For many years, plant pathology was divided into two schools of thought. It was clear that purified molecules or crude extracts from microbes or plants (referred to as general elicitors) could induce activation of general defense responses (Boller, 1995). Geneticists instead were studying plant resistance triggered by the recognition of a given pathogen Avirulence gene product by the corresponding plant Resistance (R) gene product (Dangl and Jones, 2001). This resistance followed the gene-for-gene hypothesis, was often associated with a hypersensitive response, and was widely used in breeding programs. It was however confusing why a pathogen would produce Avirulence products that would cause its recognition and subsequent host resistance.
GENERAL ELICITORS = PATHOGEN-ASSOCIATED MOLECULAR PATTERNS
By the end of 1990s, most classical general elicitors were oligosaccharides or glycoproteins (Boller, 1995). The identification of protein elicitors and their corresponding epitopes allowed a more thorough analysis of their conservation among the increasing amount of genomic information. For example, bacterial flagellin (the main building block of the flagellum) activated defense responses in tomato (Solanum lycopersicum) and Arabidopsis (Arabidopsis thaliana) and a peptide of 22 amino acid (flg22) conserved in many bacterial species was sufficient to trigger the full set of defense responses (Boller and Felix, 2009). Together with the demonstration that nonprotein pathogen-associated molecular patterns (PAMPs), e.g. lipopolysaccharides, classical activators of innate immune responses in animals, also induce defense responses in plants, it became evident that general elicitors resembled PAMPs (Nürnberger and Brunner, 2002). These were defined as conserved molecules present in whole classes of microbes (nonself) with an essential function for these microbes (Medzhitov and Janeway, 1997).
RECOGNITION OF PAMPS RELIES ON PLANT-ENCODED RECEPTORS
Although ample evidences based on binding studies on plant membranes existed (Boller, 1995), the identification of the respective plant-encoded PAMP receptors, or pattern-recognition receptors (PRRs) was required to convince geneticists that this perception was indeed specific. Flagellin Sensing2 (FLS2) is a Leu-rich repeat receptor kinase (LRR-RK) that binds flg22 and confers recognition specificity (Boller and Felix, 2009). Despite the number of known PAMPs recognized by plants, the number of cognate receptors is however still limited (Zipfel, 2009). The Arabidopsis LRR-RK EFR binds bacterial EF-Tu, the tomato LRR receptor-like proteins EIX1/2 recognize fungal xylanase, the rice (Oryza sativa) LysM receptor-like protein CEBiP binds fungal chitin, and the Arabidopsis LysM-RK CERK1/RLK1 is required for responses to chitin and unknown bacterial PAMP(s). Retrospectively it is interesting that the first plant PRR ever identified was actually the rice LRR-RK XA21 (Lee et al., 2009). However, the orphan XA21 was initially classified as an R protein due to its extreme narrow distribution and its dominant resistance against Xanthomonas oryzae pv oryzae bacteria. It is now clear that XA21 recognizes the PAMP Ax21 (or its eliciting epitope axYS22), a type I-secreted sulfated protein that is conserved across Xanthomonas species and a few related species.
PAMP PERCEPTION AS A KEY COMPONENT OF DISEASE RESISTANCE
With the identified PRRs it finally became feasible to address the importance of PAMP perception in plants. Treatment with PAMPs induces local and systemic resistances to several unrelated virulent pathogens. For example, flg22 treatment induced resistance to the bacterium Pseudomonas syringae pv tomato DC3000, as well as to the fungus Botrytis cinerea (Zipfel, 2009). Additionally, FLS2 loss of function leads to hypersusceptibility to adapted and nonadapted P. syringae strains, demonstrating that perception of a single PAMP quantitatively contributes to basal and nonhost resistances. The best manifestation that recognition of PAMPs is key to plant immunity is the fact that pathogens must suppress this level of resistance to cause disease. Pathogenic bacteria secrete several effectors inside plant cells, which target PAMP receptors or downstream components of PAMP-triggered immunity to achieve full virulence (Göhre and Robatzek, 2008; Hann et al., 2010).
Altogether, these findings have shaped the ground for a synthesis of plant innate immunity, in which the first layer of microbe recognition occurs via PRRs leading to PAMP-triggered immunity (Jones and Dangl, 2006). As a result, pathogenic microbes evolved mechanisms to avoid recognition or to suppress defense responses through secreted virulence effectors. In turn, plants evolved R proteins to specifically recognize these effectors or their action, which results in effector-triggered immunity. In this model, often referred to as the zig-zag model, PAMP perception acts as an evolutionary driving force in the constant arms race occurring between plants and their surrounding would-be pathogens.
