Abstract
The aim of this study was to validate in vivo the accuracy of a reconstruction-based partial volume correction (PVC), which takes into account the point spread function of the imaging system. The NEMA NU2 Image Quality phantom and five healthy volunteers (using [11C]flumazenil) were scanned on both HR+ and high-resolution research tomograph (HRRT) scanners. HR+ data were reconstructed using normalization and attenuation-weighted ordered subsets expectation maximization (NAW-OSEM) and a PVC algorithm (PVC-NAW-OSEM). HRRT data were reconstructed using 3D ordinary Poisson OSEM (OP-OSEM) and a PVC algorithm (PVC-OP-OSEM). For clinical studies, parametric volume of distribution (VT) images were generated. For phantom data, good recovery was found for both OP-OSEM (0.84 to 0.97) and PVC-OP-OSEM (0.91 to 0.98) HRRT reconstructions. In addition, for the HR+, good recovery was found for PVC-NAW-OSEM (0.84 to 0.94), corresponding well with OP-OSEM. Finally, for clinical data, good correspondence was found between PVC-NAW-OSEM and OP-OSEM-derived VT values (slope: 1.02±0.08). This study showed that HR+ image resolution using PVC-NAW-OSEM was comparable to that of the HRRT scanner. As the HRRT has a higher intrinsic resolution, this agreement validates reconstruction-based PVC as a means of improving the spatial resolution of the HR+ scanner and thereby improving the quantitative accuracy of positron emission tomography.
Keywords: high-resolution PET, high-resolution research tomograph, HRRT, PET, reconstruction-based partial volume correction, resolution recovery
Introduction
Positron emission tomography (PET) is a medical imaging technique that is used to study tissue function in vivo by imaging and measuring regional tracer concentrations of radiopharmaceuticals labelled with a positron emitter. A tracer model is needed to translate these measurements into a quantitative assessment of the tissue function under study. Tracer kinetic modelling, however, is less accurate for structures that have sizes smaller than or comparable to the resolution of the scanner, because these structures are affected by partial volume effects (spill-in and spill-out) (Soret et al, 2007). Partial volume effects are attributed to the fact that activity in small structures is always spread out over a resolution element. The resolution of clinical PET scanners is still rather limited– approximately 6.4 mm full-width at half-maximum (FWHM) (Brix et al, 1997b).
The amount of partial volume depends on the spatial resolution of the PET camera that is used. State-of-the-art high-resolution PET scanners (spatial resolution: ∼2.7 mm FWHM) have a higher resolution than standard clinical PET scanners, and therefore these high-resolution scanners are less affected by partial volume effects. Although such state-of-the-art PET scanners have a higher resolution, there is still the need for partial volume correction (PVC), and therefore, for both types of scanners, a PVC method would be advantageous.
For human brain studies, image-based PVC methods can roughly be divided into region- and pixel-based methods (Soret et al, 2007). Region-based PVC methods (Hoffman et al, 1979; Kessler et al, 1984) do not correct the whole PET image, but only operate at a regional level. In contrast, pixel-based PVC methods perform corrections pixel by pixel, resulting in an entire PVC corrected image. The latter methods often use co-registered anatomical information from CT or MRI images (Boussion et al, 2006). Although pixel-based PVC methods are clearly more interesting, they also have several disadvantages. First, an additional anatomical scan, usually an MRI scan, is required. More importantly, the accuracy of pixel-based PVC methods is dependent on the accuracy of the co-registration of this MRI scan to the PET scan. Inaccurate co-registration of MRI and PET is possible because of MRI image distortions (Sumanaweera et al, 1993) and/or difference in patient positioning between both scans. Finally, many PVC methods require segmentation of gray and white matter, and the final results depend on the (limited) accuracy of this segmentation (Kloet et al, 2006).
An alternative approach is to correct for partial volume during reconstruction. Recently, reconstruction-based PVC methods have been proposed (Brix et al, 1997a; Mourik et al, 2008a, 2009; Reader et al, 2003). These reconstruction-based PVC methods improve the resolution of a PET image by taking the point spread function (PSF) of the scanner into account. This approach has the advantage that no additional anatomical information is needed and it allows for the accurate modelling of the physics of the partial volume problem into the reconstruction. Previous studies have shown that the use of a reconstruction-based PVC method makes it possible to obtain image-derived input functions from the carotid artery, even when using a PET scanner with a spatial resolution of ∼6.4 mm FWHM (Mourik et al, 2008a, 2009).
The aim of this study was to assess the accuracy of reconstruction-based PVC in vivo. To this end, normal subjects were subjected to repeat [11C]flumazenil scans on two different scanners with different spatial resolutions, and acquired data were reconstructed with and without PVC. A dedicated high-resolution PET scanner (Wienhard et al, 2002) was used as a gold standard to validate PVC for standard (low-resolution) clinical PET scanners.
Materials and methods
Scanner Descriptions
Two different PET scanners were used for this study. The first scanner is a standard whole-body PET scanner, the ECAT EXACT HR+ (CTI/Siemens, Knoxville, TN, USA), with a spatial resolution of 4.1 to 7.8 mm FWHM in 3D mode (Brix et al, 1997b). A detailed description of the HR+ scanner and its performance can be found elsewhere (Brix et al, 1997b). The second scanner is the ECAT high-resolution research tomograph (HRRT, CTI/Siemens). The HRRT is the first commercially available human brain PET scanner that uses a double layer of lutetium oxyorthosilicate/lutetiumyttrium oxyorthosilicate (LSO/LYSO) crystals, thereby enabling the use of depth-of-interaction information, resulting in a spatial resolution of 2.3 to 3.4 mm FWHM. A detailed description of the scanner and its performance has been reported previously (de Jong et al, 2007).
