Abstract
CTF7 is an essential gene in yeast that is required for the formation of sister chromatid cohesion. While recent studies have provided insights into how sister chromatid cohesion is established, less is known about how specifically CTF7 facilitates the formation of cohesion, and essentially nothing is known about how sister chromatid cohesion is established in plants. In this report, we describe the isolation and characterization of CTF7 from Arabidopsis (Arabidopsis thaliana). Arabidopsis CTF7 is similar to Saccharomyces cerevisiae CTF7 in that it lacks an amino-terminal extension, exhibits acetyltransferase activity, and can complement a yeast ctf7 temperature-sensitive mutation. CTF7 transcripts are found throughout the plant, with the highest levels present in buds. Seeds containing T-DNA insertions in CTF7 exhibit mitotic defects in the zygote. Interestingly, the endosperm developed normally in ctf7 seeds, suggesting that CTF7 is not essential for mitosis in endosperm nuclei. Minor defects were observed in female gametophytes of ctf7+/2 plants, and plants that overexpress CTF7 exhibited female gametophyte lethality. Pollen development appeared normal in both CTF7 knockout and overexpression plants. Therefore, proper levels of CTF7 are critical for female gametophyte and embryo development but not for the establishment of mitotic cohesion during microgametogenesis or during endosperm development.
The proper formation of sister chromatid cohesion and its subsequent release at the metaphase-to-anaphase transition is essential for the proper segregation of genetic material during cell division. It is critical for the compaction of chromosomes and their bipolar attachment to the spindle, DNA double-strand break repair, and the regulation of gene expression (for review, see Nasmyth and Haering, 2009). Sister chromatid cohesion is controlled by the cohesin complex, which consists of a heterodimer of Structural Maintenance of Chromosome (SMC) proteins, SMC1 and SMC3, Sister Chromatid Cohesion (SCC) protein SCC3, and SCC1, an α-kleisin protein. Perhaps the most widely accepted model of how cohesin functions has been referred to as the ring model, where the cohesin ring encircles the replicated sisters, holding them together until SCC1 cleavage by separase at the metaphase-to-anaphase transition opens the ring, allowing the release of the sister chromatids (for review, see Nasmyth and Haering, 2009).
The establishment of sister chromatid cohesion in yeast involves a multistep process that begins during telophase, when cohesin complexes associate with chromatin in a process that requires the Scc2/Scc4 complex (Ciosk et al., 2000; Gillespie and Hirano, 2004) and appears to involve the binding and then opening of the SMC hinge domain (Gruber et al., 2006; Hirano and Hirano, 2006). Cohesion is established during S-phase in a Ctf7/Eco1-dependent process (Skibbens et al., 1999; Toth et al., 1999). Ctf7 interacts with a number of replication factors and appears to facilitate cohesion formation in the context of the passing replication fork (Kenna and Skibbens, 2003; Lengronne et al., 2006; Moldovan et al., 2006). An antiestablishment complex consisting of Wings Apart-Like protein (WAPL)/Rad61 and Pds5 is required to maintain cohesion during G2/M by stabilizing the interaction between cohesin and the chromosomes (Panizza et al., 2000; Losada et al., 2005; Gandhi et al., 2006). While the specific details of how Ctf7, WAPL/Rad61, and Pds5 function together to first establish and then maintain cohesion still need to be clarified, recent results indicate that Ctf7 acetylates conserved Lys residues in SMC3, which inhibits the antiestablishment function of the Wpl1-Pds5 complex and promotes cohesion establishment (Ben-Shahar et al., 2008; Unal et al., 2008; Zhang et al., 2008; Rowland et al., 2009; Sutani et al., 2009). Ctf7 is also involved in the postreplicative induction of cohesion induced by DNA double-strand breaks (Unal et al., 2007). In this instance, acetylation of the α-kleisin subunit, Mcd1, by Ctf7 is required for cohesion establishment (Heidinger-Pauli et al., 2009).
The structure of CTF7 proteins along with the effects of ctf7 mutations vary considerably between organisms (Skibbens et al., 1999; Toth et al., 1999; Tanaka et al., 2000; Bellows et al., 2003; Williams et al., 2003; Hou and Zou, 2005). Saccharomyces cerevisiae ctf7 mutations cause a delay in mitosis and cells with a 2C DNA content, separated DNA masses, and partially elongated spindles (Skibbens et al., 1999; Toth et al., 1999). The fission yeast eso1-H17 mutant displays a spindle assembly checkpoint-dependent mitotic delay and abnormal chromosome segregation (Tanaka et al., 2000). Mutation of Drosophila DECO affects the distribution of cohesin proteins, and cells progress prematurely into anaphase, where the spindle checkpoint is activated (Williams et al., 2003). The small, interfering RNA-directed depletion of ESCO1 or ESCO2 in HeLa cells caused defects in sister chromatid cohesion, misalignment of chromosomes on the spindle, and abnormal chromosome segregation; however, it did not appear to affect the distribution of cohesin proteins (Hou and Zou, 2005). Therefore, it is possible that CTF7 proteins may have multiple roles in organisms and/or have unique properties in different organisms.
The Arabidopsis (Arabidopsis thaliana) genome contains orthologs for many of the cohesion-associated genes that have been identified in yeast, and studies have shown that the cohesion machinery is generally conserved in Arabidopsis (Bai et al., 1999; Bhatt et al., 1999; Dong et al., 2001; Chelysheva et al., 2005; Lam et al., 2005; da Costa-Nunes et al., 2006; Liu and Makaroff, 2006; Jiang et al., 2007). However, essentially nothing is known about how sister chromatid cohesion is established in plants. Therefore, we were interested in determining if Arabidopsis contains a functional ortholog of CTF7, and if so, what role it plays in plant growth and development.
In this report, we present the results of an analysis of Arabidopsis CTF7, the Cohesion Establishment Factor, and the characterization of plants containing mutations in CTF7. Seeds containing T-DNA insertions in CTF7 exhibit embryo lethality early in development, while the endosperm develops normally. Minor defects were also observed in female gametophytes and pollen germination of CTF7+/2 plants; however, overexpression of CTF7 blocked female gametophyte development at FG1. Therefore, variations in CTF7 levels have very different effects in different cell types.
RESULTS
Arabidopsis Contains a Single CTF7 Gene That Lacks an N-Terminal Extension
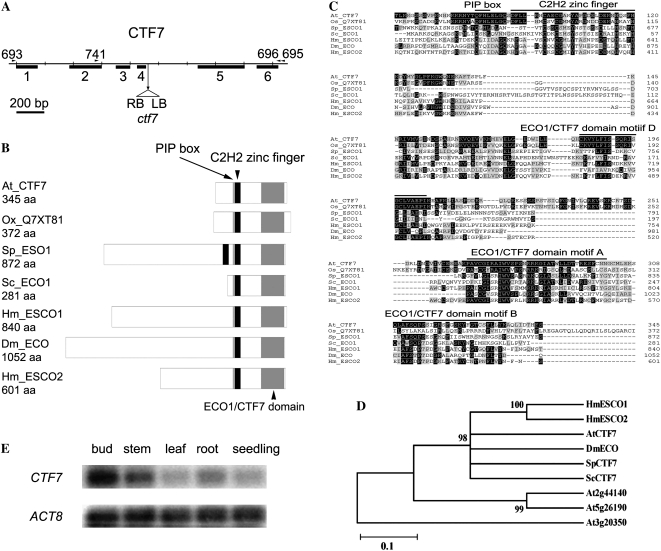
BLAST searches of the Arabidopsis protein database identified one gene (At4g31400) with high sequence similarity with both the human (E value = 3e−21) and S. cerevisiae (E value = 3e−05) Eco1 proteins. Other genes (At3g20350 and At5g26190) were identified with significantly lower similarity levels (E values = 0.62 and 1.7, respectively) that were missing one or more highly conserved residues found in all Ctf7/Eco1 proteins studied to date (see below). Therefore, the Arabidopsis genome appears to contain one putative CTF7 gene (At4g31400), hereafter called CTF7. A CTF7 cDNA was isolated and compared with the published genomic sequence of At4g31400. CTF7 consists of six exons and five introns (Fig. 1A). While the predicted and actual gene structures are similar, differences in the positions of splice sites were found for exons 2, 3, and 4.
Figure 1.
