Abstract
The concept of Receptor Mosaic (RM) is discussed; hence the integrative functions of the assemblage of G-protein coupled receptors physically interacting in the plane of the plasma membrane. The main focus is on a heterotrimer of G-protein coupled receptors, namely the A2A-D2-CB1 receptor trimer. A bioinformatics analysis was carried out on the amino acid sequence of these receptors to indicate domains possibly involved in the receptor-receptor interactions. Such a bioinformatic analysis was also carried out on the RM formed by mGLU R5, D2 and A2A.
The importance of topology, i.e., of the reciprocal localisation of the three interacting receptors in the plan of the membrane for the RM integrative functions is underlined. However, it is also pointed out that this fundamental aspect still waits techniques capable of an appropriate investigation. Finally, it is discussed how RM topology can give hints for a structural definition of the concept of hub receptor. Thus, just as in any network, the receptor operating as a hub is the one that in the molecular network formed by the receptors has the highest number of inputs.
Keywords: Receptor mosaic, A2A-D2-CB1 receptor trimer, mGLU5-D2-A2A receptor trimer, receptor topology, receptor bioinformatics analysis
1. INTRODUCTION: THE CONCEPT OF RECEPTOR MOSAIC
In opposition to the view, which was currently accepted in the Eighties, we maintained that cell signalling became a branched process beginning at the level of receptor recognition in the plasma membrane where G-Protein Coupled Receptors (GPCRs) can directly modify the ligand recognition, G protein coupling and signalling of other receptors via Receptor-Receptor Interactions (RRIs), which occur thanks to direct physical interactions between GPCRs at plasma membrane level [1, 2]. The demonstration and the theoretical and functional consequences of the existence of Receptor-Receptor Interactions as a new integrative mechanism at plasma membrane level [3] nowadays is one of the main areas of investigation for the clarification of recognition/decoding processes by GPCRs as well as for the development of new therapeutic approaches [4]. Already in the Eighties indirect in vitro and in vivo proofs of RRIs were given through the analysis of the effects of neuropeptides on the binding characteristics of monoamine receptors in membrane preparations from discrete brain regions, and by means of in vivo studies on the sleep-wakefulness cycle and arterial blood pressure control in experimental rats [5-9].
Only in the following decades several groups have provided evidence for RRI based on molecular biology techniques. While these proofs have been of extraordinary importance, it should be noted that the fundamental findings are those obtained in physiological and physio-pathological experiments since as clearly stated by the Nobel Laureate Sir James Black: “... the benefits that molecular biology will bring to pharmacology, I believe, to be circumscribed by the state of physiological knowledge, models, and concepts...” and as a consequence he foresees that we will assist to “The progressive triumph of physiology over molecular biology” [10].
Actually, from the functional evidence obtained by in vivo studies in experimental rats and by means of binding experiments on neuronal membrane preparations, our group put forward the hypothesis of the existence of assemblies of multiple receptors of various types at plasma membrane level and introduced the term receptor mosaic (RM) [11-15].
The following definition has been proposed: an RM is a cluster of receptors, which can bind and decode intra- and/or extra-cellular signals to give out an integrated input to one or more molecular networks, which represent the biochemical effectors. A cluster of receptors works as a RM only if at least one of the receptors of the cluster directly modulates by means of allosteric interactions the biochemical/functional features of at least another receptor of the cluster.
The term RM is based on the analogy of the assemblage of receptors in physical contact in the plane of the membrane with the mosaic work, which can be defined as the process of making pictures or designs by inlaying small bits of coloured stones (tesserae) (Webster's New Twentieth Century Dictionary, Prentice Hall Press, New York, 1983). We introduced such a term since it points out one of the most fundamental features of receptor assemblage, namely the spatial organization of single receptors within the cluster.
Two structural aspects and three functional aspects should be considered as fundamental features to characterize a RM:
Structural aspects
-
-
Topology, i.e., spatial distribution of the receptors within the RM
-
-
Type of receptors that are assembled in the RM.
