Abstract
Essential hypertension is a complex disorder, caused by the interplay between many genetic variants, gene-gene interactions, and environmental factors. Given that the renin-angiotensin system (RAS) plays an important role in blood pressure (BP) control, cardiovascular regulation, and cardiovascular remodeling, special attention has been devoted to the investigation of single-nucleotide polymorphisms (SNP) harbored in RAS genes that may be associated with hypertension and cardiovascular disease. MicroRNAs (miRNAs) are a family of small, ∼21-nucleotide long, and nonprotein-coding RNAs that recognize target mRNAs through partial complementary elements in the 3′-untranslated region (3′-UTR) of mRNAs and inhibit gene expression by targeting mRNAs for translational repression or destabilization. Since miRNA SNPs (miRSNPs) can create, destroy, or modify miRNA binding sites, this review focuses on the hypothesis that transcribed target SNPs harbored in RAS mRNAs, that alter miRNA gene regulation and consequently protein expression, may contribute to cardiovascular disease susceptibility.
1. Introduction
Identifying the genes and mutations that contribute to disease is a central aim in human genetics. Single nucleotide polymorphisms (SNPs) are mutations that occur at genome positions at which there are two distinct nucleotide residues (alleles) that each appear in a significant portion (i.e., a minor allele frequency greater than 1%) of the human population [1]. There are some estimated 14 million SNPs [2] in the human genome that occur at a frequency of approximately one in 1,200–1,500 bp [3]. SNPs can affect protein function by changing the amino acid sequences (nonsynonymous SNP) or by perturbing their regulation (e.g., affecting promoter activity [4], splicing process [5], and DNA and pre-mRNA conformation). When SNPs occur in 3′-UTRs, they may interfere with mRNA stability and translation by altering polyadenylation and protein/mRNA regulatory interactions. Recently, a new layer of posttranscriptional miRNA-mediated gene regulation has been discovered and shown to control the expression levels of a large proportion of genes (reviewed in [6]). Importantly, SNPs in microRNA (miRNA) target sites (miRSNPs) represent a specific class of regulatory polymorphisms in the 3′-UTR that may lead to the dysregulation of posttranscriptional gene expression. Thus, for miRNAs acting by this mechanism, the miRSNPs may lead to heritable variations in gene expression. Given that the renin angiotensin system (RAS) is intricately involved in the pathogenesis of cardiovascular disease [7–12], we review and discuss the presently available evidence for miRSNPs-mediated RAS gene regulation and its importance for phenotypic variation and disease.
2. Current View of the Renin Angiotensin System
The RAS plays a critical role in regulating the physiological processes of the cardiovascular system [reviewed in [7–14]]. The primary effector molecule of this system, angiotensin II (Ang II), has emerged as a critical hormone that affects the function of virtually all organs, including heart, kidney, vasculature, and brain, and it has both beneficial and pathological effects [7–14]. Acute stimulation with Ang II regulates salt/water homeostasis and vasoconstriction, modulating blood pressure, while chronic stimulation promotes hyperplasia and hypertrophy of vascular smooth muscle cells (VSMCs). In addition, long-term exposure to Ang II also plays a pathophysiological role in cardiac hypertrophy and remodeling, myocardial infarction, hypertension, atherosclerosis, in-stent restenosis, reduced fibrinolysis, and renal fibrosis [7–14].
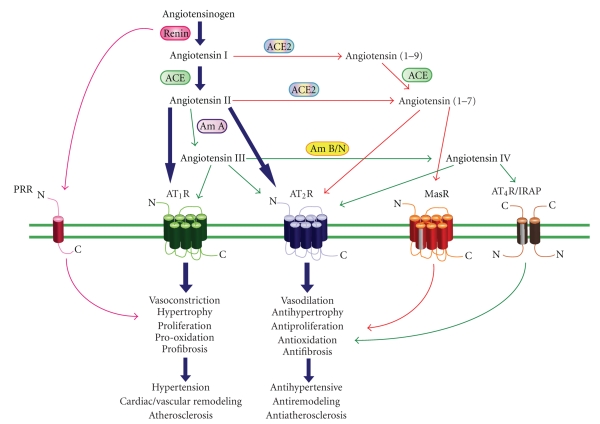
Ang II, an octapeptide hormone, is produced systemically via the classical RAS and locally via the tissue RAS [7–14]. In the classical RAS, circulating renal-derived renin cleaves hepatic-derived angiotensinogen to form the decapeptide angiotensin I (Ang I), which is converted by angiotensin-converting enzyme (ACE) in the lungs to the biologically active Ang II (Figure 1). Alternatively, a recently identified carboxypeptidase, ACE2, cleaves one amino acid from either Ang I or Ang II [15–18], decreasing Ang II levels and increasing the metabolite Ang 1–7, which has vasodilator properties. Thus, the balance between ACE and ACE2 is an important factor controlling Ang II levels [15–18]. Ang II is also further degraded by aminopeptidases to Ang III (Ang 2–8) and Ang IV (Ang 3–8) (Figure 1) [7]. Although the RAS was originally regarded as a circulating system, many of its components are localized in tissues, including the heart, brain, blood vessels, adrenal, kidney, liver and reproductive organs, indicating the existence of local tissue RASs [19]. In addition to ACE-dependent pathways of Ang II formation, non-ACE pathways have also been described. Chymotrypsin-like serine protease (chymase) may represent an important mechanism for conversion of Ang I to Ang II in the human heart, kidney, and vasculature and may be particularly important in pathological conditions such as coronary heart disease [20].
Figure 1.
Summary of the RAS incorporating the Ang peptide family and physiological effects mediated via ATR subtypes. Under the classical RAS schema, Ang II is produced, via renin and ACE, to act with equal affinity on two ATR subtypes, AT1R and AT2R (large arrows). However, it is now appreciated that a number of breakdown products of Ang II, namely, Ang (1–7), Ang III, and Ang IV exert their own unique effects that are distinct (and often opposite) to those of Ang II. Such effects are often mediated via newly recognized receptors such as MasR for Ang (1–7) and AT4R (also known as IRAP) for Ang IV, or additionally via AT2R stimulation. ACE2 is also a new pathway for the formation of Ang (1–7). Newly identified Ang receptor binding proteins associated with different ATR subtypes may also modify ATR activation. Thus, overstimulation of AT1R (and PRR) by Ang II, which can contribute to a plethora of cardiovascular disease processes, may be counter-regulated by a number of non-AT1R mechanisms. Most notably, AT2R stimulation usually causes opposing effects to AT1R, as indicated. It is also likely that the MasR exerts a similar counter-regulatory role whereas the evidence is more preliminary and speculative for AT4R/IRAP. In terms of mediators, Ang II itself stimulates AT2R whereas the shorter Ang peptides stimulate their cognate receptors and possibly also AT2R.
The biological responses to Ang II are mediated by its interaction with two distinct high-affinity G protein-coupled receptors (GPCRs) designated AT1R and AT2R (Figure 1) [7]. Both AT1R and AT2R possess similar affinity for Ang II [21]; however, pharmacologically, these receptors can be distinguished according to inhibition by specific antagonists. For example, AT1R are selectively antagonized by biphenylimidazoles such as losartan (angiotensin receptor blockers, ARB) [21] whereas tetrahydroimidazopyridines such as PD123319 specifically inhibit AT2R [21, 22]. Interestingly, all of the classical actions of Ang II, including vasoconstriction, effects on fluid and electrolyte homeostasis, and influences on cellular growth and differentiation, have been shown to be due to stimulation of AT1R located on the plasma membrane of cells [7–12]. Additionally, the majority of the pathophysiological effects (i.e., cardiac hypertrophy and remodeling, myocardial infarction, hypertension, etc.) of Ang II are also mediated via the AT1R [7–12]. In contrast, it is thought that the AT2R counter-regulates AT1R function (reviewed in [13, 14]). It is also speculated that during cardiovascular disease, AT2R upregulation and activation by Ang II, or angiotensin peptide fragments (i.e., Ang III, Ang IV, and/or Ang 1–7) may limit AT1R-mediated overactivity and cardiovascular pathologies [13, 14].
