Abstract
Development of small-molecule activators of p53 is currently focused on malignancies containing a wild-type p53 genotype, which is present in most leukemias. JNJ-26854165 is one of such p53-activating agents, but its mechanism of action remains to be elucidated. Here, we report the effects of JNJ-26854165 in acute leukemias. JNJ-26854165 treatment induced p53-mediated apoptosis in acute leukemia cells with wild-type p53, in which p53 rapidly drives transcription-independent apoptosis followed by activation of a transcription-dependent pathway. JNJ-26854165 accelerated the proteasome-mediated degradation of p21, and antagonized the transcriptional induction of p21 by p53. Interestingly, JNJ-26854165 induced S-phase delay and upregulated E2F1 expression in p53 mutant cells, resulting in apoptosis preferentially of S-phase cells. E2F1 knockdown blocked apoptosis induced by JNJ-26854165 in p53 mutant cells. Apoptotic activity of JNJ-26854165 against primary acute leukemia cells was maintained in leukemia/stroma cocultures, unlike doxorubicin, which has reduced cytrotoxicity in coculture systems. JNJ-26854165 synergizes with AraC or doxorubicin to induce p53-mediated apoptosis. Our data suggest that JNJ-26854165 may provide a novel therapeutic approach for the treatment of acute leukemias. The presence of p53-independent apoptotic activity in addition to p53-mediated apoptosis induction, if operational in vivo, may prevent the selection of p53 mutant subclones during therapy.
Keywords: AML, ALL, p53, E2F1, apoptosis
Introduction
p53-induced apoptosis plays an important role in preventing cancer development (1). p53 is the most frequently inactivated protein in human cancer, and more than 50% of all solid tumors carry TP53 mutations (2). TP53 mutations rarely occur in leukemias, but inactivation of wild-type p53 frequently occurs through binding to its principal cellular regulator HDM2 (3, 4). HDM2 binds p53 at the transactivation domain of p53 and blocks its ability to activate transcription, serves as a ubiquitin ligase that promotes p53 degradation, and mediates the nuclear export of p53 (5). HDM2 has been found to be overexpressed in approximately 50% of acute leukemias (3, 4), a process that can actively enhance tumorigenicity and resistance to apoptosis.
There is evidence that transformed cells are more sensitive to p53-induced apoptosis than their normal counterparts, leading to the suggestion that activation of p53 may cause tumor-specific cell killing (6, 7). Activation or restoration of p53 response therefore becomes an attractive therapeutic goal (6–11). Small molecule inhibitors have been described that disrupt HDM2–p53 binding, which liberate p53 from its inhibitor and enable p53 activation (9–11). One of these compounds, Nutlin-3, binds HDM2 in the p53-binding pocket, effectively dislodging p53 from HDM2 and inducing p53 response, which inhibits growth and induces p53- mediated apoptosis in leukemias (4, 11–14). Based on the preclinical data, phase I trials are undergoing to determine the maximally-tolerated dose and activity of HDM2 inhibitor R7112 (Nutlin-3 analog) in leukemia (NCT00623870) and soild tumor (NCT00559533) patients. Another such compound is MI-63, which is quite similar in its mechanism of action (10).
JNJ-26854165 is a novel p53-activating tryptamine derivative that was initially thought to act as a HDM2 ubiquitin ligase antagonist (15, 16). Preclinical data have shown potent anti-proliferative activity in various p53 wild type and mutant tumor models, implying p53-independent activities. JNJ-26854165 entered a phase I study of to determine the safety and dose in patients with advanced stage or refractory solid tumors (NCT00676910).
In this study, we investigated the potential therapeutic utility of p53 activation by JNJ-26854165 in acute leukemias. We found that (1) JNJ-26854165 treatment induces p53-mediated apoptosis in acute leukemia cells, (2) p53 rapidly drives transcription-independent apoptosis before the activation of the transcription-dependent pathway in the cells, (3) JNJ-26854165 induces S-phase delay and E2F1-mediated apoptosis in p53 mutant cells, and (4) JNJ-26854165 synergizes with AraC or doxorubicin to induce p53-mediated apoptosis.
Materials and methods
Reagents
JNJ-26854165 was provided by Ortho Biotech Oncology Research & Development (Beerse, Belgium). Nutlin-3 and pifithrin-α were purchased from Cayman Chemical (Ann Arbor, MI), and pifithrin-μ from Tocris Bioscience (Ellisville, MO). In some experiments, cells were cultured with 70 μM cycloheximide, 10 μM MG132 or 100 μM Z-VAD-FMK (Axxora, San Diego, CA), that were added to the cells 1 hour before drug administration. Cells were synchronized at the G1-S boundary by 18-hour treatment with 1 mM hydroxyurea (Sigma, St. Louis, MO).
Cell lines, primary samples and cultures
U937, K562 and REH were purchased from the American Type Culture Collection. MOLM-13, NB4 and NALM-6 were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen. OCI-AML-3 cells were kindly provided by Dr MD Minden (Ontario Cancer Institute, Toronto, Ontario, Canada), and P12-ICHIKAWA and PF-382 by Dr AA Ferrando (Columbia University Medical Center, New York, NY). U937 and K562 were validated in August 2009, by STR DNA fingerprinting using the AmpFࡁSTR Identifiler kit according to manufacturer instructions (Applied Biosystems cat 4322288). Cell lines were maintained in RPMI 1640 medium containing 10% heat-inactivated fetal calf serum (FCS). OCI-AML-3, MOLM-13, NB4 and U937 cells are derived from acute myelogenous leukemia (AML) patients, K562 from a chronic myelogenous leukemia (CML) patient in blast crisis, and NALM-6, REH, P12-ICHIKAWA and PF-382 from acute lymphoblastic leukemia (ALL) patients. OCI-AML-3, MOLM-13, NALM-6 and REH cells have wild-type p53, while NB4, U937, K562, P12-ICHIKAWA and PF-382 have mutant p53 (4, 17). Cell lines were harvested in log-phase growth, seeded at a density of 2 × 105 cell/mL and exposed to JNJ-26854165. Heparinized peripheral blood or bone marrow samples were obtained from leukemia patients with more than 70% leukemia cells after informed consent, according to institutional guidelines per Declaration of Helsinki. Mononuclear cells were purified by Ficoll-Hypaque (Sigma Chemical Co., St. Louis, MO) density-gradient centrifugation, and nonadherent cells were resuspended in RPMI 1640 medium supplemented with 10% FCS at a density of 5 × 105 cell/mL. Normal bone marrows were obtained after informed consent in accordance with institutional guidelines set forth by M. D. Anderson Cancer Center and the Declaration of Helsinki. Normal bone marrow-derived stromal cells were seeded as a layer at 3 × 104 cells/cm2 in 12-well plates in MEM-α medium 24 hours before addition of AML cells. To study the effect of bone marrow stroma on primary leukemia cells, 4 × 105 AML cells were cultured with or without the layer of stromal cells. In combination experiments of JNJ-26854165 and AraC, AML cell lines were treated with JNJ-26854165 at 0, 2, 5 or 10 μM. The concentration ratio of JNJ-26854165 to AraC was 1:1 in OCI-AML-3 and 10:1 in other cells. The two agents were added simultaneously to cells and cultured for 48 hours. In experiments involving combination of JNJ-26854165 and doxorubicin, the two agents were added simultaneously to cells and cultured for 48 hours. Cells were treated with doxorubicin at 0, 25, 50 or 100 nM (0, 100, 200 or 400 nM in OCI-AML-3 cells and primary leukemia cells) and the concentration ratio of doxorubicin to JNJ-26854165 was 1:10 (1:25 in OCI-AML-3 cells and primary leukemia cells). Cell viability was evaluated by triplicate counts of trypan blue dyeexcluding cells.
