Abstract
MicroRNAs (miRNAs) regulate gene expression and have a critical role in many biologic and pathologic processes. We hypothesized that miRNA expression profiles in injured brain (hippocampus) would show common as well as unique profiles when compared with those of blood. Adult, untouched, control rats were compared with rats with sham surgeries, ischemic strokes, brain hemorrhage (lysed blood, fresh blood, or thrombin), and kainate-induced seizures. Brain and whole-blood miRNA expression profiles were assessed 24 h later using TaqMan rodent miRNA arrays. MicroRNA response profiles were different for each condition. Many miRNAs changed more than 1.5-fold in brain and blood after each experimental manipulation, and several miRNAs were upregulated or downregulated in both brain and blood after a given injury. A few miRNAs (e.g., miR-298, miR-155, and miR-362-3p) were upregulated or downregulated more than twofold in both brain and blood after several different injuries. The results show the possible use of blood miRNAs as biomarkers for brain injury; that selected blood miRNAs may correlate with miRNA changes in the brain; and that many of the mRNAs, previously shown to be regulated in brain and blood after brain injury, are likely accounted for by changes in miRNA expression.
Keywords: blood, brain, epilepsy, hemorrhage, ischemia, microRNA (miRNA)
Introduction
RNA expression studies offer unique opportunities for diagnosis and prognosis, as well as for understanding the pathogenesis of many medical and neurologic diseases (Sharp et al, 2006, 2007). Our previous studies of mRNA expression profiles have shown that a large number of genes are induced or suppressed in brain and blood after cerebral ischemia, intracerebral hemorrhage, and kainate-induced seizures (Lu et al, 2006; Tang et al, 2001, 2002). However, the specific factors that lead to the regulation of large numbers of genes (mRNAs) in each condition remain unclear.
The recent discovery of microRNAs (miRNAs) as key regulators of gene function has introduced a new level and mechanism of gene regulation (Bartel, 2004; Chen and Rajewsky, 2007; Filip, 2007). Although there are only hundreds of miRNAs, each of them can potentially regulate hundreds of target genes (Guarnieri and DiLeone, 2008), with the prediction that more than one-third of all human genes may be regulated by miRNAs (Esquela-Kerscher and Slack, 2006). Functionally, miRNAs are implicated in fundamental cellular processes including cell-cycle regulation (Matsubara et al, 2007), hematopoietic differentiation (Hatfield and Ruohola-Baker, 2008), immune responses (Bi et al, 2009), and cellular metabolism (Gauthier and Wollheim, 2006).
MicroRNA expression has been examined in various tumors (Guarnieri and DiLeone, 2008; Lu et al, 2005; Nicoloso and Calin, 2008), in Alzheimer's disease (Hebert et al, 2008), Parkinson's disease (Kim et al, 2007), Down's syndrome (Kuhn et al, 2008), schizophrenia (Beveridge et al, 2008), and stroke (Dharap et al, 2009; Jeyaseelan et al, 2008). These miRNA expression profiles may be as diagnostically useful as mRNA expression profiles (Guarnieri and DiLeone, 2008; Lu et al, 2005). Therefore, we postulated that miRNAs alone could be adequate biomarkers and that miRNA expression profiles could explain, at least in part, the mRNA regulation in brain and blood after different neurologic diseases. To begin to address this idea, adult untouched control rats were compared with rats with sham surgeries, ischemic strokes, intracerebral hemorrhage (lysed blood, fresh blood, thrombin), and kainate-induced seizures. Total RNA (plus miRNA) was isolated 24 h later from brain and blood, and miRNA expression profiles were assessed using TaqMan Rodent miRNA Arrays. The results show unique miRNA profiles for brain, blood, and for each experimental condition, with some miRNAs being regulated in both brain and blood for a given injury. Moreover, many of the mRNA targets of regulated miRNAs have previously been reported to be regulated in brain and blood after stroke, hemorrhage, and seizures.
Materials and methods
Experiments were carried out in accordance with National Institutes of Health guidelines and were approved by the Institutional Animal Care and Use Committee (University of California at Davis). Male Sprague–Dawley rats (n=48 total), weighing 300 to 320 g, were used in this study. The animals were allowed free access to food and water and were maintained on a 12-h light/dark cycle. Rats were anesthetized with isoflurane (Minrad, New York, NY, USA) and their body temperature was maintained at 37.0°C±0.2°C. They were allowed to fully recover after a procedure in an incubator maintained at 37°C, and were then returned to their home cages with free access to food and water.
Brain Ischemia
Rats (n=6) were anesthetized with isoflurane, and the neck skin and muscle were incised to isolate the left common carotid artery. A 3-0 monofilament nylon suture was threaded through the external carotid artery stump into the internal carotid artery and up to the stem of the middle cerebral artery (MCA). The suture was anchored in place to produce a permanent MCA occlusion. This ‘suture'-induced MCA occlusion produces reliable infarction in the MCA distribution (Liu et al, 2005). Only animals with ipsilateral hemispheric infarction were selected for this study.
Brain Hemorrhage
Rats were anesthetized and placed in a stereotaxic frame (Kopf Instruments, Tujunga, CA, USA). They received intraventricular (−0.9 mm posterior, 1.4 lateral to bregma, and 4.6 mm deep) injections (intracerebroventricular) of 50 μL lysed blood (n=6), 50 μL fresh autologous tail vein blood (n=6), or thrombin (n=6) (20 U per animal in 50 μL of saline) (Paxinos and Watson, 1998). All three models have been used to study various aspects of the pathophysiology of intracerebral hemorrhage (Liu et al, 2008; Tang et al, 2001, 2002).
