Abstract
RNA-Seq is a recently developed approach to transcriptome profiling that uses deep-sequencing technologies. Studies using this method have already altered our view of the extent and complexity of eukaryotic transcriptomes. RNA-Seq also provides a far more precise measurement of levels of transcripts and their isoforms than other methods. This article describes the RNA-Seq approach, the challenges associated with its application, and the advances made so far in characterizing several eukaryote transcriptomes.
The transcriptome is the complete set of transcripts in a cell, and their quantity, for a specific developmental stage or physiological condition. Understanding the transcriptome is essential for interpreting the functional elements of the genome and revealing the molecular constituents of cells and tissues, and also for understanding development and disease. The key aims of transcriptomics are: to catalogue all species of transcript, including mRNAs, non-coding RNAs and small RNAs; to determine the transcriptional structure of genes, in terms of their start sites, 5′ and 3′ ends, splicing patterns and other post-transcriptional modifications; and to quantify the changing expression levels of each transcript during development and under different conditions.
Various technologies have been developed to deduce and quantify the transcriptome, including hybridization-or sequence-based approaches. Hybridization-based approaches typically involve incubating fluorescently labelled cDNA with custom-made microarrays or commercial high-density oligo microarrays. Specialized microarrays have also been designed; for example, arrays with probes spanning exon junctions can be used to detect and quantify distinct spliced isoforms1. Genomic tiling microarrays that represent the genome at high density have been constructed and allow the mapping of transcribed regions to a very high resolution, from several base pairs to ~100 bp2–5. Hybridization-based approaches are high throughput and relatively inexpensive, except for high-resolution tiling arrays that interrogate large genomes. However, these methods have several limitations, which include: reliance upon existing knowledge about genome sequence; high background levels owing to cross-hybridization6,7; and a limited dynamic range of detection owing to both background and saturation of signals. Moreover, comparing expression levels across different experiments is often difficult and can require complicated normalization methods.
In contrast to microarray methods, sequence-based approaches directly determine the cDNA sequence. Initially, Sanger sequencing of cDNA or EST libraries8,9 was used, but this approach is relatively low throughput, expensive and generally not quantitative. Tag-based methods were developed to overcome these limitations, including serial analysis of gene expression (SAGE)10,11, cap analysis of gene expression (CAGE)12–14 and massively parallel signature sequencing (MPSS)15–17. These tag-based sequencing approaches are high throughput and can provide precise, ‘digital’ gene expression levels. However, most are based on expensive Sanger sequencing technology, and a significant portion of the short tags cannot be uniquely mapped to the reference genome. Moreover, only a portion of the transcript is analysed and isoforms are generally indistinguishable from each other. These disadvantages limit the use of traditional sequencing technology in annotating the structure of transcriptomes.