Cell cycle and apoptosis analysis
For cell cycle analysis, cells were fixed in ice-cold ethanol (70% vol/vol) and stained with propidium iodide solution (25 μg/ml propidium iodide, 180 U/ml RNase, 0.1% Triton X-100, and 30 mg/ml polyethylene glycol in 4 mM citrate buffer, pH 7.8; Sigma Chemical). The DNA content was determined using a FACSCalibur flow cytometer (Becton Dickinson Immunocytometry Systems, San Jose, CA). Cell cycle distribution was analyzed using ModFit LT software (Verity Software House, Topsham, ME). Cells with a hypodiploid DNA content were counted as apoptotic based on DNA fragmentation. Cell debris was defined as events in the lowest 10% range of fluorescence and eliminated from analysis. For Annexin V binding studies, cells were washed twice with binding buffer (10 mM HEPES, 140 mM NaCl, and 5 mM CaCl2 at pH 7.4) and incubated with fluorescein isothiocyanate (FITC)-conjugated Annexin V (Roche Diagnostic, Indianapolis, Indiana, USA) or allophycocyanin (APC)-conjugated Annexin V (BD Biosciences, San Jose, CA). Stained cells were analyzed by flow cytometry, while membrane integrity was simultaneously assessed by propidium iodide exclusion. In some experiments on primary cells, cells were counter-stained with CD45 APC (BD Biosciences) or respective isotype control antibody. To measure mitochondrial membrane potential (Δψm), cells were loaded with MitoTracker Red CMXRos (300 nM) and MitoTracker Green (100 μM, both from Molecular Probes, Eugene, OR) for 1 hour at 37°C. The Δψm was then assessed by measuring CMXRos retention (red fluorescence) while simultaneously adjusting for mitochondrial mass (green fluorescence). All experiments were conducted in triplicate.
Transfection of small interfering RNA
U937 cells were transfected by the Amaxa electroporator Nucleofector I, using the Nucleofector Kit C (program W-001) according to the manufacturer’s instructions (Amaxa Biosystem, Cologne, Germany). To evaluate the transfection efficiency, cells were transfected with the BLOCK-iT Fluorescent Oligo (Invitrogen, Carlsbad, CA); efficiency of transfection was estimated to be approximately 70%, with approximately 90% cell viability. E2F1 and HDM2 expression was respectively down-regulated by using the ON-TARGETplus SMART pool L-003259 and L-003279 (Dharmacon, Lafayette, CO). Nonspecific control pool (Dharmacon) served for a negative control. Fourty-eight hours posttransfection, E2F1, HDM2 and β-actin expression were analyzed by Western blot analysis.
Antibodies
The following antibodies were used: rabbit polyclonal anti-p53 (Santa Cruz Biotechnology, Santa Cruz, CA); mouse monoclonal anti-p53 (DO-1; Santa Cruz Biotechnology, Santa Cruz, CA); mouse monoclonal anti-p21 (EMD Biosciences, San Diego, CA); mouse monoclonal anti-HDM2 (Santa Cruz Biotechnology); rabbit polyclonal anti-HDMX (Bethyl Laboratories, Montgomery, TX); mouse monoclonal anti-Noxa (EMD Biosciences); rabbit polyclonal anti-Puma (EMD Biosciences); mouse monoclonal anti-E2F1 (Santa Cruz Biotechnology); mouse monoclonal anti-p73 (Santa Cruz Biotechnology); rabbit polyclonal anti-phospho- retinoblastoma (Rb) (Ser608) (Cell Signaling Technology, Beverly, MA); mouse monoclonal anti-Rb (Cell Signaling Technology); and mouse monoclonal anti-β-actin (Sigma, St. Louis, MO).
Western blot analysis
Equal amounts of protein lysate were separated by SDS-PAGE (12% gel) for 2 hours at 80 V. Proteins were transferred to Hybond-P membranes (Amersham Biosciences, Piscataway, NJ), immunoblotted with appropriate antibodies and reacted with enhanced chemiluminescence reagent (Amersham Biosciences). Signals were detected by phosphoimager Storm 860 (Molecular Dynamics, Sunnyvale, CA). An anti-β-actin blot was used in parallel as a loading control. Visualized blots were analyzed by the ImageJ 1.38 software (National Institutes of Health, Bethesda, MD).
Immunofluorescence and confocal microscopy
Cells were incubated with MitoTracker Red CMXRos (300 nM) for 30 minutes at 37°C, washed twice with PBS, fixed with 2% paraformaldehyde and permeabilized with ice-cold 100% methanol. The cells were blocked in 5% normal goat serum for 30 min, followed by incubation overnight at 4C° with Alexa Fluor 488-conjugated rabbit polyclonal anti-p53 antibodies FL-393 (Santa Cruz Biotechnology). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Images were acquired using an Olympus DSU spinning disk confocal microscope. The degree of colocalization between proteins was determined by the Pearson’s correlation coefficient which was calculated from multiple areas by SlideBook 5.0 software (Intelligent Imaging Innovations, Denver, CO).
Quantitative real-time PCR using the TLDA
The TaqMan® Low Density Array (TLDA) is a 96-well microfluidic card that enables the performance of 96 simultaneous real-time PCRs. For the TLDA, the primers and probe sets were preloaded in triplicate into each of the 96 wells of the array and 100-μl reaction volume consisting of cDNA corresponding to 100 ng total RNA combined with 1 × TaqMan Universal Master Mix was loaded into each array port. According to the manufacturer’s guidelines, the level of expression was calculated based upon the PCR cycle number (Ct) at which the exponential growth in fluorescence from the probe passes a certain threshold value. For each sample, relative gene expression level was determined by subtracting the Ct value of the housekeeping gene 18S rRNA to the Ct value of the target gene (ΔCt = CtTarget gene – Ct18S rRNA). Relative quantification (fold change) between different samples (e.g., treated vs. control) was then determined according to the 2–ΔΔCt method (ΔΔCt = ΔCttreated sample – Ctcontrol sample) (18).
TP53 mutation analysis
PCR for p53 gene expression followed by direct sequencing was performed as described previously (4).
Statistical analysis
Statistical analysis was performed using the 2-tailed Student’s t test. Results were considered statistically significant at p-values < .05. Unless otherwise indicated, average values were expressed as mean ± SD. Synergism, additive effects, and antagonism were assessed, as previously described. The combination index (CI), a numerical description of combination effects, was calculated using the more stringent statistical assumption of mutually non-exclusive modes of action (19). When CI = 1, this equation represents the conservation isobologram and indicates additive effects. CI values less than 1.0 indicate a more than expected additive effect (synergism), while CI values greater than 1.0 indicate antagonism between the 2 drugs.
