Abstract
[11C]P943 is a new radioligand recently developed to image and quantify serotonin 5-Hydroxytryptamine (5-HT1B) receptors with positron emission tomography (PET). The purpose of this study was to evaluate [11C]P943 for this application in humans, and to determine the most suitable quantification method. Positron emission tomography data and arterial input function measurements were acquired in a cohort of 32 human subjects. Using arterial input functions, compartmental modeling, the Logan graphical analysis, and the multilinear method MA1 were tested. Both the two tissue-compartment model and MA1 provided good fits of the PET data and reliable distribution volume estimates. Using the cerebellum as a reference region, BPND binding potential estimates were computed. [11C]P943 BPND estimates were significantly correlated with in vitro measurements of the density of 5-HT1B receptors, with highest values in the occipital cortex and pallidum. To evaluate noninvasive methods, two- and three-parameter graphical analyses, Simplified Reference Tissue Models (SRTM and SRTM2), and Multilinear Reference Tissue Models (MRTM and MRTM2) were tested. The MRTM2 model provided the best correlation with MA1 binding-potential estimates. Parametric images of the volume of distribution or binding potential of [11C]P943 could be computed using both MA1 and MRTM2. The results show that [11C]P943 provides quantitative measurements of 5-HT1B binding potential.
Keywords: brain, evaluation of new radiotracers, 5-HT1B serotonin receptors, human, positron emission tomography (PET), tracer kinetic modeling
Introduction
Serotonin mediates numerous effects in the central nervous system by binding to a large family of receptors (Hoyer et al, 2002) with various structures, functions, and pharmacology. Six families of metabotropic receptors and one family of ionotropic receptors are known. The 5-Hydroxytryptamine (5-HT1) receptor family includes three subtypes of receptors that inhibit the formation of cAMP (cyclic adenosine monophosphate) and have been shown to be functional in vivo: the 5-HT1A, 5-HT1B, and 5-HT1D receptors.
The 5-HT1B receptor has been shown to be involved in several physiologic functions, behaviors, and psychiatric diseases (for a review, see Sari, 2004). In particular, 5-HT1B receptors are expressed as auto-receptors on serotoninergic neurons (Engel et al, 1986), and as hetero-receptors on nonserotoninergic neurons (Maura and Raiteri, 1986). Several studies have indicated that 5-HT1B receptors are located on presynaptic terminals (Boschert et al, 1994; Sari et al, 1999), where they inhibit the release of neurotransmitters, including serotonin (Martin et al, 1992) and others (e.g., acetylcholine, Maura and Raiteri, 1986). 5-HT1B receptors have been shown to be involved in locomotor activity, drug abuse reinforcement, migraine, anxiety states (Lin and Parsons, 2002), aggressive behavior, and depression (Moret and Briley, 2000). In particular, 5-HT1B antagonists have been analyzed as anxiolytics and antidepressants (Hudzik et al, 2003; Dawson et al, 2006), and 5-HT1B/5-HT1D agonists are used for the treatment of migraine (Arulmozhi et al, 2005). 5-HT1B receptors have also been investigated to explain the antidepressant action of lithium (Chenu and Bourin, 2006).
A 5-HT1B receptor radioligand suitable for positron emission tomography (PET) imaging would be useful to study the function of these receptors in vivo and to investigate their role in pathophysiology. Only recently, two selective 5-HT1B radioligands, [11C]P943 (Nabulsi et al, 2007) and [11C]AZ10419369 (Pierson et al, 2008), have been developed and evaluated in vivo as PET imaging agents.
P943 (R-1-[4-(2-methoxy-isopropyl)-phenyl]-3-[2-(4-methyl-piperazin-1-yl)benzyl]-pyrrolidin-2-one) is a potent 5-HT1B antagonist in vitro, which binds with high affinity to the human 5-HT1B receptors expressed in HeLa cells (Ki=0.77 nmol/L) (McCarthy et al, 2007; US patent application number 20080206137; see Supplementary data). In the aforementioned study, the affinity of P943 for other 5-HT and non-5-HT receptors was at least 100-fold lower, making this a highly 5-HT1B-selective ligand. In a subsequent study, P943 affinity was confirmed to be at least 1,000-fold higher for 5-HT1B receptors (Ki=1.2 nmol/L) than for 5-HT2A, 5-HT2B, 5-HT2C, and 5-H7 receptors (Ki>1 μmol/L), more than 100 times higher than for 5-HT3 receptors (Ki=157 nmol/L), and more than 50 times higher than for 5-HT1A receptors (Ki=62 nmol/L) (data obtained from the NIMH Psychoactive Drug Screening Program; see Supplementary data). P943 affinity for 5-HT1D receptors was only 10 times lower than for 5-HT1B receptors (Ki=12 nmol/L); however, the density of 5-HT1D receptors is also at least fivefold lower than that of 5-HT1B receptors (Varnäs et al, 2001); therefore, the fraction of [11C]P943 in vivo binding due to 5-HT1D receptors is expected to be negligible. The in vivo specificity of [11C]P943 binding was confirmed in rhesus monkey during a blockade study using the 5-HT1B/1D antagonist, GR127935 (see Supplementary data). P943 behaves as an antagonist in vitro, blocking the reduction in adenylate cyclase activity elicited in the isolated guinea pig substantia nigra by a 5-HT1B agonist (McCarthy et al, 2007).
The aims of this study were to evaluate the suitability of [11C]P943 to quantify the binding potential of 5-HT1B receptors in humans, and to determine the most accurate and precise quantification method. Dynamic PET images and arterial input functions were measured in human subjects to model [11C]P943 brain kinetics. Noninvasive methods avoiding arterial input function measurements and allowing high-resolution parametric imaging were also investigated.
Materials and methods
Subjects
A total of 32 subjects were included in the study to evaluate the kinetic modeling methods over a broad range of subjects. Among these subjects, 8 were healthy normal subjects and 24 were noncontrol subjects. Noncontrol subjects had psychiatric disorders, such as depression or post-traumatic stress disorders (PTSD), but did not have neurodegenerative disorders. As part of the screening procedure, magnetic resonance (MR) images were acquired for each subject to exclude those with brain abnormalities. Psychiatric evaluation included a clinical interview and application of the SCID (Structured Clinical Interview for Diagnostic) for DSM-IV (Statistical Manual of Mental Disorders, 4th Edition) axis I disorders. Patients with depression and PTSD were all medication-naive, and no use of medications was allowed during the study. The absence of recent substance use was determined by self-report and confirmed by urine toxicology and Breathalyzer on both the day of screening and on the PET scan day, before tracer injection. Moreover, this study only includes subjects whose arterial input function was successfully measured.
This study was conducted under protocols approved by the Yale School of Medicine Human Investigation Committee, the Human Subjects Subcommittee of the Veterans Affairs Connecticut Healthcare System, and the Yale Radiation Safety Committee. Subjects were recruited by public advertisement. The study protocol described methods, procedures, and prespecified inclusion/exclusion criteria. Written informed consents were obtained from all participants after a complete explanation of study procedures.
Radiochemistry
[11C]P943 was prepared by N-methylation of the precursor with [11C]methyl triflate, using the PETtrace cyclotron, MeI Microlab, and a TRACERLab FxC automated synthesizer (GE Healthcare, Chalfont St Giles, UK) as previously described (Nabulsi et al, 2007). Both the P943 standard and the N-desmethyl precursor were provided by Pfizer (Groton, CT, USA). See Supplementary data for additional details.
Positron Emission Tomography Imaging
In preparation for the scan, the antecubital vein and the radial artery were cannulated for radiotracer injection and blood sampling, respectively. Positron emission tomography imaging was performed using the high-resolution research tomograph (Siemens/CTI, Knoxville, TN, USA), which acquires 207 slices (1.2 mm slice separation) with a reconstructed image resolution of ∼3 mm. After a transmission scan, 585±139 MBq of [11C]P943 was injected over 1 min using an infusion pump (Harvard PHD 22/2000, Harvard Apparatus, Holliston, MA, USA). The tracer-specific activity at the time of injection was 150±79 MBq/nmol. The injected mass was 2.3±1.7 μg (max 9.6 μg) or 0.028±0.017 μg/kg (max 0.092 μg/kg). List mode data were acquired for 120 mins.
Arterial Input Function Measurement
Whole Blood and Plasma
Continuous and sequential discrete blood samples were obtained. For the first 7 mins, arterial blood was collected continuously using a peristaltic pump (4 mL/min), and radioactivity in whole blood was measured using a cross-calibrated radioactivity monitor (PBS-101, Veenstra Instruments, Joure, The Netherlands). During this period, the pump was stopped to collect samples at 3 and 5 mins. Subsequently, discrete blood samples were collected at 7, 10, 15, 20, 25, 30, 40, 50, 60, 75, 90, 105, and 120 mins after injection. The plasma was separated from blood cells by centrifugation (3,900 g for 5 mins at 4°C; Allegra X-22R Centrifuge, Beckman Coulter, Fullerton, CA, USA). Whole blood and plasma samples were counted in a cross-calibrated well counter (Wizard 1480, Perkin-Elmer, Waltham, MA, USA). The plasma time–activity curve (TAC) during the first 7 mins was estimated from the continuous whole blood TAC. The ratio of the whole-blood-over-plasma concentration was calculated for each sample collected between 3 and 30 mins, fitted to a linear function, and then extrapolated between 0 and 7 mins.