Reconstruction Methods
For both scanners, three different reconstructions were performed. For the HR+, data were reconstructed using normalization and attenuation-weighted ordered subsets expectation maximization (NAW-OSEM (Hudson and Larkin, 1994; Johnson et al, 1996; Shepp and Vardi, 1982), 4 iterations, 16 subsets (Boellaard et al, 2001a)). NAW-OSEM images were used without post-smoothing and with post-smoothing using a Gaussian kernel of 5 mm FWHM (NAW-OSEM-G5). The latter is the standard clinical reconstruction setting in our institute. In addition, data were reconstructed using PVC-NAW-OSEM (4 iterations, 16 subsets). A 4.5-mm FWHM Gaussian kernel, representing the PSF, was used for the PVC-NAW-OSEM reconstruction. Use of these settings for PVC-NAW-OSEM provided optimal convergence and recovery for high-activity regions (hot spots). Note that the impact of convergence also depends on the specific implementation of the recovery correction. In a previous study, it was observed that our implementation of PVC-NAW-OSEM (using the PSF in forward projection) converged after 4 iterations and that no further improvement in recovery was found after more than 4 iterations and 16 subsets (Mourik et al, 2008a). Therefore, in this study, the same settings were applied as used in a previous study (Mourik et al, 2008a). To further validate these settings, a phantom study was included in this study. More details on the implementation and optimization of PVC-NAW-OSEM and determination of the PSF can be found in Mourik et al. (2008a).
For the HRRT, data were reconstructed using the 3D ordinary Poisson (OP) OSEM algorithm with 8 iterations and 16 subsets. In addition, after reconstruction, the ordinary Poisson OSEM (OP-OSEM) images were smoothed with a 6-mm Gaussian kernel (OP-OSEM-G6) to approximate the spatial resolution of the HR+ scanner (NAW-OSEM). Finally, a PVC-OP-OSEM reconstruction (Comtat et al, 2008; Mourik et al, 2008a; Sureau et al, 2008) was used (16 iterations, 16 subsets; Mourik et al, 2008b). Default parameters were used for the PSF, similar to the study performed by Varrone et al (2009), with PSF=G1+0.05G2, where G1 represents a Gaussian function with an FWHM of 2.1 mm and G2 an FWHM of 5.9 mm.
All data were normalized and corrected for attenuation, random coincidences, scattered radiation (van Velden et al, 2008; Watson, 2000), dead time, and decay. Emission scans were reconstructed using the various reconstruction techniques mentioned above. For HR+, reconstructed images consisted of 63 planes of 256 × 256 voxels with a voxel size of 1.29 × 1.29 × 2.43 mm3. For HRRT, images consisted of 207 planes of 256 × 256 voxels with a voxel size of 1.22 × 1.22 × 1.22 mm3. The VINCI software (Max Planck Institute for Neurological Research, Cologne, Germany, http://www.mpifnf.de/vinci/) was used to co-register the HR+ scan onto the HRRT scan, using the HRRT image matrix dimensions to minimize loss in resolution.
Phantom Study
Differences between the HR+ and HRRT scanners and various reconstruction algorithms were first assessed using a phantom study. To this end, the NEMA NU2 Image Quality phantom (National Electrical Manufacturers Association NU2-2001, Rosslyn, VA, USA) with six spheres of varying internal diameter (10, 13, 17, 22, 28, and 37 mm) was scanned on both the HR+ (60 mins, 3D) and HRRT (120 mins) scanners. The NEMA NU2 Image Quality phantom itself was filled with an 18F solution of ∼2 kBq/mL and all spheres with ∼24 kBq/mL. The mean activity for each sphere was determined by deriving its standardized uptake value using a 3D isocount contour at 70% of the maximum uptake relative to the local background (Boellaard et al, 2004; Krak et al, 2005). The recovery was calculated as measured divided by true activity concentration, with the latter obtained using a calibrated well counter.
Clinical Study
For the clinical study, data were taken from the same study as previously published by van Velden et al (2009a). In that study, five healthy volunteers, with age ranging from 21 to 65 years (mean±s.d.=46±16), were scanned on both the ECAT EXACT HR+ and HRRT (in random order). Dynamic emission scans with a duration of 60 mins were acquired immediately after administration of [11C]flumazenil (366±34 MBq), with a specific activity of 54±24 GBq/mL. Administered doses and specific activities were not statistically significantly different between both scans (two-sided paired t-test, P=0.45 and P=0.46, respectively).
For the HR+, dynamic emission scans were acquired in 3D mode and histogrammed online in 16 time frames with variable frame lengths (4 × 15, 4 × 60, 2 × 150, 2 × 300 and 4 × 600 secs). For the HRRT, dynamic 64-bit list-mode emission scans were histogrammed into 16 time frames with the same variable frame lengths as for the HR+. Before emission scan and tracer administration, a 10 (HR+) or 6 (HRRT)-minute transmission scan was acquired for attenuation and scatter correction purposes.
During the entire emission scan, the arterial whole blood concentration was measured online using a continuous flow-through automatic blood sampling device (Boellaard et al, 2001b). At set times (5, 10, 15, 20, 30, 40, and 60 mins after injection), continuous withdrawal was interrupted briefly for the collection of manual samples and, after each sample, the arterial line was flushed with heparinized saline. These manual samples were used for calibrating the (online) blood sampler (BS), for measuring plasma to whole blood ratios, and for determining the plasma metabolite fractions.
