Abstract
Eph receptors and their ephrin ligands are important mediators of cell-cell communication. They are divided in two subclasses based on their affinities for each other and on sequence conservation. Receptor-ligand binding within each subclass is fairly promiscuous, while binding cross the subclasses happens rarely. EphA4 is an exception to this general rule, since it has long been known to bind both A- and B-class ephrin ligands but the reason for this exceptional behavior has not been worked out at molecular level. Recent structural and biochemical studies on EphA4 ligand-binding domain alone and in complex with its ligands have addressed this question. However, the published structures of EphA4/ephrin complexes differ considerably from each other and strikingly different explanations for the exceptional promiscuity of EphA4 were proposed. To address these contradictory findings, we have determined a crystal structure of the EphA4 ligand-binding domain at 2.3 Å resolution and show that the receptor has an unprecedented ability to exist in two very different, well-ordered conformations even in the unbound state. Our results suggest that the ligand promiscuity of the Ephs is directly correlated with the structural flexibility of the ligand-binding surface of the receptor.
Keywords: Eph, Receptor Tyrosine Kinase, Ephrin, Loop Flexibility, X-ray Crystallography
Introduction
The Erythropoietin-producing hepatocellular (Eph) receptors are the largest family of receptor tyrosine kinases and their interaction with the corresponding membrane-bound ephrin ligands at the site of cell-cell contact initiates a unique bidirectional signaling cascade [1]. Eph receptors are important mediators of cell-cell interactions, axonal pathfinding, and boundary formation. Their signaling networks have been implicated in the pathogenesis of many diseases, for example in the progression of cancer and metastasis, making them important therapeutic targets [2]. The Eph receptors are type-I transmembrane proteins with an N-terminal ectodomain, which consists of an ephrin-ligand binding domain (LBD), a cysteine rich region, and two fibronectin type III repeats. The ectodomain is separated by a single transmembrane spanning helix from the intracellular region comprising of a juxtamembrane segment, a tyrosine kinase domain, a sterile α motif (SAM), and a PDZ-binding motif. Eph receptors and their ephrin ligands are divided in two subclasses based on their affinities for each other and on sequence conservation [3; 4]. While receptor-ligand binding within the subclass is fairly promiscuous, binding cross the subclass happens rarely [5]. The most well known exception to this general rule is EphA4 that has long been known to bind both A- and B-class ligands [6; 7]. Because of this, EphA4 is an attractive target for studying the structural details of subclass specificity.
Structural studies on the binding-domains of Eph receptors and ephrins have increased considerably our understanding of the details of receptor-ligand recognition [8; 9]. The initial binding force comes from a penetration of a long, hydrophobic ephrin loop into a hydrophobic cavity on the surface of the receptor but the exact principles for the subclass specificity remain elusive. The ligand-binding cavity of the receptor consists of loops D-E, F-G, and J-K that have been shown to exhibit various amounts of conformational flexibility, depending on the subclass and the individual receptor. Several recent structures of the EphA4 ligand-binding domain alone and in complex with its ligands have addressed this conundrum [10-12]. However, the two reports of EphA4/ephrin complexes give dramatically different explanations for the exceptional promiscuity of EphA4 [10; 12]. To address these contradictory findings, we have determined a crystal structure of EphA4 ligand-binding domain at 2.3 Å resolution and show that the receptor has an unprecedented ability to adopt two distinct, well-ordered structures even in the unbound state.