Results
JNJ-26854165 inhibits cell growth and induces apoptosis in leukemia cell lines
We first examined the effect of JNJ-26854165 on the growth of cultured leukemia cell lines. The results showed that JNJ-26854165 exhibited antiproliferative activity in a variety of leukemia cell lines (Supplementary Table S1). The IC50 values at 72 hours, defined as the concentration of JNJ-26854165 causing 50% growth inhibition, were in general lower in p53 wild-type cells than p53 mutant cell lines, though the differences were small when compared to Nutlin-3. To clarify if the antiproliferative activity of JNJ-26854165 was associated with induction of apoptosis, Annexin V-positive fractions and cellular DNA content were determined. Treatment of p53 wild-type cells with JNJ-26854165 induced apoptosis in a time and dose dependent manner, as evidenced by high Annexin V positivity (Fig. 1A; Supplementary Fig. S1A) and increased sub-G1 DNA content (Supplementary Fig. S1B). JNJ-26854165 induced delayed apoptosis in p53 mutant cells, which was seen after 72 or 96 hours of exposure (Fig. 1A; Supplementary Fig. S1B). The delayed apoptosis occurred in a dose dependent manner (Supplementary Fig. S1C). Consistent with these findings, 48-hour treatment with JNJ-26854165 resulted in a significant increase of trypan blue-positive cells in wild-type p53 cells but only minimally affected the viability of p53 mutant cells (figure not shown). These results suggest that JNJ-26854165 induces early apoptosis in p53 wild-type cells while it inhibits cellular proliferation followed by delayed apoptosis in the absence of functional p53.
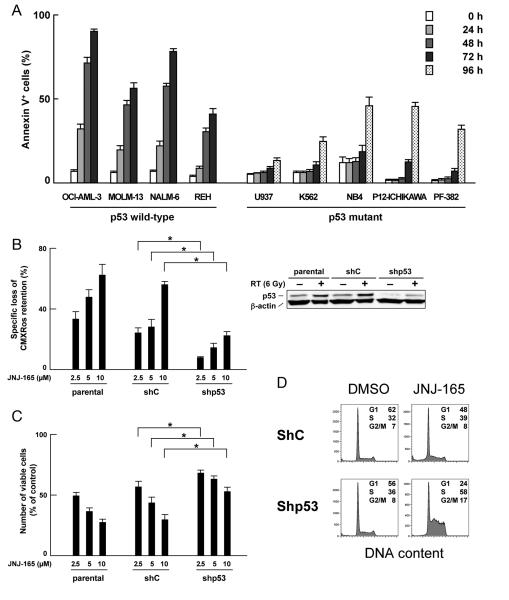
Figure 1. Functional p53 expression is required for maximal induction of apoptosis by JNJ-26854165.
(A) Cells were treated with 5 μM JNJ-26854165 for the indicated times, and the Annexin V-positive fractions were measured by flow cytometry. (B–C) Parental and lentivirally transduced OCI-AML3 cells [virus encoding either scrambled shRNA (shC) or p53-specific shRNA (shp53)] were incubated with the indicated concentrations of JNJ-26854165 for 48 hours, and CMXRos retention (B) or the number of viable cells (C) were assessed. Results are expressed as the percentage of the numbers of viable cells in an untreated group. Asterisk (*) indicates significance at P < .05. p53 expression was determined by western blot analysis in untreated or γ-irradiated cells 5 hours after 6 Gy irradiation. (D) Cells were incubated with 5 μM JNJ-26854165 for 48 hours, and stained for DNA content. Cell cycle distribution was analyzed using ModFit LT software. Results are representative of three independent experiments.
To further define the observed cell growth inhibition and cell death induced by JNJ-26854165, we investigated the effect of JNJ-26854165 in OCI-AML-3 cells infected with lentivirus encoding either scrambled shRNA or p53-specific shRNA (20). We have reported that knockdown of p53 rendered OCI-AML-3 cells resistant to Nutlin-induced apoptosis. As shown in Fig. 1B, p53 knockdown cells were less sensitive to JNJ-26854165-induced Δψm loss than parental or cells expressing scrambled shRNA, suggesting that functional p53 expression is required for full induction of apoptosis by JNJ-26854165. p53 selectivity was less pronounced for the antiproliferative effect of JNJ-26854165 (Fig. 1C), suggesting p53-independent antiproliferative activity. It has been reported that p53 activation strongly inhibits G1/S transition of cycling cells through p21 induction. Interestingly, JNJ-26854165 induced apoptosis in the absence of G1 accumulation in parental and scrambled shRNA-expressing OCI-AML-3 cells (Supplementary Fig. S1B; Fig. 1D) while in p53 knockdown cells JNJ-26854165 accumulated cells in S-phase (Fig. 1D). Absence of G1-phase arrest in p53 wild-type cells and S-phase arrest in p53 mutant cells were consistently found in a variety of leukemia cell lines treated with JNJ-26854165 (Supplementary Fig. S1B).
JNJ-26854165 increases HDM2, p53 and p21 levels in a time-dependent manner
As shown in Fig. 2A and reported previously (4), Nutlin-3 induced increased expression of p53-related proteins in OCI-AML-3 cells in a time-dependent fashion. p53 accumulated 1 hour after exposure to Nutlin-3, followed by increased levels of HDM2, p21 and Noxa. The protein expression time course after JNJ-26854165 exposure was different from that of Nutlin-3: HDM2 accumulated 4 – 6 hours after exposure to JNJ-26854165, followed by increase in p53 levels starting at 6 hours. p21 and Noxa levels were not changed significantly at 8 hours of treatment (Fig. 2A), but their increases were evident after 18 hours (Fig. 2B). Intriguingly, p21 levels were not proportional to those of induced p53 (Fig. 2B), suggesting a translational block or degradation process of p21 which could negate its transcriptional induction by p53. To elucidate if JNJ-26854165 has the ability to reduce p21 levels, OCI-AML-3 cells were treated with 10 μM JNJ-26854165 for 4 hours, either as individual agents or in combination after 1-hour preincubation with 70 μM cycloheximide, 10 μM MG132 or 100 μM Z-VAD-FMK. Cycloheximide affected the rate of decrease in p21 levels after JNJ-26854165 treatment (Fig. 2C), implying that there are mechanism(s) other than inhibition of new protein synthesis to reduce p21. The proteasome inhibitor MG132 blocked p21 reduction after JNJ-26854165 treatment (Fig. 2C), suggesting proteasome-dependent degradation of p21. p21 is a substrate for caspase-mediated degradation. However, the pancaspase inhibitor Z-VAD-FMK did not block p21 reduction significantly, indicating that the mechanism(s) is independent of caspase-mediated degradation. Our data suggest that JNJ-26854165 transcriptionally induces p21 via p53 but on the other hand, enhances p21 degradation by activation of the proteasome. The degradation process of p21 may explain why G1-phase arrest was inconspicuous in p53 wild-type cells treated with JNJ-26854165.
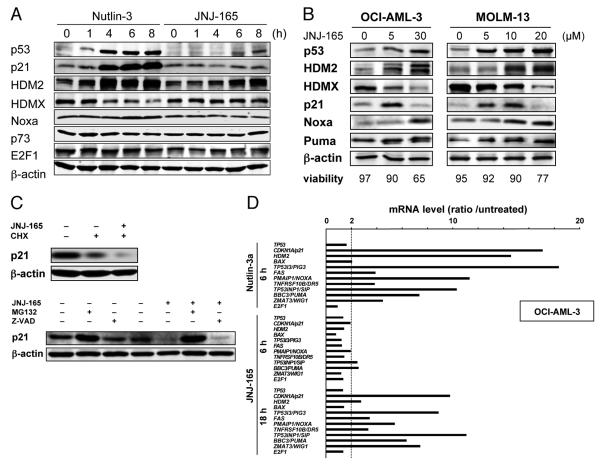
Figure 2. JNJ-26854165 induces p53 resulting in transcriptional activation of p53-regulated genes but enhances p21 degradation by proteasome activation.
