Abstract
Rapid assessment of the effect of reduced levels of gene products is often a bottleneck in determining how to proceed with an interesting gene candidate. Additionally, gene families with closely related members can confound determination of the role of even a single one of the group. We describe here an in vivo method to rapidly determine gene function using transient expression of artificial microRNAs (amiRNAs) in Arabidopsis (Arabidopsis thaliana) mesophyll protoplasts. We use a luciferase-based reporter of circadian clock activity to optimize and validate this system. Protoplasts transiently cotransfected with promoter-luciferase and gene-specific amiRNA plasmids sustain free-running rhythms of bioluminescence for more than 6 d. Using both amiRNA plasmids available through the Arabidopsis Biological Resource Center, as well as custom design of constructs using the Weigel amiRNA design algorithm, we show that transient knockdown of known clock genes recapitulates the same circadian phenotypes reported in the literature for loss-of-function mutant plants. We additionally show that amiRNA designed to knock down expression of the casein kinase II β-subunit gene family lengthens period, consistent with previous reports of a short period in casein kinase II β-subunit overexpressors. Our results demonstrate that this system can facilitate a much more rapid analysis of gene function by obviating the need to initially establish stably transformed transgenics to assess the phenotype of gene knockdowns. This approach will be useful in a wide range of plant disciplines when an endogenous cell-based phenotype is observable or can be devised, as done here using a luciferase reporter.
A 24-h biological rhythm is a distinctive feature of many organisms on earth. Time keeping by a circadian clock increases fitness under cyclic environmental conditions, and the clock is a well-conserved mechanism among most eukaryotes from uni- to multicellular organisms (Ouyang et al., 1998; Dodd et al., 2005). In plants, the circadian clock controls various output rhythms ranging from cellular and organ-level processes to whole-plant functions (McClung, 2006). Regulation by the clock can autonomously function within a single cell (Kim et al., 2003; Nakamichi et al., 2004) and many of the main components appear to be conserved among cells in different tissues (James et al., 2008).
The Arabidopsis (Arabidopsis thaliana) circadian clock is maintained by a wide range of molecules that have been identified in forward genetic screens through altered hypocotyl elongation (LATE ELONGATED HYPOCOTYL [LHY], SENSITIVITY TO RED LIGHT REDUCED1, and LUX ARRHYTHMO; Schaffer et al., 1998; Staiger et al., 2003; Hazen et al., 2005), bioluminescence-based period length assays (TIMING OF CAB1 [TOC1], ZEITLUPE [ZTL], TEJ, TIME FOR COFFEE, LIGHT INSENSITIVE PERIOD1, and XAP5 CIRCADIAN TIMEKEEPER; Somers et al., 2000; Strayer et al., 2000; Panda et al., 2002; Hall et al., 2003; Kevei et al., 2007; Martin-Tryon and Harmer, 2008), and flowering time (GIGANTEA [GI], EARLY FLOWERING3 [ELF3], ELF4, and FIONA1; Hicks et al., 1996; Park et al., 1999; Doyle et al., 2002; Kim et al., 2008). Although these efforts have been successful, network modeling (Locke et al., 2006; Zeilinger et al., 2006) and quantitative trait loci studies (Swarup et al., 1999; Michael et al., 2003; Darrah et al., 2006) predict that additional clock components are likely in the Arabidopsis circadian clock. However, forward screening approaches in whole plants have some limitations for further isolation of clock molecules since strong mutations of essential genes that contribute to clock function might be lethal, and functional redundancy within gene families might result in weak or undetectable phenotypes in single gene mutations.
In mammalian systems, the circadian clock can function cell autonomously and the use of serum-induced cultured cell lines has provided another avenue to probe the intracellular mechanisms of the mammalian circadian clock (Balsalobre et al., 1998). Recently, genome-wide reverse genetic screens using RNAi libraries have identified a large number of possible modifiers of the circadian clock using human cell lines (Maier et al., 2009; Zhang et al., 2009).
Here, we describe a rapid in vivo assay to investigate circadian clock function using Arabidopsis mesophyll protoplasts. Transient expression of artificial microRNA (amiRNA) in Arabidopsis protoplasts can reduce the expression level of an endogenous target gene or gene family, which can facilitate very rapid determination of function. This approach will be useful in a wide range of plant disciplines when an endogenous cell-based phenotype is observable.
RESULTS
Arabidopsis Protoplasts Exhibit Robust Circadian Rhythms under Constant Conditions
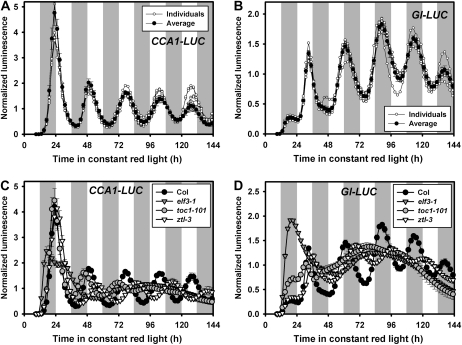
Transient gene expression in Arabidopsis mesophyll protoplasts has been used extensively for functional assays at the cellular level (Sheen, 2001). However, protoplast systems have only been exploited for studies involving relatively short times (less than 48 h), and there have been no reports indicating their viability over longer time courses. To validate the feasibility of this system for chronobiological studies, we monitored clock-driven luminescence rhythms in Arabidopsis mesophyll protoplasts by examining expression of two differently phased clock-controlled reporters after transient transfection (optimization of these conditions is described in Supplemental Results and Discussion S1 and Supplemental Materials and Methods S1). In whole plants, transcription of the myb transcription factor CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and the unique gene GI is strongly clock regulated, with peak expression in the early morning and early evening, respectively (Wang and Tobin, 1998; Fowler et al., 1999; Park et al., 1999). Protoplasts from Arabidopsis leaves transfected with either the CCA1 promoter driving the firefly (Photinus pyralis) luciferase reporter (CCA1-LUC) or the GI promoter-luciferase reporter (GI-LUC) were assayed under constant red light. CCA1-LUC and GI-LUC expression peaked in the early morning and early evening, respectively, and both maintained robust oscillations with approximately 24-h periods for over 6 d (25.8 h, Table I; Fig. 1, A and B). While traces from individual sample wells were quite similar and robust for both reporters, luminescence from the CCA1-LUC reporter was typically much higher during the first 24 to 30 h, relative to later time points (Fig. 1A). Hence, period estimates were assessed using a 36- to 144-h window when this reporter was used.
Table I. Estimates of free-running period in transiently transfected Arabidopsis protoplasts under constant light.