Although the AT1R and AT2R have been intensively investigated it is now clear that angiotensin fragments can bind to and activate other receptor subtypes. For example, Ang 1–7 acts on the Mas GPCR (MasR) and has vasodilatory and antiproliferative effects. This arm of the RAS is also thought to counterbalance the effects of Ang II acting on the AT1R (Figure 1) (reviewed in [18]). Additionally, Ang IV can bind to the angiotensin II type 4 receptor (AT4R) or the membrane-bound, insulin-regulated aminopeptidase (IRAP) (Figure 1) and mediate the enhancement of cognitive function, modulate blood flow, increase natriuresis, inhibit cardiomyocyte hypertrophy, and improve endothelial function in animal models of atherosclerosis [23, 24].
Finally, recent studies now suggest that renin, the aspartyl protease that cleaves angiotensinogen into Ang I, and prorenin, its proenzyme inactive form, can bind to what is now designated as the (pro)renin receptor (PRR) (reviewed in [25–27]). Interestingly, the binding of renin/prorenin to PRR has been shown to have two major consequences. First, the binding of renin to its receptor increases angiotensinogen conversion to Ang I by five-fold, and prorenin, which is virtually inactive in solution, also displays enzymatic activity following receptor binding [25–27]. Second, receptor-bound renin/prorenin activates the MAP kinases ERK1/2 and p38 pathways, which in turn, leads to the upregulation of profibrotic and cyclooxygenase-2 genes independent of Ang II generation [25–27]. Therefore, the activation and potentiation of renin/prorenin enzymatic activity, together with specific PRR-mediated signaling, could have striking effects on cardiovascular regulation. Taken together, these studies suggest that RAS is unexpectedly complex and multilayered. New components and functions of the RAS are still being unraveled and the physiological significance, and ultimately the clinical relevance, of these factors remain largely undefined.
3. Overview of miRNA Biology
MicroRNAs (miRNAs) are endogenous, short (20–23 nucleotide), and single-stranded nonprotein-coding RNA molecules that regulate gene expression (reviewed in [28]). These molecules act by binding to their target mRNAs, preferentially to the 3′-UTR, using a partial base-pairing mechanism. In order for a miRNA to give rise to functional consequences, the 7-8 nucleotides (nt) at the most 5′ end must have exact complementarity to the target mRNA, generally referred to as the “seed” region [29]. The current model for inhibition of expression by a miRNA suggests that a miRNA either inhibits translation or induces degradation of its target mRNA, depending upon the overall degree of complementarity of the binding site, number of binding sites, and the accessibility of those binding sites [30–32].
In mammals, computational predictions indicate that miRNAs may regulate 60% of all human protein coding genes [33], and have been increasingly implicated in the control of various biological processes, including cell differentiation, cell proliferation, development and apoptosis, and many pathological processes such as cancer, Alzheimer's disease, and cardiovascular disease [34–36]. There are estimated to be >1,000 miRNAs encoded by the human genome [37, 38], each of which can act on multiple target mRNAs. Conversely, individual mRNAs are commonly targeted by multiple miRNAs, which results in a combinatorial repression of gene expression more robust than the suppression that results from a single miRNA [39, 40]. Although miRNAs are known to mediate posttranscriptional gene silencing in the cytoplasm, recent evidence indicates that at least some fraction of mammalian miRNAs may also activate or inhibit gene expression at the transcriptional level [41, 42]. Taken together, these miRNA phenomena allow for enormous combinatorial complexity and regulatory potential.
4. miRNA Biogenesis
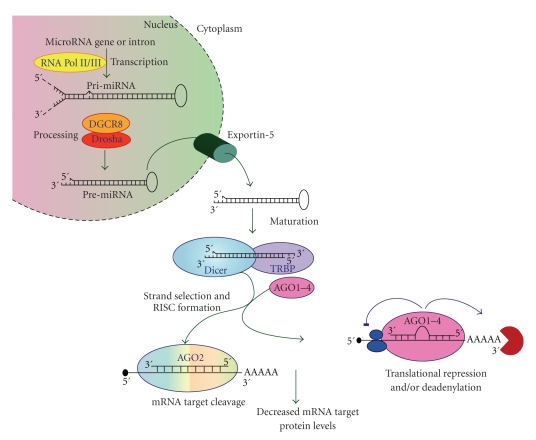
Mature miRNAs are processed from primary miRNA transcripts (pri-miRNAs), which are either transcribed from independent miRNA genes or are portions of introns of protein-coding RNA polymerase II transcripts (Figure 2) [43–45]. miRNAs tend to cluster throughout the genome and many of these clusters are likely transcribed as polycistrons [46–48]. Although little is known regarding the regulation of miRNA transcription, it is recognized that miRNA expression is usually regulated by established transcriptional mechanisms. Interestingly, however, it has been shown that each miRNA located within the same genomic cluster may be transcribed and regulated independently [49].
Figure 2.
Schematic representation outlining miRNA biogenesis including transcription, maturation, and miRNA/mRNA targeting. This diagram also outlines two potential mechanisms for miRNA/mRNA silencing. The specific details discussing these processes are included in the text.
During the transcriptional process, pri-miRNAs fold into hairpin structures containing imperfectly base-paired stems and are endonucleolytically cleaved by the nuclear microprocessor complex formed by the RNase III type endonuclease Drosha and the DiGeorge critical region 8 (DGCR8) protein [50]. The Drosha/DGCR8 complex processes pri-miRNAs into ~70-nucleotide hairpins known as pre-miRNAs (Figure 2) [28, 51]. In animals, pre-miRNAs are exported from the nucleus to the cytoplasm via Exportin-5, where they are cleaved by Dicer complexed with the TAR RNA binding protein (TRBP), to yield ~20-bp miRNA duplexes. In principle, the miRNA duplex could give rise to two different mature miRNAs. However, in a manner similar to siRNA duplexes, only one strand is usually incorporated into miRNA-induced silencing complexes (miRISCs) and guides the complex to target mRNAs; the other strand is degraded (the complementary miRNA* strand) [52]. This functional asymmetry depends on the thermodynamic stability of the base pairs at the two ends of the duplex, with the miRNA strand, which has the least stable base pair at its 5′ end in the duplex, being loaded into the miRISC [53]. Still, recent data suggest that both arms of the pre-miRNA hairpin can give rise to mature miRNAs [28, 45, 51].
5. miRNA/mRNA Silencing
Although the details are not well understood, pre-miRNA processing by Dicer is coupled with the assembly of miRNAs into ribonucleoprotein complexes called micro-RNPs (miRNPs) or miRISCs [28, 51, 54]. One of the key components of miRNPs is the Argonaute (AGO) protein family, AGO1 to AGO4, and while all four AGO proteins function in miRNA repression only AGO2 functions in mRNA target cleavage (Figure 2) [54, 55]. Once the miRNA is processed, the mature miRNA acts as an adaptor for miRISC to specifically recognize and regulate particular mRNAs. It is currently thought that the miRNA-loaded RISC is targeted to a given mRNA by a mechanism where the miRISC binds many sites nonspecifically until the correct target site is found [56].
With few exceptions, miRNA binding sites in animal mRNAs are present in the 3′-UTR and mature miRNAs base pair with their target mRNAs imperfectly, following a set of rules that have been identified by experimental and bioinformatics analyses [29, 57–60]. First, miRNA/mRNA target recognition involves Watson-Crick base pairing that must be perfect and contiguous at the 5′-end of the miRNA from nucleotides 2 to 8. This section represents the “seed” region and nucleates the miRNA-mRNA association. Second, G:U wobble pairing in the seed sequence is highly detrimental to miRNA function despite its favorable contribution to RNA:RNA duplexes. Third, an A residue across position 1 of the miRNA, and an A or U across position 9, improve the site efficiency, although they do not need to base pair with miRNA nucleotides. Fourth, it has been established that miRNAs that have suboptimal 5′ Watson-Crick base pairing need substantial complementarity to the miRNA 3′ half to stabilize the interaction and to be functional. Finally, the context of the miRNA's binding sites harbored in the 3′-UTR of target mRNAs, also influence the functional importance of these sites [61]. For example, miRNA site efficacy can be improved if the site is positioned at least 15 nt downstream from the stop codon, away from the center of long 3′-UTRs, and near AU-rich nucleotide regions. These factors can make the 3′-UTR regions less structured and hence more accessible to miRNP recognition.