Materials and Methods
Protein Expression and Purification
The PCR amplified cDNA sequence corresponding to the ligand-binding domain of human EphA4 (residues Gly22-Lys203) was cloned into a modified pET32b vector (Novagen). The vector was transformed into Escherichia coli Rosetta-gami (DE3) cells (Novagen). Cells were grown in Luria-Bertani medium supplemented with ampicillin at 37°C to an OD600 of 0.5-0.6 and were induced overnight at 18°C with 1 mM IPTG. The cells were harvested and lysed using a buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 150 mM Imidazole, 1 mM TCEP, 1 mM PMSF, 10% glycerol, 0.2% Igepal CA-630, 2.5mg DNase I (Roche), 12.5 mg Lysozyme (Sigma), and protease inhibitor cocktail tablets (Roche). The recombinant protein was purified by affinity chromatography on a His-Trap HP Nickel Sepharose column (GE Healthcare) and dialyzed into thrombin cleavage buffer containing 20 mM Tris-HCl (pH 8.4), 150 mM NaCl, and 2.5 mM CaCl2. The histidine-containing amino terminal fusion sequences were removed by overnight incubation with thrombin (1U/mg of fusion protein) at 4°C. The released EphA4 was further purified on the Nickel column followed by size exclusion chromatography (SEC) using a Superdex-200 10/30 gel-filtration column (GE Healthcare) pre-equilibrated with a buffer containing 20 mM Hepes (pH 7.2) and 150 mM NaCl. The fractions corresponding to the major peak were resolved on SDS-PAGE. The pure EphA4 LBD protein fractions were pooled and dialyzed into a buffer containing 5 mM Hepes (pH 7.0), 10 mM KCl, and 2 mM MgCl2. The ectodomain of human ephrin-A5 (residues-1-228) was cloned and expressed as a fusion to the Fc region of IgG1 as previously described[13]. The secreted ephrin-A5-Fc fusion protein was purified by affinity chromatography using Fast Flow Protein-A Sepharose (GE Healthcare).
Pull-Down Assay
The EphA4 LBD obtained after purification was incubated with ephrin-A5-Fc ligand in a 1:1 molar ratio. The pull-down experiments were performed as previously described [13].
Crystallization and Data Collection
The purified EphA4 LBD was concentrated to 15 mg/ml and was crystallized in a hanging drop by vapor diffusion at room temperature against a reservoir containing 0.1 M MES sodium salt pH 6.5 and 1.8 M ammonium sulfate. The droplet was formed by mixing 1μl of protein solution and 1μl of reservoir solution supplemented with one-tenth of the volume of 30% v/v (+/-)-2-Methyl-2,4-pentanediol (Hampton additive screen solution No. 75). The crystals were protected by cryoprotectant containing 25% glycerol, 0.1 M MES sodium salt pH 6.5, and 1.8 M ammonium sulfate. Data was collected at the Argonne Advanced Photon Source beamline 24IDE. The crystals belong to the space group P212121 and diffract to 2.3 Å resolution. Data collection statistics are presented in Table 1.
Table 1. Summary of crystallographic analysis.
| Resolution, Å | 30.0-2.3 (2.4-2.3) |
| Wavelength, Å | 1.033 |
| Completeness, % | 99.9 (99.8) |
| Redundancy, fold | 7.5 (5.9) |
| I/σI | 22.8 (3.6) |
| Rmerge, % | 11.5 (46.4) |
| Space Group | P212121 |
| Cell Dimensions, Å | a=43.09, b=62.33, c=135.93 |
| Refinement | |
| Reflections working / test | 18002 / 980 |
| Residues | 360 |
| Solvent | 240 |
| Rcrys/Rfree | 19.8/28.5 (22.3/32.3) |
| R.m.s. deviations | |
| Bonds, Å | 0.019 |
| Angles, ° | 1.80 |
| Ramachandran Analysis | |
| Most favored regions | 92.4 % |
Values in parentheses correspond to the high-resolution shell.
Rmerge = Σ|I−<I>|/ΣI, where I = observed intensity and <I> = average intensity obtained from multiple observations of symmetry-related reflections.
The r.m.s. deviations in bond lengths and angles are the respective root-mean-square deviations from ideal values
Structure Determination and Refinement
The structure was solved by molecular replacement using Phaser (CCP4 suite) [14] and the A chain of PDB entry 3CKH as the search model [11]. The best molecular replacement model was refined using Refmac5 [15], manual fitting was performed with O [16], adding solvent with Arp-Warp [17]. The final model contains two polypeptide chains of EphA4 (residues A24-A204 and B24-B204; Ala 204 coming from the expression vector) and 240 solvent molecules. Refinement statistics is presented in Table 1.