OPEN QUESTIONS IN THE PAMP PERCEPTION PATHWAYS
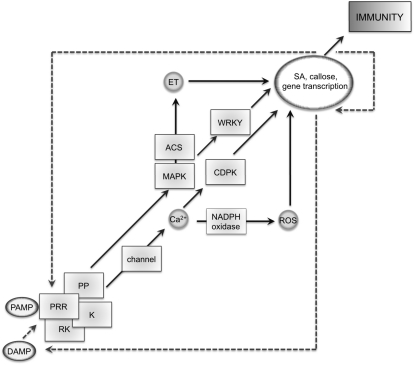
Today’s knowledge about the molecular components of PAMP perception remains a large puzzle with many missing pieces (Fig. 1). Although an increasing number of PAMP responses have been described to date, we lack understanding of the molecular mechanisms linking PAMP binding to downstream signaling. Responses to some PAMPs like flagellin and EF-Tu but not chitin depend on BAK1/SERK3, an LRR-RK initially identified in brassinolide signaling (Chinchilla et al., 2009). BAK1 is dispensable for ligand binding but required for signal transduction. It forms ligand-inducible complexes with the brassinolide receptor BRI1 and FLS2, thereby stimulating auto- and trans-phosphorylation events, leading to activation of downstream signaling (Wang et al., 2008; Schulze et al., 2010). BAK1 also plays roles in resistances to Verticillium and Alternaria fungi, and the cell death response (Chinchilla et al., 2009). As it is involved in several signaling pathways, it will be interesting to test how signal specificity is ensured and whether BAK1 is limiting. BAK1 is a member of a small family of LRR-RKs with partly redundant functions, and it is possible that PRRs are also regulated by other SERKs. Besides some evidence for protein phosphatases and the cytoplasmic kinase BIK1, which interacts with FLS2 and possibly BAK1 (Lu et al., 2010; Zhang et al., 2010), the dynamics, composition, stoichiometry of, and interactions within unstimulated and activated PRR complexes remain elusive.
Figure 1.
Molecular components of PAMP-triggered immunity and their interactions. PAMP, DAMP, PRR, regulatory RK, cytoplasmic kinase (K), phosphatase (PP), CDPKs, MAPKs, 1-aminocyclopropane-1-carboxylate synthase (ACS), WRKY transcription factors (WRKY), reactive oxygen species (ROS), ethylene (ET), and SA; black arrows indicate downstream interactions, dashed arrows possible amplifications.
One of the earliest PAMP responses detected is changes in ion fluxes across the plasma membrane, which lead to a rapid and transient extracellular alkalinization and an increase in cytosolic Ca2+ (Boller and Felix, 2009). Although the presence of respective plasma membrane channels was demonstrated, their molecular identity as well as their regulation is still unknown. Ca2+ acts as an important second messenger and could therefore provide the missing link for activating a subset of PAMP responses. The NADPH oxidase responsible for the PAMP-triggered oxidative burst is regulated by calcium, and calcium-dependent protein kinases (CDPKs) were recently described to contribute to transcriptional reprogramming upon PAMP perception (Boller and Felix, 2009; Boudsocq et al., 2010).
PAMP-induced transcriptional changes require in addition to CDPKs signaling via mitogen-activated protein kinases (MAPKs; Boudsocq et al., 2010). Despite the role of the MAPKs 3, 4, and 6, the involvement of the upstream MAPKK and MAPKKK is less clear and a link to the upstream receptor complexes is missing. This could involve cytoplasmic kinases such as BIK1, which is phosphorylated upon flg22 elicitation and transphosphorylates FLS2 and BAK1 (Lu et al., 2010; Zhang et al., 2010), but interaction with the MAPK cascade has not been demonstrated. The MAPKs 3, 4, and 6 are prominent signaling kinases in many stress-related pathways mediating specific transcriptional changes. How this is achieved and which transcriptional regulators are involved remain to be addressed. There is good evidence for a role of WRKY transcription factors in PAMP-induced transcriptional reprogramming, and a link between MAPKs was shown (Zipfel, 2009). However, to which extent members of the WRKY family and other transcriptional regulators contribute to changes in gene expression has to be demonstrated. WRKY transcription factors are also possible components linking PAMP-triggered and effector-triggered immunity (Shen et al., 2007).