Functional aspects
-
-
Presence of a so called “hub receptor” [16-19] within the RM. That is if the receptors of the RM are all equivalent or there is one receptor, which is in a privileged position and hence can strongly affect the conformations (and function) of the other receptors of the mosaic. Thus, as in any network the hub receptor is the one with the highest number of inputs [21].
-
-
Presence of cooperativity in some of the allosteric interactions, which allow the interactions between receptors [17]
-
-
The order of activation of receptors inside the RM.
The present paper will deal mainly with the first functional aspect mentioned above, hence with the topology of the RM and on some theoretical aspects, which are implicated in the topological organization.
2. THE CHEMICAL BASIS OF R ECEPTOR-RECEPTOR INTERACTIONS
Interactions between proteins underlie practically all biological processes and are fundamental for the function of the molecular networks. Thus, proteins are endowed with the so called ‘Lego Property’, i.e. with the capability to participate in protein-protein interactions, which however occur with high safety factors to allow only the necessary interactions [see, e.g., 20]. This holds true also for GPCRs, which nowadays are described as functionally ductile proteins not only capable of recognizing transmitters, but also capable of several different interactions with other small ligands (allosteric modulators) and other proteins. Actually, as discussed in the Introduction, the first description of GPCR recognition/decoding process was based on the assumption: one transmitter (or more general one signal), one receptor, one G-protein. Now the field is completely changed and it is well possible to have: more than one transmitter, which can interact with one and the same receptor; one and the same GPCR, which can interact with different G-proteins and/or decoding systems [22-24]. Furthermore, as stated above GPCRs can also interact in the plane of the membrane with other GPCRs or they can be inserted in the plane of the membrane as a multimeric complex [25]. Hence, to fully understand the biochemical processes involved in the GPCR-mediated recognition/decoding of signals, we need to unravel much more phenomena than it was thought in the beginning and in particular the principles of GPCR-GPCR interactions.
The search of these principles, as in the case of other interacting proteins, has to be focused rather than on the individuation of the entire interacting interfaces on the individuation of the binding “hot spots,” that is of the few of the interface residues, which are essential for the recognition and binding to other proteins. The limited overlap between interface residues and hotspots has been demonstrated for several cases, for example for the interaction between the human growth hormone and its receptor [26]. As a matter of fact, there are 31 residues on the receptor that are in physical contact with the hormone; however, mutation experiments indicate that only six of these residues are energetically crucial for the interaction [26].
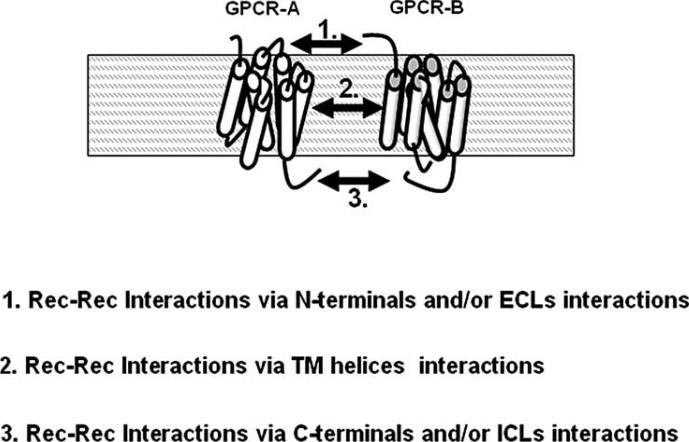
The search of principles for GPCR-GPCR interactions can start from some heuristic hypotheses on the basic aspects of this class of protein-protein interactions, which should be considered as a special class, since the two interacting proteins are embedded in three distinct environments: the extracellular space, the intra-membrane space, the intra-cellular space. Already in 1995, we proposed that interactions between two GPCRs could occur in any of the three micro-environments (see Fig. 1), with potentially different types of chemical interactions and hence different functional implications [27].
Fig. (1).
Schematic representation of the sites of GPCR-GPCR interactions [27]. Recent evidence [29, 30] has given a support to all these types of interactions, which could have different chemical characteristics, e.g., different stability and rigidity and hence different functional meanings. For further details see text.