When an endogenous miRISC programmed with miRNA binds to a recognition site that is perfectly complementary, this target mRNA will be cleaved by the miRISC (Figure 2) [56, 62–65]. In contrast, miRISCs that are imperfectly matched with target mRNAs can repress translation initiation at either the cap-recognition stage [66–70] or the 60S subunit joining stage [71]. Alternatively, binding of miRISCs can induce deadenylation and decay of target mRNAs [62].
6. Computational Algorithms to Predict miRNA/mRNA Targets
Computational miRNA/mRNA target programs remain the only source for rapid prediction of miRNA recognition sites harbored within the 3′-UTR of target mRNAs. Therefore, the development of reliable computational target prediction programs is critical in advancing our understanding of miRNA function. Given that miRNA functionality usually requires seed sequence complementarity [28, 61] the main prediction feature used in most of these programs is the sequence alignment of the miRNA seed to the 3′-UTR of candidate target genes. Additionally, many current algorithms also utilize conservation of miRNA/mRNA target sites across species as an important parameter for the identification of bona fide targets; notably however, the conservation of a miRNA binding site harbored in a given mRNA target is not a requirement for a functional miRNA.
A recent review article [72] comparing eight of the most commonly used algorithms for miRNA target prediction for the human and mouse genome programs demonstrated that the four top algorithms, DIANA-microT 3.0 (http://microrna.gr/microT) [73], TargetScan 5.0 (http://www.targetscan.org/) [74], Pictar (http://pictar.org/) [75], and ElMMo (from http://www.mirz.unibas.ch) [76] all have a precision of ~50% with a sensitivity that ranges from 6% to 12%. Of the top four performing programs, it is important to note that TargetScan is the most up-to-date regarding the number of miRNAs and genes used and Pictar is least updated [72]. Most investigators assume that mRNA targets predicted by more than one algorithm are more accurate than other targets thus leading to higher prediction precision. However, Alexiou et al. [72] demonstrated that many of the algorithm combinations performed worse than the prediction of a single algorithm. These investigators reason that the better specificity of a combination is achieved by a higher price for the sensitivity. Taking this into account, our laboratory has analyzed all of the classical and nonclassical RAS components for putative miRNA binding sites by the TargetScan algorithm (Table 1). Importantly, this analysis suggests that miRNAs may play a major role in regulating the expression of RAS proteins.
Table 1.
TargetScan* algorithm-predicted RAS putative miRNA/mRNA target sites.
| RAS Component | Total Conserved miRNA Targets | Total Poorly Conserved miRNA Targets | Overall Total miRNA Targets |
|---|---|---|---|
| AGTR1 (AT1R) | 0 | 56 | 56 |
| AGTR2 (AT2R) | 2 | 96 | 98 |
| ACE | 0 | 26 | 26 |
| ACE2 | 1 | 57 | 58 |
| AGT | 1 | 31 | 32 |
| REN | 0 | 15 | 15 |
| ATP6AP2 (PRR) | 4 | 49 | 53 |
| MAS1 (MasR) | 0 | 12 | 12 |
| LNPEP (AT4R) | 5 | 42 | 47 |
*Source: TargetScan (April, 2009). The number of potential targets is dramatically dependent upon the algorithm utilized.
Although many programs are available online for the prediction of individual mRNA targets of miRNAs (see above), the identification of authentic mRNA targets remains problematic. Mammalian miRNAs bind to the mRNA with imperfect complementarity thus how binding sites are recognized is only partially understood. Therefore, some bioinformatically predicted targets turn out to be false and others are entirely overlooked. Experimental validation of targets is therefore an important step in defining the functions of individual miRNAs (for review, see [77]).
7. Experimentally Validated miRNA/RAS Targets
Although classical and nonclassical RAS components harbor putative miRNA binding sites, very few of these sites have been experimentally validated. Our laboratory has demonstrated that miR-155 specifically interacted with the algorithm-predicted binding site harbored in the 3′-UTR of the human AT1R (hAT1R) mRNA (Table 1) [78, 79]. Additionally, miR-155 gain-of-function experiments (i.e., cells were transfected with partially double-stranded RNAs that mimic the Dicer cleavage product and are subsequently processed into their respective mature miRNAs) inhibited the expression of the hAT1R and also attenuated Ang II-induced signaling via the hAT1R in fibroblasts and vascular smooth muscle cells (VSMCs) [78, 79]. These results also demonstrated that transfection with miR-155 did not significantly decrease hAT1R steady state mRNA levels, suggesting that miR-155 can decrease hAT1R expression by inhibiting translation of the mRNA, rather than targeting it for degradation. In contrast, loss-of-function experiments (i.e., cells were transfected with miRNA inhibitors; antisense single-stranded chemically-enhanced oligonucleotides, ASO) demonstrated that transfection of anti-miR-155 not only increased hAT1R expression but also enhanced Ang II-induced signaling via the hAT1R, indicating that miR-155 plays a physiological role in regulating the expression of hAT1Rs in human fibroblasts and VSMCs [78, 79]. Recently, our laboratory also demonstrated that hAT1R expression can be regulated by miR-802 [80].
In support of the miR-155/hAT1R studies described above, trisomy 21 (Ts21) mediated overexpression of miR-155 (the bic/miR-155 gene is located on human chromosome 21 and is triplicated in Down syndrome [DS] individuals) [81, 82] resulted in the attenuation of hAT1R protein levels in fibroblasts isolated from one monozygotic twin with DS when compared to fibroblasts isolated from the unaffected euploid twin [83]. Interestingly, individuals with DS have significantly lower systolic and diastolic blood pressures [84–87], a reduced risk of vascular anomalies [88], and a low prevalence of coronary artery disease [89–93] when compared with the general population. Given that the over-expression of miR-155 in Ts21 results in attenuated hAT1R protein levels, we speculate that this may be one mechanism which contributes to the lack of cardiovascular disease observed in individuals with DS [84–87].
Finally, Boettger et al. [94] demonstrated by genomic, proteomic, and transcriptional analyses that mouse ACE mRNA is a miR-145 target. The miR-143/-145 gene cluster includes miR-143 and miR-145, which lie within a 1.7-kb highly conserved region of mouse chromosome 18 [94]. miRNA microarray hybridization experiments revealed that miR-143 and miR-145 are enriched in murine vascular smooth muscle cells (VSMCs) [94, 95]. In the mouse embryo, miR-143/-145 expression is restricted to heart, vascular, and visceral SMCs [94, 95]. During late fetal and postnatal development, miR-143/-145 expression is downregulated in the heart but persists in vascular and visceral SMCs. Homozygous miR-143/-145 knockout mice were viable but exhibited thinning of the arterial tunica media (muscular layer), with a reduction in the number of contractile VSMCs and a concomitant increase in the number of proliferative VSMCs [94]. Physiological characterization of miR-143/-145 deficient mice and arterial segments from these animals revealed defects in Ang II-induced VSMC contractility and homeostatic control of blood pressure. Consistent with this observation, pharmacological inhibition of ACE or the AT1R partially reversed vascular dysfunction and normalized gene expression in the mutant mice [94]. Since miR-145 regulates the expression of ACE, when the miR-143/-145 gene cluster is mutated, the levels of membrane-bound ACE in VSMCs increase, causing the chronic stimulation of VSMCs by Ang II, which in turn results in desensitization and “angiotensin resistance” of VSMCs. Given that miR-143/145 mutant mice developed neointimal lesions in the absence of hyperlidemia, lipid depositions, and foam cells also highlights the potential role of VSMCs in the pathogenetic process leading to atherosclerosis. The enhancement of Ang II signaling due to the increased expression of ACE would certainly contribute to this process, since increased levels of Ang II have been shown to promote atherosclerotic lesions in apoE-deficient mice [96].