Finally, for each subject, a structural T1-weighted MRI scan was acquired on a SONATA 1.5-T MRI scanner (Siemens Medical Solutions, Erlangen, Germany). PET and MRI scans of each subject were co-registered using the software package VINCI. Gray and white matter segmentation of the co-registered MRI scan was performed using Statistical Parametric Mapping (SPM5; Wellcome Department of Cognitive Neurology, University College London, UK, http://www.fil.ion.ucl.ac.uk/spm/). MRI scans were used for region of interest (ROI) definition purposes. PET and MRI scans were acquired within the same week.
The study was approved by the Medical Ethics Review Committee of VU University Medical Center, Amsterdam, and all subjects gave written informed consent before inclusion.
Plasma Input Function
BS whole blood curves were calibrated on the basis of manual samples and corrected for delay. BS whole blood curves were corrected for plasma to whole blood ratios and metabolites. The metabolite-corrected plasma input function was used during the tracer kinetic analysis.
Tracer Kinetic Analysis
Parametric volume of distribution (VT) images were generated from each reconstruction using a basis function implementation (Boellaard et al, 2005; Gunn et al, 1997) of the plasma input single-tissue compartment model, using the same settings as described by van Velden et al (2009a). Parametric images were generated using the in-house-developed software package PPET (Boellaard et al, 2006).
Using the software package DISPLAY (Montreal Neurological Institute, Montreal, QC, Canada, http://www.bic.mni.mcgill.ca/software/Display/Display.html), a total of 15 ROIs were drawn manually on individually co-registered T1-weighted MRI images (voxel size: 1.30 × 1.21 × 1.21 mm3) and resampled onto the HRRT image grid (voxel size: 1.22 × 1.22 × 1.22 mm3), in anatomical areas with varying levels of [11C]flumazenil uptake (frontal, temporal, occipital and parietal cortex, thalamus, caudate, putamen, and pons). ROIs, together with gray and white matter segmentations, were projected onto VT images to derive gray and white matter mean VT values for all individual anatomical regions. For all regions, gray matter data were used for further analysis, except for the pons, for which gray plus white matter data were used.
Per scanner, the mean VT values derived from the different reconstruction methods were compared with each other. In addition, the mean VT values derived from HR+ reconstructions were compared with those of HRRT reconstructions. The slope, intercept, and squared Pearson's correlation coefficient (R2) between VT values of different reconstructions were calculated. In addition, the slope and R2 were calculated with the intercept fixed to the origin. Finally, intra-subject test–retest variability for 15 ROIs per subject was calculated as the absolute difference between HR+ and HRRT reconstruction-based VT values divided by the mean.
Results
Phantom Study
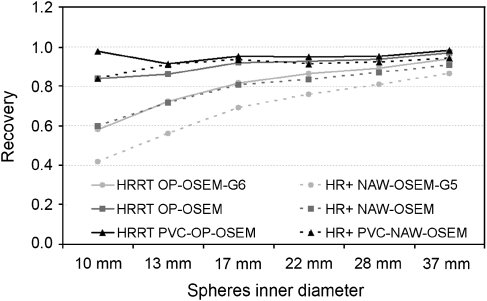
Results of the phantom study for both HR+ (dashed lines) and HRRT (straight lines) are shown in Figure 1. In case of HRRT, low recovery, ranging from 0.58 (10-mm-diameter sphere) to 0.94 (37-mm-diameter sphere), was found for the lowest resolution, reconstructed with OP-OSEM and smoothed with a Gaussian kernel of 6 mm. Comparable recovery (0.60–0.91) was found for the standard reconstruction (NAW-OSEM) of the HR+. Good recoveries were found for both standard (OP-OSEM: 0.84–0.97) and high-resolution (PVC-OP-OSEM: 0.91–0.98) HRRT reconstructions. In addition, for HR+, good recovery was found for the PVC-NAW-OSEM reconstruction (0.84–0.94), which is similar to the results found for standard HRRT OP-OSEM reconstructions.
Figure 1.
Recovery (measured/true activity) for hot spheres of different sizes. Data were acquired on both the HR+ (dashed lines) and high-resolution research tomograph (HRRT) (straight lines) scanners and reconstructed using the methods indicated. Ordinary Poisson OSEM (OP-OSEM)-G6, PVC, partial volume correction; NAW-OSEM, normalization and attenuation-weighted ordered subsets expectation maximization.
Tracer Kinetic Analysis
HR+ reconstructions
Supplementary Figure S1 shows the comparison of regional VT values obtained using NAW-OSEM and PVC-NAW-OSEM reconstructions with those obtained using the standard NAW-OSEM-G5 algorithm. Although the differences between the reconstructions are not very clear, higher VT values were found for NAW-OSEM (slope: 1.06) and especially PVC-NAW-OSEM (slope: 1.13). Very good intercept and R2 values were found (Table 1).
Table 1. Slope, intercept, and R2 derived from linear regression analyses of VT values obtained with HR+ reconstructions indicated against those obtained with HR+ NAW-OSEM-G5.
| Fixed intercept to origin | |||||
|---|---|---|---|---|---|
| Slope | Intercept | R2 | Slope | R2 | |
| NAW-OSEM | 1.06 | −0.01 | 1.00 | 1.06 | 1.00 |
| PVC-NAW-OSEM | 1.13 | −0.09 | 0.99 | 1.11 | 0.99 |
NAW-OSEM, normalization and attenuation-weighted ordered subsets expectation maximization; PVC, partial volume correction; VT, volume of distribution.