Recently, the development of novel high-throughput DNA sequencing methods has provided a new method for both mapping and quantifying transcriptomes. This method, termed RNA-Seq (RNA sequencing), has clear advantages over existing approaches and is expected to revolutionize the manner in which eukaryotic transcriptomes are analysed. It has already been applied to Saccharomyces cerevisiae, Schizosaccharomyces pombe, Arabidopsis thaliana, mouse and human cells18–24. Here, we explain how RNA-Seq works, discuss its challenges and provide an overview of studies that have used this approach, which have already begun to change our view of eukaryotic transcriptomes.
RNA-Seq technology and benefits
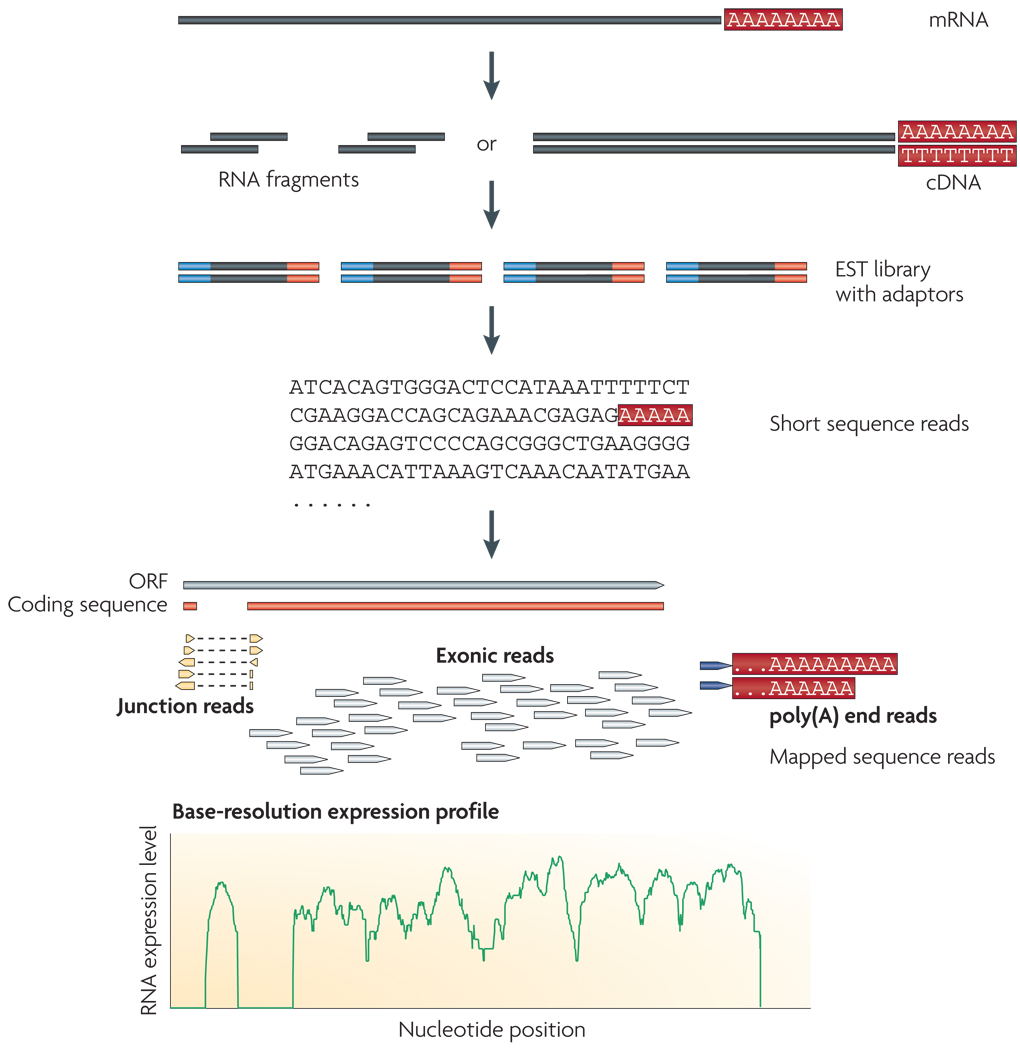
RNA-Seq uses recently developed deep-sequencing technologies. In general, a population of RNA (total or fractionated, such as poly(A)+) is converted to a library of cDNA fragments with adaptors attached to one or both ends (FIG. 1). Each molecule, with or without amplification, is then sequenced in a high-throughput manner to obtain short sequences from one end (single-end sequencing) or both ends (pair-end sequencing).The reads are typically 30–400 bp, depending on the DNA-sequencing technology used. In principle, any high-throughput sequencing technology25 can be used for RNA-Seq, and the Illumina IG18–21,23,24, Applied Biosystems SOLiD22 and Roche 454 Life Science26–28 systems have already been applied for this purpose. The Helicos Biosciences tSMS system has not yet been used for published RNA-Seq studies, but is also appropriate and has the added advantage of avoiding amplification of target cDNA. Following sequencing, the resulting reads are either aligned to a reference genome or reference transcripts, or assembled de novo without the genomic sequence to produce a genome-scale transcription map that consists of both the transcriptional structure and/or level of expression for each gene.
Figure 1. A typical RNA-Seq experiment.
Briefly, long RNAs are first converted into a library of cDNA fragments through either RNA fragmentation or DNA fragmentation (see main text). Sequencing adaptors (blue) are subsequently added to each cDNA fragment and a short sequence is obtained from each cDNA using high-throughput sequencing technology. The resulting sequence reads are aligned with the reference genome or transcriptome, and classified as three types: exonic reads, junction reads and poly(A) end-reads. These three types are used to generate a base-resolution expression profile for each gene, as illustrated at the bottom; a yeast ORF with one intron is shown.
Although RNA-Seq is still a technology under active development, it offers several key advantages over existing technologies (Table 1). First, unlike hybridization-based approaches, RNA-Seq is not limited to detecting transcripts that correspond to existing genomic sequence. For example, 454-based RNA-Seq has been used to sequence the transcriptome of the Glanville fritillary butterfly27. This makes RNA-Seq particularly attractive for non-model organisms with genomic sequences that are yet to be determined. RNA-Seq can reveal the precise location of transcription boundaries, to a single-base resolution. Furthermore, 30-bp short reads from RNA-Seq give information about how two exons are connected, whereas longer reads or pair-end short reads should reveal connectivity between multiple exons. These factors make RNA-Seq useful for studying complex transcriptomes. In addition, RNA-Seq can also reveal sequence variations (for example, SNPs) in the transcribed regions22,24.
Table 1.