(A) Protein expression in OCI-AML-3 cells, treated with 5 μM Nutlin-3 or 5 μM JNJ-26854165 for the indicated times. β-actin was used to confirm equal loading of proteins. (B) OCI-AML-3 and MOLM-13 cells were incubated with a range of concentrations of JNJ-26854165 for 18 hours. (C) OCI-AML-3 cells were preincubated for 1 hour with 70 μM cycloheximide, 10 μM MG132 or 100 μM Z-VAD-FMK, and p21 levels were determined after a 4-hour treatment with 10 μM JNJ-26854165. Results are representative of three independent experiments. (D) p53-target gene activation in response to 5 μM Nutlin-3 and 10 μM JNJ-26854165 in OCI-AML-3 cells. Ratios represent Nutlin-3 or JNJ-26854165 values divided by untreated values. Each value represents the mean of two independent experiments (each assayed in duplicate). JNJ-26854165 treatment did not alter p53 mRNA levels, suggesting that JNJ-26854165 has little effect on p53 synthesis.
In accordance with the time course of protein expression, transcriptional activation of p53-regulated genes was not seen at 6 hours but was clearly detectable at 18 hours after exposure to JNJ-26854165 in OCI-AML-3 (Fig. 2D) and MOLM-13 (Supplementary Fig. S2) cells. The p53-regulated gene expression patterns after JNJ-26854165 treatment were similar to those after Nutlin-3. JNJ-26854165 treatment did not alter p53 mRNA levels, suggesting that JNJ-26854165 has little effect on p53 synthesis. The time course of protein and transcript expression was similar in the two other studied p53 wild-type cell lines NALM-6 and REH. These findings suggest that JNJ-26854165 accumulates p53 and thereby evokes p53-mediated responses.
Wild-type p53 rapidly translocates to mitochondria in response to JNJ-26854165
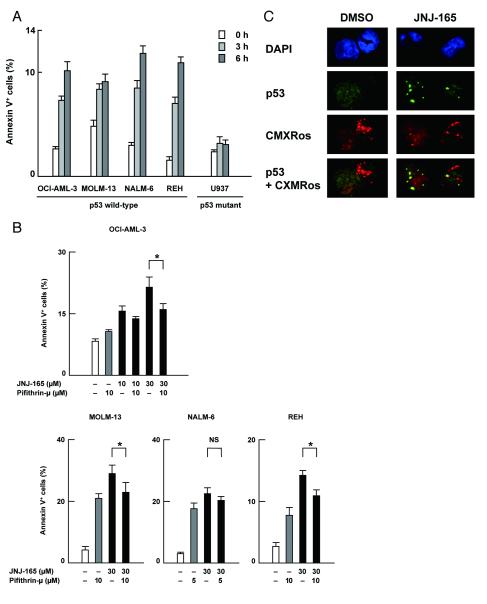
JNJ-26854165 induced apoptosis after as early as 6 hours in p53 wild-type cells (Fig. 3A) when transcriptional activation of p53-regulated genes was absent (Fig. Fig. 2D; Supplementary Fig. S2). Such early induction of apoptosis was not seen in p53 mutant cells (Fig. 1A, 3A). It has been reported that a cytoplasmic pool of p53 can induce apoptosis through a transcription-independent mechanism (21). Pifithrin-μ specifically inhibits p53 binding to mitochondria by reducing its affinity to antiapoptotic proteins Bcl-xL and Bcl-2 but has no effect on p53-dependent transactivation (22). As shown in Fig. 3B, 6-hour co-treatment with pifithrin-μ protected p53 wild-type cells partially from JNJ-26854165-induced apoptosis. Since 24-hour exposure to pifithrin-μ resulted in massive apoptosis, it remains unknown if this partial protection implies the presence of pathways distinct from p53-mediated and transcription-independent mechanism in JNJ-26854165-induced apoptosis or the interference of pifithrin-μ toxicity. We also used pifithrin-α, a transcriptional inhibitor of p53, to investigate the possible relevance of transcription-dependent p53 function in the early response of cells to JNJ-26854165. OCI-AML-3, MOLM-13, NALM-6 and REH cells were treated for 6 hours with 30 μM JNJ-26854165 and 10 μM pifithrin-α individually and in combination, and the Annexin V-positive fractions were measured by flow cytometry. Pifithrin-α did not reduce JNJ-26854165-induced phosphatidylserine externalization (data not shown). Transcriptional activation of target genes of p53 occurs in the nucleus, while mitochondrion-targeted p53 induces transcription-independent apoptosis (22). We determined the intracellular localization of p53 in OCI-AML-3 cells using confocal microscopy. A small amount of p53 was diffusely distributed in untreated cells (Fig. 3C). p53 did not specifically colocalize with the mitochondrial marker MitoTracker Red CMXRos (Pearson’s correlation coefficient: r = 0.02 ± 0.06 (mean ± SEM), n=54). After 8-hour treatment with 10 μM JNJ-26854165, individual cells showed either cytoplasmic (31%), cytoplasmic and nuclear (48%), or nuclear (21%) accumulation of p53. Independent of cellular p53 localization, approximately 70% of cells showed punctated cytoplasmic signals of p53. p53 frequently co-localized with the mitochondrial marker MitoTracker Red CMXRos (r = 0.42 ± 0.08, n=48). p53 did not show positive or negative colocalization with DAPI (r = − 0.04 ± 0.07), arguing against the idea that accelerated nuclear export of p53 contributed to mitochondrial accumulation of p53.
Figure 3. JNJ-26854165 induces p53-mediated transcription-independent apoptosis.
(A) Cells were treated with 10 μM JNJ-26854165 for the indicated times, and the Annexin V-positive fractions were measured by flow cytometry. (B) Cells were treated for 6 hours with JNJ-26854165 and pifithrin-μ either as individual agents or in combination, and the Annexin V-positive fractions were measured by flow cytometry. Asterisk (*) indicates significance at P < .05. NS, not statistically different. (C) mitochondrial relocation of p53 after JNJ-26854165 treatment. OCI-AML-3 cells were treated with 5 μM JNJ-26854165 for 8 hours. Cells were stained for p53 (green) and mitochondria (red) and visualized by confocal microscopy. Nuclei were counterstained with DAPI (blue).
JNJ-26854165 reduces the rate of DNA synthesis
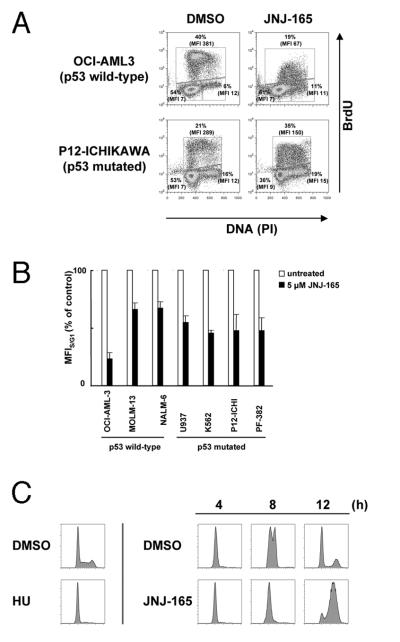
Increased S-phase percentage in p53-deficient cells was associated with a decrease in cell number, suggesting that JNJ-26854165 slowed or halted DNA synthesis. To determine if JNJ-26854165 affect DNA synthesis, leukemia cell lines were treated with 5 μM JNJ-26854165 for 24 hours and were pulse-labeled for 1 hour with BrdU. Although JNJ-26854165 treatment increased the percentage of cells incorporating BrdU, we observed that pulse labeling with BrdU of JNJ-26854165-treated cells resulted in lower levels of BrdU than control cells, as indicated by decreased mean fluorescence intensity (Fig. 4A, B). The delayed DNA synthesis was p53-independent. In p53 wild-type leukemia cells in which JNJ-26854165 induces p53-dependent apoptosis, the small fraction of cells that escaped into S-phase had significantly lower mean cellular BrdU intensity than control DMSO cells (Fig. 4A, B). To further assess the effects of JNJ-26854165 on S-phase progression, p53 mutant U937 cells were synchronized at the G1/S boundary with hydroxyurea and released into S-phase in the absence or presence of JNJ-26854165. As shown in Fig. 4C, S-phase progression was delayed over a 12-hour period.