Plasma-Free Fraction
Radiolabeled [11C]P943 (740 kBq) was added (<25 μL/mL) to a blood sample collected before injection to produce a blood standard. After mixing and centrifugation, plasma water was separated from plasma proteins using ultrafiltration tubes (Centrifree UF device number 4104, Millipore, Billerica, MA, USA) and centrifugation (1,100 g for 20 mins; IEC Medilite centrifuge, Thermo Fisher Scientific, Waltham, MA, USA). Plasma and water samples were counted in triplicate, and free fraction (fP) was estimated as the ratio of the mean concentration in water and plasma.
Determination of Ligand Metabolism in Plasma
For selected samples (10, 20, 40, 60, and 90 mins after injection) plasma proteins were precipitated using acetonitrile (1 mL/mL plasma) and centrifugation (14,000 g for 4 mins; minispin plus centrifuge, Eppendorf, Westbury, NY, USA). The supernatant was counted and analyzed by reverse-phase HPLC (high performance liquid chromatography). The HPLC system consisted of an isocratic pump (LC-20A1, Shimadzu, Kyoto, Japan), an injector (7725i, Rheodyne, Rohnert Park, CA, USA) equipped with a 2 mL sample loop, and a C18 SymmertyPrep column (7.8 × 300 mm2, 7 μm, Waters, Milford, MA, USA). The column was eluted with acetonitrile and aqueous 50 mmol/L ammonium acetate (pH 5.8 (55/45)) at a flow rate of 2.8 mL/min. The output of the HPLC column was connected to a fraction collector (CF-1 Fraction Collector, Spectrum Chromatography, Houston, TX, USA). Fractions were collected every minute and counted in a well counter.
The ratio of the radioactivity concentrations in the supernatant and plasma was obtained and fitted to a quadratic curve (fS). The unchanged fraction in the supernatant was fitted to an integrated γ-function:
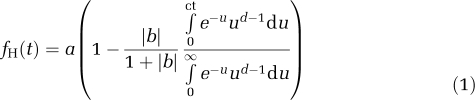 |
For improved consistency of the fits, the fourth parameter d was fixed at 2.2 (i.e. the mean value obtained with unconstrained four-parameter fits over all subjects). The unchanged fraction in the plasma was then computed as the product of functions fS and fH.
Determination of [11C]P943 Plasma Clearance and Clearance Rate
The terminal clearance rate β of [11C]P943 from the plasma was estimated by fitting the input function curve, between 30 and 120 mins, to a single exponential. Clearance was estimated by dividing the injected dose by the integral of the input function curve from 0 to ∞, where the input function curve was extrapolated on the basis of the aforementioned fit.
Magnetic Resonance Imaging
Magnetic resonance imaging was performed on a 3-T whole-body scanner Trio (Siemens Medical Systems, Erlangen, Germany) with a circularly polarized head coil. The dimension and pixel size of MR images were 256 × 256 × 176 and 0.98 × 0.98 × 1.0 mm3, respectively.
Image Reconstruction and Motion Correction
Dynamic scan data were reconstructed with all corrections (attenuation, normalization, scatter, randoms, and dead time) using the Motion-compensation Ordered subset expectation maximization (OSEM) List-mode Algorithm for Resolution-recovery Reconstruction (MOLAR) algorithm (Carson et al, 2003) with the following frame timing: 6 × 30 secs, 3 × 1 mins, 2 × 2 mins, and 22 × 5 mins. Motion correction was performed by image smoothing (Gaussian filter with a FWHM (full-width at half-maximum) of 3 pixels), followed by coregistration to an early summed image (0 to 10 mins after injection) using a six-parameter mutual information algorithm (FLIRT, FSL 3.2, Analysis Group, FMRIB, Oxford, UK). No correction was applied for misalignment between transmission and emission scans. On the basis of the estimated motion transformation matrices, the maximum displacement of each of the brain regions used in this analysis was calculated and these values were averaged across regions for each subject. The average maximum motion was 6.0±2.0 mm (n=32).
Regional Time–Activity Curve Computation
Gray matter regions-of-interest (ROIs) were taken from the AAL (Anatomical Automatic Labeling) template (Tzourio-Mazoyer et al, 2002), delineated on an MR template (Collins et al, 1998). Seven TACs were produced: the cerebellum (84 cm3 in template space), pallidum (4.6 cm3), as well as the frontal (256 cm3), occipital, parietal (64 cm3), and temporal (172 cm3) cortices. Two regions were selected in the occipital cortex: one with relatively low binding (referred to as occipital, 80 cm3) and one with relatively high binding (referred to as calcarine, 33 cm3). Nine additional ROIs were used to compare in vivo data with in vitro measurements of the density of 5-HT1B receptors: the caudate nucleus (16 cm3), putamen (17 cm3), insular cortex (29 cm3), anterior and posterior cingular cortices (22 and 6.4 cm3, respectively), raphe nucleus (1.4 cm3), substantia nigra (2.3 cm3), and the pulvinar and dorsomedial nuclei of the thalamus (5.1 and 2.8 cm3, respectively). The latter four regions were added to the template initially based on the Talairach and Tournoux atlas (Talairach and Tournoux, 1988), which is a stereotactic atlas of the thalamus (Morel et al, 1997), and the cytoarchitecture of the dorsal raphe (Baker et al, 1990). Subsequently, the position of these regions was slightly adjusted by application to spatially normalized high-resolution research tomograph PET images from tracers, which have high focal uptake in these regions.
To apply the ROIs to the PET data, a summed image (0 to 10 mins after injection) was created from the motion-corrected data and registered to the subject's T1-weighted 3-T MR image (six-parameter registration), which, in turn, was registered to the MR template using a 12-parameter affine transformation.
Kinetic Modeling
Compartmental Modeling Using the Input Function
Kinetic analysis of regional TACs was performed with the one-tissue (1T) and two-tissue (2T) models (for a review see, Gunn et al, 2001), using the following equation:
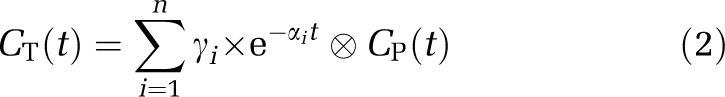 |
where n equals the number of tissue compartments. For the 1T model, γ1 is K1, the influx rate constant and α1 is k2, the efflux rate constant. For the 2T model, the eigenvalues α1 and α2 and the coefficients γ1 and γ2 are linked to the kinetic parameters K1, k2, k3 and k4 through equation set C.7 in Gunn et al (2001). The main parameters of interest are the total volume of distribution VT (computed as K1/k2 (1+k3/k4)), and the binding potential BPND (estimated as VT/VND−1, where VND is the distribution volume of the reference region, the cerebellum).
Linear Graphical Analyses
Three linear graphical analyses were tested. The first is the original graphical analysis with input function (GAIF) (Logan et al, 1990) using the following equation:
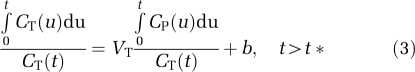 |
The remaining two graphical analyses use a reference region TAC. The first analysis (GAREF1) uses the following a two-parameter equation (Logan et al, 1996):
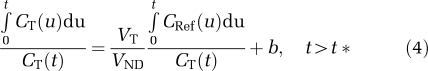 |
and the second analysis (GAREF2) uses the following three-parameter equation (Ichise et al, 1996; Logan et al, 1996):
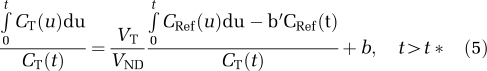 |
where b′ is the y-intercept (i.e., b) of the regression line in the GAIF analysis of the reference region TAC. In GAREF2, b′ was set to the average (across all 32 subjects) estimate of b obtained using MA1 (see below) in the cerebellum.
Multilinear Graphical Analyses
Two multilinear analyses were evaluated: MA1 (Ichise et al, 2002), using input function data, and the second version of the Multilinear Reference Tissue Model (MRTM) analysis (Ichise et al, 2003), using the cerebellum as a reference region. These two multilinear methods are based on the same hypotheses as the corresponding linear graphical analyses (GAIF for MA1 and GAREF2 for MRTM), but reduce the noise-induced bias on the parameter estimates by rearranging Equations (3) and (5). For MA1, Equation (3) is rearranged as follows:
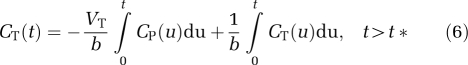 |
and for MRTM, Equation (5) is rearranged as:
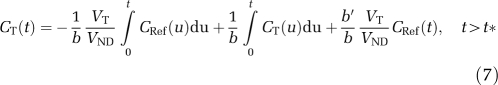 |
As b′ is a parameter linked only to the reference region TAC, it should ideally be the same in all regions (Ichise et al, 2003). Thus, two approaches were tested to reduce the noise in the parameter estimates: first, all TACs from a single subject were fitted simultaneously with a common b′ (MRTM2 method). In the second approach, b′ was fixed to the global average value obtained with MA1, as in the GAREF2 method (MRTM2* method).
Compartmental Modeling Without Input Function
The Simplified Reference Tissue Model (SRTM) (Lammertsma and Hume, 1996) was also evaluated using the following equation:
where k2 and k′2 are the efflux rates from the target region and the cerebellum, respectively, and R1 is equal to the ratio of the influx rates in the target region K1 and in the cerebellum K′1. This model assumes that a 1T model can describe both the target and the reference region TACs. Similar to b′ in MRTM, k′2 is linked only to the cerebellum. Thus, we also attempted to reduce the noise by fitting all target TACs simultaneously with a common k′2 (SRTM2: Wu and Carson, 2002).