Results and Discussion
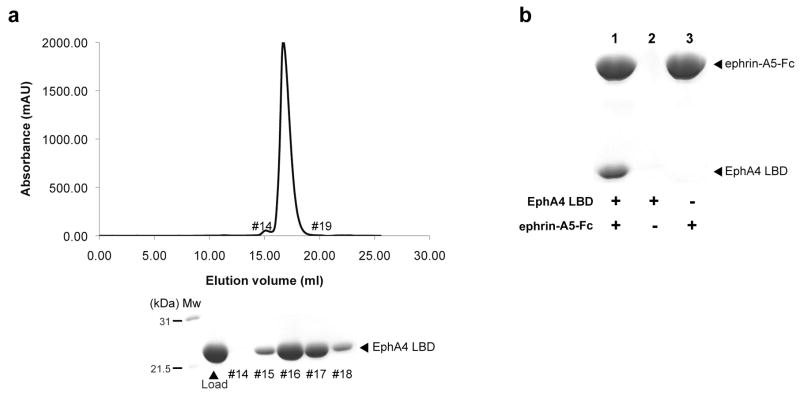
The EphA4 LBD was expressed in an E.coli strain Rosetta-gami (materials & methods) that allows for more efficient formation of disulfide bonds and expression of eukaryotic proteins. The recombinant protein was purified by affinity chromatography on His-Trap and size exclusion chromatography (SEC) on Superdex200 where EphA4 LBD elutes as a monomer (Mw ∼ 22 kDa; Fig. 1a). The EphA4 LBD obtained after SEC was incubated with Fc-tagged ephrin-A5 ligand and pulled down on Protein-A Sepharose beads demonstrating that it maintained the correct fold and its native biological ligand binding activity (Fig. 1b).
Figure 1.
Characterization of the recombinant EphA4 LBD. (a) The SEC elution profile of the EphA4 LBD after final purification. The fractions under the major peak were resolved on SDS-PAGE and correspond to the pure protein. (b) The purified EphA4 LBD retains its biological activity. The SDS-PAGE shows that the EphA4 LBD forms a specific complex with the Fc-tagged ephrin-A5 ligand (ephrin-A5-Fc) that can be pulled down by Protein-A Sepharose beads (lane 1). The EphA4 LBD by itself does not bind to the beads (lane 2).
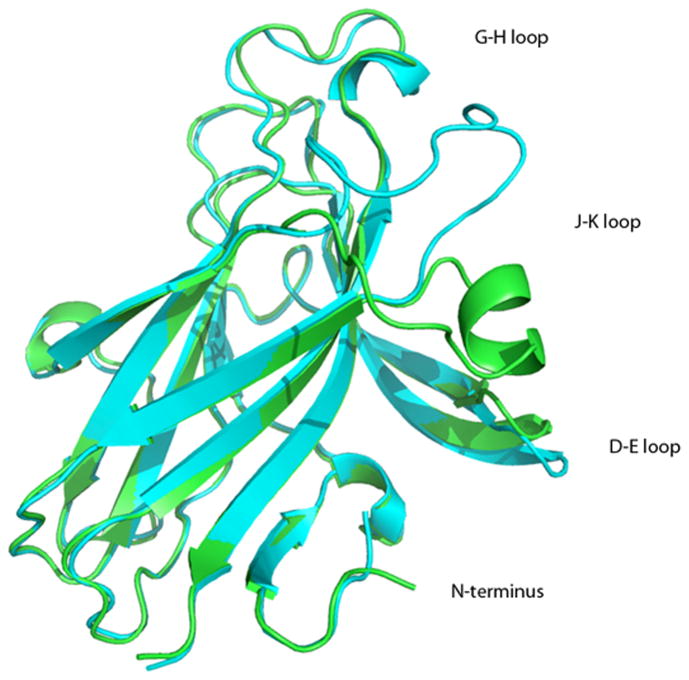
We determined the crystal structure of EphA4 LBD and refined it to 2.3 Å resolution. The overall fold of our EphA4 LBD is similar to that reported in earlier studies [10; 11]. It's a β-sandwich consisting of 11 antiparallel β-strands arranged into two β-sheets, 5 loops, and two disulphide bonds stabilizing the structure (Fig. 2). There are two molecules in the asymmetric unit (AU) that display significant variations in the conformation of D-E and J-K loops that play a fundamental role in ligand binding [1]. In addition to the crystal structures of the uncomplxed EphA4 LBD, crystal structures of the EphA4 LBD in complex with ephrin-B2 and ephrin-A2 have also been published [10; 12]. The comparison of all the available crystallographic models of EphA4 reveals unusual flexibility of D-E and J-K loops that can be either in contact (closed conformation) or pointing away from each other (open conformation).
Figure 2.
Overall structure of the EphA4 LBD. The two molecules found in the asymmetric unit are superimposed. The A chain is shown in green and the B chain in cyan.