Advantages of RNA-Seq compared with other transcriptomics methods
| Technology | Tiling microarray | cDNA or EST sequencing | RNA-seq |
|---|---|---|---|
| Technology specifications | |||
| Principle | Hybridization | Sanger sequencing | High-throughput sequencing |
| Resolution | From several to 100 bp | Single base | Single base |
| Throughput | High | Low | High |
| Reliance on genomic sequence | Yes | No | In some cases |
| Background noise | High | Low | Low |
| Application | |||
| Simultaneously map transcribed regions and gene expression | Yes | Limited for gene expression | Yes |
| Dynamic range to quantify gene expression level | Up to a few-hundredfold | Not practical | >8,000-fold |
| Ability to distinguish different isoforms | Limited | Yes | Yes |
| Ability to distinguish allelic expression | Limited | Yes | Yes |
| Practical issues | |||
| Required amount of RNA | High | High | Low |
| cost for mapping transcriptomes of large genomes | High | High | Relatively low |
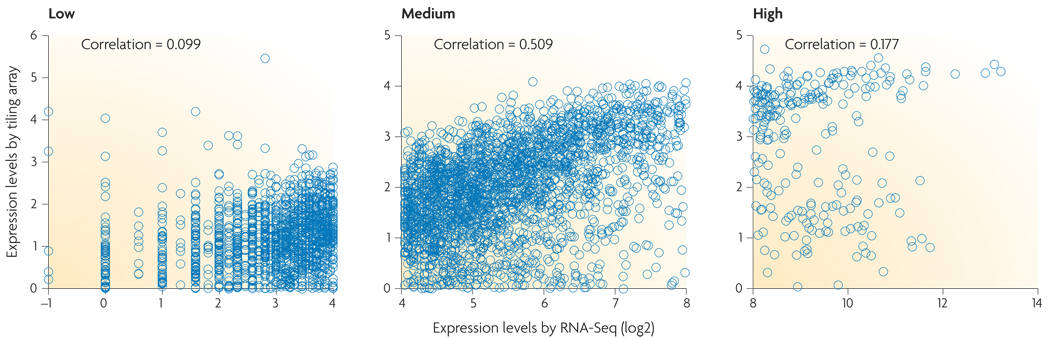
A second advantage of RNA-Seq relative to DNA microarrays is that RNA-Seq has very low, if any, background signal because DNA sequences can been unambiguously mapped to unique regions of the genome. RNA-Seq does not have an upper limit for quantification, which correlates with the number of sequences obtained. Consequently, it has a large dynamic range of expression levels over which transcripts can be detected: a greater than 9,000-fold range was estimated in a study that analysed 16 million mapped reads in Saccharomyces cerevisiae18, and a range spanning five orders of magnitude was estimated for 40 million mouse sequence reads20. By contrast, DNA microarrays lack sensitivity for genes expressed either at low or very high levels and therefore have a much smaller dynamic range (one-hundredfold to a few-hundredfold) (FIG. 2). RNA-Seq has also been shown to be highly accurate for quantifying expression levels, as determined using quantitative PCR (qPCR)18 and spike-in RNA controls of known concentration20. The results of RNA-Seq also show high levels of reproducibility, for both technical and biological replicates18,22. Finally, because there are no cloning steps, and with the Helicos technology there is no amplification step, RNA-Seq requires less RNA sample.
Figure 2. Quantifying expression levels: RNA-Seq and microarray compared.
Expression levels are shown, as measured by RNA-Seq and tiling arrays, for Saccharomyces cerevisiae cells grown in nutrient-rich media. The two methods agree fairly well for genes with medium levels of expression (middle), but correlation is very low for genes with either low or high expression levels. The tiling array data used in this figure is taken from REF. 2, and the RNA-Seq data is taken from REF. 18.
Taking all of these advantages into account, RNA-Seq is the first sequencing-based method that allows the entire transcriptome to be surveyed in a very high-throughput and quantitative manner. This method offers both single-base resolution for annotation and ‘digital’ gene expression levels at the genome scale, often at a much lower cost than either tiling arrays or large-scale Sanger EST sequencing.
Challenges for RNA-Seq
Library construction
The ideal method for transcriptomics should be able to directly identify and quantify all RNAs, small or large. Although there are only a few steps in RNA-Seq (FIG. 1), it does involve several manipulation stages during the production of cDNA libraries, which can complicate its use in profiling all types of transcript.
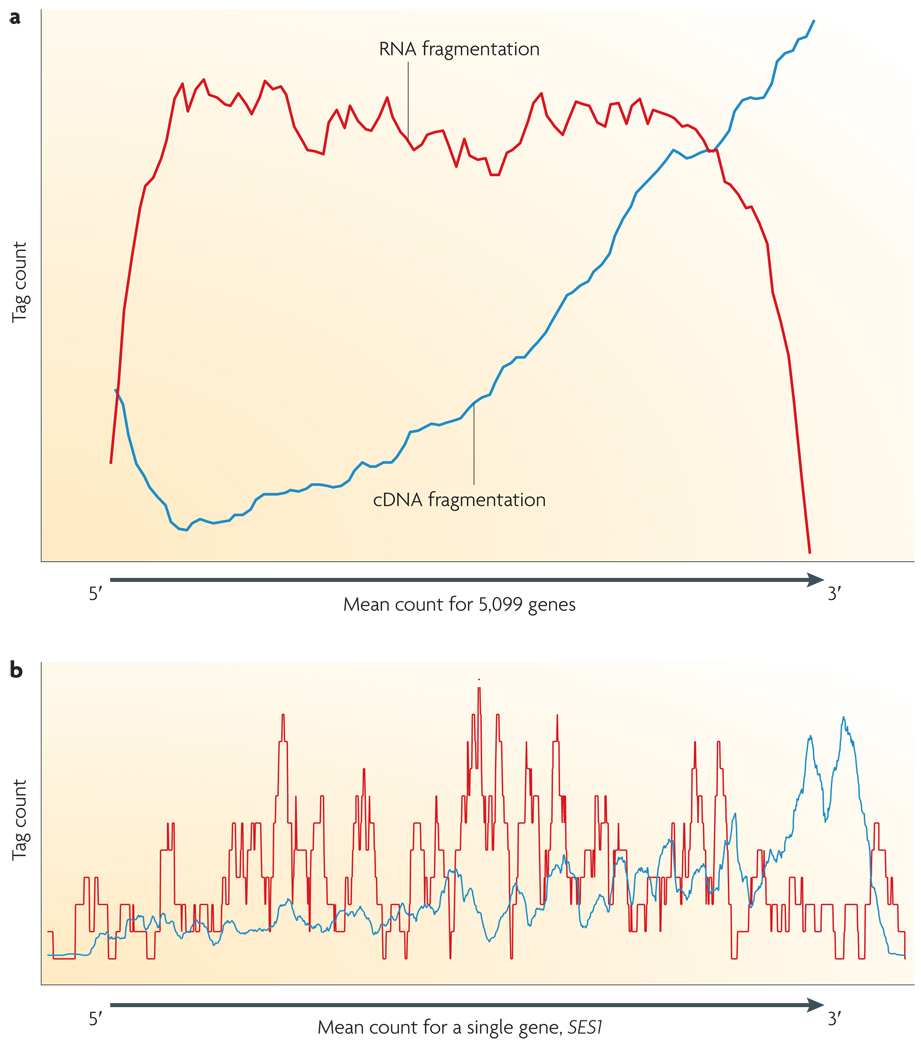
Unlike small RNAs (microRNAs (miRNAs), Piwi-interacting RNAs (piRNAs), short interfering RNAs (siRNAs) and many others), which can be directly sequenced after adaptor ligation, larger RNA molecules must be fragmented into smaller pieces (200–500 bp) to be compatible with most deep-sequencing technologies. Common fragmentation methods include RNA fragmentation (RNA hydrolysis or nebulization) and cDNA fragmentation (DNase I treatment or sonication). Each of these methods creates a different bias in the outcome. For example, RNA fragmentation has little bias over the transcript body20, but is depleted for transcript ends compared with other methods (FIG. 3). Conversely, cDNA fragmentation is usually strongly biased towards the identification of sequences from the 3′ ends of transcripts, and thereby provides valuable information about the precise identity of these ends18 (FIG. 4).