Normalization by the Free Fraction
Two others binding potentials, BPP and BPF (Innis et al, 2007), were also evaluated as VT−VND and (VT−VND)/fP, respectively.
Parametric Imaging
For parametric imaging, the MA1, SRTM, SRTM2, MRTM, and MRTM2 methods were tested. First, the dynamic images were smoothed using a Gaussian filter (FWHM=3.7 mm) before computing parametric images. Both SRTM2 and MRTM2 are two-step procedures that reduce statistical noise in parametric images by fixing k′2 (in SRTM) or b′ (in MRTM) to a common value in all brain voxels. b′ (or k′2) was estimated by first computing MRTM (or SRTM) images, then by computing the median of b′ (or k′2) estimates from selected brain voxels (4>BPND>0.5, and −1,000<b<−1 for MRTM or 1>k2>0.0001 for SRTM).
Data Weights and Parameter Estimation
Parameters were estimated using weighted least squares with weights on the basis of the noise-equivalent counts in each frame. For compartmental modeling of regional TACs, nonlinear parameter estimation was performed using a Marquardt–Levenberg algorithm (Marquardt, 1963) and custom software for IDL 6.2 (ITT Visual Information Solutions, Boulder, CO, USA). For parametric imaging using the SRTM and SRTM2 methods, a basis function approach was used. For linear and multilinear analyses, a noniterative method was used. Standard errors on the parameter estimates were estimated by computing the theoretical covariance matrix (Beck and Arnold, 1977, chapter 7.7.1) and the error propagation equation (Bevington and Robinson, 2003, chapter 3). Values are reported as rSE (relative s.e.), as a percentage of the parameter value.
Effect of the Scan Duration on VT and BPND Estimates
The effect of the scan duration on VT and BPND parameter estimates was investigated for MA1 and MRTM2*. The fit interval was reduced from 0 to 120 mins to 0 to 60 mins, by steps of 10 mins. t* was kept at 40 mins. For each healthy subject, region, and scan duration d, the distribution volume VTd and binding potential BPNDd were estimated. Two statistics were then computed across subjects: first, the relative mean values of VTd (and BPNDd, respectively), i.e., the average of VTd (BPNDd, respectively) divided by the average of VT120 (BPND120, respectively); then, the coefficients of variation, i.e., the s.d. of VTd (BPNDd, respectively) divided by the average of VTd (BPNDd, respectively). Both statistics were expressed as percentages.
Statistical Analyses
To compare methods, correlations and relative differences between sets of parameter estimates were computed using all 32 subjects. For the presentation of normal parameter values, means and s.d. were computed using only healthy normal subjects, unless otherwise specified.
Results
Input Function
Radioactivity concentration in whole blood was always lower than that in the plasma. The whole blood to plasma concentration ratio was 0.79±0.06 and 0.85±0.05 at 3 and 120 mins after injection, respectively. The free fraction of [11C]P943 in the plasma fP was 5.8±0.5% (n=8 healthy controls). [11C]P943 was slowly metabolized: unchanged [11C]P943 represented 89±4% of the total plasma radioactivity at 20 mins after injection, and 53±9% at 90 mins. Radioactivity recovered in the supernatant after plasma protein precipitation slowly decreased to 89±1% at 90 mins after injection. Overall, 94±14% of the radioactivity was recovered at the output of the HPLC system. All metabolites were more polar than the parent compound. [11C]P943 plasma clearance was 1.6±0.2 L/min in healthy controls (range=1.2 to 2.0 L/min, n=8) and 1.7±0.3 L/min when all subjects were included (range=0.8 to 2.3 L/min, n=32). The clearance rate β was 0.010±0.004 per min in healthy controls (range=0.006 to 0.020 per min, n=8) and 0.012±0.005 per min in the full cohort (range=0.005 to 0.022 per min, n=32).
Brain Time–Activity Curves
The average brain uptake of [11C]P943 peaked at 0.30±0.05 percent of injected dose per 100 mL (%ID/100 mL) at 20 mins after injection, and then slowly decreased, reaching 0.16±0.03%ID/100 mL at 120 mins after injection. Figure 1 shows selected TACs measured for one healthy normal subject. The radioactivity concentration peaked at ∼10 mins after injection in the cerebellar cortex, a region devoid of 5-HT1B receptors (Varnäs et al, 2001), and between 20 and 40 mins in cortical areas. In the pallidum, a small region with a high density of 5-HT1B receptors, the TAC peaked at 20 mins for some subjects, and then slowly decreased, whereas in other subjects, no clear peak was seen and the TAC reached a plateau after 50 mins.
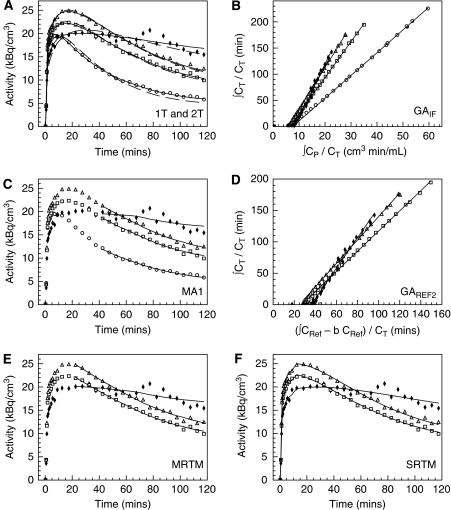
Figure 1.
Time–activity curves, Logan plots, and fits obtained in selected ROIs: cerebellum (circles), parietal (squares), calcarine cortex (triangles), and pallidum (solid diamonds). (A) Fits obtained with the one-tissue (1T) compartment model (dashed line) or the two-tissue (2T) compartment model (solid line). (B) Graphical analysis with input function (GAIF). (C) Fits obtained with the multilinear analysis using the input function (MA1). (D) Graphical analysis using reference region data GAREF2. (E) Fits obtained with multilinear analyses using reference region data (MRTM). (F) Fits obtained with the simplified reference tissue model (SRTM). t* was set to 40 mins for panels B and C, and to 10 and 20 mins for panels D and E, respectively. 313 × 352 mm2 (600 × 600 d.p.i.).
Transient equilibrium was not reached between the plasma and any brain region. Moreover, the pallidum-to-cerebellum concentration ratio steadily increased during the scan (2.9±0.2 at 120 mins after injection, n=8). The ratio of the cortex to cerebellum was also increasing throughout the scan, but reached 95% of its 120-min value (2.0±0.2, n=8) by 80 mins.
Compartmental Modeling Using the Input Function
To compare the quantification methods, seven TACs were fitted for each subject: the cerebellum, pallidum, and five cortical regions. The 2T model provided better fits of the data than did the 1T model in all regions. The residual sum of squares, χ2, Akaike information criterion, and Schwartz criterion statistics were significantly higher for 1T than for 2T in all regions (P<0.001, uncorrected paired t-test). F-tests indicate that the χ2 is significantly higher for 1T than for 2T, for all 224 TACs except 1. Figure 1A show typical fits obtained in one subject for the 1T and 2T models.
For both models, the K1 estimates in the cortex and cerebellum were relatively uniform (0.22±0.03 mL/min per cm3 for 2T, n=8) and higher than in the pallidum (0.18±0.04 mL/min per cm3 for 2T, n=8). In all regions, 1T K1 estimates were 18±6% (n=32) lower than those of the 2T model.
The average VT estimates for the 1T and 2T models in healthy normal subjects are listed in Table 1. The 2T VT values ranged from 5.4±1.1 mL/cm3 in the cerebellum to 15.6±4.1 mL/cm3 in the pallidum, and BPND estimates ranged from 0.62±0.09 in the parietal cortex to 1.81±0.32 in the pallidum.
Table 1. [11C]P943 volumes of distribution and binding potentials estimates in healthy controls (n=8).
| Parameters | Volume of distribution VT (mL/cm−3) | Binding potential BPND (unitless) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Methods | 1T | 2T | GAIFa | MA1a | 2T | MA1a | GAREF2a,b | MRTMa | MRTM2*a,b |
| Cerebellum | 4.9±1.0 (19%) | 5.4±1.1 (21%) | 5.4±1.1 (21%) | 5.4±1.1 (21%) | |||||
| Pallidum | 13.2±3.6 (27%) | 15.6±4.1 (27%) | 14.0±3.8 (27%) | 15.2±3.9 (26%) | 1.81±0.32 (17%) | 1.80±0.28 (16%) | 1.56±0.19 (12%) | 1.90±0.47 (25%) | 1.80±0.25 (14%) |
| Calcarine | 10.2±2.1 (20%) | 10.6±2.2 (21%) | 10.6±2.2 (21%) | 10.6±2.2 (21%) | 0.97±0.14 (14%) | 0.98±0.15 (15%) | 0.99±0.15 (15%) | 0.94±0.14 (15%) | 0.99±0.15 (15%) |
| Occipital | 9.0±2.1 (23%) | 9.5±2.2 (24%) | 9.4±2.3 (24%) | 9.4±2.2 (24%) | 0.75±0.22 (30%) | 0.75±0.23 (31%) | 0.76±0.24 (31%) | 0.72±0.23 (32%) | 0.76±0.24 (32%) |
| Parietal | 8.3±1.5 (18%) | 8.7±1.6 (19%) | 8.6±1.6 (19%) | 8.7±1.6 (19%) | 0.62±0.09 (15%) | 0.62±0.10 (16%) | 0.62±0.11 (18%) | 0.59±0.10 (18%) | 0.62±0.11 (18%) |
| Frontal | 9.2±1.8 (19%) | 9.7±2.0 (20%) | 9.7±2.0 (21%) | 9.6±2.0 (20%) | 0.79±0.11 (14%) | 0.80±0.12 (15%) | 0.80±0.13 (17%) | 0.76±0.13 (17%) | 0.81±0.14 (17%) |
| Temporal | 8.7±1.8 (20%) | 9.2±1.9 (21%) | 9.1±1.9 (21%) | 9.1±1.9 (21%) | 0.69±0.08 (11%) | 0.70±0.09 (12%) | 0.70±0.10 (14%) | 0.66±0.09 (14%) | 0.71±0.10 (14%) |
MRTM, Multilinear Reference Tissue Model; 1T, one tissue model; 2T, two-tissue model.