Forward genetic screens to date have largely failed in the identification of molecular components downstream of PRR activation. Instead, components of the endoplasmic reticulum quality control and protein glycosylation were isolated, which affect abundance and maturation of PRRs (Saijo, 2010). This is reminiscent to two decades of genetic screening for downstream components involved in effector-triggered immunity, mainly identifying chaperones important for R protein stability (Shirasu, 2009), and points to a vital role and/or genetic redundancy within components of PAMP signaling. Since such components are likely targeted by pathogen effectors, new approaches using biochemical techniques are needed to resolve their molecular identities.
EMERGING CONCEPTS IN PAMP-TRIGGERED IMMUNITY
PAMPs trigger early responses (seconds to minutes; e.g. ion fluxes, oxidative burst), intermediate responses (minutes to hours; e.g. MAPK/CDPK activation, ethylene production, stomatal closure, transcriptional reprogramming), and late responses (hours to days; e.g. salicylic acid [SA] accumulation, callose deposition). Intriguingly, many of these responses include the production of molecules that potentially can act as second messengers (calcium, reactive oxygen species, ethylene, SA) and we may predict roles for each of the signaling pathways in PAMP-triggered immunity. Recent data suggest that glucosinolate metabolism is required for PAMP-induced callose deposition (Clay et al., 2009). Yet, it is not evident, what is the contribution of individual PAMP responses to the outcome of PAMP-triggered immunity. This is in particular questionable for the early rapid and transient responses. A recent study reported enhanced susceptibility of rsw3 mutants, caused by a failure to elicit a sustained PAMP response (Saijo, 2010). This points to amplification mechanisms of the initial PAMP responses. More importantly, we lack knowledge on how recognition of PAMPs ultimately leads to pathogen proliferation arrest.
Like in animals, host endogenous molecules released upon wounding and infection, called damage-associated molecular patterns (DAMPs), are capable of inducing immune reactions in plants. Only recently, the first plant DAMP receptor has been identified. The LRR-RKs PEPR1 and PEPR2 are responsible for the detection of the peptidic DAMP AtPep1 (Krol et al., 2010; Yamaguchi et al., 2010). Interestingly, AtPep1 triggers similar responses as PAMPs that appear to be as well BAK1 dependent, illustrating common steps between PAMP and DAMP signaling. In addition, it is proposed that PAMP and DAMP responses may be connected in a positive feedback loop (Huffaker and Ryan, 2007), but this model has yet to be proven.
CHALLENGES AND FUTURE DIRECTIONS
Recognition of different PAMPs may play distinct roles during the infection process of diverse pathogens. FLS2-mediated immunity is highly effective at the level of preinvasion, but also acts in postinvasive immunity as shown by flg22-induced local and systemic resistance (Zipfel, 2009; Zeng and He, 2010). A better time-wise and tissue-specific resolution is required to help understanding the contribution of perception of individual PAMPs and elicited responses. Close inspection of PAMP-induced inhibition of seedling growth revealed that flagellin rather acts on leaf and root tissues, while EF-Tu is most effective on leaves. This indicates potential tissue-specific differences in the recognition of flagellin and EF-Tu, although at a global scale both PAMPs elicit an almost identical set of responses. Tissue- or cell-type specificities of PAMP responses have to be addressed to better reflect different modes of infections and colonization used by different pathogens. For example, P. syringae pv tomato DC3000 bacteria enter leaf tissues and form colonies between mesophyll cells, while Ralstonia invade roots, passage across the endodermis, and populate vessels.
Not every microbe displays all PAMPs and not every plant recognizes all PAMPs. For example, flg22 is detected by most plant species, but some pathogens evade recognition through mutation of key residues (Boller and Felix, 2009). In addition, EF-Tu is only sensed by Brassicaceae, and recognition of Ax21 is restricted to specific rice cultivars. However, EF-Tu perception can be transferred across plant families and importantly confers resistance to bacteria belonging to several classes, indicating that all necessary components downstream of EFR are conserved (Lacombe et al., 2010). These examples suggest that there is a dynamic evolution in the display of PAMPs by microbes and in the recognition of PAMPs by plants (Boller and Felix, 2009), but also illustrate that novel PRRs will provide useful tools for engineering sustainable quantitative broad-spectrum disease resistance in the field.