Figure 3. DNA library preparation: RNA fragmentation and DNA fragmentation compared.
a | Fragmentation of oligo-dT primed cDNA (blue line) is more biased towards the 3′ end of the transcript. RNA fragmentation (red line) provides more even coverage along the gene body, but is relatively depleted for both the 5′ and 3′ ends. Note that the ratio between the maximum and minimum expression level (or the dynamic range) for microarrays is 44, for RNA-Seq it is 9,560. The tag count is the average sequencing coverage for 5,000 yeast ORFs18. b | A specific yeast gene, SES1 (seryl-tRNA synthetase), is shown.
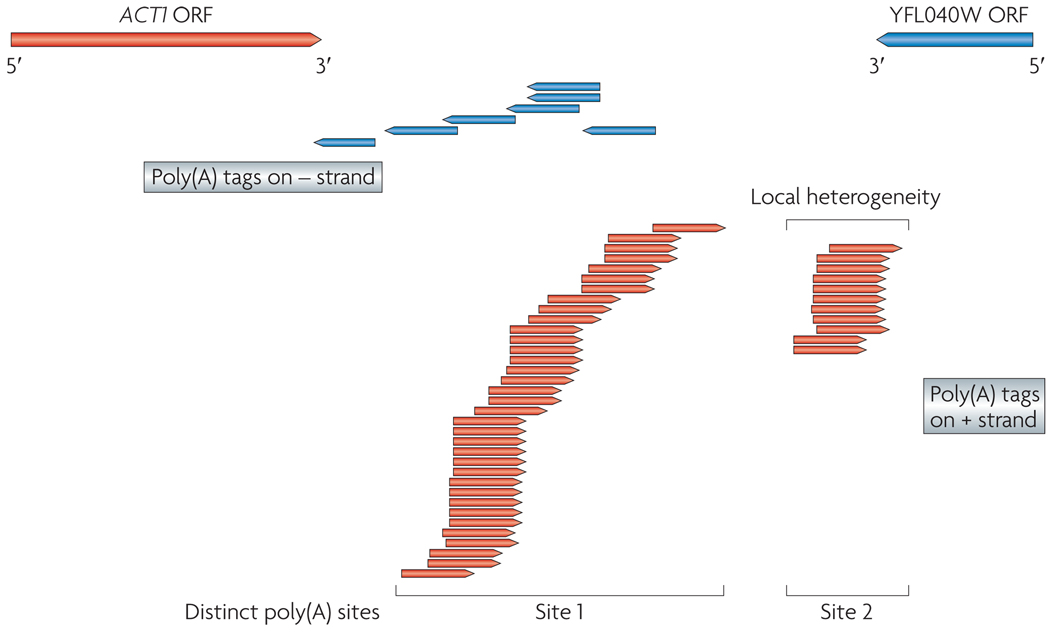
Figure 4. Poly(A) tags from RNA-Seq.
A region containing two overlapping transcripts (ACT1, from the actin gene, and YFL040W, an uncharacterized ORF) from the Saccharomyces cerevisiae genome is shown. Arrows point to transcription direction. The poly(A) tags from RNA-Seq experiments are shown below these transcripts, with arrows indicating transcription direction. The precise location of each locus identified by poly(A) tags reveals the heterogeneity in poly(A) sites, for example, ACT1 has two big clusters, both with a few bases of local heterogeneity. The transcription direction revealed by poly(A) tags also helps to resolve 3′-end overlapping transcribed regions18.
Some manipulations during library construction also complicate the analysis of RNA-Seq results. For example, many shorts reads that are identical to each other can be obtained from cDNA libraries that have been amplified. These could be a genuine reflection of abundant RNA species, or they could be PCR artefacts. One way to discriminate between these possibilities is to determine whether the same sequences are observed in different biological replicates.