Kainate Seizures
Rats (n=6) were injected subcutaneously with 10 mg/kg of kainic acid (Sigma, St Louis, MO, USA) dissolved in saline. This kainic acid model produces status epilepticus that injures neurons in the cortex and other brain regions (Zhang et al, 1996, 1997). Animals that had severe, prolonged generalized seizures were selected for this study.
Sham Surgeries
For sham ischemia, rats (n=6) were anesthetized, the suture inserted into the internal carotid artery, and then removed without being advanced to the stem of the MCA. For sham brain hemorrhage, rats (n=6) were anesthetized and 50 μL of saline was injected into the left cerebral ventricle.
Untouched Controls
Rats (n=6) that had not been handled in any way were used as controls. All experimental and control animals were housed in the same room for 1 week before the study and then during the study.
Total RNA (Plus microRNA) Isolation from Rat Brain and Blood Samples
At 24 h (1 day) after each operation or assignment as an untouched control, all subjects were anesthetized with isoflurane. Blood (0.5 mL) was drawn from the tail vein into EDTA tubes (Becton Dickinson and Company, Franklin Lakes, NJ, USA), immediately mixed with 1.3 mL RNA (Ambion, Austin, TX, USA), and stored at −80°C. Animals were then killed, brain was removed, and the hippocampus was dissected as rapidly as possible, frozen, and stored at −80°C. For ischemia and hemorrhage rats, the hippocampus adjacent to the hemispheric infarction and the hippocampus ipsilateral to ventricular injections were dissected, respectively. Hippocampus was chosen because it should sustain some cell injury or cell death in most of the models without including any areas of infarction.
Total RNA (plus miRNA) was isolated from hippocampus using a miRNeasy Mini Kit (Qiagen, Valencia, CA, USA) and a Qiacube (Qiagen). Total RNA was isolated from blood using the Mouse RiboPure-Blood RNA Isolation Kit (Ambion). The concentration and integrity of total RNA was determined using a NanoDrop-1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). RNA samples were stored at −80°C before TaqMan miRNA array studies.
TaqMan miRNA Arrays
TaqMan MicroRNA Arrays (384-Well Fluidic Cards) (Applied Biosystems, Foster City, CA) were used to profile 381 mature miRNAs. U6 was used as the endogenous control in quadruplicate and one assay not related to rat was included as a negative control. Total RNA (800 ng each sample) was converted into cDNA using Megaplex RT Primers (Applied Biosystems) and TaqMan miRNA RT Kits (Applied Biosystems). cDNAs were mixed with TaqMan Universal PCR Master Mix (Applied Biosystems) and then loaded into TaqMan rodent miRNA fluidic cards (Applied Biosystems). The cards were run using an Applied Biosystems 7900HT real-time PCR instrument equipped with a heating block for the fluidic card (Applied Biosystems).
Statistical Analysis
As cycle threshold (CT) scores >35 are considered to be nonspecific (undetectable) (Schmittgen et al, 2008), miRNAs in which 95% of individual observations had a raw CT score >35 were excluded from the final data analysis. Using these filtering criteria, 97 miRNAs were removed from the brain analysis, and 153 miRNAs were removed from blood analyses. Partek Genomics Suite (Partek, St Louis, MI, USA) was used to carry out statistical analyses. Fold change filters were applied to select the miRNAs that were regulated more than 1.5- or 2-fold. A t-test with or without FDR (false discovery rate of 5%=no more than 5% false positives) correction for multiple comparisons was used to examine the significance of miRNAs regulated in each experimental condition compared with that of the untouched control group. MicroRNAs that changed ≥1.5-fold and had a P<0.05 were subjected to hierarchical clustering using Euclidean distance on the basis of their relative mean expression.
Biologic Functional Analyses of MicroRNAs Regulated in Both Brain and Blood
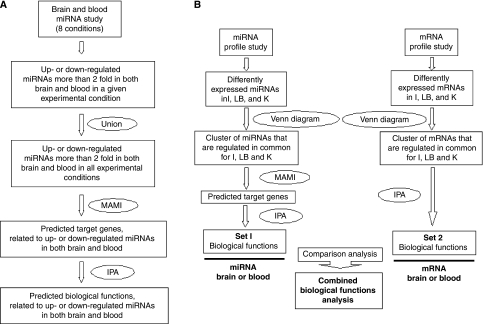
The miRNAs used for biologic functional analyses were significantly regulated (P<0.05) more than twofold in both brain and blood, as compared with those in untouched controls, in at least one experimental condition (unless stated otherwise). Predicted target genes (mRNAs) of miRNAs regulated in brain and blood were determined using publicly available software suites MetA Mir:target Inference (MAMI) (http://mami.med.harvard.edu) (Gusev, 2008). This makes composite predictions using several different algorithms, including TargetScanS, miRanda, microT, miRtarget, and PicTar. The miRNA and predicted mRNA target genes in both brain and blood were then subjected to biologic function analyses using Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Redwood City, CA USA) (Gusev, 2008). The procedure for the prediction of biologic functions of regulated miRNAs in both brain and blood is shown in Figure 1A.