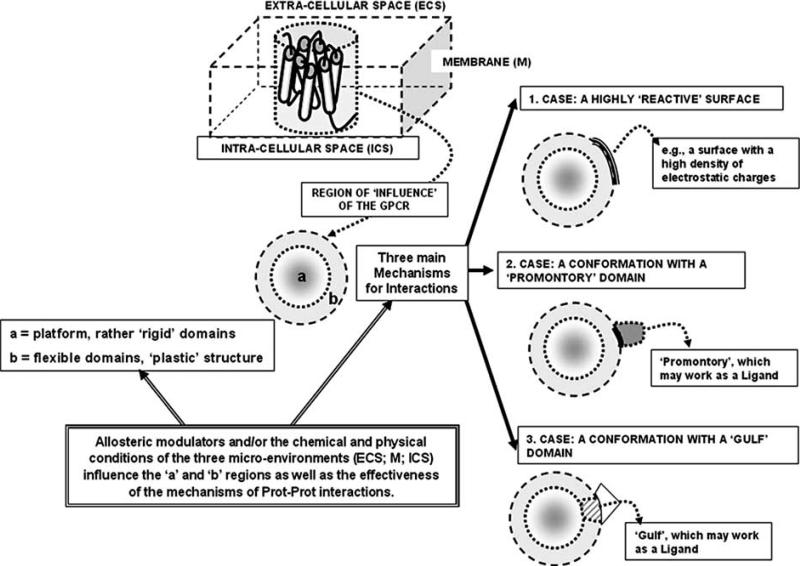
A basic aspect of interactions between GPCRs is certainly the 3D-conformation of the interacting receptors, which depend on the relative topological arrangement of their respective domains. In this context it should be mentioned that domain-domain interactions can take place within one and the same GPCR mainly by interactions between transmembrane (TM) helixes. It has been surmised that intra-receptor interactions occurring inside one and the same receptor could allow the formation of a rather rigid structure (the GPCR ‘core region’ or ‘platform’ [28]) mainly formed by transmembrane helixes, but also probably by Extra-Cellular Loop 2 (ECL2; see below). Around this core region, a more flexible region, which is mainly formed by extra- and intra-cellular loops and N- and C-terminals, can easily walk in its energy landscape by assuming different conformations (Fig. 2). Thus, the latter region may be of the highest importance for the adaptation of the receptor to the micro-environments with which it interacts. It may also be surmised that the two regions are involved in different types of interactions, in particular the more rigid ones (hence likely mainly the TM helixes) are involved in rather stable RRIs [29, 30], while the more plastic one (hence likely mainly the ECLs, the Intra-Cellular Loops (ICLs) and especially the C-terminals) may be the region mainly involved in the formation of rather flexible heteromers [31]. Furthermore, this plastic region is likely the most affected by small allosteric ligands as demonstrated by our studies on Homocysteine allosteric modulation of D2 receptors [32].
Fig. (2).
Schematic representation of a GPCR as a multi-domain protein, which is embedded into three micro-environments: the extra-cellular space (ECS), the plasma membrane (M) and the intra-cellular space (ICS). It is proposed that two regions (‘a’ and ‘b’) can be distinguished in a GPCR , namely a rather rigid core and a more plastic surrounding, respectively. The possible mechanisms for the RRI are also indicated. The chemico-physical features of the three environments as well as the action of allosteric modulators can affect both the conformation and/or the size of ‘a’ and ‘b’ regions and the mechanisms of RRI.
In agreement with such a proposal, differences between GPCRs have been demonstrated in helix orientation, helix-helix interactions, and helix topology [33]. Thus, it has been suggested the formation of a “platform”, whose core is probably formed by TM3,4,5 since a disulfide bond links TM3 and ECL2 and this link is conserved in 91.8% of GPCRs [28, 33]. The plasticity of ECLs is also suggested by the observation that they usually provide the crucial contacts in the recognition process since a direct ligand-TM domain interaction is not always necessary. The functional relevance of ECLs is also supported by the demonstration that about 52% of inactivating mutations are located in ECL2 and 3 [34].