Another key consideration concerning library construction is whether or not to prepare strand-specific libraries, as has been done in two studies21,22. These libraries have the advantage of yielding information about the orientation of transcripts, which is valuable for transcriptome annotation, especially for regions with overlapping transcription from opposite directions2,19,29; however, strand-specific libraries are currently laborious to produce because they require many steps22 or direct RNA–RNA ligation21, which is inefficient. Moreover, it is essential to ensure that the antisense transcripts are not artefacts of reverse transcription30. Because of these complications, most studies thus far have analysed cDNAs without strand information.
Bioinformatic challenges
Like other high-throughput sequencing technologies, RNA-Seq faces several informatics challenges, including the development of efficient methods to store, retrieve and process large amounts of data, which must be overcome to reduce errors in image analysis and base-calling and remove low-quality reads.
Once high-quality reads have been obtained, the first task of data analysis is to map the short reads from RNA-Seq to the reference genome, or to assemble them into contigs before aligning them to the genomic sequence to reveal transcription structure. There are several programs for mapping reads to the genome, including ELAND, SOAP31, MAQ32 and RMAP33 (information about these can be found at the Illumina forum and at SEQanswers). However, short transcriptomic reads also contain reads that span exon junctions or that contain poly(A) ends — these cannot be analysed in the same way. For genomes in which splicing is rare (for example, S. cerevisiae) special attention only needs to be given to poly(A) tails and to a small number of exon–exon junctions. Poly(A) tails can be identified simply by the presence of multiple As or Ts at the end of some reads. Exon–exon junctions can be identified by the presence of a specific sequence context (the GT–AG dinucleotides that flank splice sites) and confirmed by the low expression of intronic sequences, which are removed during splicing. Transcriptome maps have been generated in this manner for S. cerevisiae18. For complex transcriptomes it is more difficult to map reads that span splice junctions, owing to the presence of extensive alternative splicing and trans-splicing. One partial solution is to compile a junction library that contains all the known and predicted junction sequences and map reads to this library19,20. A challenge for the future is to develop computationally simple methods to identify novel splicing events that take place between two distant sequences or between exons from two different genes.
For large transcriptomes, alignment is also complicated by the fact that a significant portion of sequence reads match multiple locations in the genome. One solution is to assign these multi-matched reads by proportionally assigning them based on the number of reads mapped to their neighbouring unique sequences20,22. This method has been successful for low-copy repetitive sequences20. Short reads that have high copy numbers (>100) and long stretches of repetitive regions present a greater challenge. Obtaining longer sequence reads, for example using 454 technology, should help alleviate the multi-matching problem. Alternatively, a paired-end sequencing strategy, in which short sequences are determined from both ends of a DNA fragment25,34,35, extends the mapped fragment length to 200–500 bp and is expected to be useful in the future. Sequencing errors and polymorphisms can present mapping problems for all genomes, not just for repetitive DNA. Generally, single base differences are not problematic, because most mapping algorithms accommodate one or two base differences. However, resolving larger differences will require better reference genome annotation for polymorphisms and deeper sequencing coverage.
Coverage versus cost
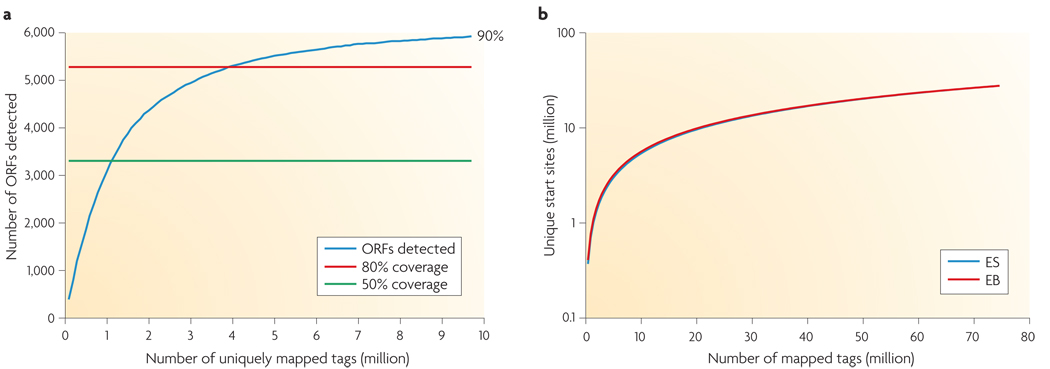
Another important issue is sequence coverage, or the percentage of transcripts surveyed, which has implications for cost. Greater coverage requires more sequencing depth. To detect a rare transcript or variant, considerable depth is needed. In simple transcriptomes, such as yeast (both S. pombe and S. cerevisiae) for which there is no evidence of alternative splicing, 30 million 35-nucleotide reads from poly(A) mRNA libraries are sufficient to observe transcription from most (>90%) genes for cells grown under a single condition (that is, in nutrient-rich medium)18. This depth is probably more than sufficient for most purposes, as the number of expressed genes detected by RNA-Seq reaches 80% coverage at 4 million uniquely mapped reads, after which doubling the depth merely increases the coverage by 10% (FIG. 5). The remaining genes are presumably either not expressed under this condition (for example, sporulation genes18) or do not have poly(A) tails. Analyzing many different conditions can further increase the coverage; in S. pombe 122 million reads from six different growth conditions detected transcription from >99% of annotated genes19.