Figure 1.
Flow diagrams showing how biologic functional analyses were carried out. (A) Prediction of target genes and biologic functions of miRNAs that changed more than twofold in both brain and blood in at least one experimental condition. (B) Comparison between miRNAs and mRNAs that were regulated 24 h after brain ischemia (I), brain hemorrhage (lysed blood, LB), and kainate seizures (K).
Comparison of Biologic Functions of MicroRNAs and mRNAs Regulated in Brain and Blood
For miRNA–mRNA biologic function analyses, miRNAs regulated more than twofold in either brain or blood after brain ischemia, brain hemorrhage (lysed blood), and kainate-induced seizures were entered into MAMI to obtain the predicted target genes (Figure 1B, left panel). The exported target genes were then entered into IPA to obtain a set of biologic functions (Figure 1B, left panel). Similarly, regulated mRNAs, obtained from our previously published studies (Lu et al, 2006; Tang et al, 2001, 2002), were input into IPA to yield another set of biologic functions (Figure 1B, right panel). We then compared the two sets of data to derive the functional pathways that were common to the miRNAs and mRNAs regulated for each of the injuries in both brain and blood (Figure 1B).
Results
Endogenous Control MicroRNAs in Brain and Blood
Five potential endogenous control miRNAs (U6, U87, Y1, snoRNA135, and snoRNA202) were used. In the brain, U6, U87, Y1, and snoRNA135 were stable (regulated <1.5-fold) in each of the seven experimental conditions, whereas snoRNA202 decreased more than 1.5-fold in one or more experimental conditions (Supplementary Table 1). In blood, U6, U87, snoRNA135, and snoRNA202 were stable (regulated <1.5-fold) in each of the seven experimental conditions, whereas Y1 was upregulated more than 1.5-fold in one or more experimental conditions (Supplementary Table 1). Thus, U6, U87, and snoRNA134 were the most stable in both brain and blood across experimental conditions. U6 was used as the endogenous control for all of the data reported in this study because it was the most stably expressed miRNA across all subjects in the control and experimental groups.
Brain and Blood MicroRNA Response
Many miRNAs changed more than 1.5-fold in brain and blood after each experimental condition as compared with those in untouched controls (Table 1; Supplementary Table 1). A number of miRNAs that changed 1.5-fold on an average were significantly changed (P<0.05), but relatively few of these miRNAs passed the false discovery rate multiple comparison correction threshold (FDR <5%) (Table 1; Supplementary Table 1). Several miRNAs were upregulated or downregulated in both brain and blood after a given injury (Table 1; Supplementary Table 1). A few miRNAs, such as miR-298, miR-155, and miR-362-3p, were upregulated or downregulated in both brain and blood after several different injuries (Table 1; Supplementary Table 1).
Table 1. Number of miRNAs that changed in brain and blood 24 h after each experimental condition as compared to untouched controls (C).
| Comparison | Up- or downregulated | Sample | No. of miRNA detectable | miRNAs regulated in both brain and blood (fold change >1.5) | No. of regulated miRNA | ||
|---|---|---|---|---|---|---|---|
| Fold change >1.5 | Fold change >1.5 and P<0.05 | Fold change >1.5, P<0.05 and FDR <0.05 | |||||
| SI versus C | Up | Brain | 198 | miR-298, miR-503, miR-672 | 43 | 5 | 0 |
| Blood | 49 | 19 | 3 | 0 | |||
| Down | Brain | 85 | miR-155,a miR-362-3p | 26 | 1 | 0 | |
| Blood | 103 | 44 | 1 | 0 | |||
| I versus C | Up | Brain | 168 | miR-10a, miR-182, miR-200b, miR-298 | 43 | 6 | 1 |
| Blood | 33 | 16 | 3 | 1 | |||
| Down | Brain | 115 | miR-155,a miR-362-3p,a miR-223, miR-210 | 22 | 5 | 2 | |
| Blood | 119 | 92 | 38 | 19 | |||
| SH versus C | Up | Brain | 143 | miR-148b, miR-503 | 36 | 3 | 0 |
| Blood | 76 | 19 | 3 | 0 | |||
| Down | Brain | 140 | miR-10b,a miR-203, miR-362-3p | 27 | 8 | 4 | |
| Blood | 76 | 36 | 3 | 2 | |||
| B versus C | Up | Brain | 163 | miR-298,a miR-200b,a miR-205,a miR-345, miR-423-5p | 34 | 6 | 0 |
| Blood | 104 | 39 | 4 | 1 | |||
| Down | Brain | 120 | None | 31 | 7 | 2 | |
| Blood | 48 | 20 | 1 | 0 | |||
| LB versus C | Up | Brain | 150 | miR-298,a miR-423-5p,a miR-10a, miR-345-5p, miR-674 | 34 | 6 | 0 |
| Blood | 87 | 30 | 6 | 1 | |||
| Down | Brain | 133 | miR-155, miR-20b-3p, miR-200a | 28 | 6 | 1 | |
| Blood | 65 | 33 | 6 | 0 | |||
| T versus C | Up | Brain | 234 | miR-107,a miR-200b, miR-331-5p, miR-672 | 69 | 12 | 0 |
| Blood | 29 | 17 | 2 | 0 | |||
| Down | Brain | 49 | miR-155,a miR-188-5pa | 19 | 3 | 3 | |
| Blood | 123 | 63 | 10 | 0 | |||
| K versus C | Up | Brain | 104 | miR-298a | 21 | 1 | 0 |
| Blood | 47 | 15 | 2 | 0 | |||
| Down | Brain | 179 | miR-155,a miR-29c, miR-34b-3p, miR-98, miR-122, miR-203, miR-450a | 39 | 9 | 4 | |
| Blood | 105 | 43 | 1 | 0 | |||
miRNA, microRNA; RT-PCR, real-time PCR.