Arabidopsis CTF7 locus and protein structure. A, Gene map of CTF7. The exon positions and T-DNA insertion site are shown. Primers used in this study are shown above the map. LB, Left border; RB, right border. B, Schematic representations of ECO1/CTF7 proteins from different organisms. The ECO1/CTF7 domain (gray box), C2H2 zinc finger domain (black box), and PIP box (thin gray line) are shown. Accession numbers are as follows: Arabidopsis CTF7 (EU077499), O. sativa CTF7 (Q7XY81), S. pombe Eso1 (O42917), S. cerevisiae Eco1 (P43605), Homo sapiens Esco1 (Q5FWF5), Drosophila Eco (Q9VS50), H. sapiens Esco2 (Q56NI9). aa, Amino acids. C, Alignment of ECO1/CTF7 proteins in different organisms. The conserved PIP box, C2H2 zinc finger motif, and acetyltransferase domain are overlined. Identical and similar amino acids are shaded black and gray, respectively. D, Phylogenetic tree of characterized ECO1/CTF7 proteins. E, CTF7 transcripts in different tissues of Arabidopsis. ACTIN8 (ACT8) transcripts were used as a control.
Arabidopsis CTF7 is capable of encoding a 345-amino acid protein. Similar to S. cerevisiae Eco1 (281 amino acids), Arabidopsis CTF7 lacks a large N-terminal extension that is found in most other Eco1 proteins (Fig. 1B). A PIP box (QxxL/I, QFHL) and a C2H2 zinc finger motif are present at residues 82 to 86 and 92 to 130, respectively, while an acetyltransferase domain is present between amino acids 184 and 335 (Fig. 1, B and C). Arabidopsis CTF7 exhibits between 24% and 58% similarity with CTF7 proteins from yeast, fruit fly, and vertebrates, which is the same level of similarity observed between other CTF7 homologs (Fig. 1C). Overall, Arabidopsis CTF7 shows the highest sequence similarity to human Eco1 (22% identity, 58% similarity).
CTF7 transcript levels were analyzed in different tissues using reverse transcription (RT)-PCR. Low but detectible levels of CTF7 transcripts were found in all tissues examined, including buds, roots, leaves, stems, and young seedlings, with buds exhibiting the highest signal and mature leaves the lowest (Fig. 1E). Expression profiling data for CTF7 are consistent with these results and show that CTF7 transcript levels are highest in bolting plants, buds, and young flowers. CTF7 transcripts are also elevated in E2FaDPa transgenic plants, which overexpress S-phase genes (Vlieghe et al., 2003). Therefore, CTF7 expression patterns are consistent with a predicted role in nuclear division.
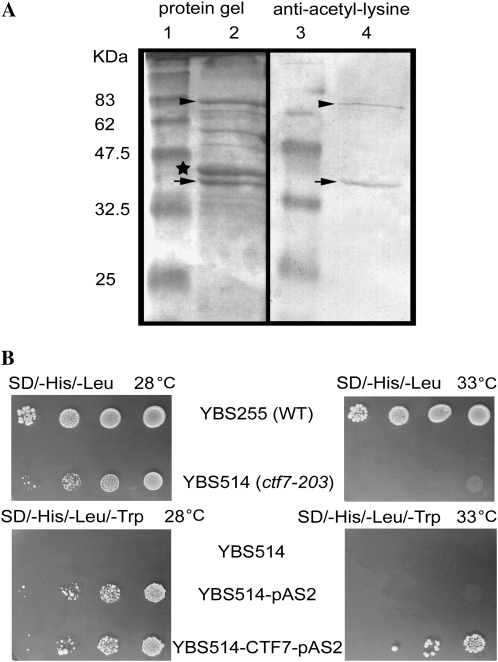
Yeast, fly, and human CTF7 proteins exhibit acetyltransferase activity (Zou et al., 1999; Ivanov et al., 2002; Bellows et al., 2003; Williams et al., 2003). In order to determine if this is also the case for Arabidopsis CTF7, the protein was overexpressed in Escherichia coli and characterized. Soluble CTF7 was produced when expressed as a maltose-binding protein (MBP) fusion in the presence of pGroESL. Acetyltransferase activity was measured by autoacetylation in crude E. coli extracts and with purified CTF7 protein. Western blotting with an anti-acetyl-Lys antibody and matrix-assisted laser-desorption ionization time of flight-mass spectrometry demonstrated that CTF7 is acetylated during the in vitro autoacetylation assays (Fig. 2A). Therefore, Arabidopsis CTF7 exhibits acetyltransferase activity in vitro and is able to undergo autoacetylation.
Figure 2.
CTF7 is the Arabidopsis cohesion establishment factor. A, Arabidopsis CTF7 exhibits acetyltransferase activity. Lanes 1 and 3, Molecular mass markers; lane 2, Coomassie Brilliant Blue-stained SDS-PAGE gel of affinity-purified CTF7-MBP; lane 4, Western blot of affinity-purified CTF7-MBP treated with anti-acetyl-Lys antibodies. Full-length CTF7-MBP is marked with arrowheads. A cleaved version of CTF7 is marked with arrows, and MBP is labeled with a star. B, Arabidopsis CTF7 can substitute for S. cerevisiae CTF7. Cultures of wild-type (WT) and temperature-sensitive ctf7-203 cells containing the pAS2 control plasmid or an Arabidopsis CTF7 complementation plasmid were grown on the appropriate selection plates at either 28°C or 33°C.
The question of whether the Arabidopsis protein can substitute for yeast Eco1 was investigated by transforming the yeast ctf7 temperature-sensitive mutant, ctf7-203 (YBS514), with the Arabidopsis CTF7 cDNA. A pAS2-CTF7 construct was able to restore the ability of YBS514 cells to grow at 33°C, while YBS514 cells containing the pAS2 vector alone were not (Fig. 2B). Therefore, Arabidopsis CTF7 is able to substitute for S. cerevisiae Eco1. Taken together, these results confirm that CTF7 is the Arabidopsis cohesion establishment factor.
Genetic Analysis of Arabidopsis ctf7 Plants
The role of CTF7 in plant growth and development was investigated by characterizing the SALK_059500 (ctf7-1) and SAIL_1214G06 (ctf7-2) T-DNA insertion lines. PCR amplification followed by DNA sequence analysis confirmed that the ctf7-1 and ctf7-2 T-DNA insertions are located in intron 4 and exon 3, respectively. Both mutations are predicted to disrupt the gene upstream of the acetyltransferase domain.
No homozygous ctf7-1 or ctf7-2 plants were identified in initial screens. Therefore, seeds from self-pollinated, heterozygous ctf7+/2 plants were collected and resown, and the progeny were screened by PCR. Fifty-nine percent (223 of 379) of the progeny from heterozygous ctf7-1 plants were heterozygous for the ctf7 T-DNA insertion, whereas 41% of the plants were wild type. No homozygous mutant plants were identified. Likewise no homozygous plants containing the ctf7-2 insertion were identified, with 54% (143 of 263) of the progeny being heterozygous for the T-DNA insertion. These results indicated that CTF7 is essential and suggested that its disruption may cause embryo lethality. Furthermore, the observation that self-pollinated ctf7-1+/2 and ctf7-2+/2 plants consistently produced fewer heterozygous progeny than expected (combined genetic data for the two lines: 336:276 [1.2:1] heterozygous:wild type; χ2 = 0.03, P > 0.05) also suggested a gametophytic effect of the mutations.
To investigate this possibility, ctf7-1+/2 plants were backcrossed with wild-type plants as both the male and female parents. When ctf7-1+/2 plants were used as the female parent, 144 wild-type and 146 ctf7-1 plants were obtained, while 120 wild-type and 91 ctf7-1 plants were obtained when ctf7-1+/2 plants were used as the male parent. Similar results were obtained in backcross experiments with ctf7-2+/2. These results indicated that the mutation is transmitted through both the male and female gametophytes but that the transmission efficiency is reduced through male gametophytes.
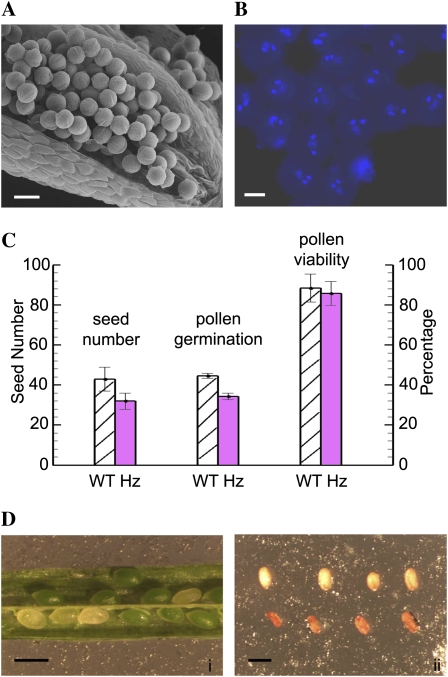
Similar phenotypes were observed for ctf7-1 and ctf7-2 plants in all of our analyses. Therefore, only the results obtained from our analysis of ctf7-1 plants are presented, and the mutant will be referred to generally as ctf7 from here on. A light microscope analysis of semithin sections through stage 11 and 12 anthers (Sanders et al., 1999) failed to identify abnormalities in pollen development in ctf7+/2 plants (data not shown). Pollen from ctf7+/2 plants appeared uniform in size and normal when examined by scanning electron microscopy (SEM; Fig. 3A). The large majority, if not all, of the pollen from ctf7+/2 plants contained two sperm nuclei and a vegetative nucleus, confirming that pollen mitosis is not disrupted by the mutation (Fig. 3B). No significant difference in pollen viability was observed between wild-type and ctf7+/2 plants (Fig. 3C). However, pollen from ctf7+/2 plants did exhibit a slightly reduced in vitro germination frequency (73% of wild-type levels; Fig. 3C).