Although the hAT1R and mouse ACE are experimentally validated targets of miRNAs, it is important to note that none of these recognition sites are conserved across species (Table 1). Therefore, although miR-155 and -802 repress hAT1R expression in humans, these miRNAs will not lead to the repression of AT1R levels in mice or rats. Likewise, miR-145 will repress ACE expression in mice but will not regulate ACE levels in humans.
8. miRSNPs
Since a large number of miRNA binding sites are harbored in the 3′-UTRs of RAS component mRNAs (Table 1), there is a high probability that SNPs will occur within miRNA target sites. By definition, miRSNPs have the potential to create, destroy, or modify the efficiency of miRNA binding, if the SNP occurs in the seed region (i.e., the region of base-pairing between nucleotides 2 and 8 of the miRNA and complementary nucleotides in the target mRNA) [97–99]. Based on this definition, there are two mechanisms by which miRSNPs can be functionally important: as a gain- or as a loss-of-function variation. A gain-of-function effect would result if the SNP enhances the targeting of the miRNA or creates a new target site in the 3′-UTR of the mRNA. In this scenario, protein expression of the target mRNA would be attenuated. In contrast, a loss-of-function effect would result when the SNP decreases or abolishes the interaction of the miRNA with its mRNA target, thus resulting in an augmentation of protein expression. In support of this conclusion, miRSNPs have been implicated in Tourette syndrome [100], papillary thyroid cancer [101], muscularity in sheep [102], hereditary spastic paraplegia type 31 [103], methotrexate resistance [104], breast cancer [105], and tumor susceptibility [106]. These examples include both gain- and loss-of-function miRSNPs.
9. The Human AT1R Gene and miRSNPs
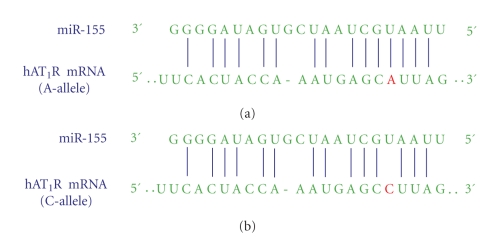
The hAT1R gene has been found to be highly polymorphic [107]. In particular, a SNP has been described in which there is an A/C transversion at position +1166 (i.e., 1166 base-pairs downstream from the start codon, dsSNP# rs5186) located in the 3′-UTR of the hAT1R gene. The increased frequency of the +1166 C-allele has been associated with hypertension [108–115], cardiac hypertrophy [116–118], aortic stiffness [119–121], myocardial infarction [122], heart failure [123–125], abdominal aortic aneurysms [126, 127], and increased oxidative stress levels in human heart failure [128]. However, the physiological relevance of this polymorphism is uncertain because of its location within the noncoding region of the hAT1R gene. Since our laboratory has previously demonstrated that miR-155 interacted with a specific cis-response element localized in the hAT1R 3′-UTR [78], we investigated whether or not there was a correlation between the +1166 A/C SNP and the miR-155-binding site [79, 83]. Importantly, computer alignment revealed that the +1166 A/C SNP occurs within the cis-response element at the site where miR-155 was shown to interact (Figure 3). The interaction between miR-155 and the hAT1R 3′-UTR harboring the A-allele fulfills the seed sequence rules [29] since there is a 7-bp region of complementarity between the 5′ end of miR-155 and the hAT1R mRNA target site (Figure 3(a)). In contrast, if a hAT1R mRNA that harbors the +1166 C-allele is expressed, the complementary seed site is interrupted (Figure 3(b)), and the thermodynamics of the miRNA:mRNA duplex would be significantly altered (i.e., a decrease in free energy) [79]. Therefore, the presence of the +1166 C-allele miRSNP would decrease the ability of miR-155 to interact with the cis-regulatory site located in the hAT1R 3′-UTR. As a consequence, it would be expected that aberrantly high levels of the hAT1R would be synthesized. In support of this hypothesis we demonstrated that when the hAT1R cis-response element harboring the C-allele was present in luciferase mRNAs, the ability of miR-155 to inhibit luciferase activity was significantly attenuated [79]. When identical experiments were performed utilizing mutant miR-155, which restored perfect Watson-Crick complementarity, luciferase activity was decreased to levels that were comparable with experiments utilizing the hAT1R cis-response element harboring the A-allele and miR-155 [79]. To further demonstrate that the presence of the +1166 C-allele can influence hAT1R density, expression constructs that produced hAT1R mRNAs containing either the A- or C-allele were cotransfected with miR-155 or mut-miR-155. These experiments again demonstrated that when seed sequence complementarity was not fulfilled, regardless of whether miR-155 or mutant miR-155 was utilized, hAT1R levels were always higher than the levels obtained when perfect complementarity was present between the miRNA and the hAT1R cis-response element [79]. Taken together, these studies provide the first feasible biochemical mechanism by which the +1166 A/C polymorphism (i.e., miRSNP) can lead to increased hAT1R densities and possibly cardiovascular disease.
Figure 3.
The human AT1R +1166 A/C SNP occurs in the miR-155-binding site. (a) Complementarity between miR-155 and the hAT1R 3′-UTR site targeted (70–90 bp downstream from the human AT1R stop codon). The +1166 A/C SNP corresponds to the nucleotide 86 bp downstream from the human AT1R stop codon (shown in red print). The binding of miR-155 to the hAT1R 3′-UTR target site fulfills the requirement of a 7-bp seed sequence of complementarity at the miRNA 5′ end when the +1166 A-allele is expressed. (b) Complementarity between miR-155 and the human AT1R 3′-UTR harboring the +1166 C-allele. If the +1166 C-allele is expressed, the seed sequence requirement would not be met and, as a consequence, it would be expected that human AT1R expression would be elevated [79].
10. miRSNPS and RAS Component Genes
To begin to investigate whether other RAS components harbored SNPs in their 3′-UTRs which created miRSNPs, the SNP Geneview Report (http://www.ncbi.nlm.nih.gov/nucleotide/) for each gene was surveyed and all of the identified SNPs were analyzed utilizing the Patrocles algorithm (http://www.patrocles.org/) [99]. Specifically the “Patrocles Finder” was utilized since it allows one to compare two sequences and subsequently determines the miRNA binding sites that are different between the two sequences. Thus, a region of the transcribed 3′-UTR which harbored the common allele was compared with the same region containing the mutated allele (select motif length: human 7 mers) and the generation of putative miRSNPs was determined (Table 2). Importantly, this analysis demonstrated that the 3′-UTRs of the RAS components harbor a number of miRSNPs some of which alter or destroy legitimate miRNA binding sites and others that create novel, illegitimate target sites. Hypothetically, if a loss-of-function miRSNP occurred in the AGTR1, ACE, AGT, REN, or ATP6AP2 gene, an increased incidence of hypertension, cardiac/vascular remodeling, and atherosclerosis would be observed (Table 3). In contrast, if a loss-of-function miRSNP occurred in the AGTR2, ACE2, MAS1, or LNPEP gene, a decreased incidence of hypertension, cardiac/vascular remodeling, and atherosclerosis would be expected (Table 3).
Table 2.