Experimental conditions included untouched control (C), sham ischemia (SI), ischemia (I), sham hemorrhage (SH), fresh blood (B), lysed blood (LB), thrombin (T) and kainate seizures (K).
The columns indicate the numbers of miRNAs that were detectable by RT-PCR; had a fold change >1.5; a fold change >1.5 and P<0.05; a fold change >1.5, P<0.05 and FDR <0.05.
Represents miRNAs regulated in both brain and blood and had a fold change >2.
Unique MicroRNA Expression Profiles in Brain and Blood
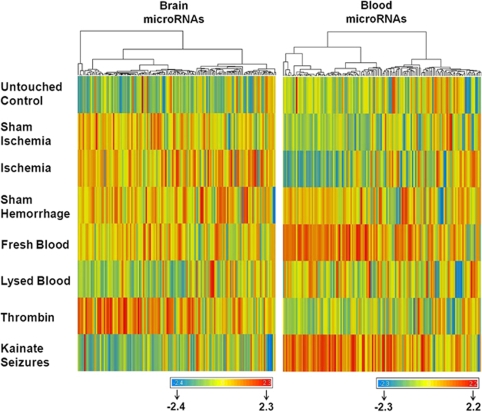
As there may not be specifically regulated miRNAs for a given disease, we looked for unique patterns of miRNA expression for each of the experimental conditions using cluster analysis. This showed that each experimental condition had a unique miRNA expression pattern in both brain (Figure 2, left panel) and blood (Figure 2, right panel). For the eight expression profiles shown, no profile was identical. The different miRNA expression patterns, including sham controls versus untouched controls, emphasize the uniqueness of the different brain and blood miRNA responses for each condition.
Figure 2.
Hierarchical clustering of miRNAs that changed more than 1.5-fold shows the unique miRNA expression profiles in brain (left panel) and blood (right panel) 24 h after each experimental condition. Red, upregulation; yellow, little change; and blue, downregulation. This experiment was performed on eight groups of rats including untouched controls (C), sham brain ischemia (SI), brain ischemia (I), sham brain hemorrhage (SH), fresh blood-induced brain hemorrhage (B), lysed blood-induced brain hemorrhage (LB), thrombin injections (T), and kainate seizures (K).
Function Analysis of MicroRNAs Regulated in Both Brain and Blood
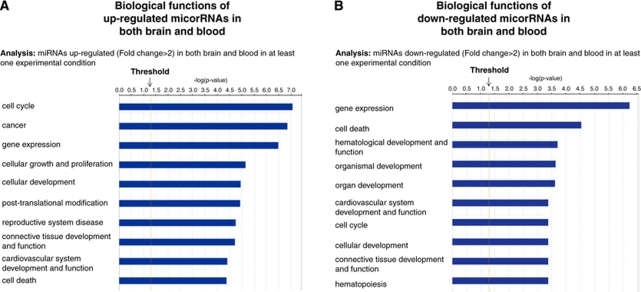
A biologic function analysis was carried out on the miRNAs regulated more than twofold in both brain and blood in at least one experimental condition. Five miRNAs (miR-298, miR-200b, miR-205, miR-107, and miR-423-5p) were upregulated in both brain and blood, and four miRNAs (miR-155, miR-362-3p, miR-10b, and miR-188-5p) were downregulated more than twofold in both brain and blood in at least one experimental condition (Table 1, marked with #). The top 10 ranked biologic functions associated with commonly upregulated miRNAs include cell cycle, cancer, gene expression, cellular growth and proliferation, cellular development, posttranslational modification, reproductive system disease, connective tissue development and function, cardiovascular system development and function, and cell death (Figure 3A, Supplementary Figure 1A). The top 10 ranked biologic functions associated with commonly downregulated miRNAs included gene expression, cell death, hematological development and function, organismal development, organ development, cardiovascular system development and function, cell cycle, cellular development, connective tissue development and function, and hematopoiesis (Figure 3B, Supplementary Figure 1B).
Figure 3.
Top 10 ranked biologic functions of miRNAs (fold change >2) regulated in both brain and blood in at least one experimental condition. (A) Biologic functions of miRNAs upregulated in both brain and blood. (B) Biologic functions of miRNAs downregulated in both brain and blood.
