Abstract
We previously reported the establishment of a rabbit (Oryctolagus cuniculus) model in which peptide immunization led to production of lupus-like autoantibodies including anti-Sm, -RNP, -SS-A, -SS-B and –dsDNA characteristic of those produced in Systemic Lupus Erythematosus (SLE) patients. Some neurological symptoms in form of seizures and nystagmus were observed. The animals used in the previous and in the present study were from a National Institute of Allergy and Infectious Diseases colony of rabbits that were pedigreed, immunoglobulin allotype-defined but not inbred. Their genetic heterogeneity may correspond to that found among patients of a given ethnicity. We extended the information about this rabbit model by microarray based expression profiling. We first demonstrated that human expression arrays could be used with rabbit RNA to yield information on molecular pathways. We then designed a study evaluating gene expression profiles in 8 groups of control and treated rabbits (47 rabbits in total). Genes significantly upregulated in treated rabbits were associated with NK cytotoxicity, antigen presentation, leukocyte migration, cytokine activity, protein kinases, RNA spliceosomal ribonucleoproteins, intracellular signaling cascades, and glutamate receptor activity. These results link increased immune activation with up-regulation of components associated with neurological and anti-RNP responses, demonstrating the utility of the rabbit model to uncover biological pathways related to SLE-induced clinical symptoms, including Neuropsychiatric Lupus. Our finding of distinct gene expression patterns in rabbits that made anti-dsDNA compared to those that only made other anti-nuclear antibodies should be further investigated in subsets of SLE patients with different autoantibody profiles.
The development of the autoimmune disease Systemic Lupus Erythematosus (SLE) is influenced by a combination of genetic (1), epigenetic (2), environmental, (3) and hormonal factors (4). The complexity of this disease has made the development of specific targeted treatments difficult, and understanding the molecular dynamics of diverse gene expression pathways that may contribute to SLE extremely challenging. The clinical manifestations of SLE are highly variable with multiple organs and organ systems affected; these include skin (5) joints (6), heart (7), kidney (8) and the nervous system (9). Presence of autoantibodies to RNA spliceosomal ribonucleoproteins and dsDNA are characteristic in this disease (10, 11). Underlying disease manifestations are a multitude of inflammatory processes and immune system dysregulation that may arise over a period of several years culminating in overt clinical disease and often marked by quiescence and flare-ups.
Combinations of several genetic defects may contribute to susceptibility to development of the complex disease processes in lupus (1, 12). Studies in mice (13-15) and human patients (16-19) have implicated individual candidate genes and genetic regions associated with development of SLE. However, few such discoveries have led to substantial improvements of clinical management. It is therefore important to continue to examine interplay of different genetic defects on pathways that become dysregulated. The collective effects may be responsible for the various manifestations of the disease.
Gene profiling microarray studies using PBMC in SLE patients have revealed overexpression of genes encoding inflammatory cytokines, chemokines and other genes that affect the immune system (20-24) including those involved in apoptosis, signal transduction, and the regulation of the cell cycle (25). The generally accepted view that gene products induced by type 1 interferons (IFN) have a role in lupus has been supported by observations of their significant upregulation in PBMC of pediatric and some adult SLE patients. DNA-containing immune complexes present in sera from lupus patients have been shown to induce genes encoding type 1 IFNs (reviewed in 26-28 and references therein). Recently a Phase I, safety and tolerability study of a human monoclonal antibody (mAb) MEDI-545 with broad specificity for type 1 IFNs utilized Affymetrix Human Genome arrays to evaluate the effects of the anti-IFN mAb treatment on IFN α/β inducible gene signatures in patients with mild SLE (28) (ClinicalTrials.gov identifier: NCT00299819A). In addition, a recent longitudinal study suggested that monitoring serum levels of IFN-regulated chemokines, most notably CXCL10 (IP-10), could greatly improve the identification of patients at risk of disease flare (29).
An important goal of biomedical research is to translate basic findings into clinical applications. Models in inbred mice that spontaneously develop SLE, along with various mutant, transgenic and knockout models have documented a variety of genetic defects leading to SLE, but from the clinical perspective, the degree to which these findings using the inbred or homogeneous artificially mutated strains apply to individuals in heterogeneous outbred human populations is open to question. Given that there is still no cure available for SLE, it is important that we continue to explore possibilities using new animal models including for neuropsychiatric lupus (NPSLE). This report extends information about the rabbit model of lupus in pedigreed, non-inbred, and genetically defined rabbits (30-32). We describe unique gene expression changes associated with lupus-like serological patterns in immunized rabbits. Our results also demonstrate that caution must be applied when choosing the structure of the carrier Multiple Antigen Peptide (MAP-peptide) for immunization. We discovered that using MAP-4 rather than MAP-8 significantly altered patterns of immune response and gene expression. Currently, microarrays specific for study of gene expression profiles are not available for rabbits. Therefore, we first conducted studies that compared identically prepared rabbit and human cRNA binding to the Affymetrix U95 microarray available for human gene expression analyses. It was determined that the human microarray could be used with rabbit cRNA to yield information on genetic pathways activated and/or suppressed in autoantibody-producing immunized rabbits. In the current report, gene expression profiles of a total of 47 rabbits, from 4 generations within a pedigreed group of control and immunized rabbits, were obtained and analyzed.
Materials and Methods
Animals
Rabbits from the NIAID colony were pedigreed, non-inbred, and allotype-defined. The animal studies described here were reviewed and approved by the animal care and use committees of NIAID/National Institutes of Health, Bethesda, MD (animal study protocol LI-6) and of the Spring Valley Laboratories (Woodbine, MD), where the NIAID allotype-defined rabbit colony was bred and housed. When introduced into a study group, the rabbits were housed in a room separate from the main colony and monitored with video cameras (30). We describe here four groups of one to two-year old rabbits totaling 47 (26 males, 21 females) that were selectively bred using high responder parents or their relatives. Rabbits in sequential Groups 2, 3 or 4 were related to Group 1 or 2 animals and/or their siblings (30). Additional rabbits from further selective breeding (31) were used for some confirmatory tests by quantitative PCR analyses. Table I summarizes the rabbits used in the microarray studies. Fourteen normal unimmunized rabbits including some that were subsequently immunized were also used for RNA extraction. The autoantibodies in Group 4 rabbits were detected as described earlier (30) and their responses summarized with their pedigree in (31). Those responses found in groups 1-3 (SM-1 through BB-31) were also previously reported (30). The pedigrees and summaries of antibody responses of animals of groups 1-4 studied by microarray expression analyses along with those in group 5 used in qPCR validations studies (31), can be accessed online, and are freely available at: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2561998/pdf/nihms51377.pdf/?tool=pmcentrez. The added pedigree and summary of autoantibody responses of group 6 described subsequently (Figure 6 in 32) is also is freely available at: http://www.plosone.org/article/slideshow.action?uri=info:doi/10.1371/journal.pone.0008494&imageURI=info:doi/10.1371/journal.pone.0008494.g006
Table 1.