Patrocles* algorithm-predicted RAS component miRSNPs.
| RAS Component | dbSNP# | Hetero | dbSNP | bp from Stop Codon | Loss of Function miRSNP | Gain of Function miRSNP |
|---|---|---|---|---|---|---|
| AGTR1 (AT1R) | rs5184 | 0.069 | A>G | 2 | miR-668 | |
| rs56343250 | N.D. | 40 | mR-1237, -1248 | |||
| rs5185 | 0.130 | T>G | 70 | miR-302c, -573 | miR-143*, -301a | |
| rs12721277 | 0.028 | G>A | 82 | miR-579 | ||
| rs5186 | 0.500 | A>C | 86 | miR-155 | ||
| rs5187 | N.D. | A>G | 93 | miR-562, -548n | miR-646 | |
| rs1799870 | 0.055 | C>T | 135 | |||
| rs5188 | 0.109 | G>A | 317 | miR-1197 | ||
| rs55707609 | N.D. | A>T | 335 | miR-299-3p | ||
| rs5189 | 0.500 | G>T | 437 | miR-570 | ||
| rs12721276 | 0.061 | C>A | 461 | miR-128, -27a | ||
| rs12721275 | 0.055 | C>T | 484 | miR-143* | ||
| rs12721274 | 0.028 | T>C | 556 | miR-641 | ||
| rs440881 | N.D. | A>C | 565 | miR-30b | ||
| rs1051649 | N.D. | C>T | 680 | miR-1197 | miR-7 | |
| rs35533650 | N.D. | A>G | 704 | |||
| rs380400 | 0.500 | A>G | 798 | |||
| rs35393661 | N.D. | ->AT | 803 | |||
|
| ||||||
| AGTR2 (AT2R) | rs34589510 | N.D. | ->T | 57 | ||
| rs5193 | 0.241 | G>T | 199 | |||
| rs5194 | 0.499 | A>G | 205 | miR-1229 | ||
| rs11091046 | 0.496 | A>C | 501 | |||
| rs17231436 | 0.131 | G>C | 581 | miR-384 | ||
| rs41312570 | N.D. | C>T | 645 | miR-301a, -130a | ||
| rs17231443 | 0.025 | C>G | 816 | miR-548f, -570 | ||
| rs17237806 | 0.050 | C>T | 837 | |||
| rs12858432 | N.D. | C>T | 906 | |||
| rs12845035 | 0.030 | C>G | 1103 | miR-361-3p | ||
| rs17237820 | 0.073 | A>T | 1111 | miR-548a-3p | ||
| rs17231478 | 0.038 | G>T | 1185 | |||
| rs17237827 | 0.025 | A>G | 1274 | |||
| rs17231450 | 0.050 | C>A | 1318 | miR-150* | ||
| rs17231457 | 0.050 | A>C | 1536 | miR-571 | ||
|
| ||||||
| ACE | None | |||||
|
| ||||||
| ACE2 | None | |||||
|
| ||||||
| AGT | rs61762526 | 0.007 | G>T | 33 | ||
| rs5042 | 0.008 | C>T | 40 | miR-486-3p | ||
| rs4753 | 0.021 | G>C | 158 | miR-337-5p | ||
| rs61751079 | 0.002 | G>A | 159 | miR-639, -539 | ||
| rs5043 | 0.021 | T>C | 167 | |||
| rs61751080 | 0.002 | T>A | 168 | |||
| rs11684 | N.D. | C>T | 184 | miR-103, -107 | ||
| rs1803103 | N.D. | G>T | 187 | miR-483-3p | ||
| rs15022 | N.D. | G>T | 188 | miR-483-3p | ||
| rs1803104 | N.D. | A>G | 316 | |||
| rs1803106 | N.D. | A>C | 338 | miR-129-5p | miR-335* | |
| rs61751081 | 0.002 | C>T | 350 | |||
| rs5044 | 0.029 | T>G | 423 | miR-1283, -606 | ||
| rs61762525 | 0.004 | C>T | 473 | miR-548l, -559 | ||
| rs7079 | 0.312 | C>A | 556 | miR-218-1*, -584 | ||
| rs55720804 | 0.035 | C>- | 573 | |||
| rs61751082 | 0.023 | C>- | 575 | |||
|
| ||||||
| REN | rs11799601 | 0.500 | C>A | 48 | miR-326, -330-5p | |
| r11571124 | 0.041 | C>T | 145 | miR-138 | miR-150* | |
|
| ||||||
| ATP6AP2 (PRR) | rs5963816 | 0.296 | A>T | 266 | miR-664 | |
| rs6609080 | 0.186 | A>G | 358 | miR-1179 | ||
| rs9062 | N.D. | G>T | 654 | miR-508-5p | miR-410 | |
| rs10536 | 0.334 | A>G | 761 | miR-802 | miR-140-3p, -497* | |
| rs1060063 | 0.340 | T>C | 809 | |||
|
| ||||||
| MAS1 (MasR) | None | |||||
|
| ||||||
| LNPEP (AT4R) | rs17087239 | 0.004 | C>T | 61 | miR-992, -15b | |
| rs39602 | 0.488 | C>G | 217 | |||
| rs75912980 | 0.180 | T>A | 364 | miR-1225-5p, -9 | ||
| rs1057808 | 0.014 | T>C | 408 | miR-922, -15b | ||
| rs3756618 | 0.028 | A>T | 501 | miR-22*, -26b* | miR-223 | |
| rs35838718 | N.D. | A>- | 561 | |||
| rs79818663 | 0.105 | G>T | 594 | |||
| rs77639920 | N.D. | C>T | 614 | miR-664* | ||
| rs62377081 | N.D. | G>T | 695 | miR-302a*, -1264 | miR-548l | |
*Source: www.patrocles.org/.
Table 3.
Physiological ramifications of RAS miRSNPs.
| RAS Component | Loss of Function miRSNP | Physiological Result | Gain of Function miRSNP | Physiological Result |
|---|---|---|---|---|
| AGTR1 (AT1R) | AT1R↑ | Hypertension Cardiac/vascular remodeling Atherosclerosis | AT1R↓ | Antihypertensive Antiremodeling Anti-atherosclerosis |
| ACE | ACE↑ | ACE↓ | ||
| AGT | AGT↑ | AGT↓ | ||
| REN | REN↑ | REN↓ | ||
| ATP6AP2 (PRR) | PRR↑ | PRR↓ | ||
| AGTR2 (AT2R) | AT2R↑ | Antihypertensive Antiremodeling Antiatherosclerosis | AT2R↓ | Hypertension Cardiac/vascular remodeling Atherosclerosis |
| ACE2 | ACE2↑ | ACE2↓ | ||
| MAS1 (MasR) | MasR↑ | MasR↓ | ||
| LNPEP (AT4R) | AT4R↑ | AT4R↓ |
11. Conclusion
miRNAs have been increasingly implicated in the control of various biological processes, including cell differentiation, cell proliferation, development and apoptosis, and many pathological processes such as cancer, Alzheimer's disease, and cardiovascular disease [34–36]. Importantly, several studies which have now demonstrated that polymorphisms at the miRNA target site in the 3′-UTRs of transcribed mRNAs can have detrimental effects given that miRSNPs can lead to the modulation of gene expression [79, 83, 100–106]. In support of this hypothesis, a recent study suggested that differences in SNP allele frequency among ethnic groups account for differences in gene expression [129]. Therefore, we speculate that miRSNPs can modulate RAS phenotypic gene expression diversities, at least in part, through alteration of miRNA target binding capability, ultimately leading to differences in the susceptibility to complex genetic disorders, such as cardiovascular disease.