R, Red light; B, blue light.
| Genotype | Effector (AmiRNA) | Reporter (Luciferase) | Light | Period | Figure |
| h | |||||
| Col | – | CCA1-LUC | R | 25.8 ± 0.1 (6/6) | Figure 1C |
| elf3-1 | – | CCA1-LUC | R | N.D. (0/6) | Figure 1C |
| toc1-101 | – | CCA1-LUC | R | 24.4 ± 0.4 (6/6) | Figure 1C |
| ztl-3 | – | CCA1-LUC | R | 27.9 ± 0.2 (6/6) | Figure 1C |
| Col | – | GI-LUC | R | 25.8 ± 0.1 (6/6) | Figure 1D |
| elf3-1 | – | GI-LUC | R | 29.7 ± 0.0 (2/6)a | Figure 1D |
| toc1-101 | – | GI-LUC | R | 18.4 ± 2.2 (6/6)b | Figure 1D |
| ztl-3 | – | GI-LUC | R | 29.2 ± 0.3 (3/5) | Figure 1D |
| Col | Control | CCA1-LUC | R | 26.3 ± 0.1 (6/6) | Figure 2A |
| Col | ELF3 | CCA1-LUC | R | 31.1 ± 1.6 (6/6) | Figure 2A |
| Col | GI | CCA1-LUC | R | 24.0 ± 0.4 (6/6) | Figure 2A |
| Col | ZTL | CCA1-LUC | R | 28.8 ± 0.1 (6/6) | Figure 2A |
| Col | Control | CCA1-LUC | B | 26.5 ± 0.1 (6/6) | Figure 2B |
| Col | ELF3 | CCA1-LUC | B | N.D. (0/6) | Figure 2B |
| Col | GI | CCA1-LUC | B | 23.4 ± 0.2 (6/6)b | Figure 2B |
| Col | ZTL | CCA1-LUC | B | 30.8 ± 0.2 (5/5) | Figure 2B |
| Col | Control | CCA1-LUC | R | 25.8 ± 0.3 (6/6) | Figure 4A |
| Col | CKBfam | CCA1-LUC | R | 29.6 ± 0.1 (6/6) | Figure 4A |
Period is not representative of the multiple trials but an artifact of the period estimation program that returned circadian range period estimates in two instances. Period estimates were generated using BRASS (http://www.amillar.org) from bioluminescence readouts shown in figures indicated. Data are shown as variance weighted mean period ± SEM (rhythmic samples/total samples). –, Without amiRNA plasmid; N.D., not detectable.
Period estimates were determined using a 60-h window (from 24–84 h).
Figure 1.
Arabidopsis protoplasts maintain a robust free-running rhythm with approximately 24-h period for over 6 d. A and C, Bioluminescence traces from protoplast cells transfected with CCA1-LUC plasmids. B and D, Bioluminescence traces from protoplast cells transfected with GI-LUC plasmids. Protoplasts were prepared from rosette leaves of 25- to 35-d Arabidopsis plants of the appropriate wild-type (Col) or mutant (elf3-1, toc1-101, and ztl-3) genotypes grown under a 12-h-light/12-h-day cycle and transfected with the reporter plasmid indicated. After transfection protoplasts were transferred to constant red light (14 μmol m−2 s−1) at ZT9 (n = 6; mean ± SEM). Image acquisition was performed every 2 h for 6 d. Each data set was normalized to the mean expression level over the 24- to 144-h sampling schedule. White and gray regions indicate subjective light and dark period. Similar results were obtained in two independent trials.
The overall oscillation patterns very closely resemble that of intact seedlings demonstrating, first, that Arabidopsis protoplasts can sustain circadian rhythms, and second, that the endogenous clock system regulates transiently introduced reporter genes and endogenous clock genes in the same manner.
Mutations of central circadian clock components can alter the endogenous period, phase, and amplitude of rhythms in plants. Mutations in TOC1 (Kevei et al., 2006; Kolmos et al., 2008) and ZTL (Somers et al., 2000; Kevei et al., 2006) shorten and lengthen periods, respectively, while mutations in ELF3 strongly and rapidly dampens circadian cycling (Hicks et al., 1996). Since transiently expressed CCA1-LUC and GI-LUC reflects the clock activity in wild-type protoplasts, we tested whether protoplasts derived from circadian clock mutants display phenotypes similar to those seen in whole plants.
CCA1-LUC and GI-LUC expression in ztl-3 protoplasts showed a robust 2- to 3-h longer period, relative to Columbia (Col) protoplasts over the 4.5-d sampling period (Fig. 1, C and D; Table I). The same reporters introduced into elf3-1 protoplasts were arrhythmic, after the first acute expression peak soon after transfection (Fig. 1, C and D; Table I). Hence, results from both mutant protoplasts closely mirror the circadian phenotype of whole seedlings.
In toc1-101 protoplasts CCA1-LUC expression showed an advanced peak phase and markedly reduced amplitude, relative to Col wild-type protoplasts, but then damped out very quickly (Fig. 1C). GI-LUC expression in toc1-101 protoplasts was severely dampened and accurate period estimates were not possible (Fig. 1D). The results using both reporters are reminiscent of a similar rapid damping or arrhythmicity seen in TOC1 RNAi and toc1-2 mutant plants assayed in red light (Mas et al., 2003).
Hence, the similarity of the reporter gene expression patterns in these mutant protoplasts to those found in the intact plants expands on our results with wild-type protoplasts by further implying that clock function in protoplasts rely on and respond to the same clock components utilized in intact leaves (e.g. ZTL, ELF3, and TOC1).
Transient Expression of AmiRNAs Targeting Clock Genes Affects Circadian Period, Phase, and Amplitude
Since our results indicate that transient expression of these reporter plasmids in protoplasts accurately reports the activity of the endogenous clock, we next tested the feasibility of this system for the functional assessment of specific clock genes by transiently reducing their endogenous gene expression. To do this we chose an amiRNA-based gene-silencing system. AmiRNAs are 21- to 27-nt single-stranded RNA molecules that are genetically designed to recognize single or multiple genes of interest by modifying a natural microRNA backbone. They regulate target gene expression by mRNA degradation or by inhibition of protein translation (Pillai et al., 2007). AmiRNAs targeting Arabidopsis genes can be generated using the amiRNA design algorithm at WMD3 Web MicroRNA Designer (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi; Schwab et al., 2006; Ossowski et al., 2008) or by employing individual amiRNA constructs (approximately 9,000) that target one-third of the Arabidopsis genome and are currently available from the Arabidopsis Biological Resource Center (ABRC; Supplemental Table S1).