Rabbits Summarya
| Group/ Backbone |
Rabbit No. | Peptide/ Rabbit Id |
Sex | ds DNA | ENA | Sm | RNP | SS-A | SS-B | ANA |
|---|---|---|---|---|---|---|---|---|---|---|
| 1/MAP-8 | XX129-3 c | SM-1 | M | 1.65 | +++ | |||||
| 2XX127-4 | SM-2 | F | ||||||||
| 2XX288-2 | SM-3 | M | 0.20 | 1.07 | ||||||
| 1XX288-4 | SM-4 | F | ||||||||
| 2XX127-2 | SM-5 | M | ||||||||
| 2XX92-6 | SM-6 | F | 0.27 | 0.93 | ||||||
| XX129-5 d | GR-7 | M | 0.33 | |||||||
| 2XX127-5 c | GR-8 | F | 1.73 | |||||||
| 1XX288-3 | GR-9 | M | 1.85 | 0.15 | + | |||||
| 2XX288-6 | GR-10 | F | 1.56 | |||||||
| 2XX127-1 | BB-11 | M | ||||||||
| 1XX78-8 | BB-12 | F | ||||||||
| 2/MAP-4 | LL191-1 | SM13 | M | 0.56 | 0.254 | 0.36 | 1.10 | +++ | ||
| LL191-2 | SM-14 | F | 0.27 | 0.49 | ||||||
| 2LL179-1 | SM-15 | M | 0.28 | 0.32 | 0.13 | +++ | ||||
| 1LL163-3 | SM-16 | F | 0.31 | |||||||
| LL164-4 | SM17 | F | ||||||||
| 1LL178-2 | GR-18 | M | ||||||||
| 1LL178-3 | GR-19 | F | 0.24 | 0.44 | ++ | |||||
| 1LL178-4 | GR-20 | F | 0.19 | ++ | ||||||
| 1LL178-5 | GR-21 | M | ||||||||
| 1LL178-6 | GR-22 | F | ||||||||
| 1LL178-8 | GR-23 | F | 0.95 | |||||||
| LL164-1 | BB-24 | M | ||||||||
| LL164-3 | BB-25 | F | ||||||||
| 2LL179-3 | BB-26 | M | ||||||||
| 1LL163-4 | BB-27 | F | ||||||||
| 3/MAP-8 | LL108-1 | GR-28 | M | 0.36 | 0.29 | 0.15 | ||||
| LL108-3 | GR29 | F | 0.87 | 0.30 | ||||||
| LL108-4 | GR-30 | F | 0.27 | 0.25 | 0.28 | 0.48 | + | |||
| 2LL179-2 | BB-31 | M | ||||||||
| 4/MAP-8b | 1QQ299-2 | SM32 | M | 0.36 | 0.36 | |||||
| 1QQ299-3 | SM-33 | F | 0.55 | 0.34 | 0.30 | 0.40 | ||||
| 6QQ299-1 | SM-34 | M | 0.43 | 0.74 | 0.31 | 0.93 | 0.31 | ++ | ||
| 6QQ299-2 | SM-35 | M | 0.56 | 0.55 | 0.47 | 0.53 | ++ | |||
| 3QQ299-1 | GR-36 | M | 0.58 | 0.74 | 0.76 | +++ | ||||
| 3QQ299-2 | GR-37 | M | 0.55 | +++ | ||||||
| 3QQ299-4 | GR-38 | M | 0.41 | ++++ | ||||||
| 4QQ299-1 | GR-39 | M | 1.09 | 0.37 | + | |||||
| 5QQ299-2 | GR-40 | F | 0.60 | ++ | ||||||
| 5QQ299-3 | GR-41 | F | 0.32 | 0.49 | 0.50 | 2.06 | + | |||
| 1QQ299-1 | BB-42 | M | ||||||||
| 5QQ299-4 | BB-43 | F | 0.43 | |||||||
| 6QQ299-3 | BB-44 | F | 0.56 | |||||||
| 1QQ173-1 | PB-45 | M | 0.45 | 0.32 | 0.71 | |||||
| 1QQ173-2 | PB-46 | M | + | |||||||
| 1QQ173-3 | PB-47 | M |
The test results on sera from groups 1-3 were previously published (Rai et al., 2006).
Group 4 relatives or offspring of groups 1-3 rabbits had high preimmune ELISA OD. The Δ OD values shown had pre-immune OD subtracted (from top to bottom): dsDNA (0.70, 0.54, 0.59, 0.16), ENA (1.24, 1.53, 1.44, 1.80, 1.87, 1.25, 2.17, 0.80); Sm (0.43, 1.05, 1.90, 0.54, 0.39, 0.49, 1.17, 0.87, 0.34); Rnp ( 0.98, 0.46, , 0.69); SS-A (0.16, 0.09, 0.66, 0.68); SS-B (0.08, 0.19, 0.08, 0.07).
Clinical signs included: seizures c ; nystagmus
(Rai et al., 2006).
FIGURE 6.
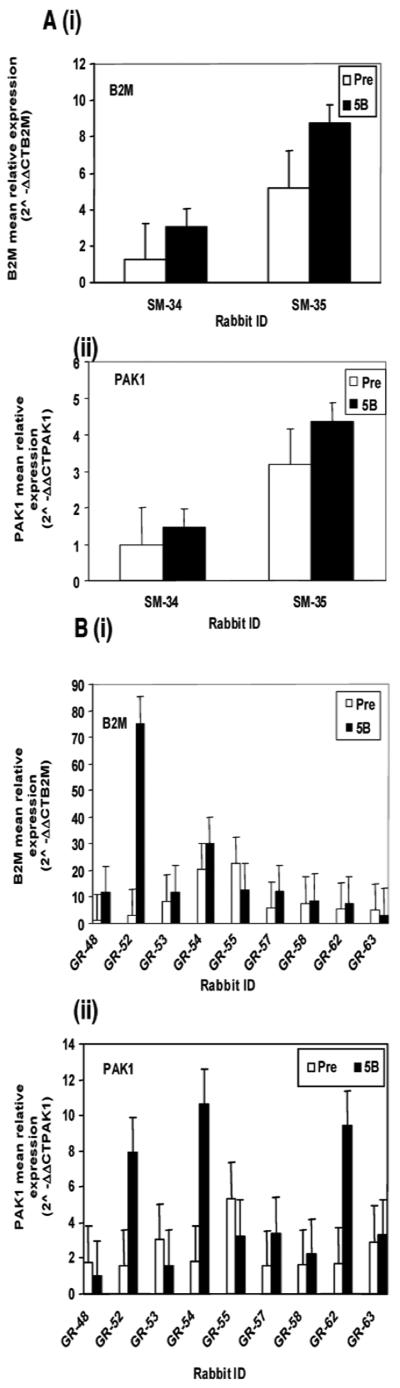
Validation of gene expression changes using real time PCR. The unit number showing relative mRNA levels in each sample was determined as a value of mRNA normalized against Peptidylprolyl isomerase A (PPIA). RT-PCR data were analyzed by using the 2−ΔΔCT method as described in (36). Results of quantitative real time PCR analyses shown in each panel were normalized relative to the lowest value set at 1. The open bars indicate expression pre-immunization (Pre) and the filled bars expression after the fifth boost (5B). Rabbit's numbers are show on the x axis. The y axis shows mean relative expression of the tested genes normalized relative to the lowest value set at 1. Increased mean relative mRNA expression of i) B2M and ii) PAK1 after the 5th boost in rabbits SM34 and SM35. B. Additional data showing increases in mean relative mRNA expression of i) B2M and ii) PAK1 in rabbits studied subsequently (31).
Immunization
Rabbits were immunized as described in (30) with one of two peptide immunogens synthesized on MAP-4 (Group 2 rabbits) or MAP-8 (Groups 1, 3, and 4) backbones (AnaSpec Inc., San Jose, CA). Rabbits were first injected s.c. with 0.5 mg MAP-peptide/0.5 ml BBS (pH8.0) + 0.5 ml CFA and then boosted at 3 week intervals with the same antigen in IFA. Peptide ‘SM’ is derived from a major antigenic region of the nuclear protein Sm B/B′ while peptide ‘GR’ corresponds to the known rabbit sequence of an extracellular epitope of the 2b subunit of neuronal postsynaptic N-methyl-D-aspartate (NMDA) receptors. Control rabbits were immunized with MAP-4 or MAP-8 backbone (BB) in identical adjuvants. Three rabbits (PB-45 through 47) that were identically handled and injected with PBS only and 14 unimmunized rabbits were also included in the microarray analysis. Sera for antibody assays and/or heparinized blood for RNA extraction were collected immediately before immunization (pre-immune) and 1 week after each boost (post-boost). Sera were stored at −20°C. Within three hours of bleeding, peripheral white blood cells (PWBC) were prepared and RNA extracted from them.
Serum autoantibodies to total extractable nuclear Ags (ENA) and to component Ags Sm, RNP, SS-A (Ro 60 and 52 kDa), SS-B (La), were assayed by ELISA using adaptations of commercially available human diagnostic kits (INOVA Diagnostics). Assays for autoantibodies to calf thymus dsDNA were adapted similarly using two different commercially available kits (Vidia, Vestec (Kit A); Zeus Scientific (Kit B)). Manufacturers' instructions were followed. Briefly, 100 μl of rabbit sera diluted 1/100 in the proprietary sample diluents were added to Ag-coated wells and incubated for 60 min at 37°C (Kit A) or 30 min at RT (Kit B). Wells were then washed, incubated for 60 min at 37°C (Kit A) or 30 min at RT (Kit B) with secondary Ab HRP-goat anti-rabbit IgG Fc (Jackson ImmunoResearch Laboratories), and developed with TMB for reading OD at 450 nm.
RNA extraction
For RNA extraction from PWBC, heparinized blood diluted 2:1 with warm 3% gelatin in PBS was incubated for 50 min at 37°C in 5% CO2. The upper layer was then removed and centrifuged at RT, 400 g for 12 min to pellet leukocytes. After discarding supernatants, leukocyte pellets were washed twice with PBS and centrifuged again for 15 min. The washed pellet was then lysed with TRIzol (Invitrogen, Carlsbad, CA) and total RNA was extracted using RNAeasy Mini columns following manufacturer's instructions (Qiagen, Valencia, CA).
Synthesis of cDNA and cRNA
The cRNA probes were prepared from mRNA using the Affymetrix gene chip eukaryotic small sample target labeling protocol assay version II (Affymetrix, Santa Clara, CA) using 2 cycles of cDNA synthesis and in vitro transcription (IVT) reactions. In the first round, cDNA was synthesized from 100 ng RNA (Superscript, Invitrogen) using a T7- Oligo (dT) promoter primer. Resulting cDNA was used in IVT reactions (T7 Megascript Ambion, Austin, Texas) to generate first cycle cRNA and cDNA synthesized from this cRNA was subsequently used in the second IVT cycle for obtaining biotinylated cRNA using CTP and UTP (EnzoBioarray, Enzo Life Sciences, Farmingdale, NY).