Figure 5. Coverage versus depth.
a | 80% of yeast genes were detected at 4 million uniquely mapped RNA-Seq reads, and coverage reaches a plateau afterwards despite the increasing sequencing depth. Expressed genes are defined as having at least four independent reads from a 50-bp window at the 3′ end. Data is taken from REF. 18. b | The number of unique start sites detected starts to reach a plateau when the depth of sequencing reaches 80 million in two mouse transcriptomes. ES, embryonic stem cells; EB, embryonic body. Figure is modified, with permission, from REF. 22 © (2008) Macmillan Publishers Ltd. All rights reserved.
In general, the larger the genome, the more complex the transcriptome, the more sequencing depth is required for adequate coverage. Unlike genome-sequencing coverage, it is less straightforward to calculate the coverage of the transcriptome; this is because the true number and level of different transcript isoforms is not usually known and because transcription activity varies greatly across the genome. One study used the number of unique transcription start sites as a measure of coverage in mouse embryonic cells, and demonstrated that at 80 million reads, the number of start sites reached a plateau22 (FIG. 5b). However, this approach does not address transcriptome complexity in alternative splicing and transcription termination sites; presumably further sequencing can reveal additional variants.
New transcriptomic insights
Despite the challenges described above, the advantages of RNA-Seq have enabled us to generate an unprecedented global view of the transcriptome and its organization for a number of species and cell types. Before the advent of RNA-Seq, it was known that a much greater than expected fraction of the yeast, Drosophila melanogaster and human genomes are transcribed2,4,36, and for yeast and humans a number of distinct isoforms have been found for many genes2,4. However, the starts and ends of most transcripts and exons had not been precisely resolved and the extent of spliced heterogeneity remained poorly understood. RNA-Seq, with its high resolution and sensitivity has revealed many novel transcribed regions and splicing isoforms of known genes, and has mapped 5′ and 3′ boundaries for many genes.
Mapping gene and exon boundaries
The single-base resolution of RNA-Seq has the potential to revise many aspects of the existing gene annotation, including gene boundaries and introns for known genes as well as the identification of novel transcribed regions. 5′ and 3′ boundaries can be mapped to within 10–50 bases by a precipitous drop in signal. 3′ boundaries can be precisely mapped by searching for poly(A) tags, and introns can be mapped by searching for tags that span GT–AG splicing consensus sites. Using these methods the 5′ and 3′ boundaries of 80% and 85% of all annotated genes, respectively, were mapped in S. cerevisiae18. Similarly, in S. pombe many boundaries were defined by RNA-Seq data in combination with tiling array data19.
These two studies led to the discovery of many 5′ and 3′ UTRs that had not been analysed previously. In S. cerevisiae, extensive 3′-end heterogeneity was discovered at two levels: first, local heterogeneity exists in which a cluster of sites are involved, typically within a 10 bp window; second, there are distinct regions of poly(A) addition for 540 genes (FIG. 4). It is plausible that these different 3′ ends confer distinct properties to the different mRNA isoforms, such as mRNA localization or degradation signals, which in turn might be responsible for unique biological functions18,19. In addition to 3′ heterogeneity, the list of upstream ORFs within the 5′ UTRs of mRNAs (uORFs) was also greatly expanded from 17 to 340 (6% of yeast genes)18; uORFs regulate mRNA translation37 or stability38, so these sequences might make a previously underappreciated contribution to the regulatory sophistication of eukaryotic genomes. Interestingly, many mRNAs with uORFs are transcription factors, suggesting that these regulators are themselves heavily regulated.
The mapping of transcript boundaries revealed several novel features of eukaryotic gene organization. Many yeast genes were found to overlap at their 3′ ends18. Using relaxed criteria similar to those employed in a recent study18 we found that 808 pairs, approximately 25% of all yeast ORFs, overlap at their 3′ ends18. Likewise, antisense expression is enriched in the 3′ exons of mouse transcripts22. These features might confer interesting regulatory properties on the affected genes. For multicellular organisms, antisense transcription could modulate gene expression through the production of siRNAs or through dsRNA editing39,40. For yeast, which seems to lack siRNA and dsRNA-editing functions, transcription from one gene might interfere with that from an overlapping gene, or coordinate gene expression through other mechanisms.
Extensive transcript complexity
RNA-Seq can be used to quantitatively examine splicing diversity by searching for reads that span known splice junctions as well as potential new ones. In humans, 31,618 known splicing events were confirmed (11% of all known splicing events) and 379 novel splicing events were discovered24. Another study of human cells found 94,241 junctions, among which 4,096 were novel, and further demonstrated that the prevalent form of alternative splicing is exon skipping41. In mice, extensive alternative splicing was observed for 3,462 genes20. In addition, 42 splicing events that join exons from multiple mouse genes were detected22.
Novel transcription
Previous studies using transposon tagging and tiling microarrays have suggested that in the genomes of yeast, D. melanogaster and humans, there are many novel transcribed regions represented in poly(A)+ RNA2,36,42,43. However, the accuracy of the tiling array results is uncertain owing to concerns about cross-hybridization (see below). RNA-Seq, which does not suffer from problems with background noise, has confirmed that at least 75% and perhaps greater than 90% of the S. cerevisiae and S. pombe genomes are expressed18,19. In addition, results from RNA-Seq suggest the existence of a large number of novel transcribed regions in every genome surveyed, including the A. thaliana21, mouse20,22, human24, S. cerevisiae18 and S. pombe19 genomes. 487 and 453 novel transcripts have been discovered in S. cerevisiae and S. pombe, respectively18,19; for S. cerevisiae half of these were not identified using microarrays. Many of these novel transcribed regions in yeast do not seem to encode any protein, and their functions remain to be determined. The current sequencing depth is not sufficient to define the boundaries of novel transcript units in mammals; however, 30–40% of reads map to unannotated regions20,22,24. These novel transcribed regions, combined with many undiscovered novel splicing variants, suggest that there is considerably more transcript complexity than previously appreciated.