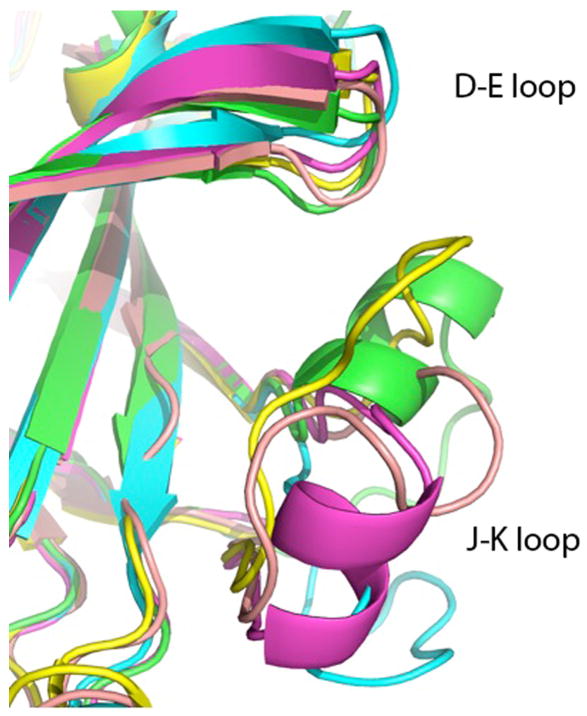
Superposition of all the EphA4 structures is summarized in Table 2 using one of the structures of free EphA4 (2WO1 chain B) as a reference. The unusually high variations in the structures, measured as root-mean-square deviation of coordinates of superimposed Cα atoms (rmsd), are unexpected for such a small protein and are indicative of exceptional conformational flexibility. The internal β-sandwich core of the molecule is rigid, as indicated by rmsd calculated with D-E and J-K loops excluded. Another region of elevated flexibility is found in the area of amino acids 107-118 (G-H loop). Moreover, even in structures of EphA4 bound to ephrin-B2, the rmsd is 1.35 Å (0.497 Å with loops omitted) and displacements of D-E and J-K loops reach 2.00 Å (residue 63) and 9.35 Å (residue 160), respectively, indicating that the remarkable flexibility is retained even in the cross-class-bound conformation. These loops also account for the dissimilarity between the structure of A and B chains of the unbound EphA4 reported here. The two chains in our EphA4 structure superimpose with an exceptionally high rmsd of 2.933 Å, as opposed to 0.873 Å when the D-E and J-K loops are omitted. The maximum displacement of the loops in question is 5.5 Å (residue 63) and 15.0 Å (residue 160), respectively. Thus, the EphA4 ligand-binding domain has the most diverse set of observed distinct structures among the Ephs, and indeed might be the only reported case of a small protein where so many different and unique loop conformations have been experimentally observed (Fig. 3).
Table 2. Comparison of conformations of different EphA4 structures.
| Structure | Conformation | rmsd | rmsd, loops omitted | Displacement D-E residue #/A (Cα) | Displacement J-K residue #/A (Cα) |
|---|---|---|---|---|---|
| Our chain A | closed | 2.523 | 1.089 | 63/8.13 | 161/10.14 |
| Our chain B | open | 3.478 | 1.268 | 61/7.64 | 159/15.39 |
| 2WO1 chain A | closed | 1.241 | 1.023 | 61/4.29 | 161/3.05 |
| 2WO1 chain B | closed | reference | reference | reference | reference |
| 3CKH chain A | open? | 3.025 | 0.969 | 64/7.98 | 156/14.12 |
| 3CKH chain B | closed? | 1.119 | 0.907 | 65/3.12 | 161/4.19 |
| 2WO2 (ephrin B2) | closed | 3.282 | 1.465 | 63/10.49 | 155/14.98 |
| 3GXU (ephrin B2) | open | 3.290 | 0.798 | 63/8.86 | 155/14.70 |
| 2WO3 (ephrin A2) | open | 3.080 | 1.019 | 63/8.13 | 155/13.01 |
(Question marks indicate that the model is incomplete in the region under consideration)
Figure 3.
Conformational flexibility of the J-K loop of EphA4. The superimposed structures are the two chains reported here (green and cyan) and three structures of ligand-bound EphA4 - 2WO2 (yellow), 3GXU (pink) and 2WO3 (magenta). A helical region is found in the J-K loop of EphA4 bound to an A class ephrin, but also in one of the structures of unbound EphA4 (chain A of our structure).