Microarray analysis
Labeled cRNA was hybridized to the Affymetrix U95A human microarray according to the manufacturer's recommended protocol. Non-normalized MAS5 signals were used to compare raw probeset intensity values between human and rabbit samples. Final rabbit study analyses were conducted with expression values summarized using dChip (33), log2 transformed and Loess normalized using an R package (http://www.elwood9.net/spike) (34). Genes with maximum expression values less than 7 for all samples were eliminated from analysis. Multi-way ANOVA along with specific contrasts, K-means clustering and gene expression visualizations were performed using Partek Genomics Suite (St. Louis, MO). Genes were identified as significantly modulated if the ANOVA contrast p-value was <0.05 and the absolute log2 fold change was >1.0.
Data were deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo).
The series entry (GSE23076) provides access to all data at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE23076
Pathway and network analyses
The gene sets were analyzed using DAVID (35) (http://david.abcc.ncifcrf.gov/knowledgebase/) and Ingenuity Pathways Analysis (IPA) (Ingenuity Systems, Mountain View, CA; www.ingenuity.com). IPA constructed hypothetical gene/protein interactions clusters between our set of differentially expressed genes and all other genes stored in the knowledge base after generating a set of networks having a network size of not more than 35 gene/proteins. IPA computes a score for each possible network according to the fit of that network to the supplied Focus Genes or the input genes. The score is derived from a p value that indicates the likelihood of Focus Genes being found together in a network due to random chance. A score >2 indicates >99% confidence that the Focus Gene network was not generated by chance alone. Biological functions are then assigned to each network.
Quantitative real time PCR (QRT-PCR)
For validation studies of the expression of genes of interest identified in the microarray studies, quantitative RT-PCR was performed using ABI Prism 7900. Total RNA extracted from rabbits' WBCs was converted to first strand cDNA. Briefly, 1 μg of total RNA was added to a 20-μL reaction mixture consisting of 0.5 μg random primers (Invitrogen, Carlsbad, CA) and RNase-free water. The reaction mixture was incubated at 65°C for 5 min and then quickly chilled on ice for 5 min. To the reaction mixture, 4 μL of 5× first strand buffer (Invitrogen, Carlsbad, CA), 1 μL of 10 mM dNTP mix (Invitrogen), and 1 μL of 0.1 M dithiothreitol (Invitrogen) was added and incubated at 25°C for 2 min. Following this one Unit of SuperScript II (Invitrogen,), was added and the reaction mixture was first incubated at 25°C for 10 min followed by 42°C for 50 min, and finally heated to 70 °C for 15 min, and then chilled on ice. Two units of DNase-free RNase H (Invitrogen) were added to the mixture that was incubated at 37°C for 20 min to remove the original RNA template. The RNase H was inactivated by heating the reaction mixture at 70 °C for 10 min. All cDNA samples were stored at −80°C until RT-PCR analyses were performed. RT-PCR analysis of mRNAs was performed on a 7900HT Sequence Detection System (Applied Biosystems). The cDNA synthesized from isolated PWBCs was directly used as template for real-time PCR by using TaqMan 2x PCR Master Mix Reagents Kit (Applied Biosystems). The primers, probes and PCR conditions used are shown in Table II. Each sample from three independent experiments was run in duplicate. The unit number showing relative mRNA levels in each sample was determined as a value of mRNA normalized against Peptidylprolyl isomerase A (PPIA). RT-PCR data were analyzed by using the 2−ΔΔCT method as described by Livak and Schmittgen (36). Based on its uniform expression among rabbit groups in the microarray analysis, rabbit peptidylprolyl isomerase A (PPIA; cyclophilin A) was selected as the housekeeping gene control and used for the calculation of ΔCT. Where rabbit sequences were unavailable, primers were designed after searching for rabbit sequences with corresponding human gene sequences in the database containing the trace archives of the whole genome shotgun sequence of the rabbit (Oryctolagus cuniculus) generated by the Broad Institute of MIT and Harvard University (NCBI trace archive: cross- species Megablast at http://www.ncbi.nlm.nih.gov/blast/tracemb.shtml) and in assemblies of rabbit scaffolds at Ensembl and UCSC (see links on the NCBI Rabbit Genome Resources site at: http://www.ncbi.nlm.nih.gov/projects/genome/guide/rabbit/.
Table II.
Primers and probes for quantitation of mRNA by Q-PCR for rabbit PPIA control, β2-microglobulin (B2M) and PAK1a
| PPIA control | |
| PPIA forward | 5′ CAACACAAATGGCTCCCAGTT 3′ |
| PPIA reverse | 5′ CATGGCTTCCACAATGCTCAT 3′ |
| PPIA probe | 5′ ATCTGCACTGCCAAGAC 3′ |
| Beta-2-microglobulin (B2M) | |
| B2M forward | 5′ TTGTTCCCCTGCCTGGAGT 3′ |
| B2M reverse | 5′TGGATGACGAGAGTACACTTGAACAT 3′ |
| B2M probe | 5′ CCAGCGTGCTCCG 3′ |
| PAK1 | |
| PAK1 forward | 5′ GAAGGCCAGATTGCAGCTGT 3′ |
| PAK1 reverse | 5′ CGAATGCAGAAACTCCAGAGC 3′ |
| PAK1 probe | 5′ AGGTCTGGGCGGATG 3′ |
The total volume of PCR reaction was 25 μl and the PCR conditions were as follows: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C, 15 s for denaturation and 60°C, 1 min for annealing and extension.
Results
Comparison of hybridization of rabbit and human cRNA to a human microarray
DNA arrays are currently available for only a limited number of species and to overcome this limitation, cross-species hybridization has been utilized as one potential solution. For example, given that only about 75% of a probe with a minimum 15 nucleotides of perfect match is needed to provide a microarray hybridization signal, a number of studies have shown successful results from hybridization of swine, bovine, canine and non-human primate RNA to human microarrays (37-43). These studies also demonstrated evolutionary conservation of basal gene expression levels within a given tissue. We conducted a controlled comparison of the hybridization of identically prepared cRNA from human and rabbit PWBC on the human Affymetrix U95A array. We hypothesized that the average probeset hybridization signal will be weaker for rabbit samples than for humans but for a large number of genes we expected individual probes within a probeset would show good hybridization signals given the level of sequence similarities between rabbit and human coding sequences (44). Comparisons of human vs rabbit in coding sequence alignments within ENCODE target regions permitted calculation of mean percent identity of rabbit and human coding sequences in these regions. Supplemental Figure 1 shows the distribution of percent identity values over all the genes within ENCODE target regions present in both rabbit and human and Supplemental Table 1 lists the data used to generate Fig. S1. Mean and median percent identities are 80% and 86% respectively. We thank Drs. E. H. Margulies and S. J. Parker for providing these analyses and calculations.
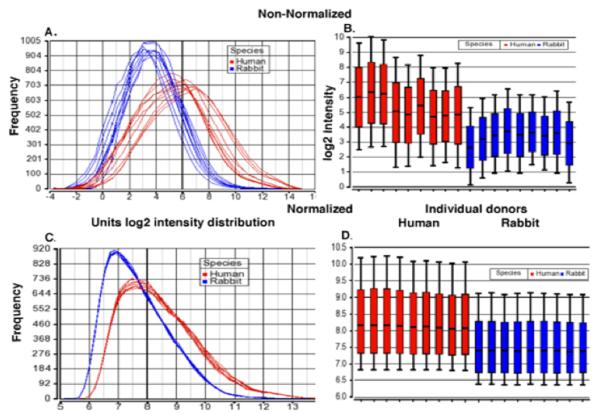
To help control for poor performing probes within a probeset, we used the dChip algorithm which gives greater weight to good performing probes when summarizing the probeset signal intensity. To test this hypothesis, we compared expression signals obtained from hybridizing Affymetrix U95A Arrays with RNA isolated from 9 different PWBC samples from rabbit and 9 from humans. Figs.1 A and B show that indeed the non-normalized MAS5 probeset intensity distribution is on average 1.7-fold (log2) dimmer for the rabbit samples than for the human samples, despite loading equal amounts of labeled cRNA onto each array. Following dChip signal summarization and within species normalization, the rabbit samples were 0.7-fold dimmer on average compared to the human samples (Figs. 1C and 1D) confirming our hypothesis that dChip should be able to better control for poor performing probes within a probeset. Confirmatory real time PCR analyses of other human microarray studies not presented here has shown a lack of detection or high false positive rates for genes expressed below a log2 signal intensity of 7 (unpublished results). Based on this result, when values below this level are considered as background, 85% of the human genes, on the U95A Array are detectably expressed in the human PWBC samples compared to about 65% of the rabbit PWBC genes (Supplemental Fig. 2A). As discussed above, previous studies have shown evolutionary conservation of gene expression levels within a given tissue. Absolute expression values for human and rabbit samples were visualized by K-means clustering (Supplemental Fig. 2B). Approximately 80% of genes had similar absolute expression values in both human and rabbits. This can be seen in Clusters 1, 4, 5, and 6 (Supplemental Figs. 2B and 2C) with >70% of the genes showing <2-fold average difference between the 9 human and 9 rabbit samples (data not shown). Clusters 2 and 3 indicate that 20% of the genes have substantially higher values in human samples compared to rabbit (Supplemental Figs. 2B and 2C), most likely due to poor hybridization of rabbit RNA to these human probesets or species-specific differences. Given that we are interested in immune-related gene expression patterns in the study of a SLE-rabbit model, the numbers of Gene Ontology “Defense Response” genes expressed above background (>7) were compared for the human and rabbit samples. Rabbit expresses >80% of the number of Defense Response genes found in human samples (Supplemental Fig. 2D). Taken together, these results give us confidence that we can use human microarrays to detect modulation of biological relevant pathways in our SLE-rabbit model.