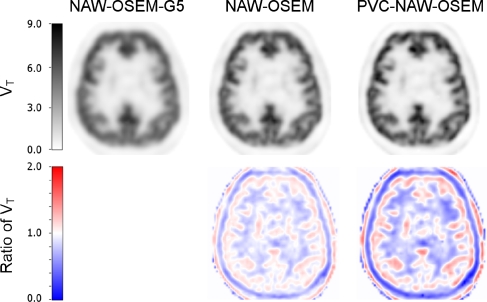
The axial parametric images of different reconstructions are shown in Figure 2 (top row). In addition, ratio images between NAW-OSEM and NAW-OSEM-G5 (bottom row, middle) and between PVC-NAW-OSEM and NAW-OSEM-G5 (bottom row, right) are also shown in Figure 2. These parametric images clearly show the differences in spatial resolution between the different reconstruction methods. Although the parametric images derived from NAW-OSEM reconstructed images (Figure 2, top row, middle) are much sharper and have a higher resolution than those derived from NAW-OSEM-G5 (Figure 2, top row, left), the highest resolution (more cortical detail (red areas) in the ratio image; Figure 2, bottom row, right) is found for PVC-NAW-OSEM reconstructions.
Figure 2.
Transaxial parametric volume of distribution (VT) images derived from dynamic [11C]flumazenil scans acquired on an HR+ scanner and reconstructed using normalization and attenuation-weighted ordered subsets expectation maximization (NAW-OSEM)-G5 (top row, left), NAW-OSEM (top row, middle), and partial volume correction (PVC)-NAW-OSEM (top row, right). Bottom row: ratio images of NAW-OSEM and NAW-OSEM-G5 (left), and PVC-NAW-OSEM and NAW-OSEM-G5 (right).
HRRT reconstructions
The comparison between the VT values obtained with OP-OSEM-G6 and those obtained with (standard) OP-OSEM and PVC-OP-OSEM reconstructions is shown in Supplementary Figure S2. The differences between OP-OSEM and PVC-OP-OSEM were small, but both reconstructions differed clearly from OP-OSEM-G6 (Supplementary Figure S2, Table 2).
Table 2. Slope, intercept, and R2 derived from linear regression analyses of VT values obtained with HRRT reconstructions indicated against those obtained with HRRT OP-OSEM-G6.
| Fixed intercept to origin | |||||
|---|---|---|---|---|---|
| Slope | Intercept | R2 | Slope | R2 | |
| OP-OSEM | 1.03 | 0.38 | 0.99 | 1.10 | 0.98 |
| PVC-OP-OSEM | 0.97 | 0.63 | 0.97 | 1.09 | 0.95 |
HRRT, high-resolution research tomograph; OP-OSEM, ordinary Poisson OSEM; PVC, partial volume correction; VT, volume of distribution.
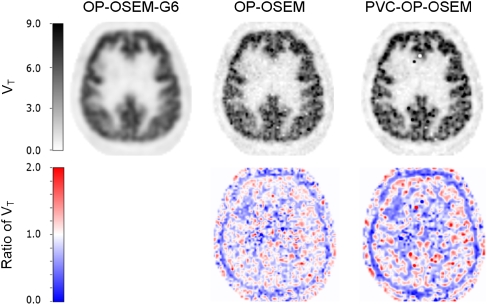
Figure 3 (top row) shows the transaxial parametric VT images derived from a dynamic [11C]flumazenil scan reconstructed with the three algorithms. Although the differences between OP-OSEM (top row, middle) and PVC-OP-OSEM (top row, right)-derived VT values were small, it is clear that the PVC-OP-OSEM images have a higher resolution than the OP-OSEM images. This is also visible in the ratio images of OP-OSEM and OP-OSEM-G6 (Figure 3, bottom row, left), and PVC-OP-OSEM and OP-OSEM-G6 (Figure 3, bottom row, right).
Figure 3.
Transaxial parametric volume of distribution (VT) images derived from dynamic [11C]flumazenil scans acquired on a high-resolution research tomograph (HRRT) scanner and reconstructed using ordinary Poisson OSEM (OP-OSEM)-G6 (top row, left), OP-OSEM (top row, middle), and partial volume correction (PVC)-OP-OSEM (top row, right). Bottom row: ratio images of OP-OSEM and OP-OSEM-G6 (left), and PVC-OP-OSEM and OP-OSEM-G6 (right).
HR+ versus HRRT reconstructions
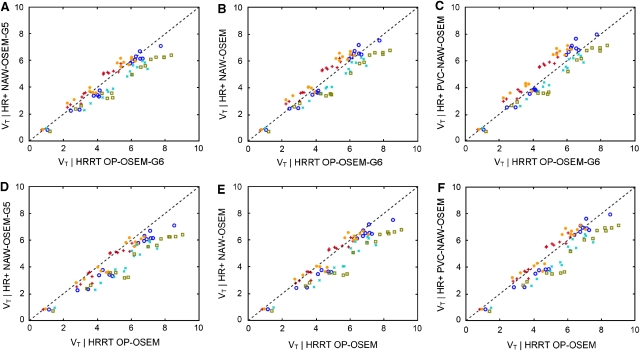
Figure 4 shows the regional VT values derived from the various HR+ reconstructions as functions of the corresponding values obtained with the HRRT OP-OSEM-G6 (top row) and OP-OSEM reconstructions (bottom row). The corresponding slope, intercept, and R2 values are shown in Supplementary Tables T1 (HRRT OP-OSEM-G6) and T2 (HRRT OP-OSEM).