Figure 4. JNJ-26854165 delays S-phase progression independent of p53.
(A–B) Quantitation of BrdU incorporation in S-phase cells. Cells were pulse-labeled with BrdU for 1 hour and analyzed by flow-cytometry for DNA content and BrdU incorporation. S-phase cells incorporating BrdU were gated and analyzed to determine the mean fluorescence intensity of BrdU staining relative to that of G1-phase cells (MFIS/G1). Changes in MFIS/G1 are reported as the ratio between the MFIS/G1 of treated cells compared to the MFIS/G1 of untreated control cells (MFIS/G1/control MFIS/G1). (C) U937 cells were treated with 1 mmol/L hydroxyurea (HU) for 18 hours, inducing a block at the G1-S boundary. Release into DMSO for the indicated time shows S-phase progression over a 12-hour period. Release into JNJ-26854165 shows a delay in S-phase progression.
JNJ-26854165 induces E2F1-mediated apoptosis in p53 mutant cells
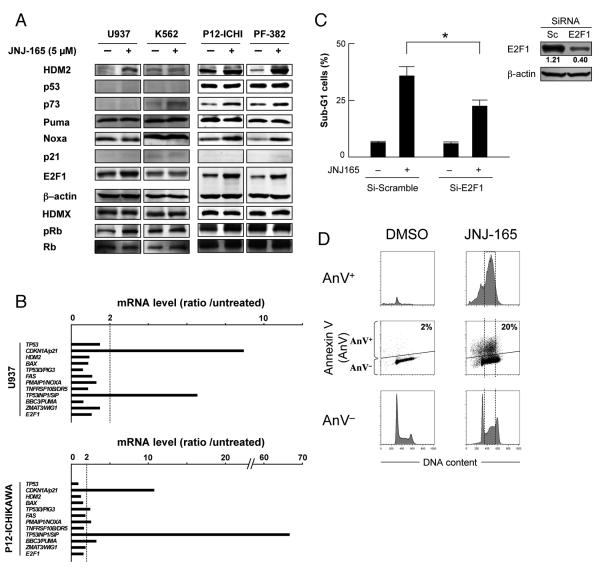
It has been reported that HDM2 stabilizes E2F1 protein in a p53-independent manner, resulting in E2F1-mediated apoptosis (23, 24). To investigate the molecular events that contribute to p53-independent apoptosis induced by JNJ-26854165 treatment, p53 mutant cells were treated with 5 μM JNJ-26854165 for 48 hours and HDM2-related protein levels were investigated (Fig. 5A). Increased levels of HDM2 and E2F1 upon JNJ-26854165 treatment were detected in 3 of 4 cell lines examined. p73 and Noxa levels were increased in 2 lines. Quantitative real-time PCR (TLDA) was performed in p53 mutant 4 leukemia cell lines including U937, K562, P12-ICHIKAWA and PF-382 cells (Fig. 5B). The transcriptional activation of p53-regulated genes was poorly detected except for CDKN1A/p21 (induced in 3 of 4 cell lines) or TP53INP1/SIP (induced in all 4 cell ines). CDKN1A/p21 and TP53INP1/SIP have been reported to be E2F1 target genes (25, 26). PMAIP1/NOXA and BC3/PUMA are also E2F1 target genes (27), and modest (2- to 6-fold mRNA) induction was detected in 2 of 4 cell lines. To elucidate if JNJ-26854165 induces E2F1-mediated apoptosis, E2F1 levels were acutely reduced in U937 cells using siRNA. E2F1 siRNA led to a significant (~ 65%) inhibition of E2F1 expression relative to cells transfected with a control siRNA, and did not interfere with β-actin synthesis (Fig. 5C). Knockdown of E2F1 in U937 cells significantly affected their susceptibility to apoptosis induced by JNJ-26854165 (35.7 ± 4.1% versus 22.4 ± 2.6% sub-G1 cells) (P = .0088), suggesting that E2F1 activation plays a role in apoptosis induction by JNJ-26854165. Consistent with this hypothesis, JNJ-26854165 induced p53-independent apoptosis predominantly in S-phase cells (Fig. 5D). Knockdown of E2F1 did not affect the increase in S-phase percentage upon JNJ-26854165 treatment (not shown), implying that p53-independent apoptosis may not be directly associated with S-phase delay. Next, HDM2 levels were similarly reduced using siRNA to test the hypothesis that HDM2 expression modulates E2F1. HDM2 siRNA led to a significant (~ 75%) inhibition of HDM2 expression relative to cells transfected with control siRNA 48 hours after transfection (Supplementary Fig. S3). However, E2F1 levels did not change significantly. Knockdown of HDM2 did not affect the susceptibility to undergo apoptosis or S-phase delay upon JNJ-26854165 treatment. These data suggest that JNJ-26854165 may upregulate E2F1 in an HDM2-independent manner.
Figure 5. JNJ-26854165 induces E2F1-mediated apoptosis in p53 mutant cells.
(A) Protein expression in p53 mutant acute leukemia cells, treated with 5 μM JNJ-26854165 for 48 hours. β-actin was used to confirm equal loading of proteins. (B) Gene activation profiles after 48-hour treatement with 5 μM JNJ-26854165 in U937 and P12-ICHIKAWA cells. Ratios represent JNJ-26854165 values divided by control values. Each value represents the mean of two independent experiments (each assayed in duplicate). (C) U937 cells were transfected with either control (Si-Scramble) or E2F1 siRNA (Si-E2F1) and incubated for 24 hours before the addition of 10 μM JNJ-26854165. After additional 72 hours incubation, DNA content was analyzed by flow cytometry and percentages of cells in sub-G1 population were determined. Intensity of the immunoblot signals was quantified by densitometry using NIH ImageJ 1.38 software, and the relative intensity of E2F1 compared to β-actin was calculated. Asterisk (*) indicates significance at P < .05. (D) JNJ-26854165 induced p53-independent apoptosis predominantly in S-phase cells. P12-ICHIKAWA cells which showed the highest ratio between the percentage of dead cells in JNJ-26854165-treated and untreated cultures among p53 mutant cell lines (see Fig. 1A), were treated with 5 μM JNJ-26854165 for 72 hours, and stained for DNA content. The Annexin V-positive fractions in correlation with DNA content were measured by flow cytometry. Data were gated on the FL2-area versus FL2-width cytogram to exclude doublets. Cells were gated on dot plots (middle row), based on Annexin staining. Annexin V-positive fraction (AnV+) corresponds to apoptotic cells, while Annexin V-negative fraction (AnV−) to viable cells. Histograms indicate the distribution of DNA content in the gated populations (AnV+: upper row, or AnV−: lower row) of the preceding dot plots. Results are representative of three independent experiments.