Figure 3.
Pollen development in ctf7+/2 plants. A, SEM micrograph of an open ctf7+/2 stamen. B, DAPI-stained mature pollen grains from a ctf7+/2 plant. C, Histogram comparing seed number, pollen germination, and pollen viability in wild-type (WT) and ctf7+/2 plants. Data represent averages of at least 20 individuals of each genotype. D, Wild-type and ctf7 seeds: i, ctf7 seeds are normal in size but white; ii, after desiccation, ctf7 seeds are orange and shrunken (bottom row). Bars = 20 μm in A, 10 μm in B, and 1 mm in D.
Further analysis of ctf7+/2 plants identified no obvious morphological alterations, with the exception that they exhibited a reduced seed set (32 ± 4 versus 43 ± 6 per silique in wild-type plants; n = 50; Fig. 3C). Approximately 25% of the seeds in the siliques of ctf7+/2 plants at the cotyledon stage were normal in size but white; later, these seeds became shrunken and orange (Fig. 3D). The presence of 25% aborted seeds inctf7+/2 siliques is consistent with a mutation that affects embryo development.
CTF7 Mutations Cause Minor Alterations in Female Gametophyte Development
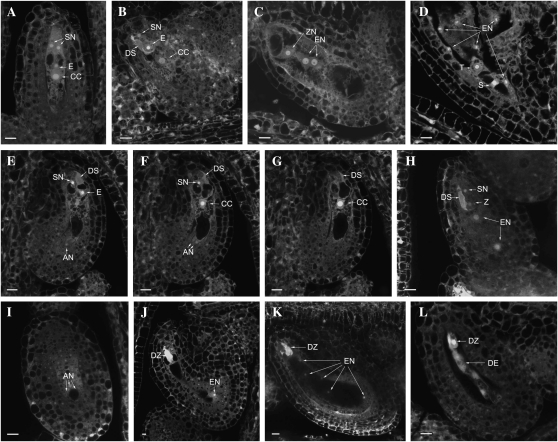
Potential effects of the ctf7 mutation on megagametogenesis were characterized by comparing ovules in wild-type and ctf7+/2 plants using laser scanning confocal microscopy. A total of 36 pistils at various stages of development from three different ctf7+/2 plants were analyzed. Female gametophyte development was defined according to Christensen et al. (1998). No detectable abnormalities were identified in embryo sacs of ctf7+/2 plants between stages FG0 and FG6 (Fig. 4, A–D). However, embryo sac development was less uniform in the siliques of ctf7+/2 plants than those of wild-type plants. In contrast to the two or three developmental stages observed in wild-type siliques (Shi et al., 2005), at least four different stages were observed in ctf7+/2 siliques (Table I). For example, both fertilized and FG5 embryo sacs were found in the same silique. This was never observed in wild-type siliques, suggesting that ctf7 female gametophytes develop slower than wild-type female gametophytes.
Figure 4.
Female gametophyte and seed development in wild-type and ctf7+/2 siliques. The images represent a single 1.5-μm optical section unless otherwise noted. A to D, Normal ovules. E to L, ctf7 ovules. A, Wild-type female gametophyte at stage FG7 containing two synergid cells (SN), an egg cell (E), and a central cell (CC). A projection of three 1.5-μm optical sections is shown. B, Female gametophyte at stage FG8 with degenerated synergid cell (DS). A projection of three 1.5-μm optical sections is shown. C, Fertilized developing seed that contains an elongated zygote (ZN) and two endosperm nuclei (EN). A projection of two 1.5-μm optical sections is shown. D, Developing seed in which the zygote has formed the suspensor (S) and terminal (T) cells. E, First optical section of an ovule with three undegenerated antipodal cells. A degenerated synergid cell, a persistent synergid cell, and the egg cell are at the micropylar pole. One of the three antipodal cells (AN) is at the bottom of the embryo sac. F, Second optical section showing two antipodal cells at the bottom of the embryo sac. G, Third optical section with the central cell in the middle of the embryo sac. H, Fertilized seed containing degenerated and persistent synergid cells and a zygote (Z). The endosperm nucleus has divided into two nuclei (EN). A projection of three 3-μm optical sections is shown. I, Optical section (3 μm) through the chalazal pole of the same seed in H. The nucleus and cell membrane of three antipodal cell nuclei are still intact. J, ctf7 seed containing a degenerated zygote (DZ) and eight endosperm nuclei. Only one of the endosperm nuclei is in the same section as the zygote. K, ctf7 seed containing a degenerated zygote and 16 endosperm nuclei. Five of the endosperm nuclei are observed. A projection of three 1.5-μm optical sections is shown. L, Degenerated ctf7 seed containing degenerated zygote and degenerated endosperm (DE) nuclei. A projection of two 1.5-μm optical sections is shown. Bars = 10 μm.
Table I. Female gametophyte development in ctf7+/2 plants.
Female gametophyte stages were defined according to Christensen et al. (1998).
| Pistil No. | No. of Female Gametophytes |
Total Female Gametophytes | |||||||
| FG1 | FG2 | FG3 | FG4 | FG5 | FG6 | FG7 | FG8 | ||
| 1 | 11 | 17 | 17 | 6 | 51 | ||||
| 2 | 13 | 17 | 10 | 6 | 46 | ||||
| 3 | 8 | 34 | 4 | 5 | 51 | ||||
| 4 | 4 | 3 | 8 | 15 | |||||
The first developmental abnormality was observed at stage FG7, when the three antipodal cells did not degenerate normally in ctf7 embryo sacs. Typically, by the time the egg and synergid cells are mature, as evidenced by the opposite positions of nuclei and vacuoles, the three antipodal cells have degenerated (Fig. 4A). However, three antipodal cells were often found along with mature FG7 egg and synergid cells in embryo sacs of ctf7+/2 siliques. Antipodal cells were also observed in recently fertilized embryo sacs and in embryo sacs containing endosperm with two or four nuclei (Fig. 4, E–G and I). Antipodal cells were never observed in fertilized embryo sacs of wild-type plants (Fig. 4, B–D). Approximately 50% (45.8% ± 1.4%) of the embryo sacs in ctf7+/2 siliques exhibited this delayed degeneration of antipodal cells, suggesting that it is linked with the ctf7 mutation. This observation, along with variations in the timing of female gametophyte development and the reduced pollen germination, indicates that the ctf7 mutation has minor effects on gametophyte development. While not severe, together they likely contribute to the reduced transmission of the mutation that we observe.
Inactivation of CTF7 Blocks Embryo Development But Has No Significant Effect on Endosperm Development
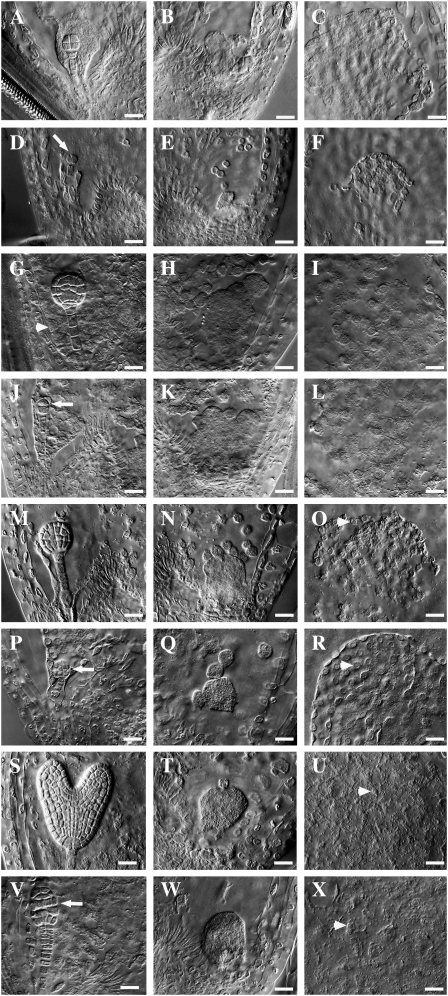
Analysis of seed development in ctf7+/2 plants identified alterations in zygote formation in approximately 25% of the seeds. In wild-type Arabidopsis plants, a single-celled zygote and a primary endosperm nucleus are normally observed along with degenerated and persistent synergid cells after fertilization of the egg and central cell. The endosperm nucleus then undergoes a series of syncytial divisions that produce an endosperm with three domains: the embryo-surrounding region or micropylar endosperm (MCE), the peripheral endosperm (PEN) in the central chamber, and the chalazal endosperm (CZE; Olsen, 2004). After the endosperm nucleus has undergone three to four rounds of nuclear division, the zygote becomes elongated and divides asymmetrically, generating a smaller apical cell and a larger basal cell (Ingouff et al., 2005). Subsequent divisions produce an embryo composed of the embryo proper and the suspensor, which is composed of an enlarged basal cell and a file of six to eight additional cells. The embryo proper undergoes successive rounds of division, forming the two-cell, quadrant, octant (Fig. 5A), dermatogen, globular (Fig. 5G), early heart (Fig. 5M), heart, torpedo (Fig. 5S), and mature embryo in turn.