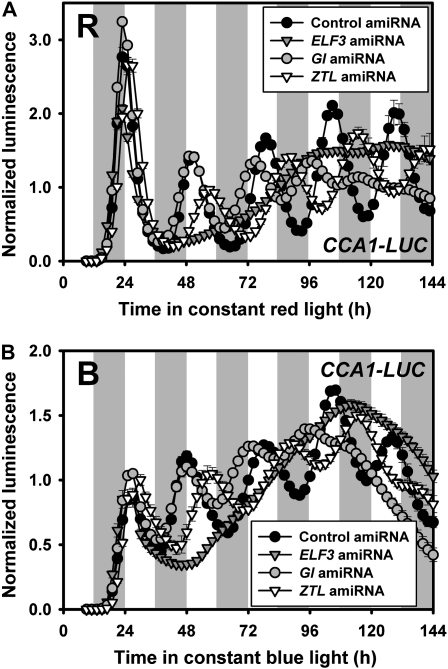
To functionally test the effectiveness of amiRNAs in Arabidopsis protoplasts, we selected amiRNAs targeting known clock genes (ELF3, GI, and ZTL) and examined their effectiveness on cyclic expression of CCA1-LUC by cotransfecting a specific amiRNA plasmid with the CCA1-LUC reporter plasmid. ELF3 and ZTL amiRNAs (amiR-ELF3; amiR-ZTL) were obtained from the ABRC amiRNA collection (see “Materials and Methods” and Supplemental Table S1), and we generated a GI amiRNA (amiR-GI) by using the Web-based amiRNA design algorithm (Ossowski et al., 2008). AmiR-ELF3 transfected cells dampened CCA1-LUC expression after the first expression peak, consistent with arrhythmicity of elf3 mutants in constant red and blue light (Fig. 2, A and B; Hicks et al., 1996). amiR-GI and amiR-ZTL transfected protoplasts had shorter (approximately 2.3 h) and longer (approximately 2.5 h) periods, respectively, than control vector-transfected cells (Table I; Fig. 2A), generally consistent with the reported effects of loss-of-function mutants in these genes (Park et al., 1999; Somers et al., 2004; Kevei et al., 2006; Martin-Tryon et al., 2007). However, some reports show a long period for some gi alleles depending on the reporter and light conditions (Park et al., 1999; Martin-Tryon et al., 2007). When tested under blue light the results were similar (Table I; Fig. 2B). Together these results show the feasibility both of using amiRNA plasmids from both the ABRC amiRNA collection and custom-designed constructs.
Figure 2.
Transient expression of amiRNAs targeting specific circadian clock components recapitulates loss-of-function mutant phenotypes. A, Bioluminescence traces from protoplast cells transfected as described in Figure 1 using the CCA1-LUC reporter and the indicated gene-specific amiRNA in constant red light (R). B, As in A but under constant blue light (B; 5 μmol m−2 s−1). Data represent mean ± SEM (n = 6). Similar results were obtained in two independent trials.
AmiRNA Affects Endogenous Expression of Target Genes
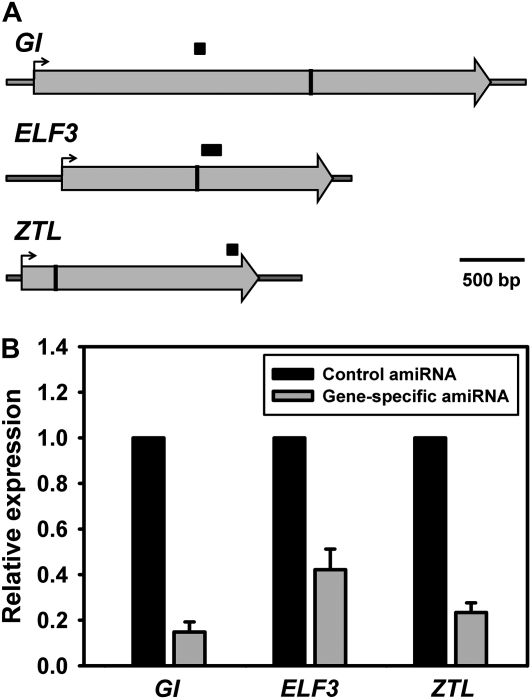
Natural miRNAs act to repress expression of target gene, either through cleavage of target mRNA in the complementary region or inhibition of translation of target protein. While most miRNAs in animal system function through translational regulation, early reports indicated that plant microRNAs mainly act through mRNA cleavage (Axtell and Bowman, 2008; Mallory and Bouche, 2008). Similarly, amiRNAs in plants are believed to regulate the transcript levels of the target genes (Schwab et al., 2006; Ossowski et al., 2008). To investigate the effect of amiRNA on the regulation of gene expression in protoplasts, we performed reverse transcription (RT)-quantitative (q)PCR in protoplasts transfected with gene-specific amiRNAs to determine the message level of the corresponding target gene (Fig. 3A). We observed strong reductions in endogenous message levels in GI, ELF3, and ZTL amiRNA-transfected protoplasts, respectively, compared to mRNA levels in control vector transfected protoplasts (Fig. 3B). The extent of transcript reduction ranged from between 5- and slightly more than 2-fold. Additionally, we tested the level of endogenous ZTL in protoplasts 24 h after transfection with amiR-ZTL. We observed reproducibly reduced levels of ZTL (data not shown), confirming that the long period in these protoplasts resulted from amiRNA effects on ZTL expression.
Figure 3.
Transient expression of amiRNA reduces expression of the endogenous target genes. A, Schematic of the GI, ELF3, and ZTL transcripts with the amiRNA targeted site (vertical bars) and the PCR-amplified region (horizontal black bars) indicated for each. B, Relative expression of GI, ELF3, and ZTL from protoplasts transfected with control amiRNA or the gene-specific amiRNA indicated and harvested at ZT12. Expression levels were determined by real-time qPCR and normalized to ACTIN2 (ACT2). Data represent mean ± SEM (n = 3).
Targeting the Casein Kinase II β-Subunit Family by AmiRNA
One potential advantage of amiRNAs is that they can be designed to regulate the expression of multiple genes as well as a single gene. The multiple numbers of gene families in Arabidopsis and the potential for functional redundancy among family members often complicates the determination of their function when only classic mutational genetics is available. To investigate a possible use of amiRNA for global knockdown of a gene family in the Arabidopsis circadian clock, we chose to test the casein kinase II β-subunits (CKBs). Casein kinase II (CK2) is a well-conserved regulator of eukaryotic circadian clocks (Sugano et al., 1999; Lin et al., 2002; Yang et al., 2002; Perales et al., 2006; Maier et al., 2009). In addition, the four β-subunits of CK2 in Arabidopsis show relatively high sequence similarity, offering the possibility of relatively easy targeting of the entire family with a single construct. Lastly, although overexpression of CKB3 and CKB4 shortens period in Arabidopsis, clock phenotypes of loss-of-function mutations within this family remain unreported (Sugano et al., 1999; Perales et al., 2006).
Using the Web-based amiRNA design algorithm, we designed a single amiRNA to target all four β-subunits as well as four amiRNAs targeting each subunit individually (Ossowski et al., 2008; Supplemental Table S1). The four-member family knockdown amiRNA (amiR-CKBfam) is highly specific for all four CKB genes without off targets. Additionally, the hybridization energies of the amiRNA to each member of the CKB family are low enough to be predicted to be functional (less than −32 kcal mol−1; Supplemental Table S1). Similarly, amiRNAs designed to affect a single CKB gene were identified (amiR-CKB1; amiR-CKB2; amiR-CKB3; amiR-CKB4), showing sufficiently low hybridization energies for the targeted gene, but much higher energies for the other three family members (Supplemental Table S1).