FIGURE 1.
Comparison of distribution of gene expression as measured with rabbit and human cRNA probes binding to human microarrays before and after dChip and within species normalization. A. and B. Non-normalized MAS5 probe set intensity distribution for log transformed gene expression of RNA isolated from 9 different PWBC samples from rabbit (blue) and 9 from human (red) on the Affymetrix huU95A array. C. and D. Following dChip summarization and within species normalization, rabbit samples average 0.7 fold dimmer. In panels B. and D., the middle lines show the median, the boxes represent 25th and 75th percentile of the data distribution, and whiskers representing 10% and 90% of the data distribution.
Microarray analysis
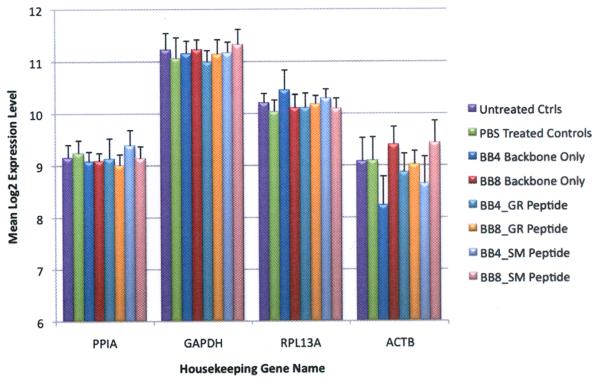
To further confirm the performance of the human arrays, we evaluated the consistency of the average expression levels of four housekeeping genes (ACTB, PPIA, RPL3A, GAPDH) across the eight different study groups listed in Fig. 2. As anticipated, these genes are well expressed and there was no significant gene expression difference between the groups for a given gene (ANOVA p>0.05).
FIGURE 2.
Average expression levels of housekeeping genes of the 8 rabbit groups studied showing consistency of the mean log2 expression levels of four housekeeping genes [PPIA (Peptidylprolyl isomerase A), GAPDH, RPL3a (ribosomal protein L13a), and ACTB]. Error bars show SEM.
The purpose of the present study was to investigate differential gene expression using our earlier described rabbit model (30-32). The rabbits chosen for this study were genetically related yet heterogeneous and therefore were more closely representative of human lupus patients than inbred mice. Immunization groups comprised rabbits immunized with one of two different peptide immunogens SM or GR synthesized on branched lysine MAP-8 (groups 1, 3, 4; 10 SM and 13 GR immunized rabbits) or MAP-4 BB (group 2; 5 SM and 6 GR immunized rabbits). Control groups included rabbits immunized with either MAP-8 or MAP-4 BB (6 and 4 rabbits respectively) without peptide. Fourteen unimmunized rabbits and three rabbits injected with PBS alone served as additional control groups.
Gene expression analyses distinguish rabbits immunized with Peptide-MAP-4, Peptide-MAP-8, MAP Backbone only, and unimmunized rabbits
Figure 3 shows peptide effects on gene expression in animals that received SM or GR on MAP-4 (A) or MAP-8 backbones (B). MAP-4 and MAP-8 backbones (BB4 or BB8) alone did not evoke striking alterations of gene expression. This is consistent with their not stimulating strong autoantibody responses to dsDNA and the other tested autoantigens (SM, RNP, SS-A and SS-B) post immunization (Table I). However, it was surprising to find that not only peptides but the nature of the MAP backbones (MAP-4 vs MAP-8) influenced the expression profiles of peptide-immunized rabbits. Immunizations with SM-and GR-peptides on MAP-4 backbone each upregulated sets of genes that differed from those genes overexpressed in rabbits immunized with the peptides on MAP-8 backbone.
FIGURE 3.
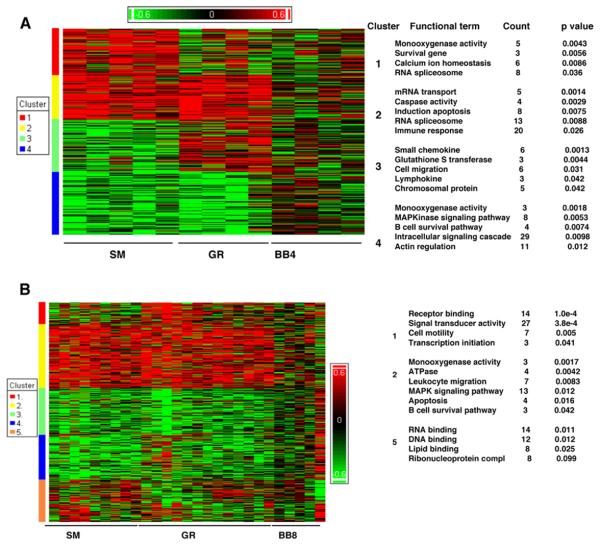
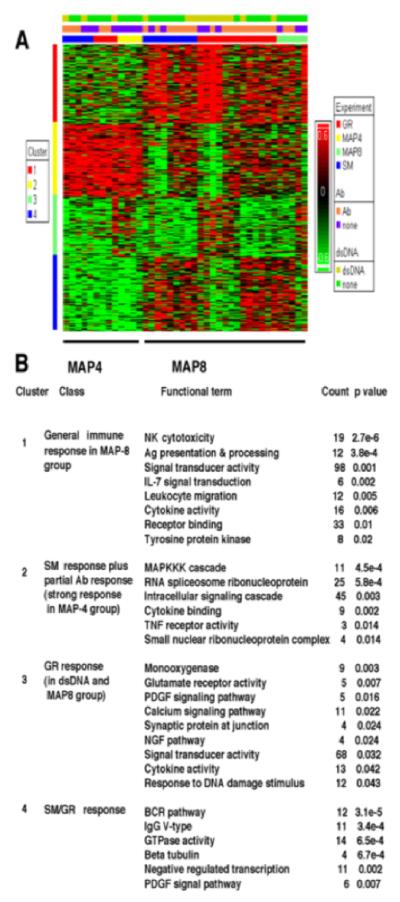
MAP-4 vs MAP-8 backbones (BB) affect gene expression patterns. At p < 0.05, and at 2≤ Filter cut-off (Fc) ≤2,768 genes passed the filter. Shades of red represent upregulation, shades of green downregulation and genes are clustered by similarity. The top functional categories for each of the clusters are indicated along the side of the figures. (A) Microarray analysis of the peptide effect of MAP-4 group reveals genes in cluster 1 showing over-representation in the SM group and genes in cluster 3 over-expressed in the GR group. Cluster 2 comprises genes, with a common elevated expression pattern in both SM and GR groups and cluster 4 genes have a common decreased expression pattern in both SM and GR groups. (B) Heat map depicting peptide effect in MAP-8 group includes 224 genes commonly overexpressed in SM and GR group. Count shows the number of different genes upregulated in expression associated with each Functional term within the cluster.