Variability is shown as s.d. across subjects (percentage s.d. in parentheses).
t* was set to 40 mins for GAIF and MA1, and to 10, 20, and 40 mins for GAREF2, MRTM, and MRTM2*, respectively.
b′ was fixed to the mean b value estimated in the cerebellum with MA1.
The rSE estimated by sensitivity analysis were ≤3% for K1 and 2% for VT with the 1T model, and ≤17% for K1 (mean rSE=3%) and ≤15% for VT (mean rSE=2%) with the 2T model.
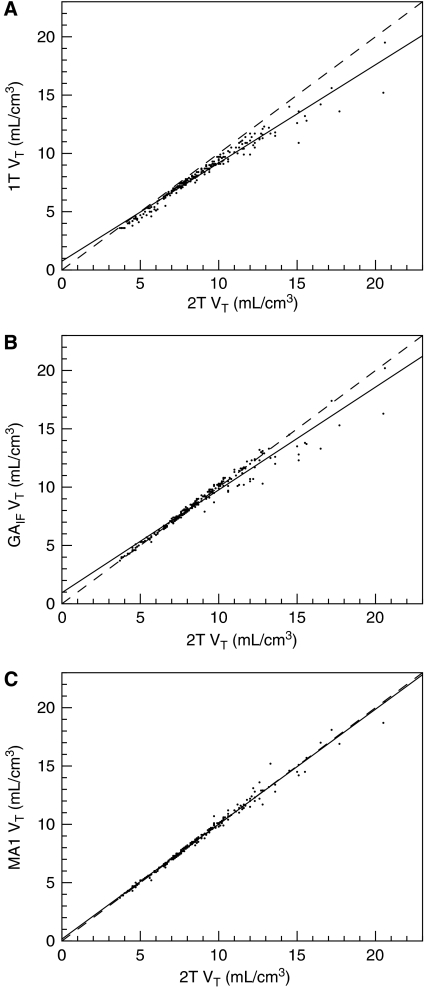
The correlations between VT estimates obtained using various methods are shown in Figure 2. The corresponding regression equations and the relative differences between methods across regions are presented in Table 2. In particular, 1T VT estimates tended to be lower than those of the 2T model in all regions, whereas 1T BPND estimates were higher than 2T estimates in cortical areas, and lower in the pallidum.
Figure 2.
Selected correlations between VT estimated with analyses using input function data. Solid line represents the line of best fit. Dashed line is the line of identity. (A) 1T versus 2T (B) GAIF versus 2T (C) MA1 versus 2T. See Table 2 for regression values. 156 × 352 mm2 (600 × 600 d.p.i.).
Table 2. Correlations and relative differences between VT or BPND estimates.
| Parameters | Method 1 | Method 2 | Correlation | Relative difference | ||||
|---|---|---|---|---|---|---|---|---|
| Slope | Intercept | r2 | Cerebellum | Cortex | Pallidum | |||
| VT | 1T | 2T | 0.843 | 0.752 | 0.966 | −9±4% | −5±2% | −12±6% |
| GAIFa | 2T | 0.882 | 0.943 | 0.964 | 2±3% | 1±2% | −9±6% | |
| MA1a | 2T | 0.984 | 0.173 | 0.989 | 0.4±3% | 0.5±1% | 0.5±5% | |
| MA1b | MA1a | 0.978 | 0.126 | 0.983 | −2±3% | −0.7±3% | −0.4±6% | |
| BPND | 1T | 2T | 0.832 | 0.200 | 0.929 | 12±8% | −5±10% | |
| GAIFa | 2T | 0.758 | 0.162 | 0.903 | −1±7% | −17±11% | ||
| MA1a | 2T | 0.964 | 0.033 | 0.966 | 0.4±6% | 0.5±9% | ||
| GAREF1a | MA1a | 0.540 | 0.254 | 0.784 | −10±4% | −35±8% | ||
| GAREF2a | MA1a | 0.783 | 0.148 | 0.925 | −0.7±3% | −15±10% | ||
| MRTMa | MA1a | 1.043 | −0.069 | 0.833 | −5±3% | −0.2±29% | ||
| MRTM2a | MA1a | 0.865 | 0.082 | 0.868 | −2±6% | −10±18% | ||
| MRTM2*a,c | MA1a | 0.965 | 0.026 | 0.970 | −0.05±3% | −2±9% | ||
| SRTM | MA1a | 0.825 | 0.070 | 0.968 | −8±3% | −15±8% | ||
| SRTM2 | MA1a | 0.617 | 0.234 | 0.859 | −5±4% | −28±9% | ||
| MRTM2b | MA1a | 0.909 | 0.105 | 0.922 | 6±11% | 2±17% | ||
| SRTM2b | MA1a | 0.610 | 0.243 | 0.857 | −4±7% | −27±10% | ||
MRTM, Multilinear Reference Tissue Model; SRTM, Simplified Reference Tissue Model; 1T, one tissue model; 2T, two-tissue model.
Correlations and relative differences calculated from the various tested methods (n=32).
Slope and intercept are determined by linear fit of (Method 1)=slope × (Method 2)+intercept.
For GAIF and MA1, t* was set to 40 mins; for GAREF1 and GAREF2, t* was set to 50 and 10 mins respectively; for MRTM, MRTM2, and MRTM2*, t* was set to 20, 30, and 40 mins, respectively.
Using mean regional values from parametric images. t* was set to 20 mins to compute the MA1 and MRTM2 parametric images.
b′ was fixed to the mean b value estimated in the reference region with MA1 (i.e., −37 mins).
Graphical Analysis with Input Function
Typical Logan (GAIF) plots and MA1 fits are shown in Figures 1B and 1C, respectively. GAIF provided similar VT and BPND estimates than did 2T in the cerebellum and cortex, but lower VT and BPND estimates than did 2T in the pallidum (−9±6% and −17±11% for VT and BPND, respectively) (Table 2). Conversely, MA1 VT and BPND estimates in the pallidum were not underestimated (mean relative differences were <2% in all regions) and were very well correlated with 2T estimates (Figure 2C, Table 2). VT and BPND estimates obtained with MA1 (t*=40 mins) in healthy controls are listed in Table 1. Owing to the excellent agreement between MA1 and 2T and because of the computational simplicity of MA1 and its suitability for parametric imaging, MA1 will be used as the method of choice for modeling with the input function (see the ‘Discussion' section).
Graphical Analysis using a Reference Region
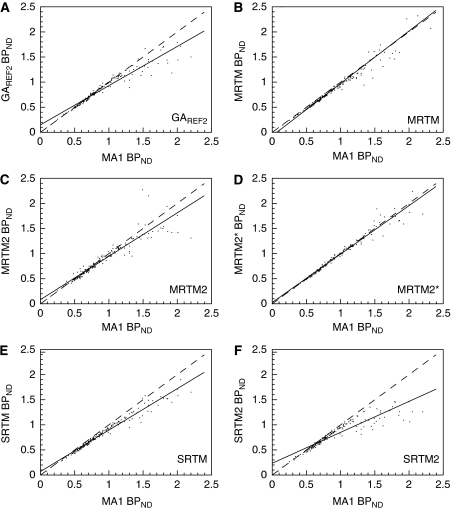
Typical Logan plots obtained using GAREF2 and fits obtained with MRTM, are shown in Figures 1D and 1E, respectively. The linear section of the GAREF1 Logan plot was not reached within the 120-min scanning duration (data not shown). Thus, GAREF1 BPND estimates were consistently lower than the 2T estimates for any t* setting. With t*=50 mins, the underestimation was −10±4% and −35±8% in the cortex and pallidum, respectively (Table 2). For GAREF2, the best correlation with MA1 BPND estimates was obtained with t*=10 mins (b′=−30 mins). GAREF2 then provided similar results to MA1 in the cortex, but underestimated BPND in the pallidum (by −15±10%, Table 2).
Stable MRTM BPND estimates could not be obtained with t* settings above 20 mins. Using MA1 as a reference, MRTM provides slightly larger underestimations of BPND values than does GAREF2 in the cortex (−5±3% with MRTM, −0.7±3% with GAREF2), but less underestimated BPND values than does GAREF2 in the pallidum (−0.2±29% with MRTM, −15±10% with GAREF2) However, pallidum MRTM BPND estimates were more variable than either MA1 or GAREF2 BPND estimates (the relative s.d. in healthy controls was 25% with MRTM, versus 16% with MA1 and 12% with GAREF2), because of the additional fitted parameter b′.