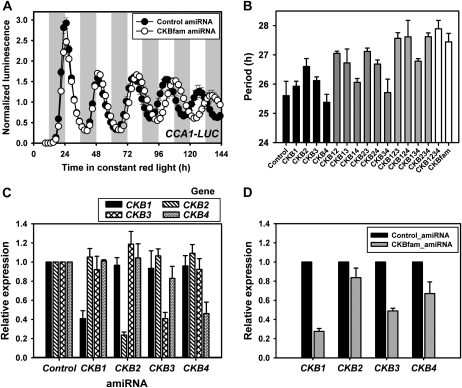
In protoplasts, expression of amiR-CKBfam had no effect on amplitude, but resulted in a 2- to 4-h period lengthening, depending on the specific transfection (Fig. 4A; Table I). These results are opposite to the effects of CKB3 and CKB4 overexpression (period shortening), and consistent with the notion that this amiRNA effectively reduced the levels of the CKB family. We also tested amiRNAs specific for each CKB subunit, individually and in all possible pairwise and multiple combinations by cotransfection (Fig. 4B). The effects of a single amiRNA on period were slight in most cases, although amiR-CKB2 had the strongest effect in lengthening period by about an hour. The period effects of additional amiRNAs were generally additive, up to the effect of all four transfected together showing a period lengthening similar to amiR-CKBfam (Fig. 4B). One exception is that amiR-CKB4 had little effect on period on its own, or when in combination with amiRNAs to the other family members. It is notable that we observed additive effects with additional plasmids although the absolute amount of each was reduced as another was included in the transfection, to keep the total amount of plasmid the same. This is consistent with dilution experiments using a single amiRNA that showed similar efficacy down to 0.25× dilution (data not shown).
Figure 4.
AmiRNA to CKB delays circadian period in Arabidopsis protoplasts. A, The average bioluminescent readout (CCA1-LUC) from protoplasts transfected with amiR-CKBfam. Experiments were performed as described in Figure 1. Data represent mean values ± SEM (n = 6). Similar results were obtained from two independent trials. B, The period length (CCA1-LUC) from protoplasts transfected with combinatorial sets of amiRNA targeting each CKB subunit. Experiments were performed as described in Figure 1 except for using different amounts of plasmid for each combination (one, two, three, and four effector plasmids: 55 μg, 22.5 μg, 11.3 μg, and 5.7 μg per each plasmid, respectively). Data represent mean ± 95% confidence interval (n = 6). Similar results were obtained in two independent trials. C, Relative expression of four CKB subunit genes from cDNA of protoplasts transfected with control amiRNA, or amiR-CKB1, -CKB2, -CKB3, or -CKB4. Protoplast harvest and determination of gene expression was performed as described in Figure 3B. Data represent mean ± SEM (n = 3). D, Relative expression of four CKB subunit genes from cDNA of protoplasts transfected with control amiRNA or amiR-CKBfam. Protoplast harvest and determination of gene expression was performed as described in Figure 3B. Data represent mean ± SEM (n = 3).
We performed RT-qPCR to test the effectiveness and specificities of the amiR-CKBs. Each gene-specific amiRNA effectively reduced the transcript level to between 25% and 40% of the wild-type level with no significant effect on closely related family members (Fig. 4C). The effectiveness of the amiR-CKBfam in reducing message levels of individual family members was variable, with the strongest effect on CKB1 and CKB3 (25%–50% reduction) and a lesser reduction on CKB2 and CKB4 transcript levels (Fig. 4D).
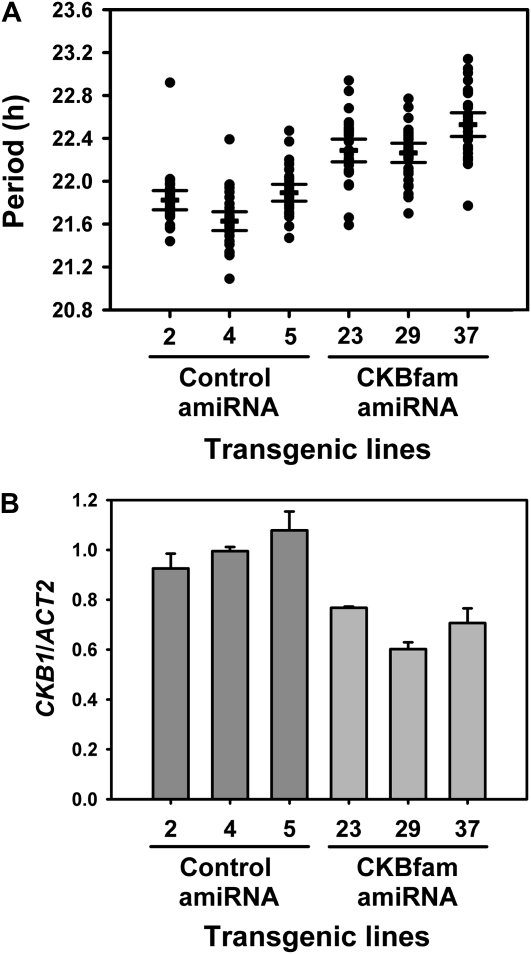
We further confirmed these results in protoplasts by generating transgenic lines expressing amiR-CKBfam in the GI-LUC reporter background (Supplemental Fig. S1). We found that the T3 transformants had slight but reproducibly longer periods, compared with those of control amiRNA expression cassette (Fig. 5A). Interestingly, these reduced effects on period correlate with the significantly lesser effects of the CKBfam amiRNA on CKB expression in the stable transformants, as compared to effects of transient expression in protoplasts (compare Fig. 4C to Fig. 5B and Supplemental Fig. S2). Nonetheless, these results demonstrate an authentic role of CK2 in regulating period length in plants and emphasize the feasibility of targeting a family or subfamily of genes to rapidly assess their clock function.
Figure 5.
Reduction of CKB gene expression by amiR-CKBfam delays circadian period in transgenic plants. A, Period estimates of GI promoter-driven luciferase expression in transgenic lines harboring control amiRNA and amiR-CKBfam. Lines with the longest period among the amiRNA control transformants and the lines with the longest period among the amiR-CKBfam transformants are shown (n = 8 for control amiRNA and 12 for CKBfam amiRNA). Luciferase activity was determined in T3 homozygous seedlings. Transgenic lines were entrained under 12-h-light/12-h-dark cycles for 7 d, transferred to constant red light, and luminescence levels were measured every 2 h for 5 d. Period lengths were determined using 24 to 120 h of free running using the BRASS program. Each symbol represents one seedling, and the lines indicate the average period value (thick) ± 95% confidence interval (thin; n = 32). Similar results were obtained in two independent trials. B, Relative expression of CKB1 in transgenic lines harboring control amiRNA and amiR-CKBfam. Transgenic lines were entrained under 12-h-light/12-h-dark cycles for 10 d and harvested at ZT9. Gene expression was determined as described in Figure 3B. Data represent mean ± SEM (n = 2).
DISCUSSION
In this report we have validated the utility of transient transfection of amiRNA constructs in conjunction with luciferase-based reporters in Arabidopsis mesophyll protoplasts to specifically reduce the expression of clock genes with known period phenotypes. In addition, we demonstrate a new approach for functional studies of gene families in the circadian clock by using amiRNA-based silencing to reduce CKB expression, thereby uncovering a novel clock phenotype in protoplasts and in whole plants.