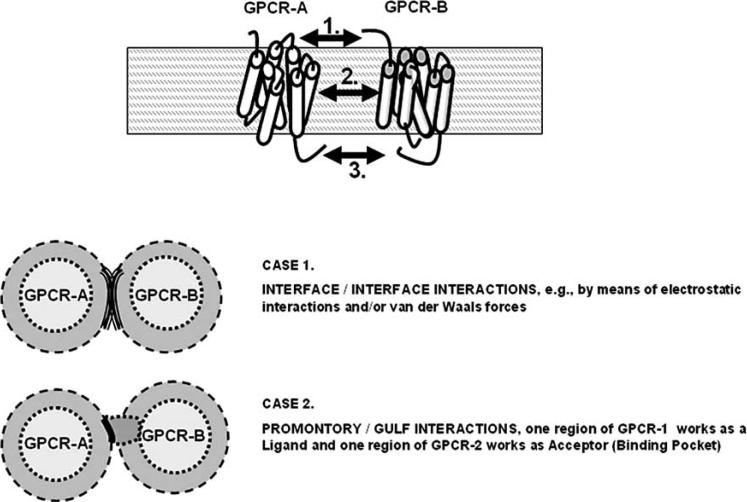
On the basis of these data, it can be surmised that RRI (let us say between receptor A. and B.) can occur via three main mechanisms (for the first two, see Figs. 2 and 3):
Contact between interfaces of receptor A. and B., which have suitable complementary “hot spots” [12-15, 20, 35]. For example, it has been demonstrated that, e.g., A2A dimerisation occurs via helix-helix interaction involving TM5 [36].
Complementary 3D-conformations, which can work as ‘Ligand-region’ (i.e., a ‘promontory-region’ of the receptor A. structure) and as suitable ‘Pocket-region’ (i.e., a ‘gulf-region’ of the receptor B. structure).
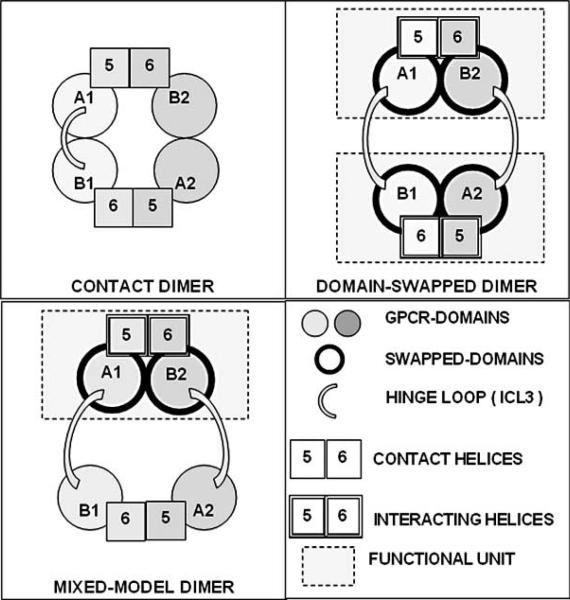
Crossing-over of the two receptor proteins; thus, it has been proposed that GPCRs have two main domains linked by a hinge loop, hence, swapping of domains is possible [12-15, 20, 35]. Gouldson suggested to distinguish two main components within a GPCR, one made by the N-terminus and loop 1-5, the other one made by loop 6-7 and the C-terminus, which could be swapped between two dimerizing receptors (Fig. 4).
These mechanisms can be based rather than in a lock-key process on an induced-fit process, which mainly involves the plastic regions of the interacting GPCRs. Furthermore, it should be considered that if more than two receptors are involved in the formation of the RM, cooperation can be present, favouring (positive cooperativity) or impairing (negative cooperativity) the assemblage of the RM [17, 37]. It should, also, be noted that all this important information does not take into account the possibility of differences in GPCR structure and function accordingly to the RM, which it belongs hence further more detailed studies are needed.
Fig. (3).
Schematic representation of the possible Receptor-Receptor Interactions. Two main case are illustrated: the interface/interface interactions and the ligand (promontory)/pocket (gulf) interactions. For further details, see text.
Fig. (4).