It has been proposed earlier [10] that the formation of a helix in the J-K loop occurs upon binding of an A-class ligand to EphA4. The structure reported here demonstrates that residues A159-A163 and B153-B155 have main chain dihedral angles characteristic of an α-helix. In case of the chain A, it is a true helical region, while in the chain B, the stretch is too short to form an α-helical pattern of hydrogen bonds. The observed conformations of the J-K loop in our structure indicate that helix formation in this region can occur in the unbound EphA4 as well.
It is worth mentioning that in our structure the J-K loop of chain B, in respect to the chain A, is found in a position similar to the loop 115-124 of ephrin-B2 in the structures of EphA4/ephrin-B2 complex [10; 12], although the pattern of interactions is different. Even though there is no direct evidence of dimerization of the EphA4 LBD in solution, this observation might be of relevance considering the proposed dimerization of the Eph receptors on cell membranes [18].
The Eph/ephrin high-affinity interface is divided in two general regions, the main being the hydrophobic ephrin G-H loop inserting into the hydrophobic channel created by the Eph D-E and J-K loops, and the second being a mostly polar docking site on the side of the Eph beta barrel. This second region of the interface is present most notably in the ephrin-B class containing structures (see e.g. [19]) and was recently elegantly studied by mutagenesis experiments [12] using EphA4 binding to ephrin-B2. These studies show that this contact region involves a network of hydrogen bonds and salt bridges between the A-C loop and the D and E strands of EphA4, and the C, G, and F strands of ephrin-B2. The authors further show that while mutations of Gln40 or Glu42 in the A-C loop of EphA4 might not significantly affect binding to A-class ligands, they do abolish the ability of EphA4 to effectively bind the B-class ligands. Consistent with this, in the EphA2/ephrin-A1 complex [9], the only previous A-class structure reported, only a weak hydrogen bond within the same interface (with a distance of about 4 Å) between Glu40 (EphA2) and Arg94 (ephrin-A1) was observed.
EphB2, has been documented to undergo fairly dramatic conformational changes in the D-E and J-K loop region, prompting the suggestions that it utilizes an “induced fit” binding mechanism [20], distinct from the “lock-and-key” mechanism proposed for EphA2 [9]. Our results presented here, together with previous structural studies, reveal that EphA4 also displays an exceptionally large conformational flexibility in the D-E and J-K loop region, even in its unbound state. These observations suggest that the ligand promiscuity of the Ephs could be directly correlated with the structural flexibility of the ligand-binding surface of the receptor.
Importantly, the reported here structures indicate that the conformational differences within the EphA4 loops in the vicinity of the hydrophobic cavity are not consequences of ligand-binding, but are preexisting in the unbound receptor, which exists in multiple conformations that can be grouped into either a ‘closed’ or an ‘open’ state. This is a rather specific and intrinsic feature of the EphA4 receptor that differentiate it from the other Eph receptors, with the possible exception of the only other cross-subclass binder, EphB2. Of course, the more Eph and Eph/ephrin structures become available, the more detailed picture of these phenomena we should be able to draw.
Research Highlights
Crystal Structure of the ligand-binding domain of the promiscuous EphA4 receptor reveals two distinct conformations (Singla et al.)