The Circadian Clock in Arabidopsis Mesophyll Protoplasts
We have established that Arabidopsis mesophyll protoplasts released from the leaves of entrained plants sustain robust rhythmic gene expression of several clock components with a near 24-h period in constant conditions (Fig. 1; Supplemental Fig. S3). These include morning-phased genes, CCA1 and LHY, as well as evening-phased genes, GI and TOC1, all of which maintain phases of expression similar to that found in intact plants. In addition, the luciferase activity driven from these clock-controlled promoters are also affected by mutant backgrounds (Fig. 1) or amiRNA-based silencing of known clock components (Fig. 2). These results support the notion that individual plant cells have a fully functional circadian clock that is maintained by plant clock components identified from studies in whole plants.
Our results substantiate the existence of a cell-autonomous circadian clock as previously described in plant cell-culture systems (Kim et al., 2003; Nakamichi et al., 2003). However, this does not exclude the possibility of additional intercellular regulation or the existence of cell- or tissue-specific clock components (Thain et al., 2002; James et al., 2008). Additional cell-based clock studies may suggest how similarly or differently the protoplast clock functions to that of intact plants. Nevertheless, the Arabidopsis protoplast system will be very useful in studies of the plant circadian clock system as a new tool complementary to molecular genetic approaches that use Arabidopsis mutant or transgenic lines. This system could also be used to rapidly probe the clock through genomic approaches and through screening systems such as chemical libraries or genome-wide plasmid collections expressing open reading frame and RNAi cassettes. One of these advantages was exemplified by testing functional activity of CKB amiRNA in protoplast circadian clock prior to a time-consuming transgenic approach. In addition, a similar approach might be devised to study the clock in other systems in which transgenesis is difficult, but protoplasting is facile.
Functional Tools of Reverse Genetic Studies Using AmiRNA in Protoplasts
The advantages and effectiveness of amiRNA technology in plant systems have been verified from several studies using transgenic approaches (Alvarez et al., 2006; Niu et al., 2006; Schwab et al., 2006; Choi et al., 2007; Ossowski et al., 2008) since the first finding of amiRNA activity in Arabidopsis (Parizotto et al., 2004). Our study extends amiRNA versatility into Arabidopsis protoplasts. We have shown the feasibility of amiRNA technology in protoplasts using both custom-designed amiRNAs (GI and CKBs) and ready-made amiRNAs from genome-wide amiRNA collection (ZTL and ELF3) by monitoring the clock activity of protoplasts transfected with each amiRNA (Figs. 2 and 4, A and B), as well as observing their gene-silencing activity (Figs. 3 and 4, C and D). In addition, we showed the possible use of amiRNA to reduce expression of gene families by showing the effect of CKB amiRNA on the protoplast clock (Fig. 4).
The individual targeting of CKB genes allows a new level of resolution on the role of CK2 in the plant circadian clock. CKB1, CKB2, and CKB3 appear to be additive in their effect in controlling period, even when taking into account the variation in the effectiveness of reducing the expression of the respective gene (Fig. 4B). Although the only demonstrated targets of CK2 are CCA1 and LHY (Sugano et al., 1998, 1999; Daniel et al., 2004), the additive nature of the reduction of these three CKBs suggests that they all contribute similarly to the control of CCA1 and LHY activity, and/or that the activity of other clock components are also affected. The latter case is very likely since ectopic expression of CCA1 or LHY, or loss-of-function CCA1/LHY alleles result in arrhythmicity or period shortening, respectively. Neither condition causes the period lengthening seen with reduced CKB levels. Previous reports indicate that CK2-mediated phosphorylation of CCA1 enhances CCA1 DNA binding (Sugano et al., 1998), suggesting that excess CKB might increase the residence time of CCA1 at promoters and possibly mimic CCA1 overexpression (arrhythmicity). Similarly, the converse (reduced CCA1 activity and period shortening) would be expected when CKB levels are reduced, and this is not observed. Instead short periods result from CKB3 and CKB4 overexpression and long periods when CKB levels are reduced. Hence, the role of CK2 in the plant circadian clock likely extends beyond its effect on CCA1 and LHY activity.
AmiRNA to CKB4 had little effect on period length despite the strong reduction in CKB4 message levels (Fig. 4, B and C). These results contrast with the previous report of period lengthening by CKB4 overexpression, which was interpreted as a connection between CKB4 activity and the clock (Perales et al., 2006). Given the close similarity between the CKBs it is likely that the overexpression of one family member can phenocopy the effects of any of the other three, as indicated by the similar clock phenotypes of the CKB3 and CKB4 ectopic expressions (Sugano et al., 1999; Perales et al., 2006). Hence, the knockdown approaches described here are likely more informative in uncovering the true in vivo roles of these closely related family members than are ectopic studies. Alternatively, the 60% reduction in CKB4 message levels may not be sufficient to affect circadian period. In general, however, these amiRNA approaches will be more helpful to understand in planta gene function by reducing endogenous gene activity than the overexpression approaches traditionally used in protoplast systems.
The amiR-CKBfam appeared to be even more effective in protoplasts than in transgenic lines (compare Fig. 4, A and D, to Fig. 5), which could be due to a higher copy number of plasmids, and/or their expression, in protoplasts than in stably transformed plants. Alternatively, severe reduction in CKB levels may be lethal or limiting to plants, only allowing the recovery of lines that reduce levels minimally. Genetic stacking of true knockouts of each of the four family members, if viable, will be the definitive way of answering this question.
CONCLUSION
Previous research indicated that gene silencing can be achieved in T87 and seedling protoplasts using in vitro synthesized double-stranded RNA interference (RNAi) through a reduction in expression of the endogenous target gene (An et al., 2005; Zhai et al., 2009). However, amiRNA technology has potential advantages over the double-stranded RNAi approach for functional studies through: (1) a more explicitly tailored design to determine specific targets or off targets among several sequence-related genes by allowing variable mismatches between miRNAs and potential targets, (2) a possible timing-dependent regulation using an inducible or a phase-specific promoter, and (3) premade clones from genome-wide collections of amiRNA-expressing plasmids.
Compared with a suspension cell-culture system, Arabidopsis mesophyll protoplasts have the following advantages: (1) differentiated and defined material with characteristic features of leaf cells, rather than undifferentiated cells maintained using phytohormones, (2) a well-established transformation method within a relatively short time, (3) the ability to use mutant and transgenic lines without the time-consuming steps of cell line establishment, and (4) no requirement for a sterile tissue culture facility and maintenance of continuous cell suspension culture. Alternatively, seedling protoplasts could be used, but these appear to be less suitable for circadian rhythm assays (Supplemental Fig. S3).