Gene expression patterns associate MAP-4 immunization with anti-ENA and MAP-8 with anti-dsDNA Ab responses
It was observed earlier (30) that immunization with MAP-4 peptide resulted in fewer anti-dsDNA positive (3 of 11) and more anti ENA (anti-Sm/RNP/SS-A/SS-B) positive responders (5 of 11) compared to MAP-8 peptide immunization where we observed higher and more consistent anti-dsDNA along with other autoantibody responses (7 of 13 each). When analyzed on the microarray, the gene expression data also reflected such a response pattern (Fig. 4). Gene expression assayed using the Affymetrix Human Genome U95A array identified 1418 differentially expressed genes. Samples were grouped by hierarchical clustering into four distinct clusters. Cluster 1 contains a set of 410 genes that were up-regulated in MAP-8 immunization groups and were involved in general immune responses including NK cytotoxicity, antigen presentation listed in Fig. 4B. This cluster was further analyzed using IPA to compare the differences in the gene expression among anti-dsDNA positive and negative rabbits. Cluster 2 indicates a set of 354 genes that were strongly upregulated in expression in the MAP4 group. This cluster contains 25 upregulated genes (p < 0.001) belonging to the RNA spliceosome ribonucleoprotein family. The genes that were upregulated were found to belong to pathways including MAP kinase, kinase, kinase (MAPKKK) cascade, RNA spliceosomal ribonucleoprotein, and intracellular signaling cascade (Fig. 4B). Cluster 3 highlights the gene expression pattern in anti-dsDNA positive MAP-8 rabbits, especially in the GR group although some BB-MAP-4 rabbits also have hit this cluster. The top functional categories of genes with up-regulated expression in cluster 3 were in the monooxygenase pathway, glutamate receptor activity, PDGF signaling pathway and others, particularly from the GR-MAP-8 group positive for anti-dsDNA. Upregulated genes in cluster 4 were mainly found among the SM/GR MAP-8 group and belonged to BCR pathway, IgG V-type, GTPase activity and others (Fig. 4B). The top pathway identified in both SM and GR-peptide responses (Cluster 4, Fig. 4) includes B-cell receptor (BCR) related genes.
FIGURE 4.
Clustering of differentially expressed genes in rabbits immunized with GR and SM peptides synthesized on MAP-4 (Group 2 rabbits) or MAP-8 (Groups 1, 3, and 4) backbones (BB) or with BB alone. Differences in relative levels of gene expression (Z-score) are indicated in color, where red indicates up-regulation and green indicates down-regulation relative to that of corresponding gene expression in controls. The legend on the right refers to the set of bars across the top. The lower bar indicates whether the MAP immunogen carried SM peptide (blue), GR peptide (red), only MAP-4 BB (bright yellow) or MAP-8 BB (pale green). The center bar indicates which rabbits made some detectable autoantibody (orange) and which did not (purple) and the upper bar which animals produced anti-dsDNA (chartreuse yellow) and which did not (dark green).
Distinctive gene expression patterns in dsDNA positive and anti- ENA positive rabbits
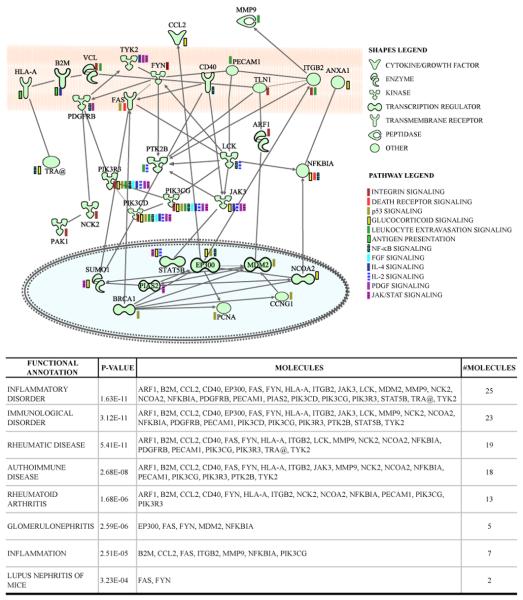
Those rabbits having anti-dsDNA Abs (SM-34, SM-35, GR-7, GR-8, GR-9, GR-10, GR-28, GR-36) and those having other autoantibodies including anti-Sm/RNP/SS-A/SS-B but negative for anti-dsDNA antibodies (SM-3, SM-6, SM-32, SM-33, GR-37, GR-38, GR-39, GR-40) (Table I) were compared using Ingenuity Pathways Analysis (IPA) in an attempt to understand the genetic basis for variation in the autoantibody profile. In order to remove the BB variable, only peptide-MAP-8 immunized rabbits of groups 1, 3 and 4 from cluster 1, Fig. 4 and not MAP-4 rabbits were included. Upon analysis, several pathways including NF-kB signaling, Death receptor signaling, Integrin signaling, Antigen presentation pathway, Leukocyte extravasation signaling, Endoplasmic reticulum stress pathway, IL-2 signalling, JAK/STAT signaling, IL-4 signaling, PDGF, Glucocorticoid receptor, p53 signaling, FGF signaling in particular, were seen to be up-regulated in the anti-dsDNA positive group as compared to the anti-dsDNA negative group.
Genes from the above pathways were selected for further analysis using IPA to identify direct interactions among these genes. Some or all genes from each of the selected pathways showed interaction with each other (Fig. 5 top). The only exception was Endoplasmic reticulum stress pathway genes (CASP9, ERN1, MBTPS1) that did not show direct relationship with the other genes and they do not appear in the interactive pathway. Fig. 5 bottom tabulates functional annotation by IPA of the disease conditions associated with these genes. IPA identifies twenty-five of these genes as involved in inflammatory processes, and subsets of these are listed in the tabulation of immunological disorders including Rheumatic Disease, Autoimmune Disease, Rheumatoid Arthritis, Glomerulonephritis, and Lupus Nephritis of Mice.
FIGURE 5.
Interactive pathway network of upregulated genes in anti-dsDNA positive rabbits. The shapes legend classifies the proteins found as transmembrane receptors, cytokines/growth factors, kinases, peptidases, other enzymes and transcriptional regulators. The pathway legend identifies of genes that were common to the listed pathways that were upregulated in the anti-dsDNA positive rabbits. The connecting lines indicate direct interactions among the products of these genes.
Validation by real time PCR
Two genes B2M (β-2-microglobulin) and PAK1 [p21-protein (Cdc42/Rac)-activated kinase 1] were selected on the basis of availability of primer sequences for validation of microarray data using quantitative real time PCR analyses of PWBC RNA. Rabbits SM 34 and 35, that both produced anti-dsDNA as well as anti-ENA, ANA and other autoantibodies (Group 4 Table 1), were chosen for validation on the basis of RNA sample availability. Fig. 6A shows increase in mRNA expression of B2M and PAK1 after the 5th boost in rabbits SM-34 and 35. Fig. 6B shows additional data from PWBC RNA prepared in an identical manner from descendants of high-responders of the earlier groups (Group 5) GR-immunized rabbits in which high incidence of autoantibody responses were found after the microarray project was completed (31). Elevated levels of expressed B2M were found in five and PAK1expression was elevated in three of the nine tested rabbits after the fifth boost.
Discussion
The gene expression study reported here was conducted to expand understanding of SLE and in particular NPSLE using a rabbit model (30-32). We reported earlier that immunization of rabbits with the SM- or GR-MAP peptides led to development of anti-nuclear autoantibodies, including anti-dsDNA, as well as neurological symptoms in the form of seizures and nystagmus in some rabbits (30). After establishing that it was possible to use the Affymetrix U95 human microarray for the rabbit gene expression studies, through comparative hybridization of identically prepared cRNA from human and rabbit PWBC, the human microarray was used due to lack of rabbit-specific microarrays.
The observation that the autoantibody responses differ depending on the backbone (BB) of the MAP conjugate used (MAP-4 or MAP-8) was unexpected. After our laboratory had good success in eliciting anti-peptide antibodies on MAP-4 backbones (45) we chose MAP-4 for immunization of Group 2 rabbits but switched back to using MAP-8 for the subsequent immunizations (Groups 3, 4 and in references (31, 32) Groups 5 and 6 because of our initial observations of the different effect of MAP-4 and MAP-8 BB on the autoantibody responses. The MAP component was expected to simply augment the Ab response and not evoke an Ab response itself (46). However, 7 of 13 rabbits of Groups 1 and 3 receiving MAP-8 peptide produced elevated anti-dsDNA levels compared with only 3 of 11of the Group 2 recipients of MAP-4 peptides and preliminary DNA microarray analyses of groups 1-3 provided additional evidence suggesting that MAP carrier influences the elicited response (30). In the original groups 1-4 (30), anti-peptide responses generally peaked by 49 days and anti-dsDNA by 70 days (after 3rd boost) with some animals responding only after the 5th to 7th boosts. The selected rabbits in the pedigree used in group 5 (31), generally developed elevated ANA above pre-immune (by 3rd boost) earlier than anti-dsDNA (5th to 7th boost).
There are reports that Toll-like receptor signaling may serve as a factor that redirects from DNA (anti-dsDNA Abs) to RNA associated (anti-Sm, -RNP, -SS-A, -SS-B etc) patterns of autoantibody production by B cells. Pisitkun et al (2006) (47), have shown that a duplication of the TLR7 gene and resultant doubling of the TLR7 gene dosage in B cells in conjunction with the lupus allele of FcγRIIB, results in increased responsiveness of B cells to TLR7 resulting in a shift in pattern of autoantibody responses.
