulatory genes (although the definition of a regulatory gene in this case was not restricted to TFs), with three out of the five genes showing strong associations between regulatory genes and major trait variation in standing populations of teosinte, encoding TFs.
IS EVOLUTION OF CIS-ACTING REGULATION PREDOMINANT, REGARDLESS OF THE GENES REGULATED?
Many scientists researching the evolution of development (EvoDevo) have been persuaded that evolution of form is primarily dependent on variation in cis-acting regulatory elements, regardless of the product that the gene encodes. The arguments used to support this case are as follows. (1) Changes in cis-acting regulatory motifs are likely less pleiotropic than changes in protein-coding sequences due to the modular nature of promoters (Stern, 2000; Wray et al., 2003; Wray, 2007). (2) Changes in cis-acting regulatory motifs are more likely to be codominant and therefore exposed more rapidly to selection than changes in coding sequences that are more likely to be recessive (Wray et al., 2003; Wray, 2007). (3) A third generalization is that these principles apply to the evolution of form but not the evolution of physiology. Although this is not a denial that physiological changes can give rise to changes in phenotype, this distinction probably arises from the inherent interest of evolutionary biologists in form. Indeed, differences in cuticle pigmentation patterns in insects (the system on which evidence supporting many of these arguments has been based; Carroll, 2008, 2000, 1995) might be viewed as a metabolic trait, rather than associated with form, by many general biologists. Similarly, changes in flowering time and responses to environmental stresses could be viewed as essentially physiological, although these processes may contribute significantly to phenotypic variation, reproductive isolation, and speciation. Hoekstra and Coyne (2007) have provided persuasive arguments against the idea that the parameters determining evolution should be any different for form than those determining evolution of any aspects of physiology, including metabolism, behavior, responses to environment, etc.
The main problem with emphasizing the evolution of cis-acting motifs regardless of the changes in the products encoded by those genes, is that there is really a lack of empirical supporting evidence. This may be because it is much more difficult to identify changes that impact the functionality of cis-acting motifs compared to those affecting coding sequence. Functionality tests are essential and in many cases are missing because of technical difficulties in undertaking them. It is also possible that gene duplication, now recognized as being far more common than previously thought, especially in plants, provides a means whereby the pleiotropic effects of changes in coding sequence can be minimized, so facilitating evolution through the accumulation of small changes in protein structure and functionality (Hoekstra and Coyne, 2007). Of course, this does not mean that evolution of cis-acting sequences is not the most important mechanism for evolutionary change—it means that the jury is still out.
The proposal of Doebley and Lukens (1998) that variation in cis-regulatory elements of genes encoding transcriptional regulators provides the predominant mechanism for the generation of novel phenotypes has been largely subsumed within the more general, cis-acting element model. Consequently, what started as a relatively robust proposal has more recently been watered down, such that the definition of regulatory genes includes genes involved in signal transduction (Weber et al., 2007). Originally Doebley and Lukens suggested that evolution was more likely to occur through changes in the expression of genes involved in transcriptional regulation than those involved in signaling pathways; an argument based on the idea that signaling components operate upstream of transcriptional regulators, so that changes in the expression of signaling components were more likely to cause pleiotropic effects (and therefore had many more ways of being severely deleterious). Consequently the more important aspect of their argument was not actually the biochemical function of the products encoded by the evolutionary hot spot genes, but their positions in regulatory hierarchies or networks. Changes affecting genes operating lower down are less likely to give rise to pleiotropic effects, and therefore more likely to provide variation that might be selected during evolution.