MicroRNAs Regulated by Multiple Conditions in either Brain or Blood
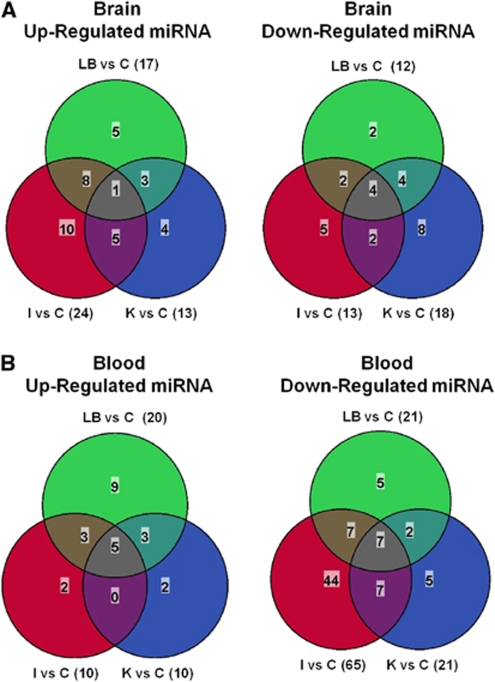
Many upregulated miRNAs were modulated in two or more of the groups in either brain or blood. In brain, 24 miRNAs were upregulated more than twofold by brain ischemia, 17 by brain hemorrhage (lysed blood), and 13 by kainate seizures (Figure 4A, left panel). In addition, 13 miRNAs were downregulated more than twofold by brain ischemia, 12 by brain hemorrhage (lysed blood), and 18 by kainate seizures (Figure 4A, right panel). A Venn diagram of these results showed that one miRNA (miR-542-3p) was upregulated (Figure 4A, left panel) and four miRNAs (miR-155, miR-362-3p, miR-122, and miR-450a-5p) were downregulated in all three conditions (Figure 4A, right panel). In blood, 10 miRNAs were upregulated more than twofold by brain ischemia, 21 by brain hemorrhage (lysed blood), and 21 by kainate seizures (Figure 4B, left panel). In addition, 65 miRNAs were downregulated more than twofold by brain ischemia, 20 by brain hemorrhage (lysed blood), and 10 by kainate seizures (Figure 4B, right panel). The Venn diagram of these results showed that five miRNAs (miR-96, miR-152, miR-298, miR-333, and miR-505) were upregulated (Figure 4B, left panel) and seven miRNAs (miR-125a-5p, miR-130b, miR-142-3p, miR-330, miR-342-5p, miR-685, and miR-347) were downregulated in all three conditions (Figure 4B, right panel).
Figure 4.
Venn diagrams showing the numbers of miRNAs that were regulated more than twofold in brain and blood 24 h after brain ischemia (I), brain hemorrhage (LB, lysed blood), and kainate seizures (K) compared with untouched controls (C). (A) MicroRNAs upregulated (left panel) and downregulated (right panel) in the brain. (B) MicroRNAs upregulated (left panel) and downregulated (right panel) in blood.
Analyses of MicroRNA Target Genes (mRNAs) and Previously Reported Regulated mRNAs in Both Brain and Blood After Ischemia, Hemorrhage, and Seizures
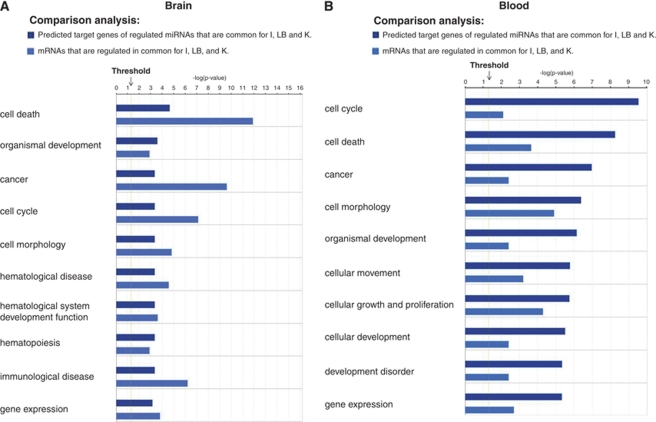
We previously reported unique mRNA expression profiles in both brain and blood after brain ischemia, brain hemorrhage (lysed blood), and kainate-induced seizures (Tang et al, 2001, 2002). To make the previously reported data comparable with the data in this study, it was re-normalized and re-analyzed using Partek software (included in Supplementary Tables 2A and B). Using the work flow in Figure 1B, the biologic functions of the mRNAs regulated in our previous studies were compared with the biologic functions of the miRNAs regulated in this study, for both brain and blood. Figures 5A and 5B show that almost all of the predicted biologic functions for regulated miRNAs and mRNAs overlapped considerably. The top 10 ranked functions in brain for regulated miRNAs and mRNA are cell death, organismal development, cancer, cell cycle, cell morphology, hematological disease, hematological system development function, hematopoiesis, immunologic disease, and gene expression (Figure 5A, Supplementary Figure 2A). The top 10 ranked functions in blood for regulated miRNAs and mRNA are cell cycle, cell death, cancer, cell morphology, organismal development, cellular movement, cellular growth and proliferation, cellular development, development disorder, and gene expression (Figure 5B, Supplementary Figure 2B). Furthermore, the top five ranked functions that overlapped for both brain and blood included cell cycle, cell death, cancer, cell morphology, and organismal development (Figures 5A and 5B).
Figure 5.
Top 10 ranked biologic functions of miRNAs and the mRNAs that were regulated in brain and blood after all three main types of brain injury: brain ischemia (I), brain hemorrhage (LB, lysed blood), and kainate-induced seizures (K). The log of the P-value is shown for each biologic function for both regulated miRNAs (dark blue) and mRNAs (light blue). Regulated miRNAs were derived from this study, and regulated mRNAs were derived from our previous studies (Lu et al, 2006; Tang et al, 2001, 2002). (A) Comparison of the biologic functions of regulated miRNAs with those of regulated mRNAs in brain. (B) Comparison of the biologic functions of regulated miRNAs with those of regulated mRNAs in blood.