Despite its many advantages, protoplasts do have some limitations since potential stresses may arise during enzyme digestion of the cell wall, and elimination of potentially important cell-to-cell communication may limit interpretation in some instances. Results using protoplasts will need to be confirmed with intact plants, but clearly, amiRNA-based silencing approaches in protoplasts will be a rapid and convenient way to reveal the biological functions of unknown genes, not only for studies of the clock, but potentially in a variety of plant research areas. While our study exploited a luciferase-based assay, it is easy to envision investigating any number of cell biological phenotypes using, for example, GFP-tagged proteins as markers of localization. Any reporter that is facile to introduce and can act as readout of a cellular function can be combined with amiRNA to probe a wide range of cellular processes.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) Col-0 wild-type and mutant seeds were imbibed at 4°C for 3 d, followed by growth on soil at 23°C under 12-h-light/12-h-dark photocycles (50–60 μmol m−1s−1). elf3-1 (Kim et al., 2008), toc1-101 (Kikis et al., 2005), ztl-3 (Somers et al., 2004), and GI-LUC (Onai and Ishiura, 2005) plants have been described previously.
Plasmid Construction and Transgenics
The PCR template vector expressing amiRNA under the cassava vein mosaic virus (CsVMV) promoter (Verdaguer et al., 1998) in protoplasts (pCsVMV-PP2C-AmiR) was generated by transferring the PP2C amiRNA foldback fragment of pAmiR-PP2C (CSHL_062516), digested with PstI and BamHI, into the pCsVMV-999 vector. The control vector (pCsVMV-AmiR) was prepared by removing the amiRNA foldback region from the pCsVMV-PP2C-AmiR by EcoRI and HindIII digestion followed by filling in and self ligation. ELF3 and ZTL amiRNA plasmids were generated by digesting pAmiR-ELF3 (CSHL_033174) and pAmiR-ZTL (CSHL_080312) with PstI and BamHI and then ligating each resulting amiRNA foldback fragment into PstI/BamHI-digested pCsVMV-PP2C-AmiR vectors. Primers for GI, CKBfam, and CKB1 to CKB4 amiRNA were designed using Web MicroRNA Designer 2 or 3 (http://wmd2 or 3.weigelworld.org). The amiRNA foldback fragments were generated by overlapping PCR using the pCsVMV-PP2C-AmiR vector as a template and primers that were specified by the Web MicroRNA Designer 3 oligo design algorithm initiated using the RS300 vector sequence and the following core amiRNA sequences of the target genes (or common gene family) to generate each specific amiRNA (GI: TATTGCCAAAATTCGGCGCCT; CKBfam: TTAACGAGCATGAATCAGACC; CKB1: TAAGTCTGATTGACGAACCGG; CKB2: TTCTTTACGGTGCTCGCTCTA; CKB3: TTTTAAGTTCCCGTAAGTCAA; CKB4: TGGAACGTAGTTTTGCGCCGT), together with two vector primers in the adjacent region that defines the amiRNA foldback (F: 5′-GGTGTAAGCTATTTTCTTTGAAGTAC-3′ and R: 5′-GCAACAGGATTCAATCTTAAGA-3′). Detailed information using overlapping PCR and each primer set is available at the Web MicroRNA Designer 2 Web site. Each resulting PCR fragment containing the full amiRNA foldback was cloned downstream of the CsVMV promoter into unique PstI and BamHI restriction sites of pCsVMV-AmiR.
The CsVMV-CKBfam-AmiR expression construct for making transgenic plants was generated by transferring a PstI/BamHI fragment as described above into pCsVMV-1300. The resulting construct was introduced into Agrobacterium tumefaciens AGL1 using electroporation and transformed in GI-LUC plants by floral dipping (Clough and Bent, 1998).
To create CCA1-LUC and GI-LUC reporters, the 5′ upstream region encompassing the CCA1 and GI promoters were amplified by PCR with Pfu DNA polymerase, using Arabidopsis Col genomic DNA as a template and the following sets of primers : CCA1 (5′-TTTGGATCCATCAAAGGAGGAAGAAGAAGAAGAAG-3′ and 5′-TTTAAGCTTCACTAAGCTCCTCTACACAACTTCA-3′) and GI (5′-TTTGGATCCAAGAGGTAGGCAAAGTAGCT-3′ and 5′-TTTAAGCTTCCAGGAACCGAAACTAAAC-3′), respectively. The underlined nucleotides indicate restriction enzyme sites. The resulting PCR products were digested with HindIII and BamHI, and ligated into pOmegaLUC_SK+ (J. Kim and H.G. Nam, unpublished data).
Protoplast Transfection with AmiRNA and Reporter Plasmids
Protoplast isolation and DNA transfection were performed according to Yoo et al. (2007) with minor modifications. All procedures were carried out in a clean hood. Protoplasts were isolated from seven to 10 leaves of 25- to 35-d-old Arabidopsis plants at ZT4 (i.e. 4 h after lights on). The detached leaves were sterilized with 70% ethanol for 30 s, followed by two sterile water rinses. Adaxial leaf surfaces were briefly abraded with sandpaper and soaked in 7 mL of an enzyme solution (400 mm mannitol, 20 mm KCl, 20 mm MES-KOH [pH 5.7], 10 mm CaCl2, 1% Cellulase R10, 0.5% Macerozyme R10, and 0.1% bovine serum albumin) in a 60-mm-diameter petri dish. This enzyme digestion was performed in the dark, with gentle shaking (35 rpm) for two and a half hours at room temperature. The released protoplasts were filtered through sterile 100 μm nylon mesh, and transferred into round-shaped culture tubes containing 5 mL of W5 solution (154 mm NaCl, 125 mm CaCl2, 5 mm KCl, 1.5 mm MES-KOH [pH 5.7], and 5 mm Glc). Protoplasts were harvested by centrifugation at 100 g for 5 min. After decanting the supernatant, the protoplast pellet was resuspended with W5 solution and held on ice for 30 min. Protoplasts were centrifuged and resuspended in 400 mm mannitol, 15 mm MgCl2, and 4 mm MES-KOH (pH 5.7), at a concentration of 1 to 1.5 × 105 mL−1.
The amiRNA and reporter plasmids for transfection were prepared by CsCl gradient purification and their DNA concentration was adjusted to 2 μg μL−1 per 4 kb DNA. For luminescence measurements, 200 μL of protolasts were transferred to a 2-mL microfuge tube containing 25 μL of amiRNA plasmid and 5 μL of the reporter plasmid. Transfections were performed by adding 230 μL of polyethylene glycol (PEG) solution [40% PEG-4000, 200 mm mannitol, 100 mm Ca(NO3)2] and mixed by inverting gently 12 times. After incubation for 8 to 15 min, the protoplast-DNA-PEG mixture was diluted with 920 μL of W5 solution and mixed by inverting gently four times. Transfected protoplasts were collected by centrifugation at 100 g for 1 min, resuspended in 650 μL of W5 solution containing 5% fetal bovine serum (F4135, Sigma), 50 μm luciferin, and 50 μg mL−1 ampicillin. For RT-PCR analysis, 800 μL of protoplasts were aliquoted into a 15-mL round-bottom tube containing 120 μL of amiRNA plasmids.