THE IMPORTANCE OF THE ROLES OF TFs IN REGULATORY NETWORKS
One criticism of the idea that evolution works primarily on TFs comes from the concept that all TFs regulate the expression of very large numbers of genes, and that when they change in functionality either through mutation of their protein sequence or through changes in the cells in which they are expressed, they are likely to have very large pleiotropic effects that are unsuitable for neo-Darwinian evolution. This concept has come largely from studies in animal development and is based on the idea that, on average, TFs regulate hundreds of target genes (Wray et al., 2003). But it must be remembered that this is an average figure. While it is possible to claim that TFs give rise to many pleiotropic/unexpected effects, this is based on the mistaken premise that TFs can be considered functionally as a single group. TFs operate within regulatory hierarchies or networks and, clearly, ectopic expression of a TF high up in such a hierarchy is likely to affect multiple genes/processes. In addition, empirical evidence shows that TFs that operate early in a regulatory hierarchy often continue to work controlling processes later in differentiation as well (Stern, 2000; Wray et al., 2003). This can be seen in the operation of some MADS domain proteins in plants, including AP1 that plays a role in determining the induction of the transition to the floral meristem and also in determining floral organ identity (Becker and Theissen, 2003), and the B-function proteins Deficiens and Globosa in Antirrhinum (equivalent to AP1 and PI, respectively, in Arabidopsis) that control later stages of petal and stamen differentiation as well as determining organ identity (Schwarz Sommer et al., 1992; Zachgo et al., 1995; Bey et al., 2004; Lauri et al., 2006). This feature of higher-order TFs adds to their potential for causing pleiotropic effects when misexpressed, but also complicates the possible outcomes of variation in their expression on phenotype.
Such difficulties with the model proposed by Doebley and Lukens (1998) might be resolved by remembering that some TFs are concerned with specialized roles, and occupy lower orders in regulatory hierarchies or networks, and consequently, they may regulate a much more limited number of target genes. In addition, some TFs may be concerned with regulating specialized processes at late stages in development, even though they may have earlier roles at a higher regulatory level. Perhaps evolution works predominantly through variation in the lower-order regulators, or the lower-order components of regulation, particularly those determining aspects of cellular specialization.
While it is easy to understand the roles and specificity of such lower-order TFs in regulating metabolic pathways—particularly pathways of secondary metabolism in plants, there are also probably TFs controlling morphological cellular specializations in similar ways, such as members of the MYB/bHLH/WD40 complex controlling trichome formation in Arabidopsis, MIXTA/ML1 regulating conical cell and trichome development in Antirrhinum flowers (Peres-Rodriguez et al., 2005), and FAMA in stomatal development in Arabidopsis (Ohashi-Ito and Bergmann, 2006). Studies looking at target genes regulated by these TFs show relatively restricted numbers of direct targets (Morohashi and Grotewold, 2009) and misexpression does not have highly pleiotropic effects but is usually restricted to the numbers of the specialized cell types that develop. GL1 has been identified as a determinant of natural variation in trichome number between Arabidopsis species and ECT as a determinant of variation in trichome number between natural accessions of Arabidopsis (Hauser et al., 2001; Hilscher et al., 2009). In Drosophila, variation in trichome/bristle number, which differs between species, is controlled by variation in the cellular activity of a similar, lower-order TF, shavenbaby (McGregor et al., 2007).
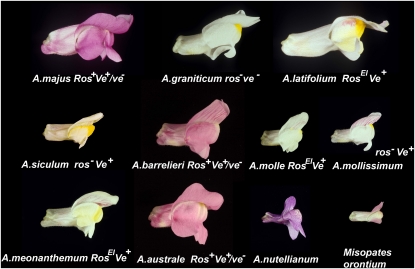
Of course, evolution through cis-acting regulation of any gene must occur in genes for which there are consequences to changes in expression, in order to have an impact on phenotype. When one considers metabolic pathways it is easy to see this as a persuasive argument. Evolution that affects primarily where or when a pathway is turned on is more likely to be of significance in speciation. Variation that expands the expression domain of an individual gene encoding an enzyme of that pathway is unlikely to have a major impact on phenotype, because no product can be made unless the earlier enzymes are also active in the same cells. It may well be that cellular morphogenesis and specialization have similar interdependencies on structural gene products, such that the primary material for evolution could be changes that involve shifting the expression of all the target genes through cis-acting variation in genes encoding TFs. Observations based on changes in flower coloration and patterning in the genus Antirrhinum support this idea (Fig. 1; Schwinn et al., 2006; Shang et al., 2010).
Figure 1.