Figure 5.
Embryo and endosperm development in wild-type and ctf7+/2 siliques. Wild-type (A–C, G–I, M–O, and S–U) and ctf7+/2 (D–F, J–L, P–R, and V–X) embryo and endosperm whole-mount images are shown. A, Wild-type octant-stage embryo. The MCE is still at the NCD stage. B, CZE of the embryo sac in A. C, PEN of the embryo sac in A. D, ctf7 embryo showing aberrant cell division. The arrow indicates the division plane. E, CZE of the embryo sac in D. F, PEN of the embryo sac in D. G, Wild-type globular embryo. The MCE shows signs of cellularization (arrowhead). H, CZE of the embryo sac in G. Endosperm nodules and chalazal cyst are visible. I, PEN of the embryo sac in G. J, ctf7 embryo showing aberrant cell division. The arrow indicates the division plane. K, CZE of the embryo sac in J. L, PEN of the embryo sac in J. M, Wild-type early-heart-stage embryo. N, CZE of the embryo sac in M. O, PEN of the embryo sac in M. The arrowhead indicates NCDs starting to cellularize. P, ctf7 embryo showing aberrant cell division. The arrow indicates abnormal cell shape and cell organization. Q, CZE of the embryo sac in P. R, PEN of the embryo sac in P. The arrowhead indicates NCDs starting to cellularize. S, Wild-type torpedo-stage embryo. T, CZE of the embryo sac in S. NCDs are still present. U, PEN of the embryo sac in S. The arrowhead shows cells that have completed cellularization. V, ctf7 embryo with an abnormal shape and disorganized cell order. The arrow indicates abnormal cell organization. W, CZE of the embryo sac in V. X, PEN of the embryo sac in V. The arrowhead shows cells that have completed cellularization. Bars = 10 μm.
Fertilization appeared to occur normally (Fig. 4, H and I), but alterations were observed in ctf7 zygotes soon after the first division, when a number of embryos arrested and subsequently degenerated at the one- or two-cell zygote stage (Figs. 4, J–L, and 5D). Some zygotes reached the four-celled proembryo stage, at which point both the suspensor and embryo proper exhibited altered division planes (Fig. 5, J and P). A small number of embryos, which exhibited dramatic alterations in division, reached the dermatogen and early globular stages before arresting (Fig. 5V). Altogether, 23.7% ± 0.6% of the seeds in ctf7+/2 siliques (n = 7) exhibited defects in early zygote and embryo development, strongly suggesting that the mutation causes embryo lethality by either blocking or altering nuclear division.
In contrast, the endosperm in ctf7 seeds appeared relatively normal throughout development. In wild-type seeds, the endosperm contains approximately 26 to 50 nuclear cytoplasmic domains (NCDs) by the time the embryo first begins to divide, while octant-stage embryos are typically surrounded by approximately 100 uncellularized NCDs (Fig. 5, B and C). The numbers and appearance of NCDs were normal in ctf7 seeds containing arrested zygotes and proembryos (Fig. 5, D–F). Cellularization of the MCE begins when the embryo is at the early globular stage in wild-type seeds (Fig. 5G). At this time, the PEN showed a gradient of cellularization stages (Fig. 5I), and endosperm nodules as well as the chalazal cyst were observed in the CZE (Fig. 5H). The endosperm of ctf7 seeds showed a similar pattern of development with no obvious abnormalities, even though the embryo had already arrested (Fig. 5, J–L). By late globular/early heart stage, the MCE had completed cellularization and the PEN had started the cellularization process in wild-type seeds (Fig. 5, M–O). Again, cellularization of the MCE and PEN and formation of the chalazal cyst occurred normally in ctf7 seeds (Fig. 5, Q and R). Finally, by the time the wild-type embryo had developed to the torpedo stage, the PEN had completed cellularization and the endosperm began to show signs of degeneration (Fig. 5, S–U). Complete cellularization, followed by degeneration of the endosperm, was also observed in ctf7 seeds (Fig. 5, W and X).
Subtle defects in the NCDs of some ctf7 seeds, however, were detected, beginning when the corresponding wild-type seeds were at approximately the 32-cell embryo stage. In particular, a small number of NCDs with enlarged nucleoli, NCDs that contained multiple nucleoli, and NCDs that contained multiple nuclei were observed (less than 5%; data not shown). However, these alterations were generally rare and did not appear to have any significant effect on endosperm development. Therefore, while inactivation of CTF7 blocks embryo development beginning at the zygote stage, it has little to no effect on endosperm development.
Overexpression of CTF7 Blocks Early Ovule Development
We next tested the effect of overexpression of Arabidopsis CTF7 on plant growth. Transgenic plants that overexpress CTF7 from the 35S promoter appeared normal during vegetative growth; however, 10 out 13 independent lines exhibited varying degrees of reduced fertility, ranging from 5% to 52% aborted seeds. Southern-blot analysis of two lines (1 and 18) that exhibited the highest levels of aborted seeds (484:311 and 568:627 normal:aborted) showed that they contain a single insert. A genetic analysis of one of these lines showed that the T2 generation segregated approximately 1:1 for BASTA resistance (47 BASTA resistant:53 BASTA sensitive), suggesting that the 35S:CTF7 construct causes gametophytic lethality.
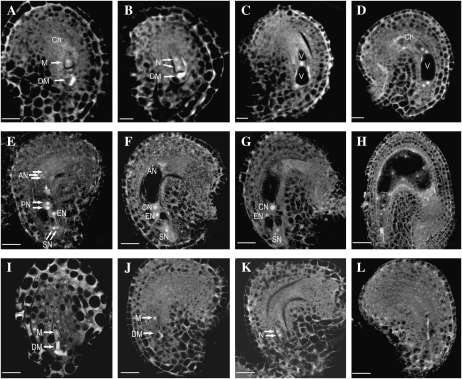
Further analysis of the 10 35S:CTF7 reduced fertility lines showed that they all produced normal levels of viable pollen but contained alterations in female gametophyte development. Detailed analyses of gametophyte development in lines 1 and 18 demonstrated that pollen development is unaffected (data not shown), while 46% (n = 795) of the ovules were found to abort early in development. Specifically, laser scanning confocal microscopy analysis of female gametophyte development in lines 1 and 18 showed that approximately half of the ovules arrested at approximately FG1 (Fig. 6, I and J). In the ovules exhibiting wild-type development, the functional megaspore (FG1; Fig. 6A) underwent mitosis to give rise to a two-nucleate embryo sac (FG2; Fig. 6B). A central vacuole formed between the two nuclei (FG3; Fig. 6C), and the two nuclei underwent a second division to give rise to a four-nucleate embryo sac at early FG4 (Fig. 6D). At this time, the division plane between the two chalazal nuclei was parallel to the chalazal-micropylar axis. Then, two of the four nuclei migrated so that the division plane between them was orthogonal to the axis. Another round of division gave rise to an eight-nucleate embryo sac. The two polar nuclei, one from each pole, then migrated toward the micropylar half of the developing female gametophyte (FG5; Fig. 6E) and eventually fused to form the central cell (FG6; Fig. 6F). The three antipodal cells degenerated just before fertilization, giving a mature female gametophyte consisting of two synergid cells, one egg cell, and one central cell (FG7; Fig. 6G).
Figure 6.