EphA4 receptor has two distinct conformations in the unbound state
Conformational plasticity of EphA4 centers around two flexible ligand-binding loops
Loop-flexibility is retained also in the ligand-bound forms of EphA4
Unusual conformational flexibility makes EphA4 able to bind all ephrin ligands
Acknowledgments
This work was supported by the National Institutes of Health grant NS38486 (to D.B.N.) The work is based upon research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. SP was financially supported by the Academy of Finland and Juselius Foundation. The authors are grateful for Rudiger Klein for cDNA and fruitful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Himanen JP, Nikolov DB. Eph signaling: a structural view. Trends Neurosci. 2003;26:46–51. doi: 10.1016/s0166-2236(02)00005-x. [DOI] [PubMed] [Google Scholar]
- 2.Pasquale EB. Eph-ephrin bidirectional signaling in physiology and disease. Cell. 2008;133:38–52. doi: 10.1016/j.cell.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Gale NW, Holland SJ, Valenzuela DM, Flenniken A, Pan L, Ryan TE, Henkemeyer M, Strebhardt K, Hirai H, Wilkinson DG, Pawson T, Davis S, Yancopoulos GD. Eph receptors and ligands comprise two major specificity subclasses and are reciprocally compartmentalized during embryogenesis. Neuron. 1996;17:9–19. doi: 10.1016/s0896-6273(00)80276-7. [DOI] [PubMed] [Google Scholar]
- 4.Himanen JP, Saha N, Nikolov DB. Cell-cell signaling via Eph receptors and ephrins. Curr Opin Cell Biol. 2007;19:534–42. doi: 10.1016/j.ceb.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Himanen JP, Chumley MJ, Lackmann M, Li C, Barton WA, Jeffrey PD, Vearing C, Geleick D, Feldheim DA, Boyd AW, Henkemeyer M, Nikolov DB. Repelling class discrimination: ephrin-A5 binds to and activates EphB2 receptor signaling. Nat Neurosci. 2004;7:501–9. doi: 10.1038/nn1237. [DOI] [PubMed] [Google Scholar]
- 6.Prevost N, Woulfe DS, Jiang H, Stalker TJ, Marchese P, Ruggeri ZM, Brass LF. Eph kinases and ephrins support thrombus growth and stability by regulating integrin outside-in signaling in platelets. Proc Natl Acad Sci U S A. 2005;102:9820–5. doi: 10.1073/pnas.0404065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith A, Robinson V, Patel K, Wilkinson DG. The EphA4 and EphB1 receptor tyrosine kinases and ephrin-B2 ligand regulate targeted migration of branchial neural crest cells. Curr Biol. 1997;7:561–70. doi: 10.1016/s0960-9822(06)00255-7. [DOI] [PubMed] [Google Scholar]
- 8.Barton WA, Himanen JP, Antipenko A, Nikolov DB. Structures of axon guidance molecules and their neuronal receptors. Adv Protein Chem. 2004;68:65–106. doi: 10.1016/S0065-3233(04)68003-X. [DOI] [PubMed] [Google Scholar]
- 9.Himanen JP, Goldgur Y, Miao H, Myshkin E, Guo H, Buck M, Nguyen M, Rajashankar KR, Wang B, Nikolov DB. Ligand recognition by A-class Eph receptors: crystal structures of the EphA2 ligand-binding domain and the EphA2/ephrin-A1 complex. EMBO Rep. 2009;10:722–8. doi: 10.1038/embor.2009.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowden TA, Aricescu AR, Nettleship JE, Siebold C, Rahman-Huq N, Owens RJ, Stuart DI, Jones EY. Structural plasticity of eph receptor A4 facilitates cross-class ephrin signaling. Structure. 2009;17:1386–97. doi: 10.1016/j.str.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin H, Shi J, Noberini R, Pasquale EB, Song J. Crystal structure and NMR binding reveal that two small molecule antagonists target the high affinity ephrin-binding channel of the EphA4 receptor. J Biol Chem. 2008;283:29473–84. doi: 10.1074/jbc.M804114200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin H, Noberini R, Huan X, Shi J, Pasquale EB, Song J. Structural characterization of the EphA4-Ephrin-B2 complex reveals new features enabling Eph-ephrin binding promiscuity. J Biol Chem. 2010;285:644–54. doi: 10.1074/jbc.M109.064824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singla N, Himanen JP, Muir TW, Nikolov DB. Toward the semisynthesis of multidomain transmembrane receptors: modification of Eph tyrosine kinases. Protein Sci. 2008;17:1740–7. doi: 10.1110/ps.035659.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 16.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 17.Lamzin VS, Wilson KS. Automated refinement of protein models. Acta Crystallogr D Biol Crystallogr. 1993;49:129–47. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- 18.Himanen JP, Yermekbayeva L, Janes PW, Walker JR, Xu K, Atapattu L, Rajashankar KR, Mensinga A, Lackmann M, Nikolov DB, Dhe-Paganon S. Architecture of Eph receptor clusters. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1004148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Himanen JP, Rajashankar KR, Lackmann M, Cowan CA, Henkemeyer M, Nikolov DB. Crystal structure of an Eph receptor-ephrin complex. Nature. 2001;414:933–8. doi: 10.1038/414933a. [DOI] [PubMed] [Google Scholar]
- 20.Goldgur Y, Paavilainen S, Nikolov D, Himanen JP. Structure of the ligand-binding domain of the EphB2 receptor at 2 A resolution. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009;65:71–4. doi: 10.1107/S1744309108043078. [DOI] [PMC free article] [PubMed] [Google Scholar]