References
- 1.Kruglyak L, Nickerson DA. Variation is the spice of life. Nature Genetics. 2001;27(3):234–236. doi: 10.1038/85776. [DOI] [PubMed] [Google Scholar]
- 2.Sherry ST, Ward M, Sirotkin K. dbSNP - database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Research. 1999;9(8):677–679. [PubMed] [Google Scholar]
- 3.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 4.Nückel H, Frey UH, Bau M, et al. Association of a novel regulatory polymorphism (−938C>A) in the BCL2 gene promoter with disease progression and survival in chronic lymphocytic leukemia. Blood. 2007;109(1):290–297. doi: 10.1182/blood-2006-03-007567. [DOI] [PubMed] [Google Scholar]
- 5.Krawczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Human Genetics. 1992;90(1-2):41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target Recognition and Regulatory Functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Gasparo M, Catt KJ, Inagami T, Wright JW, Unger TH. International union of pharmacology. XXIII. The angiotensin II receptors. Pharmacological Reviews. 2000;52(3):415–472. [PubMed] [Google Scholar]
- 8.Thomas WG, Mendelsohn FAO. Angiotensin receptors: form and function and distribution. International Journal of Biochemistry and Cell Biology. 2003;35(6):774–779. doi: 10.1016/s1357-2725(02)00263-7. [DOI] [PubMed] [Google Scholar]
- 9.Hunyady L, Catt KJ. Pleiotropic AT1 receptor signaling pathways mediating physiological and pathogenic actions of angiotensin II. Molecular Endocrinology. 2006;20(5):953–970. doi: 10.1210/me.2004-0536. [DOI] [PubMed] [Google Scholar]
- 10.Mehta PK, Griendling KK. Angiotensin II cell signaling: physiological and pathological effects in the cardiovascular system. American Journal of Physiology. 2007;292(1):C82–C97. doi: 10.1152/ajpcell.00287.2006. [DOI] [PubMed] [Google Scholar]
- 11.Elton TS, Martin MM. Angiotensin II type 1 receptor gene regulation: transcriptional and posttranscriptional mechanisms. Hypertension. 2007;49(5):953–961. doi: 10.1161/HYPERTENSIONAHA.106.070565. [DOI] [PubMed] [Google Scholar]
- 12.Oro C, Qian H, Thomas WG. Type 1 angiotensin receptor pharmacology: signaling beyond G proteins. Pharmacology and Therapeutics. 2007;113(1):210–226. doi: 10.1016/j.pharmthera.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porrello ER, Delbridge LM, Thomas WG. The angiotensin II type 2 (AT2) receptor: an enigmatic seven transmembrane receptor. Frontiers in Bioscience. 2009;14:958–972. doi: 10.2741/3289. [DOI] [PubMed] [Google Scholar]
- 14.Jones ES, Vinh A, McCarthy CA, Gaspari TA, Widdop RE. AT2 receptors: functional relevance in cardiovascular disease. Pharmacology and Therapeutics. 2008;120(3):292–316. doi: 10.1016/j.pharmthera.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crackower MA, Sarao R, Oudit GY, et al. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002;417(6891):822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- 16.Chappell MC, Gregory Modralt J, Diz DI, Ferrario CM. Novel aspects of the renal renin-angiotensin system: angiotensin-(1-7),ACE2 and blood pressure regulation. Contributions to Nephrology. 2004;143:77–89. doi: 10.1159/000078713. [DOI] [PubMed] [Google Scholar]
- 17.Raizada MK, Ferreira AJ. ACE2: a new target for cardiovascular disease therapeutics. Journal of Cardiovascular Pharmacology. 2007;50(2):112–119. doi: 10.1097/FJC.0b013e3180986219. [DOI] [PubMed] [Google Scholar]
- 18.Iwai M, Horiuchi M. Devil and angel in the renin-angiotensin system: ACE-angiotensin II-AT1 receptor axis vs. ACE2-angiotensin-(1-7)-Mas receptor axis. Hypertension Research. 2009;32(7):533–536. doi: 10.1038/hr.2009.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danser AHJ. Local renin-angiotensin systems. Molecular and Cellular Biochemistry. 1996;157(1-2):211–216. doi: 10.1007/BF00227900. [DOI] [PubMed] [Google Scholar]
- 20.Petrie MC, Padmanabhan N, McDonald JE, Hillier C, Connell JMC, McMurray JJV. Angiotensin converting enzyme (ACE) and non-ACE dependent angiotensin II generation in resistance arteries from patients with heart failure and coronary heart disease. Journal of the American College of Cardiology. 2001;37(4):1056–1061. doi: 10.1016/s0735-1097(01)01111-1. [DOI] [PubMed] [Google Scholar]
- 21.Timmermans PBMWM, Wong PC, Chiu AT, Herblin WF. Nonpeptide angiotensin II receptor antagonists. Trends in Pharmacological Sciences. 1991;12(2):55–62. doi: 10.1016/0165-6147(91)90498-h. [DOI] [PubMed] [Google Scholar]
- 22.Timmermans PBMWM, Wong PC, Chiu AT, et al. Angiotensin II receptors and angiotensin II receptor antagonists. Pharmacological Reviews. 1993;45(2):205–251. [PubMed] [Google Scholar]
- 23.Chai SY, Fernando R, Peck G, et al. The angiotensin IV/AT4 receptor. Cellular and Molecular Life Sciences. 2004;61(21):2728–2737. doi: 10.1007/s00018-004-4246-1. [DOI] [PubMed] [Google Scholar]
- 24.Vanderheyden PML. From angiotensin IV binding site to AT4 receptor. Molecular and Cellular Endocrinology. 2009;302(2):159–166. doi: 10.1016/j.mce.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 25.Danser AHJ. (Pro)renin receptors: are they biologically relevant? Current Opinion in Nephrology and Hypertension. 2009;18(1):74–78. doi: 10.1097/MNH.0b013e3283196aaf. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen G, Muller DN. The biology of the (pro)renin receptor. Journal of the American Society of Nephrology. 2010;21(1):18–23. doi: 10.1681/ASN.2009030300. [DOI] [PubMed] [Google Scholar]
- 27.Danser AHJ. The increase in renin during renin inhibition: does it result in harmful effects by the (pro)renin receptor. Hypertension Research. 2010;33(1):4–10. doi: 10.1038/hr.2009.186. [DOI] [PubMed] [Google Scholar]
- 28.Bushati N, Cohen SM. MicroRNA functions. Annual Review of Cell and Developmental Biology. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 29.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biology. 2005;3(3, article e85) doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiriakidou M, Tan GS, Lamprinaki S, De Planell-Saguer M, Nelson PT, Mourelatos Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129(6):1141–1151. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 31.Humphreys DT, Westman BJ, Martin DIK, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(47):16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bagga S, Bracht J, Hunter S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122(4):553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 33.Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Research. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dalmay T. MicroRNAs and cancer. Journal of Internal Medicine. 2008;263(4):366–375. doi: 10.1111/j.1365-2796.2008.01926.x. [DOI] [PubMed] [Google Scholar]
- 35.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. NeuroReport. 2007;18(3):297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 36.van Rooij E, Sutherland LB, Liu N, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(48):18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RHA, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120(1):21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 38.Rigoutsos I, Huynh T, Miranda K, Tsirigos A, McHardy A, Platt D. Short blocks from the noncoding parts of the human genome have instances within nearly all known genes and relate to biological processes. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(17):6605–6610. doi: 10.1073/pnas.0601688103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krek A, Grün D, Poy MN, et al. Combinatorial microRNA target predictions. Nature Genetics. 2005;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 40.Hon LS, Zhang Z. The roles of binding site arrangement and combinatorial targeting in microRNA repression of gene expression. Genome Biology. 2007;8(8, article R166) doi: 10.1186/gb-2007-8-8-r166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Place RF, Li L-C, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(5):1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim DH, Sætrom P, Snøve O, Jr., Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(42):16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO Journal. 2004;23(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10(12):1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature Reviews Genetics. 2008;9(2):102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 46.Altuvia Y, Landgraf P, Lithwick G, et al. Clustering and conservation patterns of human microRNAs. Nucleic Acids Research. 2005;33(8):2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Research. 2004;14(10):1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim Y-K, Kim VN. Processing of intronic microRNAs. EMBO Journal. 