JNJ-26854165 induces apoptosis in primary acute leukemia cells
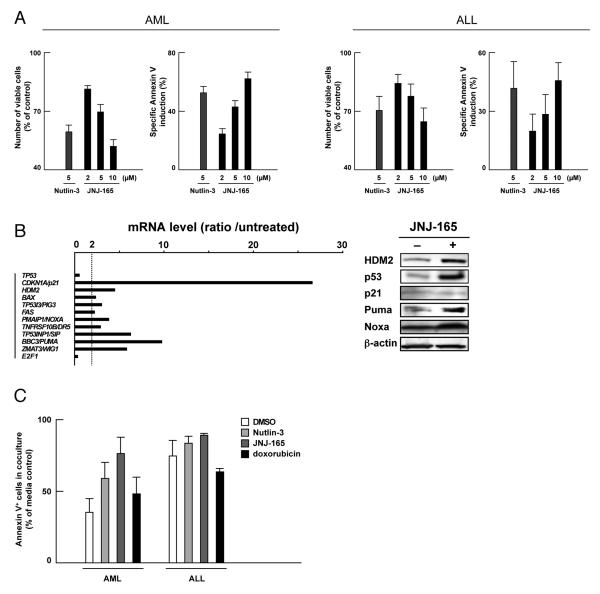
We examined the apoptotic effect of JNJ-26854165 on primary cells from 16 patients with AML and 5 patients with ALL. The percentages of spontaneous apoptosis as determined by Annexin V positivity in the series was 25.1 ± 2.8 % (mean ± SEM, 4.8 – 38.8%) in AML at 72 hours and 28.9 ± 3.4 % (20.4 – 38.6%) in ALL at 48 hours. 15 AML samples and all 5 ALL samples were sensitive to Nutlin-induced apoptosis, suggesting wild-type p53 status. The relation between p53 status and apoptotic sensitivity to Nutlins has already been established. The remaining one AML sample showed a minimal (3%) increase in the proportion of Annexin V-binding cells upon 10 μM Nutlin-3 treatment, and had a missense mutation S215G that occurred in the DNA-binding domain. The sample showed only moderate increase in the proportion of Annexin V-binding cells upon 10 μM Nutlin-3 treatment (22.1% specific apoptosis). As shown in Fig. 6A, treatment of Nutlin-sensitive samples with JNJ-26854165 caused dose-dependent increase in the percentage of Annexin V-positive cells. We correlated the extent of apoptosis induced by 10 μM JNJ-26854165 with that induced by 5 μM Nutlin-3 in the AML samples. The levels of JNJ-26854165-induced apoptosis directly correlated (r = 0.73; P < .05) with those induced by Nutlin-3. p53-related gene expression and protein levels were investigated in 2 AML samples with more than 90% blasts and wild-type p53 confirmed by sequencing analysis (Fig. 6B). Increased protein levels of HDM2, p53 and Puma and transcriptional activation of p53-regulated genes including CDKN1A/p21, HDM2, BBC3/PUMA and PMAIP1/NOXA were detected after 48-hour exposure to JNJ-26854165 in both cases. JNJ-26854165 induced Noxa at the protein level in one sample. p21 protein levels were not increased in either case despite its increased mRNA expression.
Figure 6. JNJ-26854165 induces p53-mediated apoptosis in primary acute leukemia cells.
(A) Effect of JNJ-26854165 on Nutlin-sensitive primary cells from 15 patients with AML and 5 patients with ALL. Cells were incubated with the indicated concentrations of JNJ-26854165 for 72 hours (AML) or 48 hours (ALL). The cell viability was determined by trypan blue exclusion method and the Annexin V-positive fractions were measured by flow cytometry. (B) p53-target gene activation and protein expression in response to 48-hour treatment with 10 μM JNJ-26854165 in primary AML cells. mRNA ratios represent JNJ-26854165 values divided by untreated values. β-actin was used to confirm equal loading of proteins. (C) Normal bone marrow-derived stromal cells protect primary leukemia cells from spontaneous and induced apoptosis by p53 activating agents. Primary leukemia cells were cultured in medium alone or with an adherent layer of bone marrow stromal cells for 72 hours (AML, n = 6) or 48 hours (ALL, n = 3) in the presence or absence of 5 μM Nutlin-3, 10 μM JNJ-26854165 or 400 nM doxorubicin. Cells were collected by vigorous pipetting and apoptotic cells were detected by Annexin V flow cytometry after gating on CD45+ leukemia cells. Results are expressed as the ratio of the percentage of apoptotic leukemia cells in leukemia/stroma cocultures compared to cells in medium only.
Apoptotic activity of JNJ-26854165 is enhanced by combination with AraC or doxorubicin
AraC is one of the most active chemotherapeutic agents for the therapy of acute leukemias and it remains the backbone of induction and consolidation regimens. To determine if JNJ-26854165 treatment in AML or ALL cells might potentiate the effects of AraC, we combined JNJ-26854165 and AraC in acute leukemia cell lines. An interaction study between JNJ-26854165 and AraC showed highly synergistic effects on apoptosis induction in four acute leukemia cell lines with wild-type p53 tested (Supplementary Table S2), with averaged CI values ranging from 0.2 to 0.3. Potentiation effects were not seen in p53 mutant U937, K562, P12-ICHIKAWA or PF-382 cells (not shown). We then cultured primary cells from 3 AML and 1 ALL patients with more than 90% blasts with JNJ-26854165 and/or AraC, and evaluated apoptosis after 48 hours. The cells had wild-type p53. The averaged CI values ranged from 0.4 to 0.7, confirming the synergistic nature of the JNJ-26854165/AraC interaction in primary leukemia cells (Supplementary Table S2).
Anthracyclines are another class of chemotherapeutic agents commonly used in the treatment of acute leukemias. Similar to the experiments with AraC, cells were treated for 48 hours with JNJ-26854165 and doxorubicin either as individual agents or in combination. The results indicated an additive or slightly synergistic interaction of JNJ-26854165 and doxorubicin on induction of apoptosis (Supplementary Table S3), with averaged CI values ranging from 0.3 to 1.1 (median, 0.9). Potentiation effects were not seen in p53 mutant leukemia cell lines (not shown).
Apoptotic activity of JNJ-26854165 against acute leukemia cells is relatively maintained in leukemia/stroma cocultures
A total of 9 primary acute leukemia samples (6 AML and 3 ALL) were cultured alone or cocultured with bone marrow-derived stromal cells from a normal donor. The stromal cells were resistant to 5 μM Nutlin-3, 10 μM JNJ-26854165 or 400 nM doxorubicin, as evidenced by unchanged sub-G1 DNA content after 96-hour exposure to these agents. The percentages of spontaneous apoptosis as determined by Annexin V positivity in AML series and an ALL sample were 32.6 ± 1.9 % (mean ± SEM, 27.1 – 39.5 %; at 72 hours) and 24.9 % ± 5.6 % (19.2 – 36.1 %; at 48 hours), respectively. The percentages of cells undergoing apoptosis after exposure to 5 μM Nutlin-3, 10 μM JNJ-26854165 and 400 nM doxorubicin, were 53.3 ± 3.3 %, 71.7 ± 5.5 % and 65.1 ± 4.4 % in AML and 73.3 ± 7.2 %, 75.7 ± 12.2 % and 87.3 ± 2.7 % in ALL, respectively. Co-culture of the stromal cells with acute leukemia cells protected the latter from both spontaneous apoptosis and apoptosis induced by Nutlin-3, JNJ-26854165 or doxorubicin (Fig. 6C). However, the protective effect was smaller in Nutlin-3 or JNJ-26854165 than doxorubicin. Nutlin-3, JNJ-26854165 and doxorubicin are p53 activators but the mechanisms and off-target effects are different for each compound. Our results may suggest that p53 induction by Nutlin-3 or JNJ-26854165 is more efficient in overcoming stromal cell-mediated leukemia cell resistance than wide-ranging DNA damage responses.