With MRTM2 (i.e., a coupled fit of all TACs from each subject, with a single parameter b′), only t* settings up to 30 mins could be tested. With this t* value, b′ was slightly higher than b estimated in the cerebellum with MA1 (−31±10 mins versus −34±5 mins or +8±34%). The MRTM2 BPND estimates in the pallidum were lower than MA1 estimates (−10±18% Table 2).
With MRTM2* (i.e., with b′ fixed to the mean MA1 estimate of b in the cerebellum), t* settings up to 70 mins could be used. The best correlation with MA1 BPND estimates was obtained with t*=40 mins (and b′ set to −37 mins). The mean relative difference between MA1 and MRTM2* BPND estimates was then below 2% in all regions.
BPND estimates, obtained with GAREF2, MRTM, and MRTM2* with t* set to 10, 20, and 40 mins, respectively, are listed in Table 1. The correlations between MA1 BPND estimates and these GAREF2, MRTM, MRTM2, and MRTM2* BPND estimates are presented in Figures 3A–3D.
Figure 3.
Selected correlations between BPND estimated with methods using the cerebellum as a reference region and MA1. Solid line is the line of best fit. Dashed line is the line of identity. (A) GAREF2 versus MA1. t* is 10 mins for GAREF2 and 40 mins for MA1. (B) MRTM versus MA1. t* is 20 mins for MRTM and 40 mins for MA1 (C) MRTM2, with b′ coupled across regions for each subject, versus MA1. t* is 30 mins for MRTM2 and 40 mins for MA1. (D) MRTM2*, with b′ fixed at −37 mins, versus MA1. t* is 40 mins for both methods. (E) SRTM versus MA1. (F) SRTM2, with k′2 coupled across region for each subject, versus MA1. See Table 2 for regression values. 313 × 352 mm2 (600 × 600 d.p.i.).
Simplified Reference Tissue Model
Typical fits obtained with SRTM are shown in Figure 1F. F-tests indicate that the χ2 is significantly higher for SRTM than for the 2T model for a majority of cortical TACs (95 out of 160), but only for a few pallidum TACs (4 out of 32). The χ2, Akaike information criterion, and Schwartz criterion statistics were significantly higher for SRTM than for 2T in the frontal and occipital regions, and in the calcarine region for χ2 (P<0.05, uncorrected paired t-test).
The SRTM BPND estimates were lower than MA1 estimates, especially in the pallidum (−15±8% Table 2). Estimates of k′2 were not constant across regions, being lower in the pallidum (0.035±0.004 per min, n=8) than in the cortex (0.051±0.006 per min, n=8). Moreover, attempting to reduce s.e. on BPND estimates by coupling the parameter k′2 across regions (SRTM2) leads to an even greater underestimation of pallidum BPND (−28±9% Table 2). With SRTM2, k′2 was 0.046±0.004 per min.
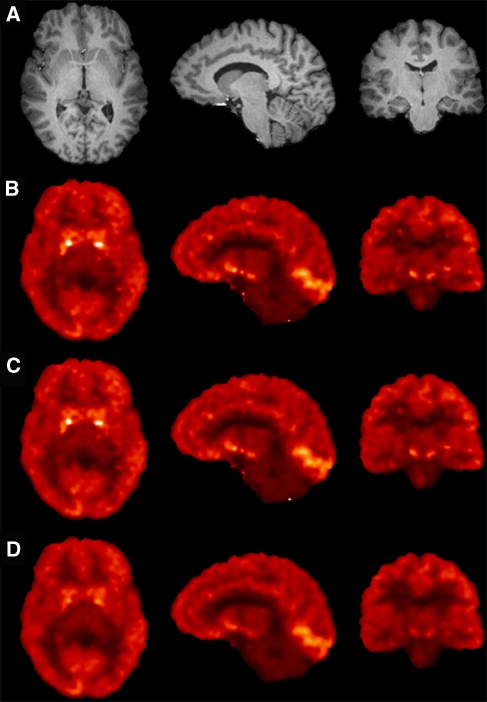
Parametric Imaging
Parametric images of VT (MA1) and BPND (MRTM2 and SRTM2) are shown in Figure 4. The value of t* was reduced to 20 mins for MA1 and MRTM2 to reduce noise. Parametric MA1 VT estimates were very well correlated with MA1 VT estimates using regional TACs (Table 2). Similarly, MRTM2 BPND estimates were highly correlated with MA1 BPND estimates using regional TACs (Table 2). Conversely, SRTM2 substantially underestimated BPND in high binding areas (Table 2).
Figure 4.
MR anatomic image (row A) and parametric images of VT obtained on the same healthy control subject with MA1 (row B), and BPND obtained with MRTM2 (row C) and SRTM2 (row D), in axial, sagittal, and coronal views (from left to right). The slices were selected to display high binding regions: pallidum (axial view), occipital (sagittal view), and substantia nigra (coronal view). The VT color scale ranges form 0 to 17.3 mL/cm3, and the BPND color scale ranges from −1 (to maximize the visual identity between VT and BPND images) to 3. Differences between the SRTM2 images and the MA1 or MRTM2 images are especially visible in the small receptor-rich regions, where SRTM2 BPND values are underestimated. 79 × 106 mm2 (300 × 300 d.p.i.).
The parameter b′, estimated from the first pass of MRTM2, was −33±5 mins, very close to the average b estimates with MA1 from cerebellum TACs with the same t* value (−32±4 mins). However, there was no significant correlation across subjects between these two sets of estimates (y=0.285x−23, r2=0.115, n=32, where y values are MRTM2 b′ estimates, and x values are MA1 b estimates in the cerebellum). Thus, as the coefficients of variation were small, estimation uncertainty seemed to be the dominant source of uncertainty. For SRTM2, k′2 was 0.045±0.006 per min.
Comparison of Binding Potential Outcome Measures
The binding potentials BPP and BPF were computed from MA1 VT estimates and [11C]P943 plasma-free fraction fP (for BPF). For healthy controls (n=8), BPP estimates ranged from 3.3±0.6 in the parietal to 9.8±2.9 in the pallidum, and BPF estimates ranged from 56±9 in the parietal to 168±47 in the pallidum (see Supplementary Table S5 in the Supplementary information file). Among the three binding potentials (i.e., BPND, BPP, and BPF), BPP estimates were the most variable (the relative s.d. is up to twice as high), and BPF estimates were slightly less variable than BPP estimates: e.g., the relative s.d. of BPND, BPP and BPF in the pallidum for healthy controls were 16, 29, and 28%, respectively (n=8). This suggests that variability in the measurement of the input function is a strong contributor to intersubject variability; this variability was minimized by the use of BPND.
Comparison of Binding Potential BPF Estimates with In Vitro Bmax Measurements
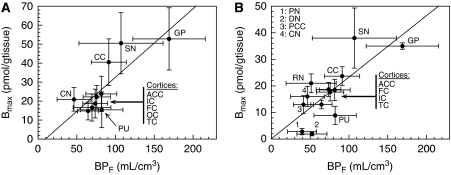
The correlations between BPF (Bmax/Kd) estimates obtained from healthy subjects (n=8) and measurements of the density of 5-HT1B receptors from two in vitro autoradiography studies (Varnäs et al, 2001, 2004) are presented in Figure 5. For this comparison, additional ROIs were included in the study as described in the ‘Materials and methods' section. BPF estimates were significantly correlated with the in vitro Bmax measurements (BPF=0.36 × Bmax−3, r2=0.671, n=10, P<0.05 for the first in vitro study and BPF=0.23 × Bmax+0.2, r2=0.576, n=13, P<0.001 for the second in vitro study). The regression slopes are estimates of the in vivo Kd, and these values are in good agreement with the reported in vitro Ki value (see the ‘Discussion' section).
Figure 5.
Correlations between BPF (Bavail/Kd) estimates from [11C]P943 regional TAC fits with MA1 (t*=40 mins) and the 5-HT1B Bmax measurements using in vitro autoradiography and [3H]GR 125743 reported by Varnäs et al (2001) (A) and Varnäs et al (2004) (B). Region labels are defined as: GP, pallidum; SN, substantia nigra; RN, raphe nucleus; PN, pulvinar nucleus; DN, dorsomedial nucleus; CN, caudate nucleus; PU, putamen; AAC, anterior cingulate cortex; PCC, posterior cingulate cortex; CC, calcarine cortex; FC, frontal cortex; IC, insular cortex; TC, temporal cortex. For cortical regions, the labels are grouped and displayed in the order of the points following the y axis (Bmax) values. In subfigure B, for the cortical regions, the in vitro Bmax values were computed as the unweighted average across cortical layers. For the pallidum, the in vitro Bmax values were computed as the unweighted average of the (internal and external) globus pallidus and ventral pallidum values. The regression line equations are BPF=0.36 Bmax−3, r2=0.671, n=10, P<0.05 (panel A) and BPF=0.23 Bmax+0.2, r2=0.576, n=13, P<0.001 (panel B). The slope of each regression line is an estimate of the in vivo Kd value, and these values are in good agreement with the reported in vitro Ki value (0.77 nmol/L; McCarthy et a, (2007); 1.2 nmol/L; NIMH Psychoactive Drug Screening Program). 355 × 133 mm2 (600 × 600 d.p.i.).