Luminescence Measurement and Circadian Rhythm Analysis
For luminescence analysis of protoplasts 300 μL of each batch of transfected protoplasts was transferred into a single well of a black 96-well microplate, covered by a transparent plastic lid, and incubated under controlled conditions (Percival E30LEDL3 growth chamber; Percival Scientific) under constant red (14 μmol m−2 s−1) or blue light (5 μmol m−2 s−1) at 23°C. Image collection and quantification were performed as described previously (Kim et al., 2008) with minor modifications. Images were collected (25 min) using a Princeton Instruments VersArrayXP:512B digital CCD camera and processed using MetaView. Data were imported into the Biological Rhythms Analysis software system (BRASS Ver. 2.14, available from http://www.amillar.org) and analyzed with the FFT-NLLS suite of programs, as described previously (Plautz et al., 1997; Somers et al., 2004). Period lengths are reported as variance-weighted periods ± SEM, which were estimated using bioluminescence data obtained from 24 to 96 h under constant conditions, unless otherwise noted.
For luminescence assays in transgenics, T3 seedlings with the GI-LUC reporter were grown on Murashige and Skoog containing 3% Suc and hygromycin under 12-h-light/12-h-dark white fluorescence light (50–60 μmol m−2 s−1) for 7 d. Luminescence measurement was performed under continuous red light (20 μmol m−2 s−1) as previously described (Somers et al., 2004). Period estimates was calculated with BRASS using bioluminescence data obtained from 24 to 120 h.
Real-Time PCR Analysis of Gene Expression in Arabidopsis Protoplasts
Protoplasts were harvested after washing once with W5 buffer around ZT 12, after 1 d of incubation in the entraining chamber post transfection. Total RNA was extracted using Trizol reagent (300 μL/105 cells; Invitrogen) and treated with DNase I (Ambion) according to the manufacturer’s instructions. A total of 0.25 μg of total RNA was reverse transcribed in a 10-μL reaction using oligo (dT20) primers and SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. RT reactions were diluted 6-fold and 3 μL of the dilutions were used for PCR reactions. Real-time PCR was carried out using iQ SYBR Green Supermix (Bio-Rad) and an iQ5 real-time PCR detection system (Bio-Rad) in 15-μL reactions. The following PCR condition used: 94°C for 2 min, followed by 40 cycles of 94°C for 15 s and 60°C for 34 s. Primers for GI and ACT2 have been described previously (Locke et al., 2005). Primers to detect other transcripts were designed using Primer Express v2.0 (Applied Biosystems) and are shown below: ELF3, 5′-ATTGCTGCATCACCGGATCT-3′ and 5′-TCACCCCTTTGTTTGACGACA-3′; ZTL, 5′-TCTTGATATTTGGCGGCTCAGT-3′ and 5′-TTGTCCTCCGTTGGGTCAAGTA-3′; CKB1, 5′-GGCAAAGGAACCGTTACGG-3′ and 5′-TTGATGTTATTCGACTGCTGCTTC-3′; CKB2, 5′-TCAGCCATCAAGTTCCTTACTACGAC-3′ and 5′-CTCACCATGCGAAGACTCCAC-3′; CKB3, 5′-CGGAAACTGATAGTGAAGGGTCTG-3′ and 5′-ATCCACGACGTATCATCACCCTC-3′; CKB4, 5′-TTCAACTGCTAAATCTCAGCTTCATTC-3′ and 5′-CACATCTGATCCTTCACTATCCGTG-3′.
Sequence data for the genes described in this article can be found in the Arabidopsis Genome Initiative and GeneBank/DDBJ/EMBL data libraries under the following accession numbers: CCA1 (At2g46830), LHY (At1g01060), GI (At1g22770), ELF3 (At2g25930), ZTL (At5g57360), TOC1 (At5g61380), ACT2 (At5g09810), CKB1 (At5g47080), CKB2 (At4g17640), CKB3 (At3g60250), and CKB4 (At2g44680).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. AmiR-CKBfam delays circadian period in transgenic plants.
Supplemental Figure S2. Relative expression of CKB2, CKB3, and CKB4 in transgenic lines harboring CKB amiRNA.
Supplemental Figure S3. Optimization of clock-controlled luminescence cycling in Arabidopsis protoplasts.
Supplemental Table S1. Calculated interaction likelihoods between amiRNAs and potential target gene(s).
Supplemental Results and Discussion S1. Assay optimization for circadian rhythm measurements in Arabidopsis protoplasts.
Supplemental Materials and Methods S1. Methods for optimized assay.
Supplementary Material
Acknowledgments
We acknowledge the assistance of the ABRC, which provided amiRNA clones. We thank Dr. Hong Gil Nam for a kind donation of vectors, including pCsVMV-999, pCsVMV-1300, pCR-CCD-F, and pOmegaLUC+_SK.
References
- Alvarez JP, Pekker I, Goldshmidt A, Blum E, Amsellem Z, Eshed Y. (2006) Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18: 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An CI, Sawada A, Kawaguchi Y, Fukusaki E, Kobayashi A. (2005) Transient RNAi induction against endogenous genes in Arabidopsis protoplasts using in vitro-prepared double-stranded RNA. Biosci Biotechnol Biochem 69: 415–418 [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Bowman JL. (2008) Evolution of plant microRNAs and their targets. Trends Plant Sci 13: 343–349 [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, Schibler U. (1998) A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93: 929–937 [DOI] [PubMed] [Google Scholar]
- Choi K, Park C, Lee J, Oh M, Noh B, Lee I. (2007) Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 134: 1931–1941 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Daniel X, Sugano S, Tobin EM. (2004) CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis. Proc Natl Acad Sci USA 101: 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrah C, Taylor BL, Edwards KD, Brown PE, Hall A, McWatters HG. (2006) Analysis of phase of LUCIFERASE expression reveals novel circadian quantitative trait loci in Arabidopsis. Plant Physiol 140: 1464–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, Hibberd JM, Millar AJ, Webb AA. (2005) Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633 [DOI] [PubMed] [Google Scholar]
- Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognar L, Nagy F, Millar AJ, Amasino RM. (2002) The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419: 74–77 [DOI] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J. (1999) GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J 18: 4679–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A, Bastow RM, Davis SJ, Hanano S, McWatters HG, Hibberd V, Doyle MR, Sung S, Halliday KJ, Amasino RM, et al. (2003) The TIME FOR COFFEE gene maintains the amplitude and timing of Arabidopsis circadian clocks. Plant Cell 15: 2719–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA. (2005) LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA 102: 10387–10392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks KA, Millar AJ, Carré IA, Somers DE, Straume M, Meeks-Wagner R, Kay SA. (1996) Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274: 790–792 [DOI] [PubMed] [Google Scholar]
- James AB, Monreal JA, Nimmo GA, Kelly CL, Herzyk P, Jenkins GI, Nimmo HG. (2008) The circadian clock in Arabidopsis roots is a simplified slave version of the clock in shoots. Science 322: 1832–1835 [DOI] [PubMed] [Google Scholar]
- Kevei E, Gyula P, Feher B, Toth R, Viczian A, Kircher S, Rea D, Dorjgotov D, Schafer E, Millar AJ, et al. (2007) Arabidopsis thaliana circadian clock is regulated by the small GTPase LIP1. Curr Biol 17: 1456–1464 [DOI] [PubMed] [Google Scholar]
- Kevei E, Gyula P, Hall A, Kozma-Bognar L, Kim WY, Eriksson ME, Toth R, Hanano S, Feher B, Southern MM, et al. (2006) Forward genetic analysis of the circadian clock separates the multiple functions of ZEITLUPE. Plant Physiol 140: 933–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikis EA, Khanna R, Quail PH. (2005) ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J 44: 300–313 [DOI] [PubMed] [Google Scholar]