Two main models have been suggested by Gouldson [35] for the formation of GPCR dimers: the “domain swapping” and the “domain contact”. According to Gouldson it is possible to distinguish two pseudo-independent units in the GPCR. Thus, the N-terminus and helices 1-5 constitute the A-GPCR domain, while helices 6 and 7 through to the C-terminus constitute the B-GPCR domain. A and B domains are connected by a hinge loop (third intracellular loop: ICL3), which is frequently the longest loop in GPCRs and therefore very well suited to allow reciprocal movements of the two A- and B-GPCR domains. Even considering only contact 5-6 dimers and swapped 5-6 dimers three basic models can occur: pure swapping, pure contact, and mixed model (contact and swapping). These models are shown in the scheme of the Figure [13].
In summary, a GPCR can take different 3D-conformations, which can affect its decoding characteristics as well as its RRIs. It may be suggested that 3D-conformations and hence RRIs depend on:
flexibility of the extra- and/or intracellular loops, which can be, in turn, modulated by other interacting molecules such as allosteric modulators, but also ions present in the extra- and intra-cellular environment, respectively;
TM conformations, which are affected by the lipid environment of the membrane (e.g. a lipid raft; [20]);
Presence of disordered sequences, which may allow intra- or inter-receptor interactions as well as interactions with multiple ligands.
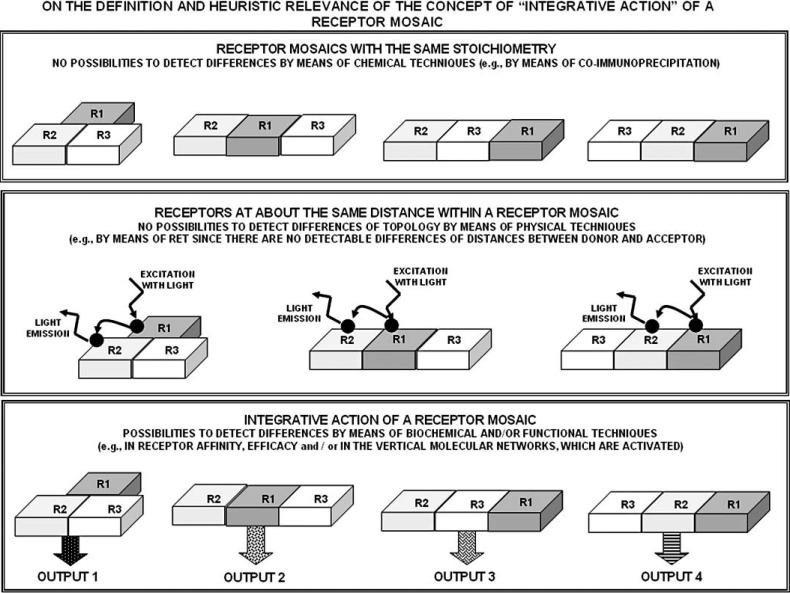
Thus, several aspects of GPCR conformations and RRIs are still to be clarified and the available techniques are insufficient above all for understanding the topology of even an hetero-trimer (see Fig. 5 two upper panels). However, a support to formulate heuristic hypotheses on RM topology can come not only from the still incomplete molecular biology data, but also, and perhaps mainly, from functional studies (e.g., in vivo studies on RM ‘integrative actions’), as Sir James Black already 15 years ago pointed out. Furthermore, theoretical considerations and models could be of a great importance, and these speculative models could, in some instances, find an indirect support from bioinformatics data.
Fig. (5).
Schematic illustration of the heuristic relevance of the integrative action of a hetero-trimer to have hints on its topology. The upper two panels illustrate the limits of the chemical and biophysical approaches in allowing a precise determination of the localisation (topology) of receptors within a RM. For further details, see text.