Figure 4.
Volume of distribution (VT) derived from normalization and attenuation-weighted ordered subsets expectation maximization (NAW-OSEM)-G5 (A, D), NAW-OSEM (B, E), and partial volume correction (PVC)-NAW-OSEM (C, F) reconstructions of HR+ data compared with VT derived from ordinary Poisson OSEM (OP-OSEM)-G6 (top row) or standard OP-OSEM reconstructions (bottom row) of high resolution research tomograph (HRRT) data. Each colored symbol represents a single subject and the same symbol per subject represents different anatomical regions. The dashed line is the line of identity.
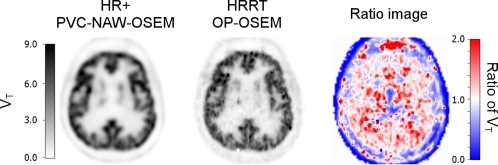
On the basis of these VT comparisons, HRRT OP-OSEM-G6 corresponds best with HR+ NAW-OSEM (Figure 4B). In addition, very good (fixed) slope and R2 values were found (Supplementary Table T1). Compared with standard HRRT OP-OSEM, best correspondence was found for HR+ PVC-NAW-OSEM (Figure 4F, slope: 1.02±0.08, see Supplementary Table T2). In Figure 5, parametric VT images are shown for HR+ PVC-NAW-OSEM and HRRT OP-OSEM, together with the ratio of both images. Although no large differences in cortical regions were observed, there were some larger differences in white matter regions (Figure 5, right image), which is in agreement with a previous study (van Velden et al, 2009a).
Figure 5.
Parametric volume of distribution (VT) images derived from dynamic [11C]flumazenil scans acquired on an HR+ scanner and reconstructed using partial volume correction (PVC)-normalization and attenuation-weighted ordered subsets expectation maximization (NAW-OSEM) (left), and on a high-resolution research tomograph (HRRT) scanner and reconstructed using ordinary Poisson OSEM (OP-OSEM) (middle). The ratio image of both images is shown at the right.
Test–retest variability
Test–retest variability between the various HR+ reconstructions and HRRT-based OP-OSEM-G6 and OP-OSEM reconstructions can be found in the last columns of Supplementary Tables T1 and T2, respectively. Test–retest variability was ⩾14% for all combinations of HR+ and HRRT reconstructions. In case of HRRT-based OP-OSEM-G6, the best test–retest results were obtained for HR+-based NAW-OSEM or PVC-NAW-OSEM (0.14±0.09). In case of standard HRRT-based OP-OSEM, the best results were found for HR+-based PVC-NAW-OSEM only (0.15±0.13). Comparable test–retest variability was found between HRRT-based PVC-OP-OSEM and HR+-based PVC-NAW-OSEM (0.14±0.13).
Discussion
Phantom Study
The phantom study was performed to determine recovery for different reconstruction algorithms in combination with both HRRT and HR+ scanners. This study showed that, for the HR+, correction for partial volume effects during reconstruction resulted in increasing recovery and thus spatial resolution (Figure 1). As mentioned previously, in case of the generally used NAW-OSEM algorithm, the spatial resolution of HR+ is approximately 4.1 to 7.8 mm FWHM in 3D mode (Brix et al, 1997b). When using the PVC-NAW-OSEM algorithm on the same scanner, however, the resolution appears to be similar to the resolution obtained with standard OP-OSEM reconstructions on the HRRT (Figure 1), which is approximately 2.3 to 3.4 mm FWHM (de Jong et al, 2007; Wienhard et al, 2002). Interestingly, this large improvement in spatial resolution was not visible for the PVC-reconstructed HRRT data. This may be explained by the intrinsic high resolution of the HRRT itself, which leaves less room for improvements. The small difference in recovery between HRRT-based OP-OSEM and PVC-OP-OSEM agrees with findings in a previous study, in which only small differences were observed in the outcome of tracer kinetic analyses using image-derived input functions extracted from OP-OSEM and PVC-OP-OSEM images (Mourik et al, 2008b).
Tracer Kinetic Analysis
HR+ reconstructions
Differences in resolution between the NAW-OSEM, PVC-NAW-OSEM, and NAW-OSEM-G5 images (Supplementary Figure S1) were not clearly visible in a correlation plot of VT values. Nevertheless, slopes between NAW-OSEM-G5 and both NAW-OSEM (1.06) and PVC-NAW-OSEM (1.13) were different. In addition, parametric images generated from the PVC-NAW-OSEM images showed more (cortical) detail compared with the NAW-OSEM images, confirming improved resolution (Figure 2B).
HRRT reconstructions
The phantom study already indicated that differences between OP-OSEM and PVC-OP-OSEM were relatively small. The same trend was observed when VT values obtained with these reconstruction algorithms were compared with those obtained with OP-OSEM-G6 (Supplementary Figure S2). Nevertheless, improved resolution could be seen in the parametric VT images (Figure 3).
Much higher differences between OP-OSEM and PVC-OP-OSEM were found in studies performed by Sureau et al (2008) and Varrone et al (2009). Both studies used dopamine transporter radioligands, which show a high contrast between the striatum and the surrounding tissue. Consequently, these structures may suffer to a high extent from spill-out effects when compared with other tracers. Although [11C]flumazenil also shows relatively high uptake, the impact on changes in resolution may be different from that seen with the dopamine transporter radioligand, as less contrast is observed overall between the structures and their surrounding tissues. Therefore, improving the tracer kinetic analysis by applying PVC might be tracer dependent and therefore needs further investigation. Note, however, that on a voxel level, VT values increased by >15% by using PVC-OP-OSEM, which is in agreement with the study performed by Varrone et al (2009).
