OPERONS AND GENE CLUSTERS IN PROKARYOTES AND EUKARYOTES
The first gene cluster for a secondary metabolic pathway was discovered in maize (Zea mays) over a decade ago (Frey et al., 1997) and was regarded as something of an oddity. However clusters of genes for secondary metabolic pathways are now an emerging theme in plant biology, and are providing some provocative insights into plant genome plasticity and evolution. Gene clusters containing nonhomologous functionally related genes are common in bacterial genomes. Most are organized as operons, in which the different genes are expressed as a single polycistronic mRNA allowing the tight coupling of transcription and translation (Zheng et al., 2002; Rocha, 2008; Koonin, 2009). Operons, in turn, may be clustered. For example in actinomycetes, which produce a prolific array of pharmaceuticals and other high-value molecules, the genes for secondary metabolite production are clustered and typically encode several transcripts, some of which are bi- or polycistronic (Osbourn, 2010). True operons are rare in eukaryotes because transcription is uncoupled from translation and mRNAs are generally monocistronic; therefore, genes under common regulation are often dispersed throughout the genome and coordinately regulated in trans. Although there are clusters of functionally related genes in eukaryotes (Hurst et al., 2004; Sproul et al., 2005; Michalak, 2008), most of these consist of paralogs that have evolved by repeated tandem gene duplication and divergence (e.g. the globin and homeobox [Hox] loci in mammals; Osbourn and Field, 2009). In filamentous fungi, however, there are numerous exceptions to this rule, most notably gene clusters for secondary metabolic pathways. These include clusters for the synthesis of important pharmaceuticals such as the β-lactam antibiotics penicillin and cephalosporin, and for the production of toxins (e.g. aflatoxin and host-selective toxins associated with virulence on plants; Hoffmeister and Keller, 2007; Turgeon and Bushley, 2010). Other examples of clusters of nonhomologous functionally related genes in eukaryotes are the GAL and DAL gene clusters in the yeast Saccharomyces cerevisiae (which enable the utilization of Gal and allantoin, respectively; Hittinger et al., 2004; Wong and Wolfe, 2005) and the major histocompatibility complex in mammals (Horton et al., 2004). In general, gene clusters of this type appear to be required for growth or survival under certain environmental conditions and can therefore be regarded as adaptive gene clusters. In particular, they relate to the exploitation of new environments or the management of interactions with other organisms (Field and Osbourn, 2010).
OPERON-LIKE GENE CLUSTERS IN PLANTS
Until recently it was thought that gene clusters in plants were restricted to tandem duplicates, for example arrays of Leu-rich repeat disease resistance genes. Computational analysis of the Arabidopsis (Arabidopsis thaliana) genome has suggested that genes for some primary metabolic pathways are more tightly linked than would be expected by chance, but most of these clusters are very loose, in some cases spanning nearly one-quarter of a chromosome (Lee and Sonnhammer, 2003). Genes for well-characterized secondary metabolic pathways such as the anthocyanin pathway are unlinked, and so it is clear that clustering of genes for plant metabolic pathways is not the rule. However five compact gene clusters for secondary metabolic pathways have now been identified in plants. These clusters are operon like in that they consist of groups of physically linked genes that are functionally related and coregulated, but that for the most part share no obvious sequence homology. Unlike operons, the genes within these clusters are transcribed separately. The five plant clusters are for the synthesis of cyclic hydroxamic acids in maize (Frey et al., 1997, 2003, 2009; von Rad et al., 2001; Jonczyk et al., 2008), triterpenes in oat (Avena sativa) and Arabidopsis (the avenacin and thalianol clusters, respectively; Papadopoulou et al., 1999; Haralampidis et al., 2001; Qi et al., 2004, 2006; Field and Osbourn, 2008; Mylona et al., 2008; Mugford et al., 2009), and diterpenes in rice (Oryza sativa; the momilactone and phytocassane clusters; Sakamoto et al., 2004; Wilderman et al., 2004; Shimura et al., 2007; Swaminathan et al., 2009). These clusters are diverse in organization and function and all appear to have evolved independently (Field and Osbourn, 2008; Frey et al., 2009; Osbourn and Field, 2009; Swaminathan et al., 2009). The four clusters from cereals are all required for the synthesis of preformed or stress-induced compounds implicated in plant defense (Papadopoulou et al., 1999; Gierl and Frey, 2001; Wilderman et al., 2004; Shimura