Defining transcription level
As RNA-Seq is quantitative, it can be used to determine RNA expression levels more accurately than microarrays. In principle, it is possible to determine the absolute quantity of every molecule in a cell population, and directly compare results between experiments. Several methods have been used for quantification. For RNA fragmentation followed by cDNA synthesis, which gives more uniform coverage of each exon, gene expression levels can be deduced from the total number of reads that fall into the exons of a gene, normalized by the length of exons that can be uniquely mapped24; for 3′-biased methods, read counts from a window near the 3′ end are used18. Gene expression levels determined by these methods closely correlate with qPCR and RNA spike-in controls.
One particularly powerful advantage of RNA-Seq is that it can capture transcriptome dynamics across different tissues or conditions without sophisticated normalization of data sets19,20,22. RNA-Seq has been used to accurately monitor gene expression during yeast vegetative growth18, yeast meiosis19 and mouse embryonic stem-cell differentiation22, to track gene expression changes during development, and to provide a ‘digital measurement’ of gene expression difference between different tissues20. Because of these advantages, RNA-Seq will undoubtedly be valuable for understanding transcriptomic dynamics during development and normal physiological changes, and in the analysis of biomedical samples, where it will allow robust comparison between diseased and normal tissues, as well as the subclassification of disease states.
Future directions
Although RNA-Seq is still in the early stages of use, it has clear advantages over previously developed transcriptomic methods. The next big challenge for RNA-Seq is to target more complex transcriptomes to identify and track the expression changes of rare RNA isoforms from all genes. Technologies that will advance achievement of this goal are pair-end sequencing, strand-specific sequencing and the use of longer reads to increase coverage and depth. As the cost of sequencing continues to fall, RNA-Seq is expected to replace microarrays for many applications that involve determining the structure and dynamics of the transcriptome.
Acknowledgements
We thank D. Raha for many valuable comments.
Glossary
- Cap analysis of gene expression (CAGE)
Similar to SAGE, except that 5′-end information of the transcript is analysed instead of 3′-end information.
- Contigs
A group of sequences representing overlapping regions from a genome or transcriptome.
- dsRNA editing
Site-specific modification of a pre-mRNA by dsRNA-specific enzymes that leads to the production of variant mRNA from the same gene.
- Genomic tiling microarray
A DNA microarray that uses a set of overlapping oligonucleotide probes that represent a subset of or the whole genome at very high resolution.
- Massively parallel signature sequencing (MPSS)
A gene expression quantification method that determines 17–20-bp ‘signatures’ from the ends of a cDNA molecule using multiple cycles of enzymatic cleavage and ligation.
- MicroRNA(miRNA)
Small RNA molecules that are processed from small hairpin RNA (shRNA) precursors that are produced from miRNA genes. miRNAs are 21–23 nucleotides in length and through the RNA-induced silencing complex they target and silence mRNAs containing imperfectly complementary sequence.
- Piwi-interacting RNAs (piRNA)
Small RNA species that are processed from single-stranded precursor RNAs. They are 25–35 nucleotides in length and form complexes with the piwi protein. piRNAs are probably involved in transposon silencing and stem-cell function.
- Quantitative PCR (qPCR)
An application of PCR to determine the quantity of DNA or RNA in a sample. The measurements are often made in real time and the method is also called real-time PCR.
- Sequencing depth
The total number of all the sequences reads or base pairs represented in a single sequencing experiment or series of experiments.
- Serial analysis of gene expression (SAGE)
A method that uses short ~14–20-bp sequence tags from the 3′ ends of transcripts to measure gene expression levels.
- Short interfering RNA (siRNA)
RNA molecules that are 21–23 nucleotides long and that are processed from long double-stranded RNAs; they are functional components of the RNAi-induced silencing complex. siRNAs typically target and silence mRNAs by binding perfectly complementary sequences in the mRNA and causing their degradation and/or translation inhibition.
- Spike-in RNA
A few species of RNA with known sequence and quantity that are added as internal controls in RNA-Seq experiments.