HR+ versus HRRT reconstructions
The phantom study showed that approximately the same resolution was obtained for the HRRT-based OP-OSEM and HR+-based PVC-NAW-OSEM reconstructions. The same was true for HRRT OP-OSEM-G6 and HR+ NAW-OSEM. These findings were confirmed by the [11C]flumazenil study (see Figure 4 and Supplementary Table T1 and T2). Again, good correspondence was found between HR+ NAW-OSEM and HRRT OP-OSEM-G6 (slope: 1.01±0.11) and between HR+ PVC-NAW-OSEM and HRRT OP-OSEM (slope: 1.02±0.08). In summary, HRRT-based OP-OSEM and HR+-based PVC-NAW-OSEM showed the best agreement.
It appears that differences in spatial resolution had different effects on the change in VT values for the HR+ than for the HRRT. For HR+, the comparison between NAW-OSEM and NAW-OSEM-G5 indicates that high VT regions are more affected by a change in resolution than low VT regions. The same observation was found for HR+ when comparing PVC-NAW-OSEM with NAW-OSEM-G5. Therefore, changes in resolution for HR+ appear as a change in the correlation slope (Supplementary Figure S1). For HRRT, a change in resolution obtained either by omitting smoothing or by using PVC-OP-OSEM resulted in an offset (overall increase) of VT values (Supplementary Figure S2). The observation that different effects on VT between both scanners were already observed by simply smoothing the data suggests that these differences are not caused by the PVC-OP-OSEM algorithm but only by the differences in resolution. The reason why differences in resolution had a different effect on VT values is not fully understood and needs further investigation.
Although there is a good agreement between HR+ and HRRT, OP-OSEM-based HRRT parametric images seem to have higher noise levels than PVC-NAW-OSEM HR+ parametric images (Figure 5). It is hypothesized that differences in image quality are explained by a number of factors. First, noise levels in reconstructed data depend not only on scanner sensitivity but also on the quality of setup and on normalization, attenuation, and scatter corrections. Second, the amount of random and scattered events for the HRRT is higher than those observed for the HR+. The latter observation might be explained by the use of a neuroshield during HR+ studies, which is at present not available for the HRRT. The neuroshield reduces the effects of the outside field of view activity, thereby lowering the amount of scattered and random events.
The relatively larger differences in white matter regions (Figure 5, right image) might be the result of small differences in the implementation of the scatter correction methods and the positive bias in OSEM reconstructions. The latter was studied for HRRT by Johansson et al (2007) and van Velden et al (2009b), and for HR+ by Boellaard et al (2001a).
In this study, the HR+ and HRRT reconstructions used the same kind of scatter correction algorithms. The scatter correction implemented for both scanners are based on the single scatter simulation algorithm by Watson (2000). This scatter correction method corrects only for photon scatter within the patient and does not correct for inter-detector photon scatter. Inter-patient scatter consists of low spatial frequencies that appear as a very smooth image. The use of a different scatter correction algorithm may, to some extent, affect the observed contrast between the high- and low-uptake regions, but has only a minor effect on the spatial resolution. The only exception might be when algorithms that correct for inter-detector scatter would have been applied. However, this is, to some extent, taken into account by the use of PSF within the PVC-OSEM algorithms. It is therefore hypothesized that the results will be similar when other patient scatter correction methods are used. However, this cannot be substantiated because different scatter correction algorithms are currently not available for both scanners.
Test–retest variability
In general, high test–retest variabilities were observed (>14%, see Supplementary Tables T1 and T2) compared with the test–retest data of HR+ itself (e.g., see Lubberink et al (2007) or Tolboom et al (2009)) or [11C]flumazenil test–retest data (Salmi et al, 2008). The high test–retest values are, at least in part, a result of differences in scanner performance attributed to, e.g., differences in attenuation (van Velden et al, 2008) and scatter (van Velden et al, 2009a) correction, even though good resolution matching was observed in the phantom study. This difference in performance of attenuation and scatter corrections is therefore seen in all low-resolution HR+ versus HRRT comparisons in this study, i.e., most correlation slopes (fixed intercepts) are <1.0. However, again, best test–retest variabilities were observed between HR+-based PVC-NAW-OSEM and HRRT reconstructions.
Conclusion
Reconstruction-based PVC HR+ data showed good agreement with non-corrected HRRT data. As the HRRT has a higher intrinsic resolution, this agreement validates reconstruction-based PVC as a means of improving the spatial resolution of the HR+ scanner and thereby improving the quantitative accuracy of PET.
Acknowledgments
This work was financially supported by the Netherlands Organisation for Scientific Research (NWO, VIDI Grant 016.066.309). We thank Nelleke Tolboom and Saskia PA Wolfensberger for executing the PET study and collecting data, the technology staff of the Department of Radiology for acquisition of the MRI data, and the PET radiochemistry and technology staff of the Division of Nuclear Medicine and PET Research for production of isotopes and acquisition of PET data.
Footnotes
Supplementary Information accompanies the paper on the Journal of Cerebral Blood Flow & Metabolism website (http://www.nature.com/jcbfm)
Conflict of interest
The authors declare no conflict of interest.