et al., 2007). The function of the Arabidopsis thalianol cluster is not known, but the high level of conservation of this cluster across different Arabidopsis accessions suggests that it is also like to have an important function in ecological interactions (Field and Osbourn, 2008). An obvious assumption is that these metabolic gene clusters have arisen by horizontal gene transfer from microbes. However there is compelling evidence to indicate this is not the case, and that the clusters are most likely to have been assembled from plant genes by recruitment of genes from elsewhere in the genome through gene duplication, neofunctionalization, and genome reorganization (Gierl and Frey, 2001; Qi et al., 2004, 2006; Field and Osbourn, 2008; Osbourn and Field, 2009; Swaminathan et al., 2009). Thus clustering appears to have occurred de novo through some form of convergent evolutionary process. These clusters represent a new paradigm in plant evolutionary biology, and provide tantalizing links with adaptive genome plasticity in microbes and animals (Field and Osbourn, 2010; Osbourn, 2010).
THE QUESTIONS
The identification of five apparently unrelated secondary metabolic gene clusters in different plant species poses some interesting questions. These are discussed below.
How Common Are Secondary Metabolic Gene Clusters in Plants?
It is not yet known how common clusters of nonhomologous but functionally related genes are throughout the plant kingdom. This will be revealed through further analysis of fully sequenced plant genomes. The maize and rice gene clusters were defined using a combination of molecular biology, biochemistry, and reverse genetics (Frey et al., 1997, 2003, 2009; von Rad et al., 2001; Sakamoto et al., 2004; Wilderman et al., 2004; Shimura et al., 2007; Jonczyk et al., 2008; Swaminathan et al., 2009). Analysis of the two rice clusters was aided by the fact that expression of these gene clusters is elicitor inducible and by the availability of rice genome sequence information. The oat avenacin cluster was initially defined using forward genetics, facilitated by a simple screen for loss of root fluorescence (Papadopoulou et al., 1999; Qi et al., 2006; Mugford et al., 2009). The thalianol cluster was predicted by searching the genome sequence of Arabidopsis for candidate gene clusters containing triterpene synthase (oxidosqualene cyclase) signature genes and then validated by functional analysis (Field and Osbourn, 2008). Genome mining for new candidate secondary metabolic pathways based on clustering and coexpression has proved to be a highly successful approach in microbes (Zerikly and Challis, 2009; Osbourn, 2010). It is relatively straightforward to predict the types of gene that one might expect to find in a gene cluster for a secondary metabolic pathway (e.g. a gene for a signature enzyme such as a terpene synthase to make the backbone, and genes for tailoring enzymes such as cytochrome P450s and other oxidoreductases, acyltransferases, methyltransferases to make further modifications). Through systematic analysis of sequenced plant genomes we will now be able to discover new secondary metabolic gene clusters and to gain an idea of their frequency and distribution. With the growing body of genome sequence information that is now available for plants and the advent of next-generation sequencing it will be possible to search for candidate secondary metabolic gene clusters in a wide range of different species.
Gene clusters for synthesis of secondary metabolites in bacteria and filamentous fungi often (but not always) contain genes for pathway-specific transporters and regulators in addition to genes for the signature enzyme and tailoring enzymes (Osbourn, 2010). This merits some comment from the plant perspective. To date, transporters for the five characterized plant secondary metabolic gene clusters have not yet been identified. However the recent demonstration that Lr34, a gene that confers broad-spectrum disease resistance in wheat (Triticum aestivum), is predicted to encode an ATP-binding cassette transporter implicated in the transport of defense compounds is intriguing, given that the immediate neighbors of this gene in the wheat genome are of unknown function but are also implicated in secondary metabolism (e.g. sugar transferase and cytochrome P450 genes; Krattinger et al., 2009). So far the only transcription factor to be reported for plant secondary metabolic gene clusters is a positive regulator of both the momilactone and phytocassane clusters in rice (Okada et al., 2009). The gene for this cluster lies outside of both of the clusters that it regulates. Thus at present it is not clear how many features plant secondary metabolic gene clusters will share in common with microbial clusters.