3. SOME THEORETICAL CONSIDERATIONS ON THE TOPOLOGICAL ARRANGEMENTS OF A TRIMERIC RECEPTOR MOSAIC
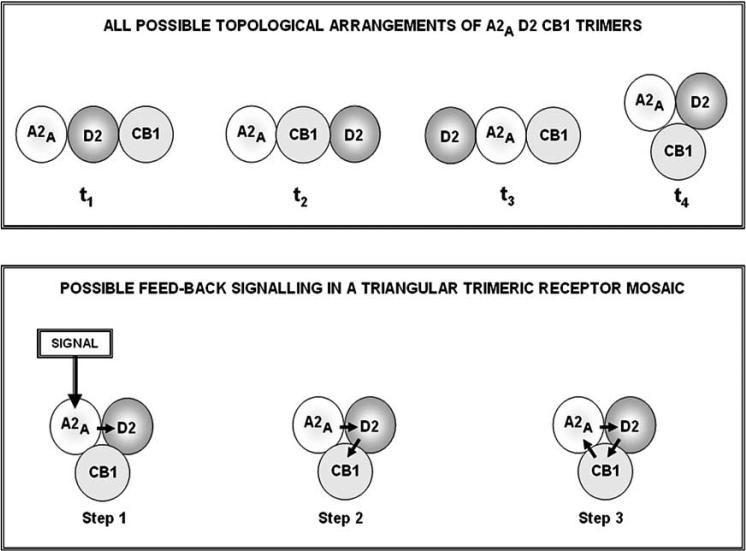
Let us consider a very simple RM, for example a trimeric RM. If we are dealing with a homo-trimer, as a first, self-evident, consideration, we must assume that each receptor has at least two binding sites available to bind other receptors in the assembled trimeric RM. Furthermore, it may be questioned whether the three originally equivalent orthosteric binding sites are equivalent in the RM or, they become not only topologically, but also functionally different. Moreover, it has also to be observed that the three receptors can be arranged according to two different topologies (linear versus triangular), which again can lead to different integrative actions of the RM (see Fig. 5, third panel). In particular, it may be surmised that a circulation of the information can take place in the triangular topology, that is that the ‘ring model’ proposed by Agnati, Fuxe, Guidolin can be in action. The ring model is of a great potential interest especially for hetero-trimeric RM since, an alteration in the conformation of one receptor after recognition/decoding of its ligand (or even after a transient change in conformation due, e.g., to temperature-induced walk of the GPCR in its energy landscape) is fed-back to the same receptor (see Fig. 6, lower panel). This phenomenon can potentiate or damper the conformational change.
Fig. (6).
Schematic illustration of the possible feed-back circulation of the information in a triangular trimeric RM. It should be noticed that all receptors have two sites allowing RRIs, hence the flow of the information. For further details, see text.
As a last consideration on this topic, it should be mentioned the Jaffe's proposal [38], which suggests a new model of allosteric regulation, the so-called morpheein model. According to this model different quaternary assemblies of the same primary protein sequence are possible in which the stochiometry of the complete assembly is dictated by different conformations of the monomeric unit. As discussed in the cited paper, the function of the possible different assemblies can be substantially different. Thus, the term morpheeins describes the differences in oligomeric multiplicity, structure and function dictated by a conformational change in the monomer. It is of substantial interest that allosteric regulators can shift the equilibrium towards alternative structures, i.e., towards assemblies with a different multiplicity and function [38]. When this model is applied to the RM, it can be surmised that not only different RM can be formed according to a transient monomer conformation, but also that possible inter-conversions of RMs can occur under the action of allosteric modulators which can bind one or more monomers leading to RMs with different multiplicity according to the conformation of the monomers and hence to different integrative actions [28].
5. A BIOINFORMATICS ANALYSIS OF THE A2ACB1-D2 TRIMERIC RECEPTOR-MOSAIC
In 2003 indications have been obtained for the existence of cannabinoid receptor CB1-D2 receptor heteromers in cotransfected HEK-293 cells based on FRET analysis and for antagonistic CB1/D2 interactions in postulated striatal CB1/D2 heteromers based on D2 binding analysis [39, 40]. In 2005 studies with co-immunoprecipitation in HEK-293 cells gave further indications for CB1/D2 heteromers with an enhanced formation after concurrent activation of the two receptors. It was noticed that in this heteromer the CB1 signalling in part switches from an inhibition of the adenylyl cyclase to a pertussis insensitive activation of the AC [41]. New evidence has now been obtained for the existence of CB1/D2 receptor heteromers based on FRET analysis in HEK-293 cells [42]. These heteromers probably exist also in native striatal tissue since CB1 agonists have been found to reduce D2 receptor affinity in dorsal striatal membrane preparations and in nuc accumbens shell [42]. Thus, these receptor-receptor interactions probably occur in CB1-D2 receptor heteromers through receptor-receptor interactions operating via allosteric mechanisms.