Female gametophyte development revealed by laser scanning confocal microscopy in wild-type and CTF7 overexpression plants. A to H, Wild-type female gametophyte development. I to L, Female gametophyte development in CTF7 overexpression lines. A, Female gametophyte stage 1 (FG1) ovule showing the functional megaspore (M). A trace of the degraded megaspore (DM) is still visible. Ch, Chalaza. B, FG2 ovule with a two-nucleate (N) embryo sac. The degraded megaspore is still visible in some cases. C, FG3 ovule showing a late two-nucleate embryo sac with an enlarged central vacuole (V) and a small chalazal vacuole. D, Ovule with a four-nucleate embryo sac at FG4. E, Ovule with an embryo sac at FG5. Cellularization and cell differentiation are complete with the formation of two synergid nuclei (SN), an egg nucleus (EN), three antipodal nuclei (AN), and the two prominent polar nuclei (PN), which have not yet fused. F, Ovule with a mature seven-celled embryo sac at FG6. The polar nuclei have fused to form a diploid central nucleus (CN). G, Ovule at FG7 in which the antipodal cells have begun to degenerate. H, Ovule after fertilization. One synergid cell is degraded and the endosperm has completed several rounds of nuclear division. I, 35S-CTF7 ovule. No difference with wild-type FG1 ovules is apparent. The degraded megaspore is visible. J, 35S-CTF7 ovule. The embryo sac is enlarged but the female gametophyte remains at FG1. K, 35S-CTF7 ovule. The female gametophyte completed one round of mitosis. L, 35S-CTF7 ovule. The female gametophyte is degraded. Bars = 5 μm.
In contrast, a significant number of the female gametophytes, which we predict overexpress CTF7, arrested at FG1 (Fig. 6, I and J). The arrested female gametophytes did not exhibit any obvious abnormalities but typically failed to divide to form a two-nucleate embryo sac. Occasionally, we observed an arrested female gametophyte at FG2 (Fig. 6K); however, this was quite rare. Obvious abnormalities were also not observed in the arrested FG2 female gametophytes, but ultimately, all of the arrested female gametophytes degraded (Fig. 6L). The size of the embryo sac continued to enlarge in seeds with arrested female gametophytes, indicating that integument growth was normal (Fig. 6, I–L).
The possibility that overexpression of CTF7 affects female gametophyte development was further investigated by analyzing transgenic plants that express CTF7-yellow fluorescent protein (YFP) from the 35S promoter. Of the 10 transgenic lines analyzed, six exhibited varying levels of reduced fertility (9%–41% aborted seeds). Analysis of the lines showed that YFP is expressed from the 35S promoter in the female gametophyte throughout ovule development and that, similar to 35S:CTF7 lines, female gametophytes arrested at FG1 and FG2 (Fig. 7, A–H). Therefore, expression of CTF7 from the 35S promoter can block female gametophyte development beginning at FG1.
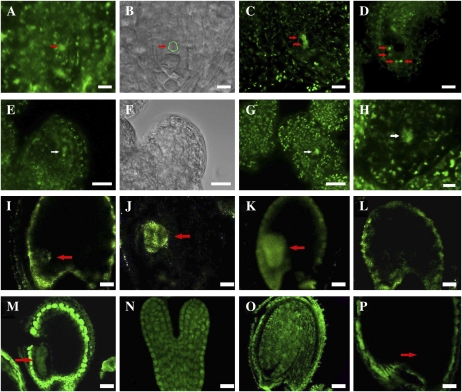
Figure 7.
Cellular distribution of CTF7 in wild-type and 35S-CTF7::YFP plants. A to D, 35S-driven CTF7::YFP localization during female gametophyte development. A, FG1 ovule. The arrow indicates the female gametophyte nucleus. B, Bright-field image of A. The position of the female gametophyte is circled. C, FG2 ovule. The arrows indicate nuclei. D, FG7 ovule. The arrows indicate nuclei. E to H, Aborted female gametophytes in 35S-CTF7::YFP plants. E, Aborted FG1 ovule. The arrow indicates degraded nucleus. F, Bright-field image of E. G, Aborted ovule at early FG1. The arrow indicates degraded nucleus. H, Aborted ovule. The arrow indicates remnants of degraded nucleus. I, Seed with globular-stage embryo from a genomic CTF7::YFP plant. J, Seed with heart-stage embryo from a genomic CTF7::YFP transgenic plant. K, Seed with torpedo-stage embryo from a genomic CTF7::YFP transgenic plant. L, Seed from a wild-type, negative control plant. M to P, CTF7 immunolocalization on whole-mount cleared seeds. M, Torpedo-stage embryo. N, Single embryo at early torpedo stage. O, Cotyledon-stage embryo. P, Negative control with no primary antibody. Arrows in I to K, M, and P denote the position of the embryo. Bars = 5 μm.
CTF7 Is Abundant in the Embryo But Present at Very Low Levels in the Endosperm
Our observation that endosperm development is not significantly affected by inactivation of CTF7 suggested that CTF7 is not required for nuclear division in the endosperm. This raised the question of whether CTF7 is actually present in the endosperm. To investigate this question, we analyzed CTF7 distribution and localization patterns in seeds using both immunolocalization and transgenic plants in which YFP was fused to the C terminus of the CTF7. The CTF7-YFP fusion protein could be readily detected in the embryo, while signals from the endosperm were barely detectable (Fig. 7, I–L). While weak fluorescence was detected in the MCE, PEN, and CZE, the signals in the endosperm were very weak, not always uniform, and often not above background levels. Immunolocalization experiments using a polyclonal antibody to full-length Arabidopsis CTF7 again revealed strong CTF7 signal in the embryo at various stages of development, while cross-reactivity in the endosperm was typically similar to that observed in negative control experiments with preimmune or secondary antibody alone (Fig. 7, M–P). Strong autofluorescence was observed in the integuments in all samples. Therefore, while CTF7 is present at high levels in the embryo, it is present at very low levels, if at all, in the endosperm. Finally, an analysis of the subcellular distribution of CTF7 showed that it is present in both the nucleus and the cytoplasm, with the nuclear signal always significantly stronger than the signal detected in the cytoplasm. A function for CTF7 outside the nucleus has not been described in any organism. Therefore, the significance, if any, of cytoplasmic CTF7 is unknown.
DISCUSSION
Ctf7 proteins possess several highly conserved domains, including a PIP box, a C2H2 zinc finger, and an acetyltransferase domain, which are all present in Arabidopsis CTF7. The PIP box is required for Proliferating Cell Nuclear Antigen binding (Moldovan et al., 2006). The C2H2 zinc finger motif appears to enhance substrate recognition (Onn et al., 2009). The C-terminal Ctf7 acetyltransferase domains of yeast Ctf7 and human ESCO1 have been shown to acetylate two conserved Lys residues on SMC3 (Ben-Shahar et al., 2008; Unal et al., 2008; Zhang et al., 2008). Furthermore, Eco1-dependent acetylation of Smc3p during S-phase and Mcd1 during DNA double-strand break repair stimulates cohesion formation by inhibiting the activity of Wpl1 (Ben-Shahar et al., 2008; Unal et al., 2008; Zhang et al., 2008; Rowland et al., 2009; Sutani et al., 2009). It will be interesting to determine if Arabidopsis CTF7 functions in a similar fashion.
CTF7 proteins in most organisms contain additional diverse N-terminal domains. In fission yeast, the N-terminal two-thirds of ESO1 is similar to DNA polymerase η of budding yeast, and deletion of this domain increases sensitivity to UV irradiation (Tanaka et al., 2000). Human ESCO1 contains an N-terminal extension with similarity to linker histone proteins, which facilitates its binding to chromosomes (Hou and Zou, 2005). Arabidopsis CTF7 is significantly shorter than the Schizosaccharomyces pombe and human proteins and similar to the S. cerevisiae protein in that it lacks an extended N-terminal domain. Predicted rice (Oryza sativa) and maize (Zea mays) CTF7 proteins also lack an N-terminal extension, suggesting that this is a common feature of plant CTF7 proteins. Arabidopsis CTF7 is able to complement a S. cerevisiae ctf7 temperature-sensitive mutant, while the human protein is not (Bellows et al., 2003). Taken together, these results suggest that CTF7 may have evolved additional different roles or may function somewhat differently in different organisms.
Proper Levels of CTF7 Are Essential for Mitosis in the Female Gametophyte
Self-pollinated ctf7+/2 plants consistently produce a 1.2:1 ratio of heterozygous:wild-type progeny, indicating that the mutation has an effect on gametophyte development. Transmission through pollen was reduced slightly in ctf7+/2 plants, with the pollen exhibiting reduced germination frequencies. However, no dramatic alterations in pollen development were observed and the generative nucleus and two sperm nuclei were present and appeared normal. Likewise, pollen development appeared normal in transgenic plants overexpressing CTF7. Therefore, changes in CTF7 levels either by mutation or overexpression do not have a dramatic effect on pollen development or function.