Discussion
The identification and characterization of a compound that activates p53 has strong implications for both the development of novel chemotherapeutics and understanding of the p53 pathway. A majority of leukemias retain wild-type TP53 and have developed mechanisms to disrupt p53 signaling, such as HDM2/HDMX overexpression or CDKN2A/ARF deletions (3, 28, 29). In such cases, use of small-molecule direct activators of p53 may induce sufficient levels of the protein to signal apoptosis (30, 31). It has been suggested that restoration of p53 activity utilizing small molecules Nutlins or RITA (reactivation of p53 and induction of tumor cell apoptosis) may be attractive chemotherapeutics in leukemias (4, 32–34). In this study, we have demonstrated that JNJ-26854165 induces p53-mediated apoptosis in leukemia cells and may be an attractive chemotherapeutic as single agents or in combination with AraC or anthracyclines. Phase I pharmacokinetic and pharmacodynamic study of JNJ-26854165 in patients with advanced refractory solid tumors showed that after consecutive doses of 300mg daily, peak plasma concentrations ranged from 2 to 3 μg/mL (6 – 9 μM), which is well within the dose range used in our studies (15, 35).
Although functional p53 protein contributed to apoptosis induction in leukemia cell lines by JNJ-26854165, p53 might not be essential for long-term cytotoxic effects. E2F1, at least partially, was found to contribute to p53-independent apoptosis induction by JNJ-26854165. Interestingly, the HDM2 antagonist Nutlin-3 has been reported to increase chemotherapy-induced apoptosis in cancer cells lacking functional p53 by activating E2F1 (36, 37). Furthermore, E2F1 transcriptional activity has been reported to be a critical determinant of Nutlin 3-induced apoptosis in human tumour cell lines (38). We think that targeting E2F1 may be exploited in the treatment of tumours without functional p53 and that the E2F1-inducing activity of JNJ-26854165 may be a potential benefit in its development as a cancer therapeutic.
p21, a transcriptional target of p53, has been shown to protect cells from p53-dependent and –independent apoptosis. It is therefore a reasonable strategy to combine p53 activators with agents which inhibit transcriptional induction of p21 and/or degrade p21 (39). Interestingly, unlike Nutlin-3, JNJ-26854165 itself induced proteasome-mediated degradation of p21, antagonizing transcriptional induction of p21 by p53. Although the precise mechanism remains unknown, Nutlin-3 which binds to HDM2 in the p53-binding pocket, has been describled to limit the binding of HDM2 to other substrates including p21 (9). Similar to the recently reported scenario in RITA (9), JNJ-26854165 may enhance p53-mediated apoptosis via degradation of p21.
We were surprised to observe that JNJ-26854165 initiates p53-mediated apoptosis before increase in cellular levels of p53 and transcriptional induction of p53 target genes, when a fraction of cytoplasmic p53 was associated with mitochondria. The conventional view of p53-mediated apoptosis has emphasized its role as a transcription factor. However, accumulating evidence indicates that p53 restoration also triggers transcription-independent apoptosis (25, 40, 41). The latter has been reportedly mediated by mitochondrial translocation of cytoplasmic p53, for which p53-dependent transcription is irrelevant (40–43). Our data shown here are compatible with the interpretation that transcription-independent mechanisms are important for p53-mediated induction of apoptosis.
In conclusion, our work suggests that JNJ-26854165 may provide a novel therapeutic tool for the therapy of acute leukemias and may be partially dependent and independent from p53 mutation status. This latter feature, if operational in vivo, could also delay or prevent the selection of p53 mutant subclones during therapy.
Supplementary Material
Acknowledgements
The authors would like to thank Erika Spaeth, Vivian Ruvolo, Teresa McQueen and Twee Tsao for valuable technical help. The authors acknowledge elucidating discussions with Dr.Tarig Bashir of Cancer Biology, Ortho Biotech Oncology Research & Development, Beerse, Belgium.
Grant Support NIH grants CA55164, CA49639, CA89346 and CA16672, the Paul and Mary Haas Chair in Genetics and research support from Johnson&Johnson (M.Andreeff). Yasuda Medical Foundation and the Japan Leukemia Research Fund (K. Kojima).
Glossary
- ALL
acute lymphoblastic leukemia
- AML
acute myelogenous leukemia
- APC
allophycocyanin
- CI
combination index
- CML
chronic myelogenous leukemia
- DAPI
4′,6-diamidino-2-phenylindole
- DMSO
dimethyl sulfoxide
- FCS
fetal calf serum
- FITC
fluorescein isothiocyanate
- HDM2
human double minute-2
- HDMX
human double minute-4
- Rb
retinoblastoma
- RITA
reactivation of p53 and induction of tumor cell apoptosis
- TLDA
TaqMan® Low Density Array
Footnotes
Disclosure of Potential Conflicts of Interest J.A. is an employee of Ortho Biotech Oncology Research & Development. The remaining authors declare no competing financial interests.
References
- 1.Hainaut P, Wiman KG. 30 years and a long way into p53 research. Lancet Oncol. 2009;10:913–9. doi: 10.1016/S1470-2045(09)70198-6. [DOI] [PubMed] [Google Scholar]
- 2.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 3.Bueso-Ramos CE, Yang Y, deLeon E, McCown P, Stass SA, Albitar M. The human MDM-2 oncogene is overexpressed in leukemias. Blood. 1993;82:2617–23. [PubMed] [Google Scholar]
- 4.Kojima K, Konopleva M, Samudio IJ, et al. MDM2 antagonists induce p53-dependent apoptosis in AML: implications for leukemia therapy. Blood. 2005;106:3150–3159. doi: 10.1182/blood-2005-02-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moll UM, Petrenko O. The MDM2-p53 interaction. Mol Cancer Res. 2003;1:1001–8. [PubMed] [Google Scholar]
- 6.Lu C, El-Deiry WS. Targeting p53 for enhanced radio- and chemo-sensitivity. Apoptosis. 2009;14:597–606. doi: 10.1007/s10495-009-0330-1. [DOI] [PubMed] [Google Scholar]
- 7.Vazquez A, Bond EE, Levine AJ, Bond GL. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov. 2008;7:979–87. doi: 10.1038/nrd2656. [DOI] [PubMed] [Google Scholar]
- 8.Lain S, Hollick JJ, Campbell J, et al. Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer Cell. 2008;13:454–63. doi: 10.1016/j.ccr.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enge M, Bao W, Hedström E, Jackson SP, Moumen A, Selivanova G. MDM2-dependent downregulation of p21 and hnRNP K provides a switch between apoptosis and growth arrest induced by pharmacologically activated p53. Cancer Cell. 2009;15:171–83. doi: 10.1016/j.ccr.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 10.Shangary S, Qin D, McEachern D, et al. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci U S A. 2008;105:3933–8. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–8. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 12.Kojima K, Konopleva M, McQueen T, O’Brien S, Plunkett W, Andreeff M. Mdm2 inhibitor Nutlin-3a induces p53-mediated apoptosis by transcription-dependent and transcription-independent mechanisms and may overcome Atm-mediated resistance to fludarabine in chronic lymphocytic leukemia. Blood. 2006;108:993–1000. doi: 10.1182/blood-2005-12-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Secchiero P, Barbarotto E, Tiribelli M, et al. Functional integrity of the p53-mediated apoptotic pathway induced by the nongenotoxic agent nutlin-3 in B-cell chronic lymphocytic leukemia (B-CLL) Blood. 2006;107:4122–9. doi: 10.1182/blood-2005-11-4465. [DOI] [PubMed] [Google Scholar]