The behavioural analysis also showed that CB1 agonists can counteract the D2 agonist induced hyper-locomotion, an effect blocked by rimonabant the CB1 antagonist, which also can enhance the action of the D2 like agonist quinpirole [42]. Furthermore, the A2A antagonist MSX-3 could counteract the ability of the CB1 agonist CP 55,940 to counteract the D2 agonist action in the behavioural analysis. Furthermore, it has been demonstrated in HEK-293 cells and in neuroblastoma cells that the CB1 signalling was entirely dependent of A2A receptor activation [43].In addtion,CB1 agonists including Δ9-tetrahydrocannabinol (THC) can increase a PKA mediated DARPP-32 phosphorylation at Thr34 position via A2A receptors in striatal slices [44, 45]. Thus, there may exist an A2A-CB1-D2 RM in striatum integrating DA, adenosine and endocannabinoid signals [42, 46]
Recently it has been shown that three GPCRs (A2A, D2 CB1) can be assembled into a hetero-trimeric RM [47]. On the bases of the current experimental evidence, no information is available on which topology (linear or triangular) is the most likely spatial arrangement. It may be surmised that, in the case of a linear arrangement, the central position could be a privileged one, since, in analogy to what has been proposed for molecular networks, the central receptor might work as a “hub receptor” (for a definition of “hub node” see, e.g., [21, 48]). Such a hub receptor could easily control the other two receptors [17].
Table 1 reports the results of bioinformatic analyses of the amino acid sequence of the three proteins. A first analysis was aimed at evaluating the degree of disorder of the various protein domains, which depends on the presence of sequences lacking a clearcut secondary structure [49]. This characteristic makes them very suitable for different and multiple protein-protein interactions, since they usually acquire an ordered structure only upon binding with partners. Several methods were recently proposed [50-54] to classify whether any given residue of a protein chain was in a disordered region. On that basis, our group developed a disorder index (DI) by pooling together the results of ten different computer programs capable of detecting with a good prediction whether an amino acid sequence is ‘disordered’ [28]. Such a parameter (ranging between 0 and 1) increases as the propensity to disorder of the analysed sequence increases. A second type of analysis was focused on the estimation of the propensity of the various domains to be involved in protein aggregation, i.e. in the formation of stable, rigid arrangements or complexes. It depends on the presence of specific motifs acting as ‘hot spots’ driving aggregation [55, 56]. Table 1 reports the Aggregation indexes (AI) obtained by using the public-domain predictor AGGRESCAN [57].