Inactivation of CTF7 also did not did not have a dramatic effect on female gametophyte development, although again, subtle alterations were observed. Specifically, inactivation of CTF7 slowed the progression of mitosis and overall development of female gametophytes (Table I); in addition, the three antipodal cells did not degenerate normally in ctf7 embryo sacs (Fig. 4, E, F, and I). However, these alterations do not appear to directly affect the function of the embryo sac or fertilization, as the mutation is transmitted normally through female gametophytes (Table I). These results are consistent with a number of reports showing that inactivation of essential genes, including several involved in the formation or release of cohesion, does not block gametophyte development but rather cause alterations in embryo development (Liu and Meinke, 1998; Lam et al., 2005; Liu and Makaroff, 2006). It has been suggested that gametophytes may have a pool of mRNA or protein that can be utilized for the successful completion of the limited number of nuclear divisions required for gametophyte development (Liu and Meinke, 1998). Evidence in support of this hypothesis comes from the observation that transgenic plants expressing a separase RNA interference construct from the meiotic DMC1 promoter exhibit defects much earlier than T-DNA insertion lines (Liu and Makaroff, 2006).
In contrast, expression of CTF7 and CTF7:YFP from the 35S promoter significantly impacted female gametophyte development, typically resulting in arrest at FG1 or FG2 (Fig. 6, I–K). The arrested female gametophytes did not exhibit any obvious abnormalities but generally failed to divide to form a two-nucleate embryo sac. The observed effect was variable, with the number of aborted ovules per silique ranging from 5% to 52%. We believe that this variability is likely due to the relatively weak and variable expression of the 35S promoter in female gametophytes of the different transgenic lines. Our results showing that the 35S promoter is active in female gametophytes of 35S-CTF7-YFP plants (Fig. 7, A–H) support this hypothesis and are consistent with another report showing that the 35S promoter is active during female gametophyte development (Liu et al., 2008). An alternative possibility, that the observed female gametophyte arrest results from chromosomal rearrangements associated with T-DNA insertion (Ray et al., 1997; Curtis et al., 2009), is less likely given the absence of pollen defects, the results of our expression studies, and the multiple independent occurrences of the phenotype in the transgenic lines.
At this time, it is not clear how overexpression of CTF7 affects female gametophyte development. However, it has recently been shown that the acetylation state of SMC3 affects the rate of clamp-loader-dependent fork progression in human cells (Terret et al., 2009) and that overexpression of Ctf7 in yeast bypasses the requirement for double-strand breaks to generate cohesion during G2/M (Heidinger-Pauli et al., 2009). Therefore, it is possible that overexpression of CFT7 may shift the balance of the acetylated cohesin complex, resulting in a slowdown or blockage of the replication fork. It is also possible that a noncohesin protein is aberrantly acetylated, causing gametophyte arrest. Our observation that CTF7 overexpression only affects the female gametophyte suggests that female gametophytes are more sensitive to these perturbations than male gametophytes or that overexpression of CTF7 in the transgenic plants may be higher in the female gametophytes.
CTF7 Is Essential for Mitosis in the Embryo But Not in Pollen or the Endosperm
CTF7 is essential for nuclear division in the embryo, with most ctf7 embryos arresting soon after fertilization and prior to the first division (Fig. 5, J and K). In contrast, the syncytial nuclear divisions of the fertilized central cell proceeded normally in ctf7 seeds. Furthermore, cellularization of the endosperm and further cell division appeared similar to the situation in wild-type endosperm (Brown et al., 1999; Sorensen et al., 2002; Ingouff et al., 2005). Therefore, while the zygote is not able to undergo nuclear division in the absence of CTF7, multiple rounds of endosperm nuclear division occur and are relatively normal in the absence of CTF7.
Our localization studies show that while CTF7 is present at high levels in the embryo, it is absent/barely detectable in the endosperm (Fig. 7). These results, along with those from our phenotypic studies, suggest that CTF7 is not required for the establishment of sister chromatid cohesion in the endosperm. In S. pombe, Eso1 becomes dispensable if Pds5 is deleted (Tanaka et al., 2001), while in S. cerevisiae, the lethality of an Eco1 deletion is suppressed by inactivation of Rad61/Wpl1 (Ben-Shahar et al., 2008). Furthermore, cohesion defects in human cells depleted of Esco1 can be rescued by codepletion of Wapl1 (Gandhi et al., 2006). Therefore, it is possible that the endosperm lacks CTF7, WAPL1, and PDS5 and instead uses a different mechanism to establish sister chromatid cohesion.
The possibility also exists that a CTF7-like protein may substitute for CTF7 in the endosperm. Several predicted proteins (At3g20350, At5g26190, and At2g44140) exhibit low-level similarity to yeast, human, and Arabidopsis CTF7. However, none of the proteins contain all of the elements conserved in CTF7 proteins (e.g. PIP box, C2H2 zinc finger, and acetyltransferase domain), and phylogenetic analyses of the Arabidopsis CTF7, At3g20350, At5g26190, and At2g44140 proteins support the conclusion that Arabidopsis contains a single CTF7 ortholog (Fig. 1D). Finally, it is possible that the early embryo abortion and continued endosperm nuclear division we observe results from different cell cycle checkpoints operating in embryos and the endosperm. It is possible that endosperm nuclei either lack cell cycle checkpoints or have altered cell cycle checkpoints that tolerate the missegregation of chromosomes. Consistent with this hypothesis are several embryo-lethal mutants that arrest at the one- or two-cell zygote stage while the endosperm continues to divide. However, very few of the mutants characterized to date appear to exhibit normal endosperm development. For example, of the 70 MEE mutants that arrest very early in embryo development, only six were found to exhibit normal endosperm development (Pagnussat et al., 2005). Therefore, our observation that inactivation of CTF7 blocks embryo development soon after fertilization, but has no apparent effect on endosperm development, is quite unusual.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown in Metro-Mix200 soil (Scotts-Sierra Horticultural Products; http://www.scotts.com) or on germination plates (Murashige and Skoog; Caisson Laboratories; www.caissonlabs.peachhost.com) in a growth chamber at 22°C with a 16-h-light/8-h-dark cycle. Arabidopsis ecotype Wassilewskija was used for transcript analysis and plant transformation studies. Crossing studies were conducted with the Columbia ecotype. The SALK_059500 and SAIL_1214G06 T-DNA insertion lines were obtained from the Arabidopsis Biological Resource Center. Seedlings and roots were collected from 7-d-old plants grown on plates. Leaves were harvested from rosette-stage plants grown on soil. Approximately 20 d after germination, buds and stems were collected. All samples were harvested, frozen in liquid N2, and stored at –80°C until needed.
Molecular Analysis of CTF7
The Arabidopsis CTF7 (At4g31400) genomic DNA sequence was identified by BLAST searching of the Arabidopsis Genome Initiative Proteins Database (http://www.arabidopsis.org/blast/) with the Saccharomyces cerevisiae Eco1, Schizosaccharomyces pombe Eso1, and human ECO1 proteins.
A CTF7 cDNA was obtained from total RNA isolated using Trizol (Invitrogen) by performing reverse transcription with primer 695 followed by PCR using primers 693 and 695 (Fig. 1). CTF7 transcript levels in buds, leaves, shoots, stems, roots, and seedlings of wild-type plants were analyzed using RT-PCR. RT was conducted on total RNA (4 μg) using a Thermoscript RT-PCR system (Invitrogen) with an oligo(dT) primer followed by 25 cycles of PCR with primer pair 741/696 (Fig. 1). Amplification products were analyzed by Southern blotting using a [32P]dATP-labeled 741/696 PCR fragment. After hybridization and washing, radioactivity was detected using a Molecular Dynamics PhosphorImager. RT-PCR was conducted using primers to ACTIN8 to standardize the amount of cDNA (An et al., 1996).
Genomic DNA of T-DNA insertion plants was genotyped by PCR with primer pairs specific for the T-DNA and wild-type loci. Southern-blot analysis on ctf7+/2 plants confirmed that there was only one T-DNA insert in the line. A 4,275-bp genomic DNA fragment containing the CTF7 coding region and 2,422 bp of 5′ flanking sequence in pFGC5941 was used to complement ctf7-1 plants using the floral dip transformation method (Clough and Bent, 1998). Transgenic plants were identified by BASTA resistance and PCR genotyped using primer pairs for the transgene, the genomic CTF7 locus, and the T-DNA insert.