Variation in floral pigmentation patterns and intensity within the genus Antirrhinum. Fully self-colored, red flowers are found in Antirrhinum majus, Antirrhinum barrelieri, and Antirrhinum australe. Other species (Antirrhinum molle, Antirrhinum mollissimum, Antirrhinum meonanthemum, Antirrhinum latifolium, and Antirrhinum siculum) are palely pigmented on the corolla, with anthocyanin only clearly visible in a veination pattern that has been suggested to act as a “honey guide” for pollinating bees. Antirrhinum granticum is also very palely pigmented in the corolla, but lacks the veination pattern of pigmentation. The differences between pale corolla pigmentation and full red are determined by differences in expression of two genes encoding MYB TFs, Rosea 1 and Rosea 2 (Schwinn et al., 2006). The differences in veination pattern are determined by differences in expression of a third gene, Venosa, encoding another MYB TF (Schwinn et al., 2006). Full red pigmentation is thought to be a derived trait in the genus (Shang et al., 2010). Full red flowers show an advantage in seed set resulting from higher levels of pollinator visits over palely pigmented morphs (Glover and Martin, 1998). Pollinators visit full red flowers preferentially over palely pigmented ones, but the presence of veination patterning is a significant attraction for pollinating bees (Shang et al., 2010). Misopates orontium is not cross fertile with Antirrhinum species, so its genotype cannot be confirmed. However, it has moderate corolla pigmentation with a veination patterning. Antirrhinum nutellianum belongs to the New World Antirrhinums. It has self-colored blue flowers (presumably due to the activity of a flavonoid 3′5′ hydroxylase, which is missing in European species). It has strong veination patterning, probably conferred by the activity of a Venosa-like TF. Therefore, significant differences in flower color and patterning between species result from differences in the expression of TFs in the genus Antirrhinum.
In line with this proposal, it is interesting that regulatory mechanisms that restrict variation in expression of many major developmental genes (often that encode higher-order transcriptional regulators) with strong pleiotropic effects have evolved. Good examples of these in plants are miRNAs limiting expression of homeodomain (HDIII-Zip) TFs determining leaf polarity (Reinhart et al., 2002; Rhoades et al., 2002) and those controlling TCP TF activity and leaf form (Palatnik et al., 2003). Indeed, the phenotypic buffering provided by such miRNAs may be the reason why they are so highly conserved between different plant species (Floyd and Bowman, 2004; Axtell and Bowman, 2008).
EVIDENCE FOR THE SPECIAL ROLES OF GENES ENCODING TFs IN PHENOTYPIC VARIATION
There is also some empirical evidence emerging for this refinement of the Doebley and Lukens model. There are individual case studies, such as those based on flower coloration in the genus Antirrhinum and in Petunia. In the case of Antirrhinum, differences in flower color patterning that result from differences in the expression of the MYB TF genes encoding Rosea1, Rosea2, and Venosa (Fig. 1; Schwinn et al., 2006; Shang et al., 2010) can impact pollinator attraction, and may have been under selection during speciation (Glover and Martin, 1998; Shang et al., 2010). In Petunia, evolutionary differences in flower color that impact pollinator attraction result from repeated loss of function of the TF, An2 (Quattrocchio et al., 1999; Hoballah et al., 2007). The fact that flower color evolution in Petunia involved loss of function of a TF, rather than changes in expression of the An2 gene, may be because, in this case, loss of color is a derived trait and consequently more likely to arise by loss of function of a regulatory TF, than through cis-acting variation in its expression. In cases where gain of color is a derived trait, as in the genus Antirrhinum (Shang et al., 2010), it is hard to imagine any alternative mechanisms to those involving changes in expression of regulatory proteins underpinning these traits.
In the Ranunculaceae it has been suggested that duplications in the B-function MADS TFs, followed by changes in expression, may have underpinned the evolution of petaloid organs (Kramer et al., 2003) in apetalous species such as Thalictrum thalictroides, as an adaptation to insect pollination. In this case, it may have been the later regulatory functions of AP3-like TFs that expanded into the first whorl of floral organs, giving rise to petaloid sepals, with specialized cell types typical of petals (Di Stilio et al., 2009).