What Is the Significance of Clustering?
Why should genes for some plant metabolic pathways be clustered while those for other pathways are not? What are the advantages associated with clustering? One explanation is regulation. Clearly dispersed genes can be coregulated through a common transcription factor, and there are some excellent examples of coordinate regulation of unlinked genes for metabolic pathways in plants (Martin et al., 2010). However, physical clustering of functionally related genes in eukaryotes has the potential to add another tier to the hierarchy of gene regulation, providing mechanisms for the coordinated regulation of gene expression at the levels of nuclear organization and/or chromatin (Hurst et al., 2004; Sproul et al., 2005; Osbourn and Field, 2009). In filamentous fungi the use of mutations and drugs that affect chromatin remodelling is proving to be a powerful means of pathway discovery, enabling cryptic clusters to be identified and activated (e.g. Bok et al., 2009; Cichewicz, 2010). Noncoding RNAs have also been implicated in the recruitment of chromatin complexes, and in animals Hox gene expression can be controlled posttranscriptionally and probably also epigenetically by noncoding RNAs and Polycomb group proteins (Yekta et al., 2008). There is evidence to indicate that regulation at the level of chromatin is likely to be important for expression of secondary metabolic gene clusters in plants. In silico data suggest chromatin-mediated regulation of the thalianol cluster in Arabidopsis (Field and Osbourn, 2008), while DNA fluorescence in situ experiments have revealed cell-type-specific chromatin decondensation of the avenacin cluster associated with gene expression in oat (Wegel et al., 2009). It is also possible that the grouping of genes into functional clusters in eukaryotes may facilitate the coordinated handling of transcripts that have arisen from physically linked genes, from transcription through processing and export to protein synthesis. A challenge going forward is to better understand how these gene clusters are regulated at multiple levels, from nuclear organization and chromatin remodelling to the synthesis and localization of functional pathway proteins.
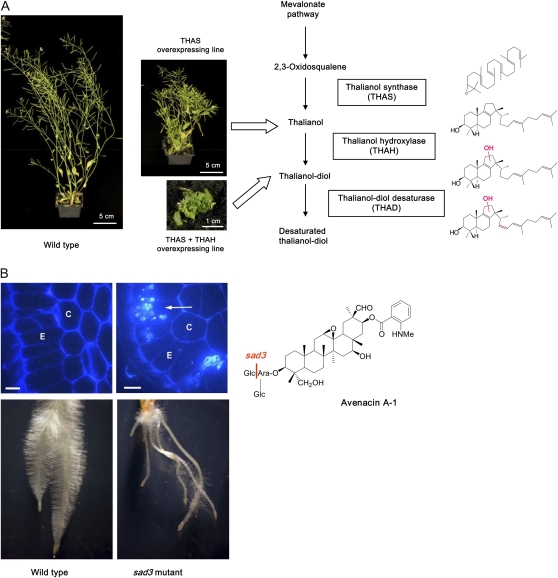
The five known plant clusters all confer the ability to synthesize secondary products that are known or are likely to have protective roles in defense against pests and pathogens, and so presumably confer a selective advantage in nature. A second reason for the clustering of functionally related genes may involve selection for the coinheritance of favorable combinations of alleles at these multigene loci. Where the fitness of an allele at one locus depends on the genotype at another locus then a selective advantage may arise for genomic rearrangements that reduce the distance between the two loci (Nei, 1967). This ratcheting effect may be enhanced where the fitness of recombinant haplotypes is low, for example where the combination of a functional and nonfunctional allele at two loci results in the premature disruption of a biochemical pathway and accumulation of toxic intermediates (Nei, 2003). There is certainly evidence to suggest that this may be the case. For example, accumulation of an intermediate in the yeast DAL pathway as a consequence of a late pathway mutation results in toxicity (Wong and Wolfe, 2005), while accumulation of toxic intermediates in the triterpene pathways in Arabidopsis and in oat leads to substantial defects in growth and development (Field and Osbourn, 2008; Mylona et al., 2008; Fig. 1).