2007;26(3):775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song G, Wang L. MiR-433 and miR-127 arise from independent overlapping primary transcripts encoded by the miR-433-127 locus. PLoS ONE. 2008;3(10) doi: 10.1371/journal.pone.0003574. Article ID e3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 51.Du T, Zamore PD. MicroPrimer: the biogenesis and function of microRNA. Development. 2005;132(21):4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 52.Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115(2):199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 53.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115(2):209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 54.Peters L, Meister G. Argonaute Proteins: mediators of RNA Silencing. Molecular Cell. 2007;26(5):611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 55.Tolia NH, Joshua-Tor L. Slicer and the argonautes. Nature Chemical Biology. 2007;3(1):36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- 56.Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nature Reviews Molecular Cell Biology. 2007;8(1):23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 57.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes and Development. 2004;18(5):504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 59.Grimson A, Farh KK-H, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Molecular Cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nielsen CB, Shomron N, Sandberg R, Hornstein E, Kitzman J, Burge CB. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007;13(11):1894–1910. doi: 10.1261/rna.768207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(11):4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes and Development. 2006;20(14):1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eulalio A, Rehwinkel J, Stricker M, et al. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes and Development. 2007;21(20):2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Giraldez AJ, Mishima Y, Rihel J, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312(5770):75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 66.Humphreys DT, Westman BJ, Martin DIK, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(47):16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pillai RS, Bhattacharyya SN, Artus CG, et al. Molecular biology: inhibition of translational initiation by let-7 microRNA in human cells. Science. 2005;309(5740):1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 68.Thermann R, Hentze MW. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447(7146):875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- 69.Mathonnet G, Fabian MR, Svitkin YV, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317(5845):1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 70.Wakiyama M, Takimoto K, Ohara O, Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes & Development. 2007;21(15):1857–1862. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chendrimada TP, Finn KJ, Ji X, et al. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447(7146):823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- 72.Alexiou P, Maragkakis M, Papadopoulos GL, Reczko M, Hatzigeorgiou AG. Lost in translation: an assessment and perspective for computational microrna target identification. Bioinformatics. 2009;25(23):3049–3055. doi: 10.1093/bioinformatics/btp565. [DOI] [PubMed] [Google Scholar]
- 73.Maragkakis M, Alexiou P, Papadopoulos GL, et al. Accurate microRNA target prediction correlates with protein repression levels. BMC Bioinformatics. 2009;10, article 1471:p. 295. doi: 10.1186/1471-2105-10-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Research. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lall S, Grün D, Krek A, et al. A genome-wide map of conserved microRNA targets in C. elegans. Current Biology. 2006;16(5):460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 76.Gaidatzis D, van Nimwegen E, Hausser J, Zavolan M. Inference of miRNA targets using evolutionary conservation and pathway analysis. BMC Bioinformatics. 2007;8(1):p. 69. doi: 10.1186/1471-2105-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kuhn DE, Martin MM, Feldman DS, Terry AV, Jr., Nuovo GJ, Elton TS. Experimental validation of miRNA targets. Methods. 2008;44(1):47–54. doi: 10.1016/j.ymeth.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martin MM, Lee EJ, Buckenberger JA, Schmittgen TD, Elton TS. MicroRNA-155 regulates human angiotensin II type 1 receptor expression in fibroblast. The Journal of Biological Chemistry. 2006;281(27):18277–18284. doi: 10.1074/jbc.M601496200. [DOI] [PubMed] [Google Scholar]
- 79.Martin MM, Buckenberger JA, Jiang J, et al. The human angiotensin II type 1 receptor +1166 A/C polymorphism attenuates microRNA-155 binding. The Journal of Biological Chemistry. 2007;282(33):24262–24269. doi: 10.1074/jbc.M701050200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 80.Sansom SE, Nuovo GJ, Martin MM, et al. MiR-802 regulates human angiotensin II type 1 receptor expression in intestinal epithelial C2BBe1 cells . doi: 10.1152/ajpgi.00120.2010. American Journal of Physiology Gastrointestinal and Liver Physiology. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kuhn DE, Nuovo GJ, Martin MM, et al. Human chromosome 21-derived miRNAs are overexpressed in down syndrome brains and hearts. Biochemical and Biophysical Research Communications. 2008;370(3):473–477. doi: 10.1016/j.bbrc.2008.03.120. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 82.Kuhn DE, Nuovo GJ, Terry AV, Jr., et al. Chromosome 21-derived microRNAs provide an etiological basis for aberrant protein expression in human down syndrome brains. The Journal of Biological Chemistry. 2010;285(2):1529–1543. doi: 10.1074/jbc.M109.033407. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 83.Sethupathy P, Borel C, Gagnebin M, et al. Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3′ untranslated region: a mechanism for functional single-nucleotide polymorphisms related to phenotypes. American Journal of Human Genetics. 2007;81(2):405–413. doi: 10.1086/519979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Murdoch JC, Rodger JC, Rao SS. Down’s syndrome: an atheroma free model? British Medical Journal. 1977;2(6081):226–228. doi: 10.1136/bmj.2.6081.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yla-Herttuala S, Luoma J, Nikkari T, Kivimaki T. Down’s syndrome and atherosclerosis. Atherosclerosis. 1989;76(2-3):269–272. doi: 10.1016/0021-9150(89)90110-x. [DOI] [PubMed] [Google Scholar]
- 86.Kapell D, Nightingale B, Rodriguez A, Lee JH, Zigman WB, Schupf N. Prevalence of chronic medical conditions in adults with mental retardation: comparison with the general population. Mental Retardation. 1998;36(4):269–279. doi: 10.1352/0047-6765(1998)036<0269:POCMCI>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 87.Draheim CC, McCubbin JA, Williams DP. Differences in cardiovascular disease risk between nondiabetic adults with mental retardation with and without Down syndrome. American Journal on Mental Retardation. 2002;107(3):201–234. doi: 10.1352/0895-8017(2002)107<0201:DICDRB>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 88.Greene AK, Kim S, Rogers GF, Fishman SJ, Olsen BR, Mulliken JB. Risk of vascular anomalies with Down syndrome. Pediatrics. 2008;121(1):e135–e140. doi: 10.1542/peds.2007-1316. [DOI] [PubMed] [Google Scholar]
- 89.Baird PA, Sadovnick AD. Causes of death to age 30 in Down syndrome. American Journal of Human Genetics. 1988;43(3):239–248. [PMC free article] [PubMed] [Google Scholar]
- 90.Brattstrom L, Englund E, Brun A. Does Down syndrome support homocysteine theory of arteriosclerosis? The Lancet. 1987;1(8529):391–392. doi: 10.1016/s0140-6736(87)91772-7. [DOI] [PubMed] [Google Scholar]
- 91.Pueschel SM, Craig WY, Haddow JE. Lipids and lipoproteins in persons with Down’s syndrome. Journal of Intellectual Disability Research. 1992;36(4):365–369. doi: 10.1111/j.1365-2788.1992.tb00535.x. [DOI] [PubMed] [Google Scholar]