Alonso-Blanco et al. (2009) have recently provided a summary of genes known to underpin natural variation in different plant species. This is very useful in attempting to define just how important variation in cis-acting regulatory motifs in genes encoding TFs may be to evolution in plants, with the proviso that it is possible that natural variation between different populations might not reflect those variations that determine the differences between species, because the former does not involve reproductive isolation. Of 75 traits defining natural phenotypic variation listed by Alonso-Blanco et al. (2009), 26 are in genes encoding TFs. If such variation were randomly distributed between genes then one would expect it to be in genes encoding TFs about 7% of the time (based on the no. of TF genes out of total no. of genes). The 36% representation of genes encoding TFs underpinning natural variation is therefore significant, although these figures could be biased by the fact that geneticists have defined those traits of evolutionary significance, often without evidence of positive selection for them. The calculation of whether variation occurs predominantly in cis-acting motifs is more difficult to make, because of the difficulties in assessing the size of regulatory domains within promoters. Most plant promoters are relatively small (up to 500 bp) but there are important examples of phenotypic variation determined by differences in regulatory motifs lying kilobase pairs away from the coding sequence even in plants (Salvi et al., 2007). A conservative estimate might place the size of the regulatory domains of genes on average as the same size as their coding sequences. If so, the reporting of 27 of the 75 traits by Alonso-Blanco as being due to changes in promoters would be close to the value expected, for example. The number of cis-acting mutations affecting TF genes out of the 75 traits is high at 10, but this is more likely a reflection of the high number of TF genes involved in phenotypic variation, rather than the result of a preponderance of variation affecting cis-acting regulatory motifs.
So the data currently available support the idea that phenotypic variation is more likely to result from variation in genes that encode TFs than from cis-acting regulatory changes, with the caveat that those traits listed by Alonso-Blanco et al. (2009) may not equate to those most important in determining differences between species.
IMPLICATIONS OF THE SPECIAL ROLES OF TFs IN PHENOTYPIC VARIATION
The possibility that phenotypic variation evolves predominantly through changes in genes encoding lower-order TFs has important repercussions not only for understanding how evolution works but also for developing strategies for how to engineer and improve plants in the future. Purugganan and Fuller (2009) have claimed that domestication of crops involves predominantly changes in transcriptional activators operating in transcriptional networks. For engineering metabolism through biotechnological approaches, it has already been shown that engineering lower-order TFs is a very effective means of increasing flux to secondary metabolites of interest without incurring massive pleiotropic effects (Grotewold et al., 1998; Bovy et al., 2002; Butelli et al., 2008; Luo et al., 2008; Zhang et al., 2009). Perhaps the most effective way to engineer other traits will be to focus on similar lower-order regulators of cell development and metabolism (Martin, 1996; Broun and Somerville, 2001).
References
- Alonso-Blanco C, Aarts MGM, Bentsink L, Keurentjes JJB, Reymond M, Vreugdenhil D, Koornneef M. (2009) What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell 21: 1877–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Bowman JL. (2008) Evolution of plant microRNAs and their targets. Trends Plant Sci 13: 343–349 [DOI] [PubMed] [Google Scholar]
- Bailey PC, Dicks J, Wang TL, Martin C. (2008) IT3F: a web-based tool for functional analysis of transcription factors in plants. Phytochemistry 69: 2417–2425 [DOI] [PubMed] [Google Scholar]
- Becker A, Theissen G. (2003) The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol Phylogenet Evol 29: 464–489 [DOI] [PubMed] [Google Scholar]