Figure 1.
Detrimental effects of accumulation of toxic metabolic intermediates on plant growth and development. A, Accumulation of elevated levels of thalianol pathway intermediates in Arabidopsis results in moderate to severe dwarfing (adapted from Field and Osbourn, 2008). B, sad3 mutants of diploid oat accumulate an incompletely glucosylated avenacin pathway intermediate that is toxic, leading to stunted roots and a root hair deficiency phenotype. The top sections show cross sections of roots of wild-type and sad3 mutant seedlings stained with Calcofluor. Callose-containing aggregates were observed in the epidermal cells of the sad3 mutant but not in the wild type (confirmed using aniline blue). The accumulation of callose is likely to be a stress response triggered by accumulation of monodeglucosyl avenacin A-1. C, Cortex; E, epidermis. Scale bars = 50 μm. Adapted from Mylona et al. (2008).
POTENTIAL APPLICATIONS
Although the fundamental research described above delves into new territories, it is also important to consider a more practical issue—how can knowledge gained from the study of plant metabolic gene clusters be applied for the development of crops for agronomic and industrial end uses, with enhanced pest and disease resistance, improved nutritional qualities, or elevated levels of high-value products. Knowledge of gene clusters facilitates the delineation and functional analysis of cluster components and new metabolic pathways, as has been demonstrated on numerous occasions in bacteria such as the actinomycetes. Physical clustering of genes can also be exploited to introgress beneficial traits such as disease resistance into other plant varieties, or alternatively to breed out metabolic gene clusters associated with undesirable traits (such as bitterness and antifeedant activity). For those species that are less closely related, components of gene clusters can be transferred individually or in combination between species using recombinant DNA technology. The latter will be enhanced by the development of improved technology for the transfer of multiple genes, or perhaps even whole gene clusters, into plants of commercial and agronomic importance.
CONCLUSION
Secondary metabolic gene clusters are among the most diverse and rapidly evolving features of plant genomes. Using plant metabolic gene clusters as readouts for metabolic diversification and, by extension, genome plasticity, it will be possible to address novel and important questions in plant biology: How widespread are such clusters, why do they exist, and how do they form? An improved understanding of plant gene clusters will enable us to establish the rules behind this phenomenon—why are some metabolic pathways represented by clusters while others are represented by dispersed genes. This could ultimately cause us to reconsider our understanding of plant metabolism. It is also intriguing to consider whether there might be other types of nonhomologous gene clusters in plants that have functions other than in secondary metabolism. Last but by no means least, can we explain the formation of nonhomologous gene clusters based on our current knowledge of plant genome dynamics or do we need to invoke new mechanisms for rapid adaptive evolution?