Effect of the Scan Duration on VT and BPND Estimates
To estimate the minimal PET data acquisition time required to derive stable VT or BPND estimates, all regional time activity curves from healthy subjects were fitted using MA1 or MRTM2* using varying total scan duration, ranging from 60 to 120 mins in 10-min increments. In the cortex and cerebellum, the coefficients of variation of VT and BPND estimates were practically constant for all scan durations tested (see Supplementary Figure S5 in Supplementary information file). When the scan duration was reduced, MA1 VT estimates very slightly decreased in the cerebellum and were almost constant in the cortex (Supplementary Figure S5A). Conversely, BPND estimates very slightly increased in the cortex when the scan duration was reduced (Supplementary Figure S5B). In the pallidum, for scan durations ≤70 mins, VT and BPND estimates were unstable (coefficient of variation ≥90%). When the scan duration was reduced from 120 to 80 mins, mean VT estimates very slightly decreased (Supplementary Figure S5A) and BPND estimates were practically constant (Supplementary Figure S5B). The coefficient of variation was practically constant for VT, but increased from 15% (for both MA1 and MRTM2*) to 29% for MA1, and 32% for MRTM2*. Consequently, using 90-min scan duration provided VT and BPND estimates within ±5% of 120-min scan estimates, in average, for all regions, but with a sizeable increase in the coefficient of variation of the BPND estimates in the pallidum (23 versus 15%).
Discussion
The primary goal of this study was to evaluate the suitability of [11C]P943 to quantify the binding potential of 5-HT1B receptors in humans, and to evaluate the most suitable quantification method. [11C]P943 is one of the first radioligands available to quantify 5-HT1B receptors in vivo, along with [11C]AZ10419369 (Pierson et al, 2008).
Regional Distribution and In Vivo Equilibrium Dissociation Constant
The distribution of [11C]P943 binding is correlated with the known density of 5-HT1B receptors from in vitro autoradiography studies (Varnäs et al, 2001, 2004). The correlation, although significant, is not perfect, with mainly two regions having higher binding than predicted by in vitro data (putamen and thalamus), and several regions having lower binding than expected from in vitro data (caudate, calcarine area when compared with data from Varnäs et al (2001), substantia nigra, raphe nucleus). These differences may be introduced by the partial volume effect, especially as the size of these regions in not similar, and spillover is different for each region. Moreover, there are surely differences in the delineation of the regions between the in vitro and in vivo studies.
However, it is worth observing than the in vivo Kd of [11C]P943 estimated using the slope of the regression line between the BPF estimates and Bmax measurements (0.36 and 0.23 nmol/L, respectively) is close to the Ki values estimated in vitro (0.77 nmol/L; McCarthy et al, 2007; 1.2 nmol/L; NIMH Psychoactive Drug Screening Program). This indicates that the theoretical prediction of binding potential values taking into account the receptor density (Bavail), radioligand affinity (Kd), plasma-free fraction (fp), and nonspecific binding in brain tissue (Innis et al, 2007) is well suited for [11C]P943.
The free fraction in brain tissue fND can be estimated to be ∼1% from the measurement of the plasma-free fraction fP (5.8±0.5%) and the cerebellum distribution volume VT (5.4±1.1 mL/cm3). Using this estimate, the lowest in vivo Kd estimate (0.23 nmol/L) and the peak uptake value observed in the cerebellum to estimate the nondisplaceable [11C]P943 concentration (0.29% ID/100 mL), the tracer injected dose limit could be estimated to be ∼40 nmol or ∼17 μg, where a tracer dose is assumed to occupy <5% of receptors. On the basis of this analysis, these studies were conducted at tracer levels.
The Cerebellum as a Reference Region
In vitro studies in rodents have shown that 5-HT1B receptors mRNA can be detected in the cerebellum, in cell bodies of the Purkinje cells in the cerebellar gray matter (Voigt et al, 1991). However, using immunoreactivity or autoradiography, 5-HT1B receptors were only detected in the deep nuclei of the cerebellum (Pazos and Palacios, 1985; Sari et al, 1999). This localization mismatch was attributed to the fact that 5-HT1B receptors are localized at the end of the projections of the Purkinje cells in the deep nuclei (Boschert et al, 1994). In vitro studies of the distribution of 5-HT1B receptor mRNA in humans did not include the cerebellum (Varnäs et al, 2005), but autoradiographic studies did not show specific binding in cerebellar gray matter (Varnäs et al, 2001, 2004), as in rodents. On the basis of these data, cerebellar gray matter may be used as a reference region, although additional pharmacological studies are necessary to validate this reference region. Moreover, care must be taken to reduce spillover from the occipital cortex, which shows high levels of specific binding, as well as from the deep nuclei. Thus, the cerebellum ROI used in this study was located in the lower half of the cerebellum gray matter.
Compartmental Modeling
Kinetic analysis of [11C]P943 brain TACs showed that all curves were better described by the 2T model. The lack of fit of the 1T model is more visually apparent for the cerebellum and pallidum TACs, and differences between 1T and 2T distribution volumes estimates were higher in these regions than in the cortex.
The need for a 2T model to accurately fit the cerebellum TAC, although it is the reference region and (putatively) is devoid of specific binding, has already been reported for other radioligands (Watabe et al, 2000). This may be caused by tissue heterogeneity as the grey and white matter are very finely interwoven in this structure. In addition, being a region with relatively low activity, the cerebellum may be more sensitive to scatter correction errors, or biases in the OSEM reconstructions at low counts (Comtat et al, 2004; van Velden et al, 2008). It is noteworthy that the latter effects can be avoided when randoms and scatters are included in the iterative steps (Comtat et al, 2004), as is the case in the MOLAR reconstruction algorithm. Indeed, with this algorithm, biases were only detected for frames with <200,000 noise-equivalent counts (Planeta-Wilson et al, 2008), which is much lower than any frame in the present data.
Similarly, the need for a 2T model in the receptor-rich pallidum is not completely intuitive as regions with higher binding potentials tend to be more 1T-like (Carson et al, 1998; Watabe et al, 2000). However, the need for a 2T model may also be due to the spill-in in this small region, especially at the beginning of the scan as the influx rate K1 is relatively lower in this region, or heterogeneity of the receptor distribution within the region as the ventral part of the pallidum has a much higher receptor density (Varnäs et al, 2001, 2004).
The equation used for the compartmental models (i.e., Equation (2)) does take into account the effect of radioactivity in the blood. However, this effect is small for [11C]P943 for three reasons: whole blood radioactivity remains lower than total plasma radioactivity throughout the scan; [11C]P943 metabolism in plasma is slow ([11C]P943 unchanged fraction is still ≥50% 90 after injection); and [11C]P943 distribution volumes are relatively high (≥5 mL/cm3). Thus, assuming that the blood volume in the brain is 0.05 mL/cm3, and that its radioactivity concentration can be approximated by the arterial blood radioactivity concentration, blood radioactivity would represents only 2% of the total activity in the cerebellum at the end of scan, and <1% in all other regions. Consequently, adding a fixed vascular fraction of 0.05 mL/cm3 to the 2T model would produce VT estimates 2±1% lower in the cerebellum and <1% lower in the other regions. The effect on the binding-potential estimates would be somewhat higher: the cortical BPND estimates would be 3±2% higher and the pallidum BPND estimates would be 4±5% higher. This small bias would be present in all tested methods, as they all neglect the blood activity; thus, ignoring the blood volume correction in the 1T and 2T fits permits a more direct comparison of all modeling methods.
Comparison of Binding Potential Outcome Measures
Among the three binding potentials, BPND estimates were the least variable across healthy controls, suggesting that variability in the measurement of the input function is a strong contributor to intersubject variability. In the measurement of the input function, the unchanged fraction estimation may be the strongest contributor to intersubject variability, as at the end of scan the relative s.d. of total plasma measurements, expressed in percentage of injected dose, is lower or equal (15%) to the relative s.d. of brain TACs (14% to 21% depending of the ROI), whereas the relative s.d. of the unchanged fraction curve is higher (26%).
Comparison of [11C]P943 and [11C]AZ10419369
At present, only qualitative comparisons between [11C]P943 and [11C]AZ10419369 (Pierson et al, 2008) in human beings can be made, as arterial input function measurements and full kinetic modeling of [11C]AZ10419369 are required for a comprehensive comparison of these two tracers. The two tracers appear to have similar brain uptake ([11C]P943 average brain uptake is 0.30±0.05% ID/100 mL of tissue at peak, which corresponds to a whole-brain uptake of 4.2% ID; this is very comparable with the brain uptake of 3.5% for [11C]AZ10419369). The time of peak uptake in the reference region (or the average brain) is higher for [11C]P943 than for [11C]AZ10419369 (<5 mins). Conversely, the washout from the reference region (or the average brain) is faster for [11C]P943 than for [11C]AZ10419369. These differences will be due to differences in both brain and plasma kinetics. Both tracers have qualitatively similar kinetics in the pallidum, with a lower initial uptake than in the cortex and no apparent transient equilibrium with the reference region. In the cortex, both tracers reach an apparent transient equilibrium between the cortical regions and the cerebellum, but [11C]P943 reached that state later (at 80 mins) than [11C]AZ10419369 (30 mins). The values of the (target) tissue to reference tissue concentration ratio at 120 mins after injection were similar for [11C]P943 and [11C]AZ10419369 (∼3 in the pallidum and ∼2 in the cortical regions).