Discussion
A major finding of this study is that different patterns of miRNA expression occur in brain and blood 24 h after brain ischemia, brain hemorrhage, kainate seizures, and even sham surgeries, compared with untouched control animals. A number of miRNAs were significantly regulated (P<0.05) more than 1.5-fold in brain and blood after each brain injury. Several miRNAs were upregulated or downregulated in both brain and blood after a given injury; and a few miRNAs, including miR-298, mir-155, mir-362-3p, etc., were upregulated or downregulated in both brain and blood after several different injuries. These results show the possible use of blood miRNAs as biomarkers for selected brain miRNAs after one or more specific types of brain injuries. They also provide a partial mechanism for specific miRNA-induced mRNA expression profiles in brain and blood after different types of brain injury.
MicroRNA expression levels can be studied using several different methods: microarray analysis, real-time PCR (RT-PCR), northern blots, in situ hybridization, and solution hybridization. Of these techniques, TaqMan miRNA assays are specific for mature miRNAs and discriminate among related miRNAs that differ by as little as one nucleotide (Chen et al, 2005). Furthermore, RT-PCR assays are not affected by genomic DNA contamination (Chen et al, 2005). Therefore, TaqMan RT-PCR miRNA assays are a sensitive and accurate method for assessing miRNA expression.
A remarkable finding in this study is that miR-298 was the only miRNA that was upregulated in both brain and blood after almost all experimental conditions. Thus, this is consistent with the finding that miR-298 was also upregulated in the brain after transient MCA occlusions in another recent study (Jeyaseelan et al, 2008). Although miR-298 has many functions, it was recently shown to recognize specific binding sites in the 3′-UTR of β-amyloid precursor protein-converting enzyme-1 (BACE1) mRNA and to regulate BACE1 protein expression in cultured neuronal cells (Boissonneault et al, 2009). This particular miRNA also emphasizes the fact that miR-298 likely regulates hundreds of other mRNAs and that the dozens of miRNAs shown to be regulated in brain and blood in this study regulate hundreds of target mRNAs in brain and blood, many of which were shown by us to be regulated in our previous studies (Lu et al, 2006; Tang et al, 2001, 2002).
Recent studies indicate that some miRNAs that are selectively and/or highly expressed in immune cells—including the miR-17-92 cluster, miR-150, miR-155, miR-181, and miR-223—have a ‘permissive' function in the maturation, proliferation, and differentiation of myeloid and lymphoid cells (Tsitsiou and Lindsay, 2009). In addition, miR-155 negatively regulates the immune response by repressing Src homology-2 domain-containing inositol 5-phosphatase 1 (SHIP1) through direct 3′-UTR interactions (O'Connell et al, 2009). In this study, miR-155 was downregulated in both brain and blood after almost all experimental conditions. This is consistent with our previous genomic studies that the expression of Lyn, one of the Src family of kinases, was upregulated 21-fold after intracerebral hemorrhage (Lu et al, 2006; Tang et al, 2002).
Many of the miRNAs upregulated or downregulated by each experimental condition were modulated in two or more of the groups. The miRNAs regulated in common by the various conditions might provide an index of common mechanisms of injury. Therefore, we used the commonly regulated miRNAs for predicting biologic functions. The results show that the five top-ranked functions predicted by the commonly regulated miRNAs and mRNAs in both brain and blood included cell cycle, cell death, cancer, cell morphology, and organismal development. This suggests that there are some functional responses of miRNAs and mRNAs that are similar in brain and blood after many types of acute brain injury. Future studies are required to determine exactly which factors regulate a similar cross talk in both brain and blood between the miRNAs and mRNAs in each. Such factors could include similar brain and blood stress responses to injury, or similar brain and blood immune responses to injury.
The ‘cell cycle' was among the top-ranked functions for miRNA regulated in both brain and blood, and for mRNAs and miRNAs that changed in brain and blood 1 day after injury. This suggests that the ‘cell-cycle' pathway might be relevant to such injuries. The miRNAs induced in blood related to the ‘cell cycle' may relate to the proliferation of white blood cells and the blood inflammatory response to acute brain injury. The miRNAs activated in brain in ‘cell-cycle' pathways could relate to glial proliferation, and could potentially relate to the possibility that cell cycle re-entry leads to postmitotic neuronal death. Cell cycle re-entry in neurons has been confirmed in many brain diseases, including stroke, brain hemorrhage, traumatic brain injury (TBI), and epilepsy (Di Giovanni et al, 2005; Imai et al, 2002; Nagy and Esiri, 1998; O'Hare et al, 2002). Inhibition of cyclin-dependent kinases increases neuronal survival and improves behavioral outcomes in animal models of these diseases (Di Giovanni et al, 2005; Hilton et al, 2008; Park et al, 2000; Verdaguer et al, 2003; Verdaguer et al, 2004). Thus, the miRNAs involved in the cell cycle in brain could relate to the birth of new cells including glia, and/or relate to neuronal re-entry into the cell cycle and death of these cells.