Two CTF7-YFP fusion protein constructs were generated. The first was generated by cloning the 2,082-bp coding sequence fragment into 35S::YFP pFGC5941. In the second construct, a 4,275-bp genomic DNA fragment containing the CTF7 coding region and 2,422 bp of 5′ flanking sequence was cloned into a pFGC5941 vector containing YFP. After confirmation by DNA sequencing, the clones were introduced into plants by Agrobacterium tumefaciens transformation and screened by BASTA. Positive plants were confirmed by PCR and further analyzed by confocal microscopy.
Analysis of CTF7 Acetyltransferase Activity
The CTF7 cDNA was cloned into pET-24b as an NdeI/EcoRI fragment and into pIADL14 as an NdeI/HindIII fragment to generate a CTF7-MBP fusion (CTF7-MBP). After DNA sequencing, pET-24b-CTF7 was transformed into BL21(DE3) pLysS Escherichia coli cells, and the overexpression and solubility of the protein were determined. The solubility of CTF7-MBP was improved by introducing pGroESL into the cell line (Goloubinoff et al., 1989). CTF7-MBP was purified on a maltose-binding resin from whole cell extracts of cells that had been grown overnight at 30°C. Fractions containing recombinant CTF7-MBP were analyzed by SDS-PAGE and confirmed by matrix-assisted laser-desorption ionization time of flight-mass spectrometry. CTF7-MBP was assayed for acetyltransferase activity as described previously (Bellows et al., 2003).
Complementation of Yeast ctf7-203 Cells
Yeast expression constructs were prepared by transferring the CTF7 protein-coding region into pAS2 (Clontech Laboratories; http://www.clontech.com). CTF7-pAS2 and pAS2 were transferred into the temperature-sensitive ctf7-203 line, YBS514, using lithium acetate transformation (Skibbens et al., 1999). The ability of the constructs to complement the ctf7-203 mutation was tested by growing YBS255 (wild-type) and YBS514 cells containing CTF7-pAS2 or pAS2 in the appropriate dropout medium. After growth at 25°C for 48 h, 10 μL of a serial dilution of YBS255, ctf7-203/YBS514, YBS514-PAS2, and YBS514-CTF7-PAS2 was plated on the selection plates and grown at 25°C, 28°C, 33°C, and 38°C for 48 h.
Morphological Analysis of ctf7+/2 and 35S-CTF7 Plants
Ovule development was analyzed in ctf7+/2 plants by laser scanning confocal microscopy essentially as described (Christensen et al., 1997). Inflorescences were collected and fixed in 4% glutaraldehyde under vacuum overnight, dehydrated in a graded ethanol series (20% steps for 1 h each), and cleared in a 2:1 mixture of benzyl benzoate:benzyl alcohol. The pistils were mounted under sealed coverslips. Images were collected and projected with Olympus Flouview 2.0 software (http://www.olympus-global.com/), analyzed with Image Pro Plus (Media Cybernetics; http://www.mediacy.com), and presented with Photoshop version 7.0 (Adobe; http://www.adobe.com).
Endosperm development was analyzed in siliques from five individual ctf7+/2 plants using laser scanning confocal microscopy essentially as described (Braselton et al., 1996). Inflorescences were fixed in 3:1 ethanol:glacial acetic acid at 4°C for 48 h, hydrolyzed with 5 mol L−1 HCl for 1 h, followed by staining using Schiff’s reagent (Sigma; www.sigmaaldrich.com) for 24 h. After dehydration in a serial ethanol series, individual seeds were dissected in methyl salicylate and viewed using an Olympus IX-81 fluorescence deconvolution microscope system. Whole-mount clearing was used to determine the embryo and endosperm phenotypes. Siliques from ctf7 heterozygous plants were dissected and cleared in Herr’s solution containing lactic acid:chloral hydrate:phenol:clove oil:xylene (2:2:2:2:1, w/w). Embryo and endosperm development was studied microscopically with a Nikon microscope equipped with differential interference contrast optics.
Pollen morphology, viability, and germination were compared in newly opened flowers of ctf7+/2 plants and wild-type siblings from the same pot. Pollen morphology was analyzed using SEM. Freshly opened flowers were fixed in formalin-acetic acid-alcohol overnight, rinsed three times in fixation buffer, and dehydrated in a graded ethanol series. Once in 85% ethanol, the anthers were dissected, further dehydrated up to 100% ethanol, critical point dried with CO2, mounted on SEM stubs with carbon tabs, sputter coated with 21-nm gold, and viewed and photographed using a Supra 35 FEG-VP scanning electron microscope (Carl Zeiss; www.zeiss.com).
Pollen viability was analyzed by soaking the grains in 2,3,5-triphenyl tetrazolium chloride (1.0% by weight in 50% Suc) at 37°C for 30 min followed by observation by fluorescence microscopy (Huang et al., 2004). Pollen mitosis was analyzed by 4′,6-diamidino-2-phenylindole (DAPI) staining (2 mg L−1) followed by observation by fluorescence microscopy. Pollen germination was tested according to Fan et al. (2001).
CTF7 Immunolocalization
Polyclonal antibodies were raised to Arabidopsis CTF7, which was overexpressed in E. coli and affinity purified using standard procedures (Harlow and Lane, 1988). For immunolocalization experiments, Arabidopsis ovules were fixed overnight in 4% paraformaldehyde in 1× phosphate-buffered saline (PBS) supplemented with 0.1% Triton X-100. The material was then squashed using a cover glass, treated for 1 h with 2% Driselase, and washed briefly with PBS and then with PBS containing 3% IGEPAL CA-630 (Sigma) plus 10% dimethyl sulfoxide for 1 h. The slides were washed three times with 1× PBS, blocked with 3% bovine serum albumin, and then treated with primary antibody overnight at 37°C. The slides were washed six times with 1× PBS treated with secondary antibody for 3 h at 37°C. After washing, the slides were stained with DAPI for 30 min, washed briefly, treated with antifade, and sealed with a cover glass. The slides were analyzed using confocal microscopy as described above.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number EU077499.
Acknowledgments
We are grateful to Richard Edelmann and Matthew L. Duley for technical support with microscopy, Robert Skibbens for providing us the yeast strains YBS255 and ctf7-203 (YBS514), Rich Jorgenson for providing the pFGC5941 plasmid, Michael Crowder for providing the MBP plasmid, and John Hawes for providing the pGroESL plasmid. We thank Meghan Holdorf and Lara Strittmatter for helpful discussions and comments on the manuscript.