Footnotes
FURTHER INFORMATION
Gerstein laboratory homepage: http://bioinfo.mbb.yale.edu
Snyder laboratory homepage: http://www.yale.edu/snyder
454 Life science: http://www.454.com
Applied Biosystems: www.appliedbiosystems.com
Helicos Biosciences: http://www.helicosbio.com
Illumina: http://www.illumina.com
Illumina forum: http://www.illumina.com/pagesnrn.ilmn?iD=245
SEQanswers: http://seqanswers.com/forums/showthread.php?t=43
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Clark TA, Sugnet CW, Ares M., Jr Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- 2.David L, et al. A high-resolution map of transcription in the yeast genome. Proc. Natl Acad. Sci. USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamada K, et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- 4.Bertone P, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 5.Cheng J, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 6.Okoniewski MJ, Miller CJ. Hybridization interactions between probesets in short oligo microarrays lead to spurious correlations. BMC Bioinformatics. 2006;7:276. doi: 10.1186/1471-2105-7-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Royce TE, Rozowsky JS, Gerstein MB. Toward a universal microarray: prediction of gene expression through nearest-neighbor probe sequence identification. Nucleic Acids Res. 2007;35:e99. doi: 10.1093/nar/gkm549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boguski MS, Tolstoshev CM, Bassett DE., Jr Gene discovery in dbEST. Science. 1994;265:1993–1994. doi: 10.1126/science.8091218. [DOI] [PubMed] [Google Scholar]
- 9.Gerhard DS, et al. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC) Genome Res. 2004;14:2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 11.Harbers M, Carninci P. Tag-based approaches for transcriptome research and genome annotation. Nature Methods. 2005;2:495–502. doi: 10.1038/nmeth768. [DOI] [PubMed] [Google Scholar]
- 12.Kodzius R, et al. CAGE: cap analysis of gene expression. Nature Methods. 2006;3:211–222. doi: 10.1038/nmeth0306-211. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura M, Carninci P. Cap analysis gene expression: CAGE. Tanpakushitsu Kakusan Koso. 2004;49:2688–2693. (in Japanese) [PubMed] [Google Scholar]
- 14.Shiraki T, et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl Acad. Sci. USA. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brenner S, et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nature Biotechnol. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- 16.Peiffer JA, et al. A spatial dissection of the Arabidopsis floral transcriptome by MPSS. BMC Plant Biol. 2008;8:43. doi: 10.1186/1471-2229-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reinartz J, et al. Massively parallel signature sequencing (MPSS) as a tool for in-depth quantitative gene expression profiling in all organisms. Brief. Funct. Genomic Proteomic. 2002;1:95–104. doi: 10.1093/bfgp/1.1.95. [DOI] [PubMed] [Google Scholar]
- 18.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilhelm BT, et al. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 20.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 21.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cloonan N, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nature Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 23.Marioni J, Mason C, Mane S, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008 Jun 11; doi: 10.1101/gr.079558.108. (doi:10.1101/gr.079558.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morin R, et al. Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. Biotechniques. 2008;45:81–94. doi: 10.2144/000112900. [DOI] [PubMed] [Google Scholar]
- 25.Holt RA, Jones SJ. The new paradigm of flow cell sequencing. Genome Res. 2008;18:839–846. doi: 10.1101/gr.073262.107. [DOI] [PubMed] [Google Scholar]
- 26.Barbazuk WB, Emrich SJ, Chen HD, Li L, Schnable PS. SNP discovery via 454 transcriptome sequencing. Plant J. 2007;51:910–918. doi: 10.1111/j.1365-313X.2007.03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vera JC, et al. Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Mol. Ecol. 2008;17:1636–1647. doi: 10.1111/j.1365-294X.2008.03666.x. [DOI] [PubMed] [Google Scholar]
- 28.Emrich SJ, Barbazuk WB, Li L, Schnable PS. Gene discovery and annotation using LCM-454 transcriptome sequencing. Genome Res. 2007;17:69–73. doi: 10.1101/gr.5145806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dutrow N, et al. Dynamic transcriptome of Schizosaccharomyces pombe shown by RNA–DNA hybrid mapping. Nature Genet. 2008;40:977–986. doi: 10.1038/ng.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu JQ, et al. Systematic analysis of transcribed loci in ENCODE regions using RACE sequencing reveals extensive transcription in the human genome. Genome Biol. 2008;9:R3. doi: 10.1186/gb-2008-9-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008 doi: 10.1101/gr.078212.108. (doi:10.1101/gr.078212.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith AD, Xuan Z, Zhang MQ. Using quality scores and longer reads improves accuracy of Solexa read mapping. BMC Bioinformatics. 2008;9:128. doi: 10.1186/1471-2105-9-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hillier LW, et al. Whole-genome sequencing and variant discovery in C. elegans. Nature Methods. 2008;5:183–188. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- 35.Campbell PJ, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nature Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manak JR, et al. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nature Genet. 2006;38:1151–1158. doi: 10.1038/ng1875. [DOI] [PubMed] [Google Scholar]
- 37.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 38.Ruiz-Echevarria MJ, Peltz SW. The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell. 2000;101:741–751. doi: 10.1016/s0092-8674(00)80886-7. [DOI] [PubMed] [Google Scholar]
- 39.Tomari Y, Zamore PD. MicroRNA biogenesis: drosha can’t cut it without a partner. Curr. Biol. 2005;15:R61–R64. doi: 10.1016/j.cub.2004.12.057. [DOI] [PubMed] [Google Scholar]
- 40.Bass BL. How does RNA editing affect dsRNA-mediated gene silencing? Cold Spring Harb. Symp. Quant. Biol. 2006;71:285–292. doi: 10.1101/sqb.2006.71.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sultan M, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 42.Ross-Macdonald P, et al. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature. 1999;402:413–418. doi: 10.1038/46558. [DOI] [PubMed] [Google Scholar]
- 43.Kumar A, des Etages SA, Coelho PS, Roeder GS, Snyder M. High-throughput methods for the large-scale analysis of gene function by transposon tagging. Methods Enzymol. 2000;328:550–574. doi: 10.1016/s0076-6879(00)28418-8. [DOI] [PubMed] [Google Scholar]