- 14.Gu L, Zhu N, Findley HW, Zhou M. MDM2 antagonist nutlin-3 is a potent inducer of apoptosis in pediatric acute lymphoblastic leukemia cells with wild-type p53 and overexpression of MDM2. Leukemia. 2008;22:730–9. doi: 10.1038/leu.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arts J, Page M, Valckx A, et al. JNJ-26854165 – a novel HDM2 antagonist in clinical development showing broad-spectrum preclinical antitumour activity against solid malignancies [abstract 1592] Proc Am Assoc Cancer Res AACR. 2008;49 [Google Scholar]
- 16.Patel S, Player MR. Small-molecule inhibitors of the p53-HDM2 interaction for the treatment of cancer. Expert Opin Investig Drugs. 2008;17:1865–82. doi: 10.1517/13543780802493366. [DOI] [PubMed] [Google Scholar]
- 17.Petitjean A, Mathe E, Kato S, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622–9. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2–ΔΔCT Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 20.Kojima K, Shimanuki M, Shikami M, et al. The dual PI3 kinase/mTOR inhibitor PI-103 prevents p53 induction by Mdm2 inhibition but enhances p53-mediated mitochondrial apoptosis in p53 wild-type AML. Leukemia. 2008;22:1728–36. doi: 10.1038/leu.2008.158. [DOI] [PubMed] [Google Scholar]
- 21.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–30. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strom E, Sathe S, Komarov PG, et al. Small-molecule inhibitor of p53 binding to mitochondria protects mice from gamma radiation. Nat Chem Biol. 2006;2:474–9. doi: 10.1038/nchembio809. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z, Wang H, Li M, Rayburn ER, Agrawal S, Zhang R. Stabilization of E2F1 protein by MDM2 through the E2F1 ubiquitination pathway. Oncogene. 2005;24:7238–47. doi: 10.1038/sj.onc.1208814. [DOI] [PubMed] [Google Scholar]
- 24.Pediconi N, Ianari A, Costanzo A, et al. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat Cell Biol. 2003;5:552–8. doi: 10.1038/ncb998. [DOI] [PubMed] [Google Scholar]
- 25.Hiyama H, Iavarone A, Reeves SA. Regulation of the cdk inhibitor p21 gene during cell cycle progression is under the control of the transcription factor E2F. Oncogene. 1998;16:1513–23. doi: 10.1038/sj.onc.1201667. [DOI] [PubMed] [Google Scholar]
- 26.Hershko T, Chaussepied M, Oren M, Ginsberg D. Novel link between E2F and p53: proapoptotic cofactors of p53 are transcriptionally upregulated by E2F. Cell Death Differ. 2005;12:377–83. doi: 10.1038/sj.cdd.4401575. [DOI] [PubMed] [Google Scholar]
- 27.Hershko T, Ginsberg D. Up-regulation of Bcl-2 homology 3 (BH3)-only proteins by E2F1 mediates apoptosis. J Biol Chem. 2004;279:8627–34. doi: 10.1074/jbc.M312866200. [DOI] [PubMed] [Google Scholar]
- 28.Han X, Garcia-Manero G, McDonnell TJ, et al. HDM4 (HDMX) is widely expressed in adult pre-B acute lymphoblastic leukemia and is a potential therapeutic target. Mod Pathol. 2007;20:54–62. doi: 10.1038/modpathol.3800727. [DOI] [PubMed] [Google Scholar]
- 29.Volanakis EJ, Williams RT, Sherr CJ. Stage-specific Arf tumor suppression in Notch1-induced T cell acute lymphoblastic leukemia. Blood. 2009;114:4451–9. doi: 10.1182/blood-2009-07-233346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–8. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 31.Enge M, Bao W, Hedström E, Jackson SP, Moumen A, Selivanova G. MDM2-dependent downregulation of p21 and hnRNP K provides a switch between apoptosis and growth arrest induced by pharmacologically activated p53. Cancer Cell. 2009;15:171–83. doi: 10.1016/j.ccr.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 32.Gu L, Zhu N, Findley HW, Zhou M. MDM2 antagonist nutlin-3 is a potent inducer of apoptosis in pediatric acute lymphoblastic leukemia cells with wild-type p53 and overexpression of MDM2. Leukemia. 2008;22:730–9. doi: 10.1038/leu.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Secchiero P, Melloni E, di Iasio MG, et al. Nutlin-3 up-regulates the expression of Notch1 in both myeloid and lymphoid leukemic cells, as part of a negative feedback antiapoptotic mechanism. Blood. 2009;113:4300–8. doi: 10.1182/blood-2008-11-187708. [DOI] [PubMed] [Google Scholar]
- 34.Nahi H, Selivanova G, Lehmann S, et al. Mutated and non-mutated TP53 as targets in the treatment of leukaemia. Br J Haematol. 2008;141:445–53. doi: 10.1111/j.1365-2141.2008.07046.x. [DOI] [PubMed] [Google Scholar]
- 35.Tabernero J, Dirix L, Schoffski P, et al. Phase I pharmacokinetic (PK) and pharmacodynamic (PD) study of HDM-2 antagonist JNJ-26854165 in patients with advanced refractory solid tumors [abstract 3514] J Clin Oncol. 2009;27:15s. [Google Scholar]
- 36.Ambrosini G, Sambol EB, Carvajal D, Vassilev LT, Singer S, Schwartz GK. Mouse double minute antagonist Nutlin-3a enhances chemotherapy-induced apoptosis in cancer cells with mutant p53 by activating E2F1. Oncogene. 2007;26:3473–81. doi: 10.1038/sj.onc.1210136. [DOI] [PubMed] [Google Scholar]
- 37.Peirce SK, Findley HW. The MDM2 antagonist nutlin-3 sensitizes p53-null neuroblastoma cells to doxorubicin via E2F1 and TAp73. Int J Oncol. 2009;34:1395–1402. [PubMed] [Google Scholar]
- 38.Kitagawa M, Aonuma M, Lee SH, Fukutake S, McCormick F. E2F-1 transcriptional activity is a critical determinant of Mdm2 antagonist-induced apoptosis in human tumor cell lines. Oncogene. 2008;27:5303–14. doi: 10.1038/onc.2008.164. [DOI] [PubMed] [Google Scholar]
- 39.Carter BZ, Mak DH, Schober WD, et al. Simultaneous activation of p53 and inhibition of XIAP enhance the activation of apoptosis signaling pathways in AML. Blood. 2010;115:306–14. doi: 10.1182/blood-2009-03-212563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vaseva AV, Marchenko ND, Moll UM. The transcription-independent mitochondrial p53 program is a major contributor to nutlin-induced apoptosis in tumor cells. Cell Cycle. 2009;8:1711–9. doi: 10.4161/cc.8.11.8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steele AJ, Prentice AG, Hoffbrand AV, et al. p53-mediated apoptosis of CLL cells: evidence for a transcription-independent mechanism. Blood. 2008;112:3827–34. doi: 10.1182/blood-2008-05-156380. [DOI] [PubMed] [Google Scholar]
- 42.Erster S, Mihara M, Kim RH, Petrenko O, Moll UM. In vivo mitochondrial p53 translocation triggers a rapid first wave of cell death in response to DNA damage that can precede p53 target gene activation. Mol Cell Biol. 2004;24:6728–41. doi: 10.1128/MCB.24.15.6728-6741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu H, Ye H, Osman NE, et al. The MDM2-binding region in the transactivation domain of p53 also acts as a Bcl-XL-binding motif. Biochemistry. 2009;48:12159–68. doi: 10.1021/bi901188s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.