References
- An Y, McDowell J, Huang S, McKinney E, Chambliss S, Meagher R. (1996) Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J 10: 107–121 [DOI] [PubMed] [Google Scholar]
- Bai X, Peirsion B, Dong F, Cai X, Makaroff C. (1999) Isolation and characterization of SYN1, a RAD21-like gene essential for meiosis in Arabidopsis. Plant Cell 11: 417–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellows AM, Kenna MA, Cassimeris L, Skibbens RV. (2003) Human EFO1p exhibits acetyltransferase activity and is a unique combination of linker histone and Ctf7p/Eco1p chromatid cohesion establishment domains. Nucleic Acids Res 31: 6334–6343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shahar TR, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F. (2008) Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science 321: 563–566 [DOI] [PubMed] [Google Scholar]
- Bhatt AM, Lister C, Page T, Fransz P, Findlay K, Jones GH, Dickinson HG, Dean C. (1999) The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J 19: 463–472 [DOI] [PubMed] [Google Scholar]
- Braselton JP, Wilkinson MJ, Clulow SA. (1996) Feulgen staining of intact plant tissues for confocal microscopy. Biotech Histochem 71: 84–87 [DOI] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE, Nguyen H, Olsen OA. (1999) Development of endosperm in Arabidopsis thaliana. Sex Plant Reprod 12: 32–42 [Google Scholar]
- Chelysheva L, Diallo S, Vezon D, Gendrot G, Vrielynck N, Belcram K, Rocques N, Marquez-Lema A, Bhatt AM, Horlow C, et al. (2005) AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J Cell Sci 118: 4621–4632 [DOI] [PubMed] [Google Scholar]
- Christensen CA, King EJ, Jordan JR, Drews GN. (1997) Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex Plant Reprod 10: 49–64 [Google Scholar]
- Christensen CA, Subramanian S, Drews GN. (1998) Identification of gametophytic mutations affecting female gametophyte development in Arabidopsis. Dev Biol 202: 136–151 [DOI] [PubMed] [Google Scholar]
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Nasmyth K. (2000) Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell 5: 243–254 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MJ, Belcram K, Bollmann SR, Tominey CM, Hoffman PD, Mercier R, Hays JB. (2009) Reciprocal chromosome translocation associated with T-DNA-insertion mutation in Arabidopsis: genetic and cytological analyses of consequences for gametophyte development and for construction of doubly mutant lines. Planta 229: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa-Nunes JA, Bhatt AM, O’Shea S, West CE, Bray CM, Grossniklaus U, Dickinson HG. (2006) Characterization of the three Arabidopsis thaliana RAD21 cohesins reveals differential responses to ionizing radiation. J Exp Bot 57: 971–983 [DOI] [PubMed] [Google Scholar]
- Dong F, Cai X, Makaroff C. (2001) Cloning and characterization of two Arabidopsis genes that belong to the RAD21/REC8 family of chromosome cohesin proteins. Gene 271: 99–108 [DOI] [PubMed] [Google Scholar]
- Fan L, Wang Y, Wang H, Wu W. (2001) In vitro Arabidopsis pollen germination and characterization of the inward potassium currents in Arabidopsis pollen grain protoplasts. J Exp Bot 52: 1603–1614 [PubMed] [Google Scholar]
- Gandhi R, Gillespie PJ, Hirano T. (2006) Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol 16: 2406–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie PJ, Hirano T. (2004) Scc2 couples replication licensing to sister chromatid in Xenopus egg extracts. Curr Biol 14: 1598–1603 [DOI] [PubMed] [Google Scholar]
- Goloubinoff P, Gatenby AA, Lorimer GH. (1989) GroE heat-shock proteins promote assembly of foreign prokaryotic ribulose bisphosphate carboxylase oligomers in Escherichia coli. Nature 337: 44–47 [DOI] [PubMed] [Google Scholar]
- Gruber S, Arumugam P, Katou Y, Kuglitsch D, Helmhart W, Shirahige K, Nasmyth K. (2006) Evidence that loading of cohesin onto chromosomes involves opening of its SMC hinge. Cell 127: 523–537 [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Heidinger-Pauli JM, Unal E, Koshland D. (2009) Distinct targets of the Eco1 acetyltransferase modulate cohesion in S phase and in response to DNA damage. Mol Cell 34: 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Hirano T. (2006) Opening closed arms: long-distance activation of SMC ATPase by hinge-DNA interactions. Mol Cell 21: 175–186 [DOI] [PubMed] [Google Scholar]
- Hou FJ, Zou H. (2005) Two human orthologues of Eco1/Ctf7 acetyltransferases are both required for proper sister-chromatid cohesion. Mol Biol Cell 16: 3908–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Zhu J, Mu X, Lin J. (2004) Pollen dispersion, pollen viability and pistil receptivity in Lenmus chinensis. Ann Bot (Lond) 93: 295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff M, Haseloff J, Berger F. (2005) Polycomb group genes control developmental timing of endosperm. Plant J 42: 663–674 [DOI] [PubMed] [Google Scholar]
- Ivanov D, Schleiffer A, Eisenhaber F, Mechtler K, Haering CH, Nasmyth K. (2002) Eco1 is a novel acetyltransferase that can acetylate proteins involved in cohesion. Curr Biol 12: 323–328 [DOI] [PubMed] [Google Scholar]
- Jiang L, Xia M, Strittmatter LI, Makaroff CA. (2007) The Arabidopsis cohesin protein SYN3 localizes to the nucleolus and is essential for gametogenesis. Plant J 50: 1020–1034 [DOI] [PubMed] [Google Scholar]
- Kenna MA, Skibbens RV. (2003) Mechanical link between cohesion establishment and DNA replication: Ctf7p/Eco1p, a cohesion establishment factor, associates with three different replication factor C complexes. Mol Cell Biol 23: 2999–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam WS, Yang XH, Makaroff CA. (2005) Characterization of Arabidopsis thaliana SMC1 and SMC3: evidence that ASMC3 may function beyond chromosome cohesion. J Cell Sci 118: 3037–3048 [DOI] [PubMed] [Google Scholar]
- Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner KP, Shirahige K, Uhlmann F. (2006) Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol Cell 23: 787–799 [DOI] [PubMed] [Google Scholar]
- Liu CM, Meinke DW. (1998) The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J 16: 21–31 [DOI] [PubMed] [Google Scholar]
- Liu J, Zhang Y, Qin G, Tsuge T, Sakaguchi N, Luo G, Sun K, Shi D, Aki S, Zheng N, et al. (2008) Targeted degradation of the cyclin-dependent kinase inhibitor ICK4/KRP6 by RING-type E3 ligases is essential for mitotic cell cycle progression during Arabidopsis gametogenesis. Plant Cell 20: 1538–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Makaroff CA. (2006) Arabidopsis separase AESP is essential for embryo development and the release of cohesin during meiosis. Plant Cell 18: 1213–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Hirano T. (2005) Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J Cell Sci 118: 2133–2141 [DOI] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S. (2006) PCNA controls establishment of sister chromatid cohesion during S phase. Mol Cell 23: 723–732 [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. (2009) Cohesin: its roles and mechanisms. Annu Rev Genet 43: 525–558 [DOI] [PubMed] [Google Scholar]
- Olsen OA. (2004) Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell 16: 214–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onn I, Guacci V, Koshland DE. (2009) The zinc finger of Eco1 enhances its acetyltransferase activity during sister chromatid cohesion. Nucleic Acids Res 37: 6126–6134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie LF, Ye D, Sundaresan V. (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614 [DOI] [PubMed] [Google Scholar]
- Panizza S, Tanaka TU, Hochwagen A, Eisenhaber F, Nasmyth K. (2000) Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr Biol 10: 1557–1564 [DOI] [PubMed] [Google Scholar]
- Ray SM, Park SS, Ray A. (1997) Pollen tube guidance by the female gametophyte. Development 124: 2489–2498 [DOI] [PubMed] [Google Scholar]
- Rowland BD, Roig MB, Nishino T, Kurze A, Uluocak P, Mishra A, Beckouet F, Underwood P, Metson J, Imre R, et al. (2009) Building sister chromatid cohesion: Smc3 acetylation counteracts an antiestablishment activity. Mol Cell 33: 763–774 [DOI] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu YC, Lee PY, Truong MT, Beals TP, Goldberg RB. (1999) Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod 11: 297–322 [Google Scholar]
- Shi DQ, Liu J, Xiang YH, Ye D, Sundaresan V, Yang WC. (2005) SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. Plant Cell 17: 2340–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens R, Corson L, Koshland D, Hieter P. (1999) Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev 13: 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen MB, Mayer U, Lukowitz W, Robert H, Chambrier P, Jurgens G, Somerville C, Lepiniec L, Berger F. (2002) Cellularisation in the endosperm of Arabidopsis thaliana is coupled to mitosis and shares multiple components with cytokinesis. Development 129: 5567–5576 [DOI] [PubMed] [Google Scholar]
- Sutani T, Kawaguchi T, Kanno R, Itoh T, Shirahige K. (2009) Budding yeast Wpl1 (Rad61)-Pds5 complex counteracts sister chromatid cohesion-establishing reaction. Curr Biol 19: 492–497 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Hao ZL, Kai M, Okayama H. (2001) Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism. EMBO J 20: 5779–5790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Yonekawa T, Kawasaki Y, Kai M, Furuya K, Iwasaki M, Murakami H, Yanagida M, Okayama H. (2000) Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol Cell Biol 20: 3459–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terret ME, Sherwood R, Rahman S, Qin J, Jallepalli PV. (2009) Cohesin acetylation speeds the replication fork. Nature 462: 231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. (1999) Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev 13: 320–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE. (2008) A molecular determinant for the establishment of sister chromatid cohesion. Science 321: 566–569 [DOI] [PubMed] [Google Scholar]
- Unal E, Heidinger-Pauli JM, Koshland D. (2007) DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7). Science 317: 245–248 [DOI] [PubMed] [Google Scholar]
- Vlieghe K, Vuylsteke M, Florquin K, Rombauts S, Maes S, Ormenese S, Van Hummelen P, Van de Peer Y, Inzé D, De Veylder L. (2003) Microarray analysis of E2Fa-DPa-overexpressing plants uncovers a cross-talking genetic network between DNA replication and nitrogen assimilation. J Cell Sci 116: 4249–4259 [DOI] [PubMed] [Google Scholar]
- Williams BC, Garrett-Engele CM, Li ZX, Williams EV, Rosenman ED, Goldberg ML. (2003) Two putative acetyltransferases, san and deco, are required for establishing sister chromatid cohesion in Drosophila. Curr Biol 13: 2025–2036 [DOI] [PubMed] [Google Scholar]
- Zhang JL, Shi XM, Li YH, Kim BJ, Jia JL, Huang ZW, Yang T, Fu XY, Jung SY, Wang Y, et al. (2008) Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol Cell 31: 143–151 [DOI] [PubMed] [Google Scholar]
- Zou H, McGarry TJ, Bernal T, Kirschner MW. (1999) Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science 285: 418–422 [DOI] [PubMed] [Google Scholar]