Supplementary Material
References
- Boellaard R, Knaapen P, Rijbroek A, Luurtsema GJ, Lammertsma AA. Evaluation of basis function and linear least squares methods for generating parametric blood flow images using 15O-water and positron emission tomography. Mol Imaging Biol. 2005;7:273–285. doi: 10.1007/s11307-005-0007-2. [DOI] [PubMed] [Google Scholar]
- Boellaard R, Krak NC, Hoekstra OS, Lammertsma AA. Effects of noise, image resolution, and ROI definition on the accuracy of standard uptake values: a simulation study. J Nucl Med. 2004;45:1519–1527. [PubMed] [Google Scholar]
- Boellaard R, van Lingen A, Lammertsma AA. Experimental and clinical evaluation of iterative reconstruction (OSEM) in dynamic PET: quantitative characteristics and effects on kinetic modeling. J Nucl Med. 2001a;42:808–817. [PubMed] [Google Scholar]
- Boellaard R, van Lingen A, van Balen SC, Hoving BG, Lammertsma AA. Characteristics of a new fully programmable blood sampling device for monitoring blood radioactivity during PET. Eur J Nucl Med. 2001b;28:81–89. doi: 10.1007/s002590000405. [DOI] [PubMed] [Google Scholar]
- Boellaard R, Yaqub M, Lubberink M, Lammertsma AA. PPET: a software tool for kinetic and parametric analysis of dynamic PET studies. Neuroimage. 2006;31 (Suppl 2:T62. [Google Scholar]
- Boussion N, Hatt M, Lamare F, Bizais Y, Turzo A, Cheze-Le Rest C, Visvikis D. A multiresolution image based approach for correction of partial volume effects in emission tomography. Phys Med Biol. 2006;51:1857–1876. doi: 10.1088/0031-9155/51/7/016. [DOI] [PubMed] [Google Scholar]
- Brix G, Doll J, Bellemann ME, Trojan H, Haberkorn U, Schmidlin P, Ostertag H. Use of scanner characteristics in iterative image reconstruction for high-resolution positron emission tomography studies of small animals. Eur J Nucl Med. 1997a;24:779–786. doi: 10.1007/BF00879667. [DOI] [PubMed] [Google Scholar]
- Brix G, Zaers J, Adam LE, Bellemann ME, Ostertag H, Trojan H, Haberkorn U, Doll J, Oberdorfer F, Lorenz WJ. Performance evaluation of a whole-body PET scanner using the NEMA protocol. National Electrical Manufacturers Association. J Nucl Med. 1997b;38:1614–1623. [PubMed] [Google Scholar]
- Comtat C, Sureau FC, Sibomana M, Hong IK, Sjöholm N, Trébossen R. Image based resolution modeling for the HRRT OSEM reconstructions software. IEEE Nucl Sci Symp Conf Rec. 2008. pp. 4120–4123.
- de Jong HWAM, van Velden FHP, Kloet RW, Buijs FL, Boellaard R, Lammertsma AA. Performance evaluation of the ECAT HRRT: an LSO-LYSO double layer high resolution, high sensitivity scanner. Phys Med Biol. 2007;52:1505–1526. doi: 10.1088/0031-9155/52/5/019. [DOI] [PubMed] [Google Scholar]
- Gunn RN, Lammertsma AA, Hume SP, Cunningham VJ. Parametric imaging of ligand-receptor binding in PET using a simplified reference region model. Neuroimage. 1997;6:279–287. doi: 10.1006/nimg.1997.0303. [DOI] [PubMed] [Google Scholar]
- Hoffman EJ, Huang SC, Phelps ME. Quantitation in positron emission computed tomography: 1. Effect of object size. J Comput Assist Tomogr. 1979;3:299–308. doi: 10.1097/00004728-197906000-00001. [DOI] [PubMed] [Google Scholar]
- Hudson H, Larkin R. Accelerated image reconstruction using ordered subsets of projection data. IEEE Trans Med Imaging. 1994;13:601–609. doi: 10.1109/42.363108. [DOI] [PubMed] [Google Scholar]
- Johansson J, Oikonen V, Teras M. Quantitative brain imaging using the new, fast iterative histogram-mode reconstruction for the HRRT PET scanner. IEEE Nucl Sci Symp Conf Rec. 2007. pp. 3463–3467.