Although only a limited number of disease conditions were studied, data show that brain and blood miRNA responses were unique for each. One example of this difference was the unique brain and blood miRNA response of sham-operated control animals to that of untouched control animals. Although there are miRNAs specifically regulated by each of the conditions, we propose that it is the composite brain and blood miRNA response of all of the regulated miRNAs that may provide miRNA fingerprints that can be distinguished for many different disease-related states.
Finally, several experimental limitations need to be mentioned. Although the functions of regulated miRNAs described in this study significantly overlap the functions of mRNAs reported in our previous studies, actual mRNA levels were not measured in this study. This must be carried out in future studies. In addition, for those mRNAs that are presumed targets of specific miRNAs, these target mRNAs must also be measured in future studies to show their actual regulation. Second, animals were not perfused at the conclusion of an experiment because of concern that this would affect miRNA expression. Thus, the miRNAs that are reported as being regulated in both brain and blood would represent the maximum number that was regulated, as some could represent contamination of brain by blood. This is likely a minor problem because of the small volume of blood compared with brain, the few shared miRNAs in brain and blood, and the comparison of hemorrhage and control showed no shared miRNAs.
Footnotes
Supplementary Information accompanies the paper on the Journal of Cerebral Blood Flow & Metabolism website (http://www.nature.com/jcbfm)
Conflict of interest
The authors declare no conflict of interest.
Supplementary Material
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Beveridge NJ, Tooney PA, Carroll AP, Gardiner E, Bowden N, Scott RJ, Tran N, Dedova I, Cairns MJ. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum Mol Genet. 2008;17:1156–1168. doi: 10.1093/hmg/ddn005. [DOI] [PubMed] [Google Scholar]
- Bi Y, Liu G, Yang R. MicroRNAs: novel regulators during the immune response. J Cell Physiol. 2009;218:467–472. doi: 10.1002/jcp.21639. [DOI] [PubMed] [Google Scholar]
- Boissonneault V, Plante I, Rivest S, Provost P. MicroRNA-298 and microRNA-328 regulate expression of mouse beta-amyloid precursor protein-converting enzyme 1. J Biol Chem. 2009;284:1971–1981. doi: 10.1074/jbc.M807530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- Dharap A, Bowen K, Place R, Li LC, Vemuganti R. Transient focal ischemia induces extensive temporal changes in rat cerebral microRNAome. J Cereb Blood Flow Metab. 2009;29:675–687. doi: 10.1038/jcbfm.2008.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giovanni S, Movsesyan V, Ahmed F, Cernak I, Schinelli S, Stoica B, Faden AI. Cell cycle inhibition provides neuroprotection and reduces glial proliferation and scar formation after traumatic brain injury. Proc Natl Acad Sci USA. 2005;102:8333–8338. doi: 10.1073/pnas.0500989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Filip A. [MiRNA—new mechanisms of gene expression control] Postepy Biochem. 2007;53:413–419. [PubMed] [Google Scholar]
- Gauthier BR, Wollheim CB. MicroRNAs: ‘ribo-regulators' of glucose homeostasis. Nat Med. 2006;12:36–38. doi: 10.1038/nm0106-36. [DOI] [PubMed] [Google Scholar]
- Guarnieri DJ, DiLeone RJ. MicroRNAs: a new class of gene regulators. Ann Med. 2008;40:197–208. doi: 10.1080/07853890701771823. [DOI] [PubMed] [Google Scholar]
- Gusev Y. Computational methods for analysis of cellular functions and pathways collectively targeted by differentially expressed microRNA. Methods. 2008;44:61–72. doi: 10.1016/j.ymeth.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Hatfield S, Ruohola-Baker H. microRNA and stem cell function. Cell Tissue Res. 2008;331:57–66. doi: 10.1007/s00441-007-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert SS, Horre K, Nicolai L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A, De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci USA. 2008;105:6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton GD, Stoica BA, Byrnes KR, Faden AI. Roscovitine reduces neuronal loss, glial activation, and neurologic deficits after brain trauma. J Cereb Blood Flow Metab. 2008;28:1845–1859. doi: 10.1038/jcbfm.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai H, Harland J, McCulloch J, Graham DI, Brown SM, Macrae IM. Specific expression of the cell cycle regulation proteins, GADD34 and PCNA, in the peri-infarct zone after focal cerebral ischaemia in the rat. Eur J Neurosci. 2002;15:1929–1936. doi: 10.1046/j.1460-9568.2002.02025.x. [DOI] [PubMed] [Google Scholar]