Transient Equilibrium State in Bolus Studies
[11C]P943 does not reach a transient equilibrium state between the plasma and any brain region within 120 mins. In particular, the tissue-to-plasma ratio increases constantly during the scan in the pallidum. Similarly, results from 2T fits indicate that the smallest eigenvalue α1 (0.009±0.002 per min) is lower than the exponential decay rate of the input function β (0.012±0.004 per min) in 18 out of the 32 subjects. In this case, no transient equilibrium state may be reached after a bolus injection (Carson et al, 1993). In addition, transient equilibrium between cortical regions and the cerebellum is biased: the cortex-to-cerebellum ratios (minus 1) at 120 mins after injection is 29±6% higher than of the binding potential BPND in the cortex. This overestimation of the cortex-to-cerebellum ratio is consistent with the results from the 2T fits. Indeed, the plasma decay rate β (0.012±0.004 per min) is not negligible compared with the smallest eigenvalue α1 in the cerebellum (0.025±0.006 min−1) and even more so in the cortex (0.018±0.002 per min). This leads to higher tissue-to-plasma ratios than the true VT (Carson et al, 1993), and higher cortex-to-cerebellum ratios (minus 1) than the true binding potentials.
Estimation of VT and BPND
Using arterial input function data, the 2T and MA1 methods can be used to analyze regional TACs. MA1 is the method of choice with input function data for parametric imaging because of its lower sensitivity to noise. For reference region analysis, the two-parameter graphical analysis is not applicable as the linear section of this Logan plot is not reached quickly enough. The SRTM provides less biased estimates than does the 1T model in cortical regions, but more biased estimates in the pallidum and high binding regions. Moreover, attempting to reduce noise by using only one k′2 value for all regions or voxels (SRTM2) noticeably increases the bias in BPND in high binding regions. Reference region compartmental models such as the Full Reference Tissue Model or Watabe's Reference Tissue Model, may be theoretically more suitable than SRTM (for a review, see Gunn et al, 2001), as all regions are better described by the 2T model than by the 1T model, although they would still not perfectly describe [11C]P943 kinetics. However, in practice, these more complex models provided BPND estimates that were less well correlated with MA1 BPND estimates than SRTM (data not shown).
Methods based on three-parameter graphical analysis are the most suitable for reference region methods. The MRTM produces unbiased BPND estimates, with a lower t* setting than does MA1 (20 versus 40 mins). However, BPND estimates were too variable in the pallidum. Three methods were tested to reduce this variability. Two methods involved fixing b′ to the mean MA1 cerebellum values: GAREF2 and MRTM2*. Both methods were applicable, putatively because of the relatively narrow range of b′ values in the present population. Between these two methods, MRTM2* was the best method, as GAREF2 has noise-induced bias in the pallidum.
Estimation of b′ in Multilinear Reference Tissue Model 2
An optimal method for reliable estimation of b′ for each subject is still under development. The MRTM2 is well adapted for ligands the brain TACs of which are well fitted with the 1T model (Ichise et al, 2008). However, when the 1T model cannot fit perfectly, the brain TACs, additional issues must be considered, as the MRTM equation is then just an approximation. For [11C]P943, the ideal b′, which maximizes the correlation of MA1 and MRTM2 BPND estimates, appears to be the MA1 b′ value obtained with the same t* value. Indeed, fixing b′ to this value obtained with MA1 in each subject leads to BPND estimates almost perfectly correlated with MA1 BPND estimates (i.e., BPND=0.986 × MA1 BPND+0.014, r2=0.989, n=32, data not shown). However, this most suitable value will not correspond to the least-squares estimate from MRTM in any region. Indeed, even in noise-free simulations, MRTM b′ estimates do not equal MA1 b′ estimates, as MRTM combines the approximations of MA1 in both reference and target regions.
On the basis of these results, MRTM2* seems to be the best method to quantify [11C]P943 binding potential BPND when arterial blood sampling is not available and when only a subset of regional TACs are used. The drawback of this approach is that it assumes an average b′ value, determined in one population, can be applied.
Conclusions
[11C]P943 is suitable to quantify 5-HT1B receptors in humans in vivo. [11C]P943 binding potentials are significantly correlated with in vitro measurements of the density of 5-HT1B receptors, and in vivo estimates of the [11C]P943 dissociation constant Kd are close to in vitro measurements. Using arterial input functions, MA1 is the method of choice, especially for parametric images computation. Among noninvasive methods, MRTM2* is the preferred method.
Acknowledgments
The authors acknowledge the excellent work of the staff of the Yale PET Center and the nursing support from Sue Kasserman, RN, for help in recruitment and patient care and Brenda Breault, RN, BSN for her contributions with patient care during the PET scans.
Footnotes
Supplementary Information accompanies the paper on the Journal of Cerebral Blood Flow & Metabolism website (http://www.nature.com/jcbfm)
Conflict of interest
The authors declare no conflict of interest.
Supplementary Material
References
- Arulmozhi DK, Veeranjaneyulu A, Bodhankar SL. Migraine: current concepts and emerging therapies. Vascul Pharmacol. 2005;43:176–187. doi: 10.1016/j.vph.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Baker KG, Halliday GM, Törk I. Cytoarchitecture of the human dorsal raphe nucleus. J Comp Neurol. 1990;301:147–161. doi: 10.1002/cne.903010202. [DOI] [PubMed] [Google Scholar]
- Beck JV, Arnold KJ. Parameter Estimation in Engineering and Science. New York, NY: Wiley; 1977. [Google Scholar]
- Bevington PR, Robinson DK. Data Reduction and Error Analysis for the Physical Sciences. New York, NY: McGraw-Hill; 2003. [Google Scholar]
- Boschert U, Amara DA, Segu L, Hen R. The mouse 5-hydroxytryptamine1b receptor is localized predominantly on axon terminals. Neuroscience. 1994;58:167–182. doi: 10.1016/0306-4522(94)90164-3. [DOI] [PubMed] [Google Scholar]
- Carson R, Channing M, Blasberg R, Dunn B, Cohen R, Rice K, Herscovitch P. Comparison of bolus and infusion methods for receptor quantitation: application to [18f]cyclofoxy and positron emission tomography. J Cereb Blood Flow Metab. 1993;13:24–42. doi: 10.1038/jcbfm.1993.6. [DOI] [PubMed] [Google Scholar]
- Carson RE, Barker WC, Liow J, Johnson CA. Design of a motion-compensation OSEM list-mode algorithm for resolution-recovery reconstruction for the HRRT. IEEE Nuclear Science Symposium Conference Record. 2003;5:3281–3285. [Google Scholar]
- Carson RE, Kiesewetter D, Jagoda Der M, Herscovitch P, Eckelman W. Muscarinic cholinergic receptor measurements with [18f]fp-tztp: control and competition studies. J Cereb Blood Flow Metab. 1998;18:1130–1142. doi: 10.1097/00004647-199810000-00010. [DOI] [PubMed] [Google Scholar]
- Chenu F, Bourin M. Potentiation of antidepressant-like activity with lithium: mechanism involved. Curr Drug Targets. 2006;7:159–163. doi: 10.2174/138945006775515392. [DOI] [PubMed] [Google Scholar]
- Collins D, Zijdenbos A, Kollokian V, Sled J, Kabani N, Holmes CJ, Evans AC. Design and construction of a realistic digital brain phantom. IEEE Trans Med Imaging. 1998;17:463–468. doi: 10.1109/42.712135. [DOI] [PubMed] [Google Scholar]
- Comtat C, Bataille F, Michel C, Jones J, Sibomana M, Janeiro L, Trebossen R. OSEM-3D reconstruction strategies for the ECAT HRRT. IEEE Nuclear Science Symposium Conference Record. 2004;6:3492–3496. [Google Scholar]
- Dawson LA, Hughes ZA, Starr KR, Storey JD, Bettelini L, Bacchi F, Arban R, Poffe A, Melotto S, Hagan JJ, Price GW. Characterisation of the selective 5-HT1B receptor antagonist sb-616234-a (1-[6-(cis-3,5-dimethylpiperazin-1-yl)-2,3-dihydro-5-methoxyindol-1-yl]-1-[2′methyl-4′-(5-methyl-1,2,4-oxadiazol-3-yl)biphenyl-4-yl]methanone hydrochloride): in vivo neurochemical and behavioural evidence of anxiolytic/antidepressant activity. Neuropharmacology. 2006;50:975–983. doi: 10.1016/j.neuropharm.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Engel G, Göthert M, Hoyer D, Schlicker E, Hillenbrand K. Identity of inhibitory presynaptic 5-hydroxytryptamine (5-ht) autoreceptors in the rat brain cortex with 5-HT1B binding sites. Naunyn Schmiedebergs Arch Pharmacol. 1986;332:1–7. doi: 10.1007/BF00633189. [DOI] [PubMed] [Google Scholar]