Although we did not detect antibodies to the MAP BB itself by our ELISA measurements, the current gene expression data including all rabbits in groups 1-4, suggest that whereas the BB alone did not induce unique changes in the gene expression pattern, immunizations with the same peptide on two different BB did lead to some distinct patterns of clustering (Figs. 3A, B). The BB-specific alterations may have been generated due to the differences in the physical structure of MAP-4 and MAP-8. Structurally, the MAP core matrix consists of heptalysine containing eight dendritic arms (MAP-8) or trilysine containing four arms (MAP-4) to which peptides are bound (46).
It is possible that the branched MAP-8 core with more branches could more strongly stimulate the immune system of the rabbits to generate anti-dsDNA Abs. Different amounts of peptide attached per core molecule and their presentation may also be affecting the Ab response pattern. In addition, it was observed that the gene expression profile of Group 2 rabbits, which received peptide on MAP-4 BB, showed 25 genes belonging to RNA spliceosomal ribonucleoprotein family (Fig. 4, cluster 2). This correlates with the serological responses of this group that tended towards production of antibodies to other auto-antigens related to the RNA spliceosomal ribonucleoprotein family (Sm, RNP, SS-A, SS-B). MAP-8 peptide immunizations are used by several laboratories in studies of animal models of SLE (48, 49). It would be of interest to know whether any differences in response patterns would be observed in other species if MAP-4 was substituted for MAP-8.
The GR peptide as discussed earlier is derived from the NMDA glutamate receptor. The GR-MAP-8 rabbits, which were anti-dsDNA positive, upregulated expression of 5 genes related to glutamate receptor activity (Fig. 4, cluster 3). This observation is consonant with reports that a pathogenic mouse anti-dsDNA Ab that bound kidney tissue recognized a peptide sequence found within NMDA NR2b and that some murine and human anti-dsDNA Abs bound the peptide or the receptor itself (49-50). Cross reactivity of some anti-dsDNA autoantibodies with NMDA glutamate receptor NR2b led to suggestions that such cross reactive anti-dsDNA play a role in neuropsychiatric manifestations of SLE (reviewed in 51), may cause brain pathology, and cognitive dysfunction (52). Finally, in some SLE patients showing signs of NPSLE, serum Abs that bind NR2a and NR2b receptors were found (53, 54). Our model that elicits autoantibodies in rabbits after immunization with a peptide from the NMDA glutamate receptor can provide a means for development of further insights into NPSLE and pursuit of therapeutic targets.
General immune function genes related to NK cytotoxicity, antigen presentation and processing, and BCR genes (Fig. 4, clusters 1 and 4, respectively) are dysregulated in SM and GR rabbits. Whereas it is well established that B cells, being a source of characteristic antinuclear autoantibodies, play a crucial role in the pathogenesis of SLE, impaired natural killer cell cytotoxicity has recently been reported in SLE (55, 56). It has also been documented that alterations in B-cell regulation are responsible for B-cell hyperactivity as seen in SLE (57).
Comparisons of anti-dsDNA positive rabbits with those only positive for other autoantibodies have indicated variation in the gene expression with differentially regulated pathways in both the groups. Recent reports from human studies have also directly or indirectly indicated upregulation of many of these pathways including NF-kB (58); death receptor signaling (59); integrin signaling (60); IL-4 signaling (61), PDGF signaling (62); and Glucocorticoid receptor signaling (63).
There were distinct direct or indirect interactions among the genes belonging to these different pathways (Fig. 5) suggesting that a select set of genes when dysregulated, can result in several distressed physiological pathways. The observation that dysregulation of such pathways is associated with high anti-dsDNA Ab responses in the rabbits underlines the fact that anti-dsDNA Abs are the hallmarks of lupus pathogenesis. No evidence of kidney damage or pathology was seen in the first four groups studied here by cross-species microarrays. This was also true of the 5th group (31) included in qPCR analyses (Fig.6) as well as the 6th group (32). In the 5th and 6th groups, hematological profiles suggested development of chronic inflammatory responses. This may have been due to selectively bred animals developing greater responses to exposure to Freund's complete and incomplete adjuvant. In addition, generalized anti-dsDNA Abs may generate inflammation, damage of multiple organs due to their ability to deposit in tissues or when present in immune complexes, to activate inflammatory cells (64). The observed gene interactions offer insight into how multiple processes may be affected as a result of anti-dsDNA Abs. Even though for the Endoplasmic reticulum stress pathway genes (CASP9, ERN1, MBTPS1), IPA did not show direct relationships with the other genes, and they do not appear in the interactive pathway (Fig. 5), Caspase 9 is a CASP9 encoded apoptosis-related cysteine protease and required for neurological development. Caspase 9 activation via APAF1 is one of the earliest in the caspase activation cascade and may affect neuronal death in Alzheimer's patients (65).
The genes in the interactive diagram generated using the knowledge base of IPA analysis tool (Fig. 5) were found to be associated with inflammation and several pathological conditions including autoimmune disease, immunological disorder, and glomerulonephritis, which further supports the role of anti-dsDNA Abs in various clinical manifestations in lupus. One of the goals of our first paper (30) was to determine whether we could confirm and extend the early report of SLE-like serology development after immunization of rabbits with peptide SM on MAP-8 (48). The original study included rabbits immunized with negative control peptides or Freund's adjuvant only (48). At the start of this work, we did not know whether peptide GR would serve as a negative control or induce autoantibodies. For our expression analyses we included unimmunized and PBS only treated animals for comparison with immunized rabbits.
Potential roles of exposure to environmental pathogens as well to Freund's adjuvant have already been discussed in (31). By this stage in development of the 5th group of pedigreed rabbits, we suggested that continued breeding to obtain offspring of selected responder parents could produce “rabbit models with spontaneous occurrence of SLE-like serology and disease phenotypes” (31). In the 6th group, controls immunized with Complete Freund's adjuvant followed by incomplete were included (32). The trend toward more consistent autoantibody production (31) continued and one of 7 adjuvant-only recipients produced increased anti-dsDNA above pre-immune background after the 5th boost. This and two other adjuvant-only recipients also expressed elevated B2M mRNA measured by qPCR (32).
The model developed in pedigreed and non-inbred rabbits remains a promising one for further genetic investigations targeted to understanding NPSLE. The differential gene expression results we report resemble those found in human SLE. The observations in the rabbits studied that differential gene expression is at the core of their different autoantibody responses, suggest that this should be further investigated in subsets of SLE patients with different autoantibody profiles.
Supplementary Material
Acknowledgements
We thank Cornelius Alexander and Barbara Newman, for technical advice and assistance, Michael Mage, Rajesh Kumar and Nicholas Beckloff for helpful comments about the manuscript, E. H. Margulies and S.J. Parker for providing the sequence analyses and calculations shown in supplemental Table 1 and Fig. S1, Mariame Quinones for advice about IPA analysis and presentation of results in Fig. 5, Les Klimczak for statistical advice, and Shirley Starnes for expert Editorial Assistance.
Abbreviations used in this paper
- ANA
anti-nuclear antibody
- BB
backbone
- B2M
β2-microglobulin
- ENA
extractable nuclear antigens
- IFN
Interferon
- IPA
Ingenuity Pathways Analysis
- MAP
multiple antigen peptide
- NMDA
N-Methyl-D-Aspartate
- NPSLE
neuropsychiatric systemic lupus erythematosus
- PAK1
p21-protein (Cdc42/Rac)-activated kinase 1
- PID
percentage identity
- PWBC
peripheral white blood cells
- SLE
systemic lupus erythematosus
Footnotes
This research was supported by the intramural research program of the NIH, NIAID (http://www3.niaid.nih.gov/) and in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U. S. Government.