- Kim J, Kim Y, Yeom M, Kim JH, Nam HG. (2008) FIONA1 is essential for regulating period length in the Arabidopsis circadian clock. Plant Cell 20: 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Geng R, Somers DE. (2003) Circadian phase-specific degradation of the F-box protein ZTL is mediated by the proteasome. Proc Natl Acad Sci USA 100: 4933–4938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmos E, Schoof H, Plumer M, Davis SJ. (2008) Structural insights into the function of the core-circadian factor TIMING OF CAB2 EXPRESSION 1 (TOC1). J Circadian Rhythms 6: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JM, Kilman VL, Keegan K, Paddock B, Emery-Le M, Rosbash M, Allada R. (2002) A role for casein kinase 2alpha in the Drosophila circadian clock. Nature 420: 816–820 [DOI] [PubMed] [Google Scholar]
- Locke JCW, Kozma-Bognar L, Gould PD, Feher B, Kevei E, Nagy F, Turner MS, Hall A, Millar AJ. (2006) Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol Syst Biol 2: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JCW, Southern MM, Kozma-Bognar L, Hibberd V, Brown PE, Turner MS, Millar AJ. (2005) Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol Syst Biol 1: 2005.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier B, Wendt S, Vanselow JT, Wallach T, Reischl S, Oehmke S, Schlosser A, Kramer A. (2009) A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev 23: 708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Bouche N. (2008) MicroRNA-directed regulation: to cleave or not to cleave. Trends Plant Sci 13: 359–367 [DOI] [PubMed] [Google Scholar]
- Martin-Tryon EL, Harmer SL. (2008) XAP5 CIRCADIAN TIMEKEEPER coordinates light signals for proper timing of photomorphogenesis and the circadian clock in Arabidopsis. Plant Cell 20: 1244–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Tryon EL, Kreps JA, Harmer SL. (2007) GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol 143: 473–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas P, Alabadi D, Yanovsky MJ, Oyama T, Kay SA. (2003) Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15: 223–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CR. (2006) Plant circadian rhythms. Plant Cell 18: 792–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Salomé PA, Yu HJ, Spencer TR, Sharp EL, McPeek MA, Alonso JM, Ecker JR, McClung CR. (2003) Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302: 1049–1053 [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Ito S, Oyama T, Yamashino T, Kondo T, Mizuno T. (2004) Characterization of plant circadian rhythms by employing Arabidopsis cultured cells with bioluminescence reporters. Plant Cell Physiol 45: 57–67 [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Matsushika A, Yamashino T, Mizuno T. (2003) Cell autonomous circadian waves of the APRR1/TOC1 quintet in an established cell line of Arabidopsis thaliana. Plant Cell Physiol 44: 360–365 [DOI] [PubMed] [Google Scholar]
- Niu QW, Lin SS, Reyes JL, Chen KC, Wu HW, Yeh SD, Chua NH. (2006) Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat Biotechnol 24: 1420–1428 [DOI] [PubMed] [Google Scholar]
- Onai K, Ishiura M. (2005) PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells 10: 963–972 [DOI] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D. (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. (1998) Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA 95: 8660–8664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Poirier GG, Kay SA. (2002) tej defines a role for poly(ADP-ribosyl)ation in establishing period length of the Arabidopsis circadian oscillator. Dev Cell 3: 51–61 [DOI] [PubMed] [Google Scholar]
- Parizotto EA, Dunoyer P, Rahm N, Himber C, Voinnet O. (2004) In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev 18: 2237–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Somers DE, Kim Y, Choy Y, Lim H, Soh M, Kim H, Kay SA, Nam HG. (1999) Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285: 1579–1582 [DOI] [PubMed] [Google Scholar]
- Perales M, Portoles S, Mas P. (2006) The proteasome-dependent degradation of CKB4 is regulated by the Arabidopsis biological clock. Plant J 46: 849–860 [DOI] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Filipowicz W. (2007) Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol 17: 118–126 [DOI] [PubMed] [Google Scholar]
- Plautz JD, Kaneko M, Hall JC, Kay SA. (1997) Independent photoreceptive clocks throughout Drosophila. Science 278: 1632–1635 [DOI] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G. (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229 [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. (2001) Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol 127: 1466–1475 [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Kim WY, Geng R. (2004) The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell 16: 769–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA. (2000) ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101: 319–329 [DOI] [PubMed] [Google Scholar]
- Staiger D, Allenbach L, Salathia N, Fiechter V, Davis SJ, Millar AJ, Chory J, Fankhauser C. (2003) The Arabidopsis SRR1 gene mediates phyB signaling and is required for normal circadian clock function. Genes Dev 17: 256–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA. (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Sugano S, Andronis C, Green RM, Wang ZY, Tobin EM. (1998) Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein. Proc Natl Acad Sci USA 95: 11020–11025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano S, Andronis C, Ong MS, Green RM, Tobin EM. (1999) The protein kinase CK2 is involved in regulation of circadian rhythms in Arabidopsis. Proc Natl Acad Sci USA 96: 12362–12366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K, Alonso-Blanco C, Lynn JR, Michaels SD, Amasino RM, Koornneef M, Millar AJ. (1999) Natural allelic variation identifies new genes in the Arabidopsis circadian system. Plant J 20: 67–77 [DOI] [PubMed] [Google Scholar]
- Thain SC, Murtas G, Lynn JR, McGrath RB, Millar AJ. (2002) The circadian clock that controls gene expression in Arabidopsis is tissue specific. Plant Physiol 130: 102–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaguer B, de Kochko A, Fux CI, Beachy RN, Fauquet C. (1998) Functional organization of the cassava vein mosaic virus (CsVMV) promoter. Plant Mol Biol 37: 1055–1067 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM. (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Yang Y, Cheng P, Liu Y. (2002) Regulation of the Neurospora circadian clock by casein kinase II. Genes Dev 16: 994–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zeilinger MN, Farre EM, Taylor SR, Kay SA, Doyle FJ III. (2006) A novel computational model of the circadian clock in Arabidopsis that incorporates PRR7 and PRR9. Mol Syst Biol 2: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z, Sooksa-nguan T, Vatamaniuk OK. (2009) Establishing RNA interference as a reverse-genetic approach for gene functional analysis in protoplasts. Plant Physiol 149: 642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang EE, Liu AC, Hirota T, Miraglia LJ, Welch G, Pongsawakul PY, Liu X, Atwood A, Huss JW, III, Janes J, et al. (2009) A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell 139: 199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.