- 92.Licastro F, Marocchi A, Penco S, et al. Does Down’s syndrome support the homocysteine theory of atherogenesis?. Experience in elderly subjects with trisomy 21. Archives of Gerontology and Geriatrics. 2006;43(3):381–387. doi: 10.1016/j.archger.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 93.Goi G, Baquero-Herrera C, Licastro F, Dogliotti G, Corsi MM. Advanced oxidation protein products (AOPP) and high-sensitive C-reactive protein (hs-CRP) in an “atheroma-free model”: Down’s syndrome. International Journal of Cardiology. 2006;113(3):427–429. doi: 10.1016/j.ijcard.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 94.Boettger T, Beetz N, Kostin S, et al. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. Journal of Clinical Investigation. 2009;119(9):2634–2647. doi: 10.1172/JCI38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cordes KR, Sheehy NT, White MP, et al. MiR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460(7256):705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Daugherty A, Manning MW, Cassis LA. Angiotensin II promotes atherosclerotic lesions and aneurysms in apolipoprotein E-deficient mice. Journal of Clinical Investigation. 2000;105(11):1605–1612. doi: 10.1172/JCI7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bao L, Zhou M, Wu L, et al. PolymiRTS Database: linking polymorphisms in microRNA target sites with complex traits. Nucleic Acids Research. 2007;35(1):D51–D54. doi: 10.1093/nar/gkl797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hariharan M, Scaria V, Brahmachari SK. dbSMR: a novel resource of genome-wide SNPs affecting microRNA mediated regulation. BMC Bioinformatics. 2009;10, article 108 doi: 10.1186/1471-2105-10-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hiard S, Charlier C, Coppieters W, Georges M, Baurain D. Patrocles: a database of polymorphic miRNA-mediated gene regulation in vertebrates. Nucleic Acids Research. 2009;38(supplement 1):D640–D651. doi: 10.1093/nar/gkp926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Abelson JF, Kwan KY, O’Roak BJ, et al. Medicine: sequence variants in SLITRK1 are associated with Tourette’s syndrome. Science. 2005;310(5746):317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- 101.He H, Jazdzewski K, Li W, et al. The role of microRNA genes in papillary thyroid carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(52):19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Clop A, Marcq F, Takeda H, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nature Genetics. 2006;38(7):813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- 103.Züchner S, Wang G, Tran-Viet K-N, et al. Mutations in the novel mitochondrial protein REEP1 cause hereditary spastic paraplegia type 31. American Journal of Human Genetics. 2006;79(2):365–369. doi: 10.1086/505361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mishra PJ, Humeniuk R, Mishra PJ, Longo-Sorbello GSA, Banerjee D, Bertino JR. A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(33):13513–13518. doi: 10.1073/pnas.0706217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Adams BD, Furneaux H, White BA. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-α (ERα) and represses ERα messenger RNA and protein expression in breast cancer cell lines. Molecular Endocrinology. 2007;21(5):1132–1147. doi: 10.1210/me.2007-0022. [DOI] [PubMed] [Google Scholar]
- 106.Nicoloso MS, Sun H, Spizzo R, et al. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Research. 2010;70(7):2789–2798. doi: 10.1158/0008-5472.CAN-09-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Baudin B. Polymorphism in angiotensin II receptor genes and hypertension. Experimental Physiology. 2005;90(3):277–282. doi: 10.1113/expphysiol.2004.028456. [DOI] [PubMed] [Google Scholar]
- 108.Bonnardeaux A, Davies E, Jeunemaitre X, et al. Angiotensin II type 1 receptor gene polymorphisms in human essential hypertension. Hypertension. 1994;24(1):63–69. doi: 10.1161/01.hyp.24.1.63. [DOI] [PubMed] [Google Scholar]
- 109.Kainulainen K, Perola M, Terwilliger J, et al. Evidence for involvement of the type 1 angiotensin II receptor locus in essential hypertension. Hypertension. 1999;33(3):844–849. doi: 10.1161/01.hyp.33.3.844. [DOI] [PubMed] [Google Scholar]
- 110.Wang WYS, Zee RYL, Morris BJ. Association of angiotensin II type 1 receptor gene polymorphism with essential hypertension. Clinical Genetics. 1997;51(1):31–34. doi: 10.1111/j.1399-0004.1997.tb02410.x. [DOI] [PubMed] [Google Scholar]
- 111.Kobashi G, Hata A, Ohta K, et al. A1166C variant of angiotensin II type 1 receptor gene is associated with severe hypertension in pregnancy independently of T235 variant of angiotensinogen gene. Journal of Human Genetics. 2004;49(4):182–186. doi: 10.1007/s10038-004-0129-4. [DOI] [PubMed] [Google Scholar]
- 112.Wang J-G, Staessen JA. Genetic polymorphisms in the renin-angiotensin system: relevance for susceptibility to cardiovascular disease. European Journal of Pharmacology. 2000;410(2-3):289–302. doi: 10.1016/s0014-2999(00)00822-0. [DOI] [PubMed] [Google Scholar]
- 113.Jiang Z, Zhao W, Yu F, Xu G. Association of angiotensin II type 1 receptor gene polymorphism with essential hypertension. Chinese Medical Journal. 2001;114(12):1249–1251. [PubMed] [Google Scholar]
- 114.Ono K, Mannami T, Baba S, Yasui N, Ogihara T, Iwai N. Lack of association between angiotensin II type 1 receptor gene polymorphism and hypertension in Japanese. Hypertension Research. 2003;26(2):131–134. doi: 10.1291/hypres.26.131. [DOI] [PubMed] [Google Scholar]
- 115.Palatini P, Ceolotto G, Dorigatti F, et al. Angiotensin II type 1 receptor gene polymorphism predicts development of hypertension and metabolic syndrome. American Journal of Hypertension. 2009;22(2):208–214. doi: 10.1038/ajh.2008.319. [DOI] [PubMed] [Google Scholar]
- 116.Osterop APRM, Kofflard MJM, Sandkuijl LA, et al. AT1 receptor A/C1166 polymorphism contributes to cardiac hypertrophy in subjects with hypertrophic cardiomyopathy. Hypertension. 1998;32(5):825–830. doi: 10.1161/01.hyp.32.5.825. [DOI] [PubMed] [Google Scholar]
- 117.Makeeva OA, Puzyrev KV, Pavlukova EN, et al. ACE and AGTR1 genes polymorphisms in left ventricular hypertrophy pathogenesis in humans. Molekulyarnaya Biologiya. 2004;38(6):990–996. [PubMed] [Google Scholar]
- 118.Smilde TDJ, Zuurman MW, Hillege HL, et al. Renal function dependent association of AGTR1 polymorphism (A1166C) and electrocardiographic left-ventricular hypertrophy. American Journal of Hypertension. 2007;20(10):1097–1103. doi: 10.1016/j.amjhyper.2007.04.023. [DOI] [PubMed] [Google Scholar]
- 119.Lajemi M, Labat C, Gautier S, et al. Angiotensin II type 1 receptor-153A/G and 1166A/C gene polymorphisms and increase in aortic stiffness with age in hypertensive subjects. Journal of Hypertension. 2001;19(3):407–413. doi: 10.1097/00004872-200103000-00008. [DOI] [PubMed] [Google Scholar]
- 120.Díez J, Laviades C, Orbe J, et al. The A1166C polymorphism of the AT1 receptor gene is associated with collagen type I synthesis and myocardial stiffness in hypertensives. Journal of Hypertension. 2003;21(11):2085–2092. doi: 10.1097/01.hjh.0000098127.00558.d2. [DOI] [PubMed] [Google Scholar]
- 121.Benetos A, Gautier S, Ricard S, et al. Influence of angiotensin-converting enzyme and angiotensin II type 1 receptor gene polymorphisms on aortic stiffness in normotensive and hypertensive patients. Circulation. 1996;94(4):698–703. doi: 10.1161/01.cir.94.4.698. [DOI] [PubMed] [Google Scholar]
- 122.Tiret L, Blanc H, Ruidavets J-B, et al. Gene polymorphisms of the renin-angiotensin system in relation to hypertension and parental history of myocardial infarction and stroke: the PEGASE study. Journal of Hypertension. 1998;16(1):37–44. doi: 10.1097/00004872-199816010-00007. [DOI] [PubMed] [Google Scholar]
- 123.Wu C-K, Tsai C-T, Hwang J-J, et al. Renin-angiotensin system gene polymorphisms and diastolic heart failure. European Journal of Clinical Investigation. 2008;38(11):789–797. doi: 10.1111/j.1365-2362.2008.02017.x. [DOI] [PubMed] [Google Scholar]
- 124.Lin J, Hu FB, Qi L, Curhan GC. Genetic polymorphisms of angiotensin-2 type 1 receptor and angiotensinogen and risk of renal dysfunction and coronary heart disease in type 2 diabetes mellitus. BMC Nephrology. 2009;10(1, article 9) doi: 10.1186/1471-2369-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Amir O, Amir RE, Paz H, Attias E, Sagiv M, Lewis BS. Relation between AT1R gene polymorphism and long-term outcome in patients with heart failure. Cardiology. 2009;112(2):151–157. doi: 10.1159/000143390. [DOI] [PubMed] [Google Scholar]
- 126.Jones GT, Thompson AR, van Bockxmeer FM, et al. Angiotensin II type 1 receptor 1166C polymorphism is associated with abdominal aortic aneurysm in three independent cohorts. Arteriosclerosis, Thrombosis, and Vascular Biology. 2008;28(4):764–770. doi: 10.1161/ATVBAHA.107.155564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.McColgan P, Peck GE, Greenhalgh RM, Sharma P. The genetics of abdominal aortic aneurysms: a comprehensive meta-analysis involving eight candidate genes in over 16,700 patients. International Surgery. 2009;94(4):350–358. [PubMed] [Google Scholar]
- 128.Cameron VA, Mocatta TJ, Pilbrow AP, et al. Angiotensin type-1 receptor A1166C gene polymorphism correlates with oxidative stress levels in human heart failure. Hypertension. 2006;47(6):1155–1161. doi: 10.1161/01.HYP.0000222893.85662.cd. [DOI] [PubMed] [Google Scholar]
- 129.Spielman RS, Bastone LA, Burdick JT, Morley M, Ewens WJ, Cheung VG. Common genetic variants account for differences in gene expression among ethnic groups. Nature Genetics. 2007;39(2):226–231. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]