It is not known to date what shapes the evolution of PAMP perception systems. Plants evidently will adapt to microbial populations of their local environment. There is considerable variation within accessions of Arabidopsis in the response to bacterial infection (Atwell et al., 2010), which may be partly a result of qualitative or quantitative variation in PAMP perception and/or responses. The identification of new PAMPs from plant pathogens, but also from symbionts, and their corresponding PRRs is required to fully understand the dynamics between plants and their microbial communities. How plants distinguish friends from foes and how symbionts cope with PAMP-triggered immunity also remain to be studied.
References
- Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, et al. (2010) Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T. (1995) Chemoperception of microbial signals in plant cells. Annu Rev Plant Physiol Plant Mol Biol 46: 189–214 [Google Scholar]
- Boller T, Felix G. (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng SH, Sheen J. (2010) Differential innate immunity signalling via Ca(2+) sensor protein kinases. Nature 464: 418–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Shan L, He P, de Vries S, Kemmerling B. (2009) One for all: the receptor-associated kinase BAK1. Trends Plant Sci 14: 535–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Jones JD. (2001) Plant pathogens and integrated defense responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Göhre V, Robatzek S. (2008) Breaking the barriers: microbial effector molecules subvert plant immunity. Annu Rev Phytopathol 46: 189–215 [DOI] [PubMed] [Google Scholar]
- Hann DR, Gimenez-Ibanez S, Rathjen JP. (2010) Bacterial virulence effectors and their activities. Curr Opin Plant Biol 13: 388–393 [DOI] [PubMed] [Google Scholar]
- Huffaker A, Ryan CA. (2007) Endogenous peptide defense signals in Arabidopsis differentially amplify signaling for the innate immune response. Proc Natl Acad Sci USA 104: 10732–10736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Krol E, Mentzel T, Chinchilla D, Boller T, Felix G, Kemmerling B, Postel S, Arents M, Jeworutzki E, Al-Rasheid KA, et al. (2010) Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J Biol Chem 285: 13471–13479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe S, Rougon-Cardoso A, Sherwood E, Peeters N, Dahlbeck D, van Esse HP, Smoker M, Rallapalli G, Thomma BP, Staskawicz B, et al. (2010) Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat Biotechnol 28: 365–369 [DOI] [PubMed] [Google Scholar]
- Lee SW, Han SW, Sririyanum M, Park CJ, Seo YS, Ronald PC. (2009) A type I-secreted, sulfated peptide triggers XA21-mediated innate immunity. Science 326: 850–853 [DOI] [PubMed] [Google Scholar]
- Lu D, Wu S, Gao X, Zhang Y, Shan L, He P. (2010) A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci USA 107: 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Janeway CA., Jr (1997) Innate immunity: the virtues of a nonclonal system of recognition. Cell 91: 295–298 [DOI] [PubMed] [Google Scholar]
- Nürnberger T, Brunner F. (2002) Innate immunity in plants and animals: emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr Opin Plant Biol 5: 318–324 [DOI] [PubMed] [Google Scholar]
- Saijo Y. (2010) ER quality control of immune receptors and regulators in plants. Cell Microbiol 12: 716–724 [DOI] [PubMed] [Google Scholar]
- Schulze B, Mentzel T, Jehle AK, Mueller K, Beeler S, Boller T, Felix G, Chinchilla D. (2010) Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J Biol Chem 285: 9444–9451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P. (2007) Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Shirasu K. (2009) The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu Rev Plant Biol 60: 139–164 [DOI] [PubMed] [Google Scholar]
- Wang X, Kota U, He K, Blackburn K, Li J, Goshe MB, Huber SC, Clouse SD. (2008) Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell 15: 220–235 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA. (2010) PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22: 508–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, He SY. (2010) A prominent role of the flagellin receptor FLAGELLIN-SENSING2 in mediating stomatal response to Pseudomonas syringae pv tomato DC3000 in Arabidopsis. Plant Physiol 153: 1188–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S, et al. (2010) Receptor-like cytoplasmic kinases integrate signalling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301 [DOI] [PubMed] [Google Scholar]
- Zipfel C. (2009) Early molecular events in PAMP-triggered immunity. Curr Opin Plant Biol 12: 414–420 [DOI] [PubMed] [Google Scholar]