References
- Bok JW, Chiang YM, Szewczyk E, Reyes-Domingez Y, Davidson AD, Sanchez JF, Lo HC, Watanabe K, Strauss J, Oakley BR, et al. (2009) Chromatin-level regulation of biosynthetic gene clusters. Nat Chem Biol 5: 462–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichewicz RH. (2010) Epigenome manipulation as a pathway to new natural product scaffolds and their congeners. Nat Prod Rep 27: 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field B, Osbourn AE. (2008) Metabolic diversification—independent assembly of operon-like gene clusters in different plants. Science 320: 543–547 [DOI] [PubMed] [Google Scholar]
- Field B, Osbourn AE. (April 19, 2010) Operons. Nat Chem Biol doi/10.1038/nchembio.359 [Google Scholar]
- Frey M, Chomet P, Glawischnig E, Stettner C, Grun S, Winklmair A, Eisenreich W, Bacher A, Meeley RB, Briggs SP, et al. (1997) Analysis of a chemical plant defense mechanism in grasses. Science 277: 696–699 [DOI] [PubMed] [Google Scholar]
- Frey M, Huber K, Park WJ, Sicker D, Lindberg P, Meeley RB, Simmons CR, Yalpini N, Gierl A. (2003) A 2-oxoglutarate-dependent dioxygenase is integrated in DIMBOA biosynthesis. Phytochemistry 62: 371–376 [DOI] [PubMed] [Google Scholar]
- Frey M, Schullehner K, Dick R, Fiesselmann A, Gierl A. (2009) Benzoxazinoid biosynthesis, a model for evolution of secondary metabolic pathways in plants. Phytochemistry 70: 1645–1651 [DOI] [PubMed] [Google Scholar]
- Gierl A, Frey M. (2001) Evolution of benzoxazinone biosynthesis and indole production in maize. Planta 213: 493–498 [DOI] [PubMed] [Google Scholar]
- Haralampidis K, Bryan G, Qi X, Papadopoulou K, Bakht S, Melton R, Osbourn AE. (2001) A new class of oxidosqualene cyclases directs synthesis of antimicrobial phytoprotectants in monocots. Proc Natl Acad Sci USA 98: 13431–13436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hittinger CT, Rokas A, Carroll SB. (2004) Parallel inactivation of multiple GAL pathway genes and ecological diversification in yeasts. Proc Natl Acad Sci USA 101: 14144–14149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmeister D, Keller NP. (2007) Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat Prod Rep 24: 393–416 [DOI] [PubMed] [Google Scholar]
- Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, Lush MJ, Povey S, Talbot CC, Jr, Wright MW, et al. (2004) Gene map of the extended human MHC. Nat Rev Genet 5: 889–899 [DOI] [PubMed] [Google Scholar]
- Hurst LD, Pal C, Lercher MJ. (2004) The evolutionary dynamics of eukaryotic gene order. Nat Rev Genet 5: 299–310 [DOI] [PubMed] [Google Scholar]
- Jonczyk R, Schmidt H, Osterrieder A, Fiesselmann A, Schullehner K, Haslbeck M, Sicker D, Hofmann D, Yalpani N, Simmons C, et al. (2008) Elucidation of the final reactions of DIMBOA-glucoside biosynthesis in maize: characterization of Bx6 and Bx7 biosynthesis in maize: characterization of Bx6 and Bx7. Plant Physiol 146: 1053–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. (2009) Evolution of genome architecture. Int J Biochem Cell Biol 41: 298–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krattinger SG, Laguda ES, Spielmeyer W, Singh RP, Huerta-Espino J, McFadden H, Bossolini E, Selter LL, Keller B. (2009) A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323: 1360–1363 [DOI] [PubMed] [Google Scholar]
- Lee J, Sonnhammer ELL. (2003) Genomic gene clustering analysis of pathways in eukaryotes. Genome Res 13: 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Ellis N, Rook F. (2010) Do transcription factors play special roles in adaptive variation? Plant Physiol 154: 506–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak P. (2008) Coexpression, coregulation and cofunctionality of neighboring genes in eukaryotic genomes. Genomics 91: 243–248 [DOI] [PubMed] [Google Scholar]
- Mugford ST, Qi X, Bakht S, Hill L, Wegel E, Hughes RK, Papadopoulou K, Melton R, Philo M, Sainsbury F, et al. (2009) A serine carboxypeptidase-like acyltransferase is required for synthesis of antimicrobial compounds and disease resistance in oats. Plant Cell 21: 2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylona P, Owatworakit A, Papadopoulou K, Jenner H, Qin B, Findlay K, Hill L, Qi X, Bakht S, Melton R, et al. (2008) Sad3 and Sad4 are required for saponin biosynthesis and root development in oat. Plant Cell 20: 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. (1967) Modification of linkage intensity by natural selection. Genetics 57: 625–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. (2003) Let’s stick together. Heredity 90: 411–412 [DOI] [PubMed] [Google Scholar]