- Johnson C, Seidel J, Carson RE, Gandler WR, Sofer A, Green MV, Daube-Witherspoon ME. Evaluation of 3D reconstruction algorithms for a small animal PET scanner. IEEE Trans Nucl Sci. 1996;44:1303–1308. [Google Scholar]
- Kessler RM, Ellis JR, Jr, Eden M. Analysis of emission tomographic scan data: limitations imposed by resolution and background. J Comput Assist Tomogr. 1984;8:514–522. doi: 10.1097/00004728-198406000-00028. [DOI] [PubMed] [Google Scholar]
- Kloet RW, Berckel BNM, Pouwels PJW, Schuitemaker A, Barkhof F, Kropholler MA, Lammertsma AA, Boellaard R. Effects of MR scanner type, scanning sequence and segmentation algorithm on MR-based partial volume corrections of [11C](R)PK11195. Neuroimage. 2006;31:T83. [Google Scholar]
- Krak NC, Boellaard R, Hoekstra OS, Twisk JW, Hoekstra CJ, Lammertsma AA. Effects of ROI definition and reconstruction method on quantitative outcome and applicability in a response monitoring trial. Eur J Nucl Med Mol Imaging. 2005;32:294–301. doi: 10.1007/s00259-004-1566-1. [DOI] [PubMed] [Google Scholar]
- Lubberink M, Luurtsema G, van Berckel BN, Boellaard R, Toornvliet R, Windhorst AD, Franssen EJ, Lammertsma AA. Evaluation of tracer kinetic models for quantification of P-glycoprotein function using (R)-[11C]verapamil and PET. J Cereb Blood Flow Metab. 2007;27:424–433. doi: 10.1038/sj.jcbfm.9600349. [DOI] [PubMed] [Google Scholar]
- Mourik JEM, Lubberink M, Klumpers UMH, Lammertsma AA, Boellaard R. Partial volume corrected image derived input functions for dynamic brain studies: methodology and validation for [11C]flumazenil. Neuroimage. 2008a;39:1041–1050. doi: 10.1016/j.neuroimage.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Mourik JEM, Lubberink M, Schuitemaker A, Tolboom N, van Berckel BN, Lammertsma AA, Boellaard R. Image-derived input functions for PET brain studies. Eur J Nucl Med Mol Imaging. 2009;36:463–471. doi: 10.1007/s00259-008-0986-8. [DOI] [PubMed] [Google Scholar]
- Mourik JEM, van Velden FHP, Lubberink M, Kloet RW, Berckel BNM, Lammertsma AA, Boellaard R. Image derived input functions for dynamic high resolution research tomograph PET brain studies. Neuroimage. 2008b;43:676–686. doi: 10.1016/j.neuroimage.2008.07.035. [DOI] [PubMed] [Google Scholar]
- Reader AJ, Julyan PJ, Williams H, Hastings DL, Zweit J. EM algorithm system modeling by image-space techniques for PET reconstruction. IEEE Trans Nucl Sci. 2003;50:1392–1397. [Google Scholar]
- Salmi E, Aalto S, Hirvonen J, Langsjo JW, Maksimow AT, Oikonen V, Metsahonkala L, Virkkala J, Nagren K, Scheinin H. Measurement of GABAA receptor binding in vivo with [11C]flumazenil: a test-retest study in healthy subjects. Neuroimage. 2008;41:260–269. doi: 10.1016/j.neuroimage.2008.02.035. [DOI] [PubMed] [Google Scholar]
- Shepp LA, Vardi Y. Maximum likelihood reconstruction for emission tomography. IEEE Trans Med Imaging. 1982;1:113–122. doi: 10.1109/TMI.1982.4307558. [DOI] [PubMed] [Google Scholar]
- Soret M, Bacharach SL, Buvat I. Partial-volume effect in PET tumor imaging. J Nucl Med. 2007;48:932–945. doi: 10.2967/jnumed.106.035774. [DOI] [PubMed] [Google Scholar]
- Sumanaweera TS, Glover GH, Binford TO, Adler JR. MR susceptibility misregistration correction. IEEE Trans Med Imaging. 1993;12:251–259. doi: 10.1109/42.232253. [DOI] [PubMed] [Google Scholar]
- Sureau FC, Reader AJ, Comtat C, Leroy C, Ribeiro M, Buvat I, Trebossen R. Impact of image space resolution modeling for studies with the high resolution research tomograph. J Nucl Med. 2008;49:1000–1008. doi: 10.2967/jnumed.107.045351. [DOI] [PubMed] [Google Scholar]
- Tolboom N, Yaqub M, Boellaard R, Luurtsema G, Windhorst AD, Scheltens P, Lammertsma AA, van Berckel BN. Test-retest variability of quantitative [(11)C]PIB studies in Alzheimer's disease. Eur J Nucl Med Mol Imaging. 2009;36:1629–1638. doi: 10.1007/s00259-009-1129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Velden FH, Kloet RW, van Berckel BN, Buijs FL, Luurtsema G, Lammertsma AA, Boellaard R. HRRT versus HR+ human brain PET studies: an interscanner test-retest study. J Nucl Med. 2009a;50:693–702. doi: 10.2967/jnumed.108.058628. [DOI] [PubMed] [Google Scholar]
- van Velden FH, Kloet RW, van Berckel BN, Lammertsma AA, Boellaard R. Accuracy of 3-dimensional reconstruction algorithms for the high-resolution research tomograph. J Nucl Med. 2009b;50:72–80. doi: 10.2967/jnumed.108.052985. [DOI] [PubMed] [Google Scholar]
- van Velden FHP, Kloet RW, van Berckel BNM, Molthoff CFM, de Jong HWAM, Lammertsma AA, Boellaard R. Impact of attenuation correction strategies on the quantification of high resolution research tomograph PET studies. Phys Med Biol. 2008;53:99–118. doi: 10.1088/0031-9155/53/1/007. [DOI] [PubMed] [Google Scholar]
- Varrone A, Sjoholm N, Eriksson L, Gulyas B, Halldin C, Farde L. Advancement in PET quantification using 3D-OP-OSEM point spread function reconstruction with the HRRT. Eur J Nucl Med Mol Imaging. 2009;36:1639–1650. doi: 10.1007/s00259-009-1156-3. [DOI] [PubMed] [Google Scholar]
- Watson CC. New, faster, image-based scatter correction for 3D PET. IEEE Trans Nucl Sci. 2000;47:1587–1594. [Google Scholar]
- Wienhard K, Schmand M, Casey ME, Baker M, Bao J, Eriksson L, Jones WF, Knoess C, Lenox M, Lercher M, Luk P, Michel C, Reed JH, Richerzhagen N, Treffert J, Vollmar S, Young JW, Heiss WD, Nutt R. The ECAT HRRT: performance and first clinical application of the new high resolution research tomograph. IEEE Trans Nucl Sci. 2002;49:104–110. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.