- Jeyaseelan K, Lim KY, Armugam A. MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke. 2008;39:959–966. doi: 10.1161/STROKEAHA.107.500736. [DOI] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A microRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn DE, Nuovo GJ, Martin MM, Malana GE, Pleister AP, Jiang J, Schmittgen TD, Terry AV, Jr, Gardiner K, Head E, Feldman DS, Elton TS. Human chromosome 21-derived miRNAs are overexpressed in Down syndrome brains and hearts. Biochem Biophys Res Commun. 2008;370:473–477. doi: 10.1016/j.bbrc.2008.03.120. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu DZ, Xie KQ, Ji XQ, Ye Y, Jiang CL, Zhu XZ. Neuroprotective effect of paeoniflorin on cerebral ischemic rat by activating adenosine A1 receptor in a manner different from its classical agonists. Br J Pharmacol. 2005;146:604–611. doi: 10.1038/sj.bjp.0706335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DZ, Cheng XY, Ander BP, Xu H, Davis RR, Gregg JP, Sharp FR. Src kinase inhibition decreases thrombin-induced injury and cell cycle re-entry in striatal neurons. Neurobiol Dis. 2008;30:201–211. doi: 10.1016/j.nbd.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Tang Y, Ran R, Ardizzone TL, Wagner KR, Sharp FR. Brain genomics of intracerebral hemorrhage. J Cereb Blood Flow Metab. 2006;26:230–252. doi: 10.1038/sj.jcbfm.9600183. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Matsubara H, Takeuchi T, Nishikawa E, Yanagisawa K, Hayashita Y, Ebi H, Yamada H, Suzuki M, Nagino M, Nimura Y, Osada H, Takahashi T. Apoptosis induction by antisense oligonucleotides against miR-17-5p and miR-20a in lung cancers overexpressing miR-17-92. Oncogene. 2007;26:6099–6105. doi: 10.1038/sj.onc.1210425. [DOI] [PubMed] [Google Scholar]
- Nagy Z, Esiri MM. Neuronal cyclin expression in the hippocampus in temporal lobe epilepsy. Exp Neurol. 1998;150:240–247. doi: 10.1006/exnr.1997.6753. [DOI] [PubMed] [Google Scholar]
- Nicoloso MS, Calin GA. MicroRNA involvement in brain tumors: from bench to bedside. Brain Pathol. 2008;18:122–129. doi: 10.1111/j.1750-3639.2007.00119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci USA. 2009;106:7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hare M, Wang F, Park DS. Cyclin-dependent kinases as potential targets to improve stroke outcome. Pharmacol Ther. 2002;93:135–143. doi: 10.1016/s0163-7258(02)00183-3. [DOI] [PubMed] [Google Scholar]
- Park DS, Obeidat A, Giovanni A, Greene LA. Cell cycle regulators in neuronal death evoked by excitotoxic stress: implications for neurodegeneration and its treatment. Neurobiol Aging. 2000;21:771–781. doi: 10.1016/s0197-4580(00)00220-7. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C.1998The rat brain in stereotatic coordinates4th edn.London: Academic Press [Google Scholar]
- Schmittgen TD, Lee EJ, Jiang J, Sarkar A, Yang L, Elton TS, Chen C. Real-time PCR quantification of precursor and mature microRNA. Methods. 2008;44:31–38. doi: 10.1016/j.ymeth.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp FR, Xu H, Lit L, Walker W, Apperson M, Gilbert DL, Glauser TA, Wong B, Hershey A, Liu DZ, Pinter J, Zhan X, Liu X, Ran R. The future of genomic profiling of neurological diseases using blood. Arch Neurol. 2006;63:1529–1536. doi: 10.1001/archneur.63.11.1529. [DOI] [PubMed] [Google Scholar]
- Sharp FR, Xu H, Lit L, Walker W, Pinter J, Apperson M, Verro P. Genomic profiles of stroke in blood. Stroke. 2007;38:691–693. doi: 10.1161/01.STR.0000247940.27518.38. [DOI] [PubMed] [Google Scholar]
- Tang Y, Lu A, Aronow BJ, Sharp FR. Blood genomic responses differ after stroke, seizures, hypoglycemia, and hypoxia: blood genomic fingerprints of disease. Ann Neurol. 2001;50:699–707. doi: 10.1002/ana.10042. [DOI] [PubMed] [Google Scholar]
- Tang Y, Lu A, Aronow BJ, Wagner KR, Sharp FR. Genomic responses of the brain to ischemic stroke, intracerebral haemorrhage, kainate seizures, hypoglycemia, and hypoxia. Eur J Neurosci. 2002;15:1937–1952. doi: 10.1046/j.1460-9568.2002.02030.x. [DOI] [PubMed] [Google Scholar]
- Tsitsiou E, Lindsay MA. microRNAs and the immune response. Curr Opin Pharmacol. 2009;9:514–520. doi: 10.1016/j.coph.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaguer E, Jorda EG, Canudas AM, Jimenez A, Sureda FX, Rimbau V, Pubill D, Escubedo E, Camarasa J, Pallas M, Camins A. 3-Amino thioacridone, a selective cyclin-dependent kinase 4 inhibitor, attenuates kainic acid-induced apoptosis in neurons. Neuroscience. 2003;120:599–603. doi: 10.1016/s0306-4522(03)00424-x. [DOI] [PubMed] [Google Scholar]
- Verdaguer E, Jimenez A, Canudas AM, Jorda EG, Sureda FX, Pallas M, Camins A. Inhibition of cell cycle pathway by flavopiridol promotes survival of cerebellar granule cells after an excitotoxic treatment. J Pharmacol Exp Ther. 2004;308:609–616. doi: 10.1124/jpet.103.057497. [DOI] [PubMed] [Google Scholar]
- Zhang X, Boulton AA, Yu PH. Expression of heat shock protein-70 and limbic seizure-induced neuronal death in the rat brain. Eur J Neurosci. 1996;8:1432–1440. doi: 10.1111/j.1460-9568.1996.tb01605.x. [DOI] [PubMed] [Google Scholar]
- Zhang X, Gelowitz DL, Lai CT, Boulton AA, Yu PH. Gradation of kainic acid-induced rat limbic seizures and expression of hippocampal heat shock protein-70. Eur J Neurosci. 1997;9:760–769. doi: 10.1111/j.1460-9568.1997.tb01424.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.