- Bey M, Stüber K, Fellenberg K, Schwarz-Sommer Z, Sommer H, Saedler H, Zachgo S. (2004) Characterization of Antirrhinum petal development and identification of target genes of the class B MADS box gene DEFICIENS. Plant Cell 16: 3197–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovy A, Ric de Vos R, Kemper M, Schijlen E, Almenar Pertejo M, Muir S, Collins G, Robinson S, Verhoeyen M, Hughes S, et al. (2002) High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes Lc and C1. Plant Cell 14: 2509–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ, Davidson EH. (1969) Gene regulation for higher cells: a theory. Science 165: 349–357 [DOI] [PubMed] [Google Scholar]
- Broun P, Somerville C. (2001) Progress in plant metabolic engineering. Proc Natl Acad Sci USA 98: 8925–8927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butelli E, Titta L, Giorgio M, Mock HP, Matros A, Peterek S, Schijlen EGWM, Hall R, Bovy AG, Luo J, et al. (2008) Induced anthocyanin biosynthesis in tomato results in purple fruit with increased antioxidant and dietary, health-protecting properties. Nat Biotechnol 26: 1301–1308 [DOI] [PubMed] [Google Scholar]
- Carroll SB. (1995) Homeotic genes and the evolution of arthropods and chordates. Nature 376: 479–485 [DOI] [PubMed] [Google Scholar]
- Carroll SB. (2000) Endless forms: the evolution of gene regulation and morphological diversity. Cell 101: 577–580 [DOI] [PubMed] [Google Scholar]
- Carroll SB. (2008) Evo-Devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36 [DOI] [PubMed] [Google Scholar]
- Di Stilio VS, Martin C, Schulfer AF, Connelly CF. (2009) An ortholog of MIXTA-like2 controls epidermal cell shape in flowers of Thalictrum. New Phytol 183: 718–728 [DOI] [PubMed] [Google Scholar]
- Doebley J. (2004) The genetics of maize evolution. Annu Rev Genet 38: 37–59 [DOI] [PubMed] [Google Scholar]
- Doebley J, Lukens L. (1998) Transcriptional regulators and the evolution of plant form. Plant Cell 10: 1075–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L. (1997) The evolution of apical dominance in maize. Nature 386: 485–488 [DOI] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci (in press) [DOI] [PubMed] [Google Scholar]
- Floyd SK, Bowman JL. (2004) Gene regulation: ancient microRNA target sequences in plants. Nature 428: 485–486 [DOI] [PubMed] [Google Scholar]
- Fu J, Keurentjes JJ, Bouwmeester H, America T, Verstappen FW, Ward JL, Beale MH, de Vos RC, Dijkstra M, Scheltema RA, et al. (2009) System-wide molecular evidence for phenotypic buffering in Arabidopsis. Nat Genet 41: 166–167 [DOI] [PubMed] [Google Scholar]
- Glover BJ, Martin C. (1998) The role of petal cell shape and pigmentation in pollination success in Antirrhinum majus. Heredity 80: 778–784 [Google Scholar]
- Grotewold E, Chamberlin M, Snook M, Siame B, Butler L, Swenson J, Maddock S, Clair GS, Bowen B. (1998) Engineering secondary metabolism in maize cells by ectopic expression of transcription factors. Plant Cell 10: 721–740 [PMC free article] [PubMed] [Google Scholar]
- Hauser MT, Harr B, Schlötterer C. (2001) Trichome distribution in Arabidopsis thaliana and its close relative Arabidopsis lyrata: molecular analysis of the candidate gene GLABROUS1. Mol Biol Evol 18: 1754–1763 [DOI] [PubMed] [Google Scholar]
- Hilscher J, Schlötterer C, Hauser MT. (2009) A single amino acid replacement in ETC2 shapes trichome patterning in natural Arabidopsis populations. Curr Biol 19: 1747–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoballah ME, Gubitz T, Stuurman J, Broger L, Barone L, Mandel T, Dell’Olivo A, Arnold M, Kuhlemeier C. (2007) Single gene-mediated shift in pollinator attraction in Petunia. Plant Cell 19: 779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA. (2007) The locus of evolution: evo devo and the genetics of adaptation. Evolution 61: 995–1016 [DOI] [PubMed] [Google Scholar]
- Kramer EM, Di Stilio VS, Schluter P. (2003) Complex patterns of gene duplication in the apetala3 and pistillate lineages of the Ranunculaceae. Int J Plant Sci 164: 1–11 [Google Scholar]
- Lauri A, Xing S, Heidmann I, Saedler H, Zachgo S. (2006) The pollen-specific DEFH125 promoter from Antirrhinum is bound in vivo by the MADS-box proteins DEFICIENS and GLOBOSA. Planta 224: 61–71 [DOI] [PubMed] [Google Scholar]
- Lukens L, Doebley J. (2001) Molecular evolution of the teosinte branched gene among maize and related grasses. Mol Biol Evol 18: 627–638 [DOI] [PubMed] [Google Scholar]
- Luo J, Butelli E, Hill L, Parr A, Niggeweg R, Bailey P, Weisshaar B, Martin C. (2008) AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: expression in fruit results in very high levels of both types of polyphenol. Plant J 56: 316–326 [DOI] [PubMed] [Google Scholar]