References
- 1.Moser KL, Kelly JA, Lessard CJ, Harley JB. Recent insights into the genetic basis of systemic lupus erythematosus. Genes Immun. 2009;10:373–379. doi: 10.1038/gene.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan Y, Sawalha AH. Epigenetic regulation and the pathogenesis of systemic lupus erythematosus. Transl. Res. 2009;153:4–10. doi: 10.1016/j.trsl.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Jönsen A, Bengtsson AA, Nived O, Truedsson L, Sturfelt G. Gene-environment interactions in the aetiology of systemic lupus erythematosus. Autoimmunity. 2007;40:613–617. doi: 10.1080/08916930701511051. [DOI] [PubMed] [Google Scholar]
- 4.Cohen-Solal JF, Jeganathan V, Hill L, Kawabata D, Rodriguez-Pinto D, Grimaldi C, Diamond B. Hormonal regulation of B-cell function and systemic lupus erythematosus. Lupus. 2008;17:528–532. doi: 10.1177/0961203308089402. [DOI] [PubMed] [Google Scholar]
- 5.Werth VP. Cutaneous lupus: insights into pathogenesis and disease classification. Bull. NYU Hosp. Jt. Dis. 2007;65:200–204. [PubMed] [Google Scholar]
- 6.Grossman JM. Lupus arthritis. Best Practice & Research Clinical Rheumatology. 2009;23:495–506. doi: 10.1016/j.berh.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Sitia S, Atzeni F, Sarzi-Puttini P, Di Bello V, Tomasoni L, Delfino L, Antonini-Canterin F, Di Salvo G, De Gennaro Colonna V, La Carrubba S, Carerj S, Turiel M. Cardiovascular involvement in systemic autoimmune diseases. Autoimmun. Rev. 8:281–286. doi: 10.1016/j.autrev.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Seshan SV, Jennette JC. Renal disease in systemic lupus erythematosus with emphasis on classification of lupus glomerulonephritis: advances and implications. Arch. Pathol. Lab. Med. 2009;133:233–248. doi: 10.5858/133.2.233. [DOI] [PubMed] [Google Scholar]
- 9.Efthimiou P, Blanco M. Pathogenesis of neuropsychiatric systemic lupus erythematosus and potential biomarkers. Mod. Rheumatol. 2009;19:457–468. doi: 10.1007/s10165-009-0198-5. [DOI] [PubMed] [Google Scholar]
- 10.McCarty GA, Rice JR, Bembe ML, Pisetsky DS. Independent expression of autoantibodies in systemic lupus erythematosus. J. Rheumatol. 1982;9:691–695. [PubMed] [Google Scholar]
- 11.Hahn BH. Antibodies to DNA. N. Engl. J. Med. 1998;338:1359–1368. doi: 10.1056/NEJM199805073381906. [DOI] [PubMed] [Google Scholar]
- 12.Zhu J, Mohan C. SLE 1, 2, 3…genetic dissection of lupus. Adv. Exp. Med. Biol. 2007;601:85–95. doi: 10.1007/978-0-387-72005-0_9. [DOI] [PubMed] [Google Scholar]
- 13.Tsao BP. An update on genetic studies of systemic lupus erythematosus. Curr. Rheumatol. Rep. 2002;4:359–367. doi: 10.1007/s11926-002-0046-5. [DOI] [PubMed] [Google Scholar]
- 14.Santiago-Raber ML, Laporte C, Reininger L, Izui S. Genetic basis of murine lupus. Autoimmun. Rev. 2004;3:33–39. doi: 10.1016/S1568-9972(03)00062-4. [DOI] [PubMed] [Google Scholar]
- 15.Vinuesa CG, Goodnow CC. Illuminating autoimmune regulators through controlled variation of the mouse genome sequence. Immunity. 2004;20:669–679. doi: 10.1016/j.immuni.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 16.Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat. Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N. Engl. J. Med. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 18.Nath SK, Han S, Kim-Howard X, Kelly JA, Viswanathan P, Gilkeson GS, et al. A nonsynonymous functional variant in integrin-alpha(M) (encoded by ITGAM) is associated with systemic lupus erythematosus. Nat. Genet. 2008;40:152–154. doi: 10.1038/ng.71. [DOI] [PubMed] [Google Scholar]
- 19.Kozyrev SV, Abelson AK, Wojcik J, Zaghlool A, Linga Reddy MV, Sanchez E, et al. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat. Genet. 2008;40:211–216. doi: 10.1038/ng.79. [DOI] [PubMed] [Google Scholar]
- 20.Rus V, Chen H, Zernetkina V, Magder LS, Mathai S, Hochberg MC, Via C. Gene expression profiling in peripheral blood mononuclear cells from lupus patients with active and inactive disease. Clin. Immunol. 2004;112:231–234. doi: 10.1016/j.clim.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 21.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, Shark KB, Grande WJ, Hughes KM, Kapur V, Gregersen PK, Behrens TW. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc. Natl. Acad. Sci. USA. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J, Pascual V. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J. Exp. Med. 2003;197:711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee PY, Li Y, Richards HB, Chan FS, Zhuang H, Narain S, Butfiloski EJ, Sobel ES, Reeves WH, Segal MS. Type I interferon as a novel risk factor for endothelial progenitor cell depletion and endothelial dysfunction in systemic lupus erythematosus. Arthritis Rheum. 2007;56:3759–3769. doi: 10.1002/art.23035. [DOI] [PubMed] [Google Scholar]
- 24.Abbas AR, Wolslegel K, Seshasayee D, Modrusan Z, Clark HF. Deconvolution of blood microarray data identifies cellular activation patterns in systemic lupus erythematosus. PLoS One. 2009;4:e6098. doi: 10.1371/journal.pone.0006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maas K, Chan S, Parker J, Slater A, Moore J, Olsen N, Aune TM. Cutting edge:molecular portrait of human autoimmune disease. J. Immunol. 2002;169:5–9. doi: 10.4049/jimmunol.169.1.5. [DOI] [PubMed] [Google Scholar]
- 26.Chaussabel D, Quinn C, Shen J, Patel P, Glaser C, Baldwin N, Stichweh D, Blankenship D, Li L, Munagala I, Bennett L, Allantaz F, Mejias A, Ardura M, Kaizer E, Monnet L, Allman W, Randall H, Johnson D, Lanier A, Punaro M, Wittkowski KM, White P, Fay J, Klintmalm G, Ramilo O, Palucka AK, Banchereau J, Pascual V. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qing X, Putterman C. Gene expression profiling in the study of the pathogenesis of systemic lupus erythematosus. Autoimmun. Rev. 2004;3:505–509. doi: 10.1016/j.autrev.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Yao Y, Richman L, Higgs B,W, Morehouse C,A, de los Reyes M, Brohawn P, Zhang J, White B, Coyle AJ, Kiener PA, Jallal B. Neutralization of interferon-alpha/beta-inducible genes and downstream effect in a phase I trial of an anti-interferon-alpha monoclonal antibody in systemic lupus erythematosus. Arthritis Rheum. 2009;60:1785–1796. doi: 10.1002/art.24557. [DOI] [PubMed] [Google Scholar]
- 29.Bauer JW, Petri M, Batliwalla FM, Koeuth T, Wilson J, Slattery C, Panoskaltsis-Mortari A, Gregersen PK, Behrens TW, Baechler EC. Interferon-regulated chemokines as biomarkers of systemic lupus erythematosus disease activity: a validation study. Arthritis Rheum. 2009;60:3098–4107. doi: 10.1002/art.24803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rai G, Ray S, Shaw RE, Degrange PF, Mage RG, Newman BA. Models of systemic lupus erythematosus: development of autoimmunity following peptide immunizations of noninbred pedigreed rabbits. J. Immunol. 2006;176:660–667. doi: 10.4049/jimmunol.176.1.660. [DOI] [PubMed] [Google Scholar]
- 31.Puliyath N, Ray S, Milton J, Mage RG. Genetic contributions to the autoantibody profile in a rabbit model of systemic lupus erythematosus (SLE) Vet. Immunol. Immunopathol. 2008;125:251–267. doi: 10.1016/j.vetimm.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Pospisil R, Ray S, Milton J, Mage RG. Investigations of a rabbit (Oryctolagus cuniculus) model of systemic lupus erythematosus (SLE), BAFF and its receptors. PLoS One. 2009;4:e8494. doi: 10.1371/journal.pone.0008494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choe SE, Boutros M, Michelson AM, Church GM, Halfon MS. Preferred analysis methods for Affymetrix GeneChips revealed by a wholly defined control dataset. Genome Biol. 2005:R16. doi: 10.1186/gb-2005-6-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biology. 2003;4:R60. [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Robert C, Hue I, McGraw S, Gagne D, Sirard MA. Quantification of cyclin B1 and p34(cdc2) in bovine cumulus-oocyte complexes and expression mapping of genes involved in the cell cycle by complementary DNA macroarrays. Biol. Reprod. 2002;67:1456–1464. doi: 10.1095/biolreprod.102.002147. [DOI] [PubMed] [Google Scholar]