- Okada A, Okada K, Miyamoto K, Koga J, Shibuya N, Nojiri H, Yamane H. (2009) OsTGAP1, a bZIP transcription factor, co-ordinately regulates the inductive production of diterpenoid phytoalexins in rice. J Biol Chem 284: 26510–26518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbourn A. (2010) Secondary metabolic gene clusters: evolutionary toolkits for chemical innovation. Trends Genet 26: 449–457 [DOI] [PubMed] [Google Scholar]
- Osbourn AE, Field B. (2009) Operons. Cell Mol Life Sci 66: 3755–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulou K, Melton RE, Leggett M, Daniels MJ, Osbourn AE. (1999) Compromised disease resistance in saponin-deficient plants. Proc Natl Acad Sci USA 96: 12923–12928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Bakht S, Leggett M, Maxwell C, Melton R, Osbourn A. (2004) A gene cluster for secondary metabolism in oat: implications for the evolution of metabolic diversity in plants. Proc Natl Acad Sci USA 101: 8233–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Bakht S, Qin B, Leggett M, Hemmings A, Mellon F, Eagles J, Werck-Reichart D, Schaller H, Lesot A, et al. (2006) A different function for a member of an ancient and highly conserved cytochrome P450 family: from essential sterol to plant defense. Proc Natl Acad Sci USA 103: 18848–18853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EPC. (2008) The organization of the bacterial genome. Annu Rev Genet 42: 211–233 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, et al. (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134: 1642–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura K, Okada A, Okada K, Jikumaru Y, Ko KW, Toyomasu T, Sassa T, Hasegawa M, Kodama O, Shibuya N, et al. (2007) Identification of a biosynthetic gene cluster in rice for momilactones. J Biol Chem 282: 34013–34018 [DOI] [PubMed] [Google Scholar]
- Sproul D, Gilbert N, Bickmore W. (2005) The role of chromatin structure in regulating the expression of clustered genes. Nat Rev Genet 6: 775–781 [DOI] [PubMed] [Google Scholar]
- Swaminathan S, Morrone D, Wang Q, Fulton B, Peters RJ. (2009) CYP76M7 is an ent-cassadiene C11α-hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell 21: 3315–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon BG, Bushley KE. (2010) Secondary metabolism. Borkovich K, Ebbole D, , Cellular and Molecular Biology of Filamentous Fungi. American Society of Microbiology, Washington, DC, pp 376–395 [Google Scholar]
- von Rad U, Hüttl R, Lottspeich F, Gierl A, Frey M. (2001) Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize. Plant J 28: 633–642 [DOI] [PubMed] [Google Scholar]
- Wegel E, Koumproglou R, Shaw P, Osbourn A. (2009) Cell-type specific chromatin decondensation of a metabolic gene cluster in oats. Plant Cell 21: 3926–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman PR, Xu M, Jin Y, Coates RM, Peters RJ. (2004) Identification of syn-imara-7,15-diene synthase reveals functional clustering of terpene synthases involved in rice phytoalexin/allelochemical biosynthesis. Plant Physiol 135: 2098–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S, Wolfe KH. (2005) Birth of a metabolic gene cluster in yeast by adaptive gene relocation. Nat Genet 37: 777–782 [DOI] [PubMed] [Google Scholar]
- Yekta S, Tabin CJ, Bartel DP. (2008) MicroRNAs in the Hox network: an apparent link to posterior prevalence. Nat Rev Genet 9: 789–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerikly M, Challis GL. (2009) Strategies for the discovery of new natural products by genome mining. ChemBioChem 10: 625–633 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Szustakowski JD, Fortnow L, Roberts RJ, Kasif S. (2002) Computational identification of operons in microbial genomes. Genome Res 12: 1221–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]