- Martin C. (1996) Transcription factors and the manipulation of plant traits. Curr Opin Biotechnol 7: 130–138 [Google Scholar]
- Martin C, Paz-Ares J. (1997) MYB transcription factors in plants. Trends Genet 13: 67–73 [DOI] [PubMed] [Google Scholar]
- McGregor AP, Orgogozo V, Delon I, Zanet J, Srinivasan DG, Payre F, Stern DL. (2007) Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature 448: 587–591 [DOI] [PubMed] [Google Scholar]
- Morohashi K, Grotewold E. (2009) A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet 5: e1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K, Bergmann DC. (2006) Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18: 2493–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Parenicova L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, et al. (2003) Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15: 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peres-Rodriguez M, Jaffe FW, Butelli E, Glover BJ, Martin C. (2005) Development of three different cell types is associated with the activity of a specific MYB transcription factor in the ventral petal of Anthirrhinum majus flowers. Development 132: 359–370 [DOI] [PubMed] [Google Scholar]
- Pérez-Rodríguez P, Riaño-Pachón DM, Guedes Corrêa LG, Rensing SA, Kersten B, Mueller-Roeber B. (2010) PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res 38: D822–D827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan MD, Fuller DQ. (2009) The nature of selection during plant domestication. Nature 457: 843–848 [DOI] [PubMed] [Google Scholar]
- Quattrocchio F, Wing J, van der Woude K, Souer E, de Vetten N, Mol JNM, Koes R. (1999) Molecular analysis of the anthocyanin2 gene of Petunia and its role in the evolution of flower color. Plant Cell 11: 1433–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. (2002) MicroRNAs in plants. Genes Dev 16: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. (2002) Prediction of plant microRNA targets. Cell 110: 513–520 [DOI] [PubMed] [Google Scholar]
- Salvi S, Sponza G, Morgante M, Tomes D, Niu X, Fengler KA, Meeley R, Ananiev EV, Svitashev S, Bruggemann E, et al. (2007) Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc Natl Acad Sci USA 104: 11376–11381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z, Hue I, Huijser P, Flor P, Hansen R, Tetens F, Lönnig WE, Saedler H, Sommer H. (1992) Characterization of the Antirrhinum floral homeotic MADS-box gene DEFICIENS: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J 11: 251–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwinn K, Venail J, Shang Y, Mackay S, Alm V, Butelli E, Oyama R, Bailey P, Davies K, Martin C. (2006) A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18: 831–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Venail J, Mackay S, Bailey PC, Schwinn KE, Jameson PE, Martin C, Davies KM. (2010) The molecular basis for venation patterning of pigmentation and its effect on pollinator attraction in flowers of Antirrhinum. New Phytol (in press) [DOI] [PubMed] [Google Scholar]
- Stern DL. (2000) Evolutionary developmental biology and the problem of variation. Evolution 54: 1079–1091 [DOI] [PubMed] [Google Scholar]
- Wang RL, Stec A, Hey J, Lukens L, Doebley J. (1999) The limits of selection during maize domestication. Nature 398: 236–239 [DOI] [PubMed] [Google Scholar]
- Weber AL, Briggs WH, Rucker J, Baltazar BM, de Jesús Sánchez-Gonzalez J, Feng P, Buckler ES, Doebley J. (2007) The genetic architecture of complex traits in teosinte (Zea mays ssp. parviglumis): new evidence from association mapping. Genetics 177: 2349–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AC. (1975) Evolutionary importance of gene regulation. Stadler Symp 7: 117–134 [Google Scholar]
- Wray GA. (2007) The evolutionary significance of cis-regulatory mutations. Nat Rev Genet 8: 206–216 [DOI] [PubMed] [Google Scholar]
- Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA. (2003) The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol 20: 1377–1419 [DOI] [PubMed] [Google Scholar]
- Zachgo S, de Andrade Silva E, Motte P, Tröbner W, Saedler H, Schwarz-Sommer Z. (1995) Functional analysis of the Antirrhinum floral homeotic DEFICIENS gene in vivo and in vitro by using a temperature-sensitive mutant. Development 121: 2861–2875 [DOI] [PubMed] [Google Scholar]
- Zhang W, Ning G, Lv H, Liao L, Bao M. (2009) Single MYB-type transcription factor AtCAPRICE: a new efficient tool to engineer the production of anthocyanin in tobacco. Biochem Biophys Res Commun 388: 742–747 [DOI] [PubMed] [Google Scholar]