- Gunn RN, Gunn SR, Cunningham VJ. Positron emission tomography compartmental models. J Cereb Blood Flow Metab. 2001;21:635–652. doi: 10.1097/00004647-200106000-00002. [DOI] [PubMed] [Google Scholar]
- Hoyer D, Hannon J, Martin GR. Molecular, pharmacological and functional diversity of 5-ht receptors. Pharmacol Biochem Behav. 2002;71:533–554. doi: 10.1016/s0091-3057(01)00746-8. [DOI] [PubMed] [Google Scholar]
- Hudzik TJ, Yanek M, Porrey T, Evenden J, Paronis C, Mastrangelo M, Ryan C, Ross S, Stenfors C. Behavioral pharmacology of ar-a000002, a novel, selective 5-hydroxytryptamine(1b) antagonist. J Pharmacol Exp Ther. 2003;304:1072–1084. doi: 10.1124/jpet.102.045468. [DOI] [PubMed] [Google Scholar]
- Ichise M, Ballinger J, Golan H, Vines D, Luong A, Tsai S, Kung H. Noninvasive quantification of dopamine d2 receptors with iodine-123-ibf SPECT. J Nucl Med. 1996;37:513–520. [PubMed] [Google Scholar]
- Ichise M, Cohen RM, Carson RE. Noninvasive estimation of normalized distribution volume: application to the muscarinic-2 ligand [18F]FP-TZTP. J Cereb Blood Flow Metab. 2008;28:420–430. doi: 10.1038/sj.jcbfm.9600530. [DOI] [PubMed] [Google Scholar]
- Ichise M, Liow J, Lu J, Takano A, Model K, Toyama H, Suhara T, Suzuki K, Innis RB, Carson RE. Linearized reference tissue parametric imaging methods: application to [11c]dasb positron emission tomography studies of the serotonin transporter in human brain. J Cereb Blood Flow Metab. 2003;23:1096–1112. doi: 10.1097/01.WCB.0000085441.37552.CA. [DOI] [PubMed] [Google Scholar]
- Ichise M, Toyama H, Innis RB, Carson RE. Strategies to improve neuroreceptor parameter estimation by linear regression analysis. J Cereb Blood Flow Metab. 2002;22:1271–1281. doi: 10.1097/01.WCB.0000038000.34930.4E. [DOI] [PubMed] [Google Scholar]
- Innis R, Cunningham V, Delforge J, Fujita M, Gjedde A, Gunn R, Holden J, Houle S, Huang S, Ichise M, Iida H, Ito H, Kimura Y, Koeppe RA, Knudsen G, Knuuti J, Lammertsma A, Laruelle M, Logan J, Maguire R, Mintun M, Morris E, Parsey R, Price JC, Slifstein M, Sossi V, Suhara T, Votaw JR, Wong D, Carson RE. Consensus nomenclature for in vivo imaging of reversibly binding radioligands. J Cereb Blood Flow Metab. 2007;27:1533–1539. doi: 10.1038/sj.jcbfm.9600493. [DOI] [PubMed] [Google Scholar]
- Lammertsma A, Hume S. Simplified reference tissue model for pet receptor studies. Neuroimage. 1996;4:153–158. doi: 10.1006/nimg.1996.0066. [DOI] [PubMed] [Google Scholar]
- Lin D, Parsons LH. Anxiogenic-like effect of serotonin(1b) receptor stimulation in the rat elevated plus-maze. Pharmacol Biochem Behav. 2002;71:581–587. doi: 10.1016/s0091-3057(01)00712-2. [DOI] [PubMed] [Google Scholar]
- Logan J, Fowler J, Volkow N, Wang G, Ding Y, Alexoff D. Distribution volume ratios without blood sampling from graphical analysis of pet data. J Cereb Blood Flow Metab. 1996;16:834–840. doi: 10.1097/00004647-199609000-00008. [DOI] [PubMed] [Google Scholar]
- Logan J, Fowler JS, Volkow ND, Wolf A, Dewey S, Schlyer D, Macgregor R, Hitzemann R, Bendriem B, Gatley S, Christman D. Graphical analysis of reversible radioligand binding from time-activity measurements applied to [n-11c-methyl]-(−)-cocaine pet studies in human subjects. J Cereb Blood Flow Metab. 1990;10:740–747. doi: 10.1038/jcbfm.1990.127. [DOI] [PubMed] [Google Scholar]
- Marquardt D. An algorithm for least square estimation of nonlinear parameters. SIAM J. 1963;11:431–441. [Google Scholar]
- Martin KF, Hannon S, Phillips I, Heal DJ. Opposing roles for 5-HT1B and 5-ht3 receptors in the control of 5-ht release in rat hippocampus in vivo. Br J Pharmacol. 1992;106:139–142. doi: 10.1111/j.1476-5381.1992.tb14306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maura G, Raiteri M. Cholinergic terminals in rat hippocampus possess 5-HT1B receptors mediating inhibition of acetylcholine release. Eur J Pharmacol. 1986;129:333–337. doi: 10.1016/0014-2999(86)90443-7. [DOI] [PubMed] [Google Scholar]
- McCarthy TJ, Hoglund U, Antoni G, Lindhe O, Sobolov S, Gray P, Schaeffer E, Poe R, Zhang L, Bergstrom M, Blomqvist G, Appel L, Langstrom B.2007Discovery and qualification of the first 5HT1B selective PET radiotracer using a novel PET radiotracer development paradigmPaper presented at the Joint Molecular Imaging Conference, Providence, Rhode Island, USA
- Morel A, Magnin M, Jeanmonod D. Multiarchitectonic and stereotactic atlas of the human thalamus. J Comp Neurol. 1997;387:588–630. doi: 10.1002/(sici)1096-9861(19971103)387:4<588::aid-cne8>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Moret C, Briley M. The possible role of 5-ht(1b/d) receptors in psychiatric disorders and their potential as a target for therapy. Eur J Pharmacol. 2000;404:1–12. doi: 10.1016/s0014-2999(00)00581-1. [DOI] [PubMed] [Google Scholar]
- Nabulsi NB, Huang Y, Ropchan JR, Cosgrove KP, Staley J, Planeta-Wilson B, McCarthy T, Carson RE, Frost JJ, Ding Y-S.2007Synthesis and evaluation of [11C]P943 for 5-HT1B receptor studies in primates and humansPaper presented at the Joint Molecular Imaging Conference, Providence, Rhode Island, USA
- Pazos A, Palacios JM. Quantitative autoradiographic mapping of serotonin receptors in the rat brain. I. Serotonin-1 receptors. Brain Res. 1985;346:205–230. doi: 10.1016/0006-8993(85)90856-x. [DOI] [PubMed] [Google Scholar]
- Pierson ME, Andersson J, Nyberg S, McCarthy DJ, Finnema SJ, Varnäs K, Takano A, Karlsson P, Gulyás B, Medd AM, Lee CM, Powell ME, Heys JR, Potts W, Seneca N, Mrzljak L, Farde L, Halldin C. [11C]Az10419369: a selective 5-HT1B receptor radioligand suitable for positron emission tomography (pet). Characterization in the primate brain. Neuroimage. 2008;41:1075–1085. doi: 10.1016/j.neuroimage.2008.02.063. [DOI] [PubMed] [Google Scholar]
- Planeta-Wilson B, Yan J, Mulnix T, Carson RE.2008Quantitative accuracy of HRRT list-mode reconstructions: effect of low statisticsPaper presented at the IEEE Nuclear Science Symposium and Medical Imaging Conference, Dresden, Germany, 5121–5124 [DOI] [PMC free article] [PubMed]
- Sari Y. Serotonin1B receptors: from protein to physiological function and behavior. Neurosci Biobehav Rev. 2004;28:565–582. doi: 10.1016/j.neubiorev.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Sari Y, Miquel MC, Brisorgueil MJ, Ruiz G, Doucet E, Hamon M, Vergé D. Cellular and subcellular localization of 5-hydroxytryptamine1b receptors in the rat central nervous system: immunocytochemical, autoradiographic and lesion studies. Neuroscience. 1999;88:899–915. doi: 10.1016/s0306-4522(98)00256-5. [DOI] [PubMed] [Google Scholar]
- Talairach J, Tournoux P. Co-Planar Stereotactic Atlas of the Human Brain: 3-Dimensional Proportional System: An Approach to Cerebral Imaging. New York, NY: Thieme Medical Publishers; 1988. [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Van Velden F, Kloet RW, Van Berckel BN, Wolfensberger S, Lammertsma AA, Boellaard R. Comparison of 3D-OP-OSEM and 3D-FBP reconstruction algorithms for high-resolution research tomograph studies: effects of randoms estimation methods. Phys Med Biol. 2008;53:3217–3230. doi: 10.1088/0031-9155/53/12/010. [DOI] [PubMed] [Google Scholar]
- Varnäs K, Hall H, Bonaventure P, Sedvall G. Autoradiographic mapping of 5-ht(1b) and 5-ht(1d) receptors in the post mortem human brain using [(3)h]gr 125743. Brain Res. 2001;915:47–57. doi: 10.1016/s0006-8993(01)02823-2. [DOI] [PubMed] [Google Scholar]
- Varnäs K, Halldin C, Hall H. Autoradiographic distribution of serotonin transporters and receptor subtypes in human brain. Hum Brain Mapp. 2004;22:246–260. doi: 10.1002/hbm.20035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnäs K, Hurd Y, Hall H. Regional expression of 5-HT1B receptor mRNA in the human brain. Synapse. 2005;56:21–28. doi: 10.1002/syn.20128. [DOI] [PubMed] [Google Scholar]
- Voigt MM, Laurie DJ, Seeburg PH, Bach A. Molecular cloning and characterization of a rat brain cDNA encoding a 5-hydroxytryptamine1b receptor. EMBO J. 1991;10:4017–4023. doi: 10.1002/j.1460-2075.1991.tb04977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe H, Channing MA, Der MG, Adams HR, Jagoda E, Herscovitch P, Eckelman WC, Carson RE. Kinetic analysis of the 5-ht2a ligand [11c]mdl 100,907. J Cereb Blood Flow Metab. 2000;20:899–909. doi: 10.1097/00004647-200006000-00002. [DOI] [PubMed] [Google Scholar]
- Wu Y, Carson R. Noise reduction in the simplified reference tissue model for neuroreceptor functional imaging. J Cereb Blood Flow Metab. 2002;22:1440–1452. doi: 10.1097/01.WCB.0000033967.83623.34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.