- 38.Grigoryev DN, Ma SF, Simon BA, Irizarry RA, Ye SQ, Garcia JG. In vitro identification and in silico utilization of interspecies sequence similarities using GeneChip technology. BMC Genomics. 2005;6:62. doi: 10.1186/1471-2164-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moody DE, Zou Z, McIntyre L. Cross-species hybridisation of pig RNA to human nylon microarrays. BMC Genomics. 2002;3:27. doi: 10.1186/1471-2164-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shah G, Azizian M, Bruch D, Mehta R, Kittur D. Cross-species comparison of gene expression between human and porcine tissue, using single microarray platform – preliminary results. Clin. Transplant. 2004;18(Suppl 12):76–80. doi: 10.1111/j.1399-0012.2004.00223.x. [DOI] [PubMed] [Google Scholar]
- 41.Caceres M, Lachuer J, Zapala MA, Redmond JC, Kudo L, Geschwind DH, Lockhart DJ, Preuss TM, Barlow C. Elevated gene expression levels distinguish human from non-human primate brains. Proc. Natl. Acad. Sci. USA. 2003;100:13030–13035. doi: 10.1073/pnas.2135499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gilad Y, Rifkin SA, Bertone P, Gerstein M, White KP. Multi-species microarrays reveal the effect of sequence divergence on gene expression profiles. Genome Res. 2005;15:674–680. doi: 10.1101/gr.3335705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ji W, Zhou W, Gregg K, Yu N, Davis S. A method for cross-species gene expression analysis with high-density oligonucleotide arrays. Nucleic Acids Res. 2004;32:e93. doi: 10.1093/nar/gnh084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Margulies EH, Cooper GM, Asimenos G, Thomas DJ, Dewey CN, Siepel A, Birney E, Keefe D, Schwartz AS, Hou M, Taylor J, Nikolaev S, Montoya-Burgos JI, Löytynoja A, Whelan S, Pardi F, Massingham T, Brow JB, Bickel P, Holmes I, Mullikin JC, Ureta-Vidal A, Paten B, Stone EA, Rosenbloom KR, Kent WJ, Bouffard GG, Guan X, Hansen NF, Idol JR, Maduro VV, Maskeri B, McDowell JC, Park M, Thomas PJ, Young AC, Blakesley RW, Muzny DM, Sodergren E, Wheeler DA, Worley KC, Jiang H, Weinstock GM, Gibbs RA, Graves T, Fulton R, Mardis ER, Wilson RK, Clamp M, Cuff J, Gnerre S, Jaffe DB, Chang JL, Lindblad-Toh K, Lander ES, Hinrichs A, Trumbower H, Clawson H, Zweig A, Kuhn RM, Barber G, Harte R, Karolchik D, Field MA, Moore RA, Matthewson CA, Schein JE, Marra MA, Antonarakis SE, Batzoglou S, Goldman N, Hardison R, Haussler D, Miller W, Pachter L, Green ED, Sidow A. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 2007;17:760–774. doi: 10.1101/gr.6034307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang G, Obiakor H, Sinha RK, Newman BA, Hood BL, Conrads TP, Veenstra TD, Mage RG. Activation-induced deaminase cloning, localization, and protein extraction from young VH-mutant rabbit appendix. Proc. Natl. Acad. Sci. USA. 2005;102:17083–17088. doi: 10.1073/pnas.0501338102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tam JP. Synthetic peptide vaccine design: synthesis and properties of a high-density multiple antigenic peptide system. Proc. Natl. Acad. Sci. USA. 1988;85:5409–5413. doi: 10.1073/pnas.85.15.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pisitkun P, Deane JA, Difilippantonio MJ, Tarasenko T, Satterthwaite AB, Bolland S. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 2006;312:1669–1672. doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- 48.James JA, Gross T, Scofield RH, Harley JB. Immunoglobulin epitope spreading and autoimmune disease after peptide immunization: Sm B/B′-derived PPPGMRPP and PPPGIRGP induce spliceosome autoimmunity. J. Exp. Med. 1995;181:453–461. doi: 10.1084/jem.181.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gaynor B, Putterman C, Valadon P, Spatz L, Scharff MD, Diamond B. Peptide inhibition of glomerular deposition of an anti-DNA antibody. Proc. Natl. Acad. Sci. USA. 1997;94:1955–1960. doi: 10.1073/pnas.94.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DeGiorgio LA, Konstantinov KN, Lee SC, Hardin JA, Volpe BT, Diamond B. A subset of lupus anti-DNA antibodies cross-reacts with the NR2 glutamate receptor in systemic lupus erythematosus. Nat. Med. 2001;7:1189–1193. doi: 10.1038/nm1101-1189. [DOI] [PubMed] [Google Scholar]; Sharma A, Isenberg D, Diamond B. Studies of human polyclonal and monoclonal antibodies binding to lupus autoantigens and cross-reactive antigens. Rheumatology. 2003;42:453–460. doi: 10.1093/rheumatology/keg161. [DOI] [PubMed] [Google Scholar]
- 51.Diamond B, Kowal C, Huerta PT, Aranow C, Mackay M, DeGiorgio LA, Lee J, Triantafyllopoulou A, Cohen-Solal J, Volpe BT. Immunity and acquired alterations in cognition and emotion: lessons from SLE. Adv. Immunol. 2006;89:289–320. doi: 10.1016/S0065-2776(05)89007-8. [DOI] [PubMed] [Google Scholar]
- 52.Kowal C, DeGiorgio LA, Nakaoka T, Hetherington H, Huerta PT, Diamond B, Volpe BT. Cognition and immunity: antibody impairs memory. Immunity. 2004;21:179–188. doi: 10.1016/j.immuni.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 53.Husebye ES, Sthoeger ZM, Dayan M, Zinger H, Elbirt D, Levite M, Mozes E. Autoantibodies to a NR2A peptide of the glutamate/NMDA receptor in sera of patients with systemic lupus erythematosus. Ann. Rheum. Dis. 2005;64:1210–1213. doi: 10.1136/ard.2004.029280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Omdal R, Brokstad K, Waterloo K, Koldingsnes W, Jonsson R, Mellgren SI. Neuropsychiatric disturbances in SLE are associated with antibodies against NMDA receptors. Eur. J. Neurol. 2005;12:392–398. doi: 10.1111/j.1468-1331.2004.00976.x. [DOI] [PubMed] [Google Scholar]
- 55.Park YW, Kee SJ, Cho YN, Lee EH, Lee HY, Kim EM, Shin MH, Park JJ, Kim TJ, Lee SS, Yoo DH, Kang HS. Impaired differentiation and cytotoxicity of natural killer cells in systemic lupus erythematosus. Arthritis Rheum. 2009;60:1753–1763. doi: 10.1002/art.24556. [DOI] [PubMed] [Google Scholar]
- 56.Green MR, Kennell AS, Larche MJ, Seifert MH, Isenberg DA, Salaman MR. Natural killer cell activity in families of patients with systemic lupus erythematosus: demonstration of a killing defect in patients. Clin. Exp. Immunol. 2005;141:165–173. doi: 10.1111/j.1365-2249.2005.02822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dolff S, Abdulahad WH, Bijl M, Kallenberg CG. Regulators of B-cell activity in SLE: a better target for treatment than B-cell depletion? Lupus. 2009;18:575–580. doi: 10.1177/0961203309102296. [DOI] [PubMed] [Google Scholar]
- 58.Okamoto T. NF-kappaB and rheumatic diseases. Endocr. Metab. Immune Disord. Drug Targets. 2006;6:359–372. doi: 10.2174/187153006779025685. [DOI] [PubMed] [Google Scholar]
- 59.Pitidhammabhorn D, Kantachuvesiri S, Totemchokchyakarn K, Kitiyanant Y, Ubol S. Partial construction of apoptotic pathway in PBMC obtained from active SLE patients and the significance of plasma TNF-alpha on this pathway. Clin. Rheumatol. 2006;25:705–714. doi: 10.1007/s10067-005-0162-5. [DOI] [PubMed] [Google Scholar]
- 60.Nakayamada S, Saito K, Nakano K, Tanaka Y. Activation signal transduction by beta1 integrin in T cells from patients with systemic lupus erythematosus. Arthritis Rheum. 2007;56:1559–1568. doi: 10.1002/art.22581. [DOI] [PubMed] [Google Scholar]
- 61.Lit LC, Wong CK, Tam LS, Li EK, Lam CW. Raised plasma concentration and ex vivo production of inflammatory chemokines in patients with systemic lupus erythematosus. Ann. Rheum. Dis. 2006;65:209–215. doi: 10.1136/ard.2005.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matsuda M, Shikata K, Makino H, Sugimoto H, Ota K, Akiyama K, Hirata K, Ota Z. Gene expression of PDGF and PDGF receptor in various forms of glomerulonephritis. Am. J. Nephrol. 1997;17:25–31. doi: 10.1159/000169067. [DOI] [PubMed] [Google Scholar]
- 63.Spies CM, Schaumann DHS, Berki T, Mayer K, Jakstadt M, Huscher D, Wunder C, Burmester G-R, Radbruch A, Lauster R, Scheffold A, Buttgereit F. Membrane glucocorticoid receptors are down regulated by glucocorticoids in patients with systemic lupus erythematosus and use a caveolin-1- independent expression pathway. Ann. Rheum. Dis. 2006;65:1139–1146. doi: 10.1136/ard.2005.048272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schiffer LE, Hussain N, Wang X, Huang W, Sinha J, Ramanujam M, Davidson A. Lowering anti-dsDNA antibodies--what's new? Lupus. 2002;11:885–894. doi: 10.1191/0961203302lu311rr. [DOI] [PubMed] [Google Scholar]
- 65.Engidawork E, Gulesserian T, Yoo BC, Cairns N, Lubec G. Alteration of caspases and apoptosis-related proteins in brains of patients with Alzheimer's disease. Biochem. Biophys. Res. Commun. 2001;281:84–93. doi: 10.1006/bbrc.2001.4306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.