Table 1.
Per Domain Analysis of the Propension to Disorder and to Aggregation of A2A, D2, CB1 and mGluR5 Receptor Proteins (In Brackets the Code Identifying them in the Swiss-Prot Database)
| Disorder index (DI)* | Aggregation Index (AI)§ | |||||||
|---|---|---|---|---|---|---|---|---|
| A2A (P29274) | D2 (P14416) | CB1 (P21554) | mGluR5 (P41594) | A2A (P29274) | D2 (P14416) | CB1 (P21554) | mGluR5 (P41594) | |
| N-term | 0.386 | 0.394 | 0.498 | 0.156 | 0.455 | 0.040 | 0.077 | 0.082 |
| TM1 | 0.000 | 0.000 | 0.008 | 0.000 | 0.637 | 0.741 | 0.543 | 0.754 |
| ICL1 | 0.023 | 0.009 | 0.000 | 0.000 | 0.457 | 0.229 | 0.253 | 0.456 |
| TM2 | 0.117 | 0.092 | 0.000 | 0.000 | 0.525 | 0.579 | 0.567 | 0.612 |
| ECL1 | 0.100 | 0.100 | 0.000 | 0.000 | 0.612 | 0.443 | 0.189 | 0.336 |
| TM3 | 0.013 | 0.054 | 0.000 | 0.145 | 0.612 | 0.430 | 0.371 | 0.226 |
| ICL2 | 0.055 | 0.119 | 0.050 | 0.255 | 0.380 | 0.288 | 0.237 | 0.172 |
| TM4 | 0.022 | 0.004 | 0.000 | 0.000 | 0.489 | 0.559 | 0.726 | 0.832 |
| ECL2 | 0.163 | 0.092 | 0.000 | 0.071 | 0.080 | 0.000 | 0.228 | 0.214 |
| TM5 | 0.000 | 0.042 | 0.000 | 0.131 | 0.773 | 0.801 | 0.754 | 0.446 |
| ICL3 | 0.272 | 0.620 | 0.391 | 0.163 | 0.199 | 0.074 | 0.187 | 0.193 |
| TM6 | 0.000 | 0.012 | 0.000 | 0.000 | 0.625 | 0.781 | 0.762 | 0.671 |
| ECL3 | 0.012 | 0.050 | 0.067 | 0.000 | 0.317 | 0.123 | 0.555 | 0.600 |
| TM7 | 0.037 | 0.067 | 0.009 | 0.000 | 0.508 | 0.459 | 0.469 | 0.572 |
| C-Term | 0.613 | 0.029 | 0.605 | 0.802 | 0.034 | 0.336 | 0.057 | 0.037 |
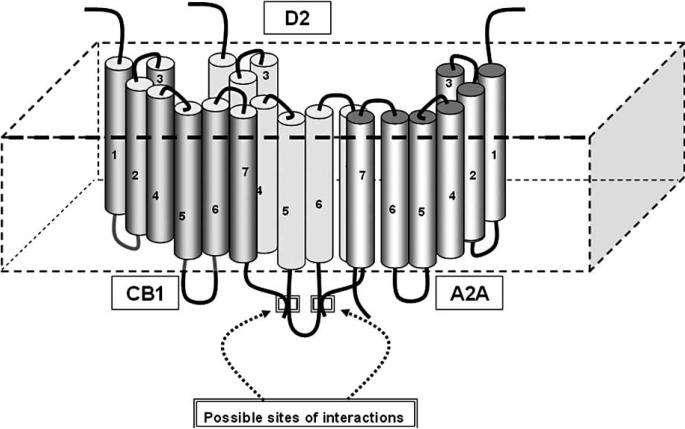
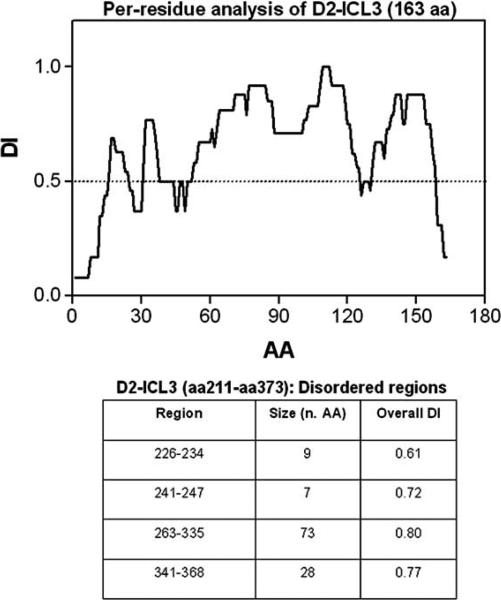
While the -helixes (especially TM5 and 6) have a high aggregation propensity (i.e. AI), the C-terminals of CB1 and A2A and the ICL3 of D2 have a high DI, which suggests a high propensity to form protein-protein interactions [28, 58]. Thus, in agreement with our previous discussion (see Section 2.) it is proposed that TM domains are mainly involved in less plastic arrangements of the GPCR 3D-conformation and in stable RRIs as indicated by several groups [29, 59-62]. On the other hand, the domains with a high DI are particularly suited for more dynamic interactions, such as those with allosteric modulators, but also possibly for more flexible RRIs. As tentative hypothesis, based also on the evidence that ICL3 has two high DI rather long amino acid sequences (namely 263-335 and 341-368) and as pointed out by Dunker high DI AA sequences can work as hubs (see Fig. 7). Thus, it can be suggested (see also the plot of Fig. 8) the C-terminal of CB1 interacts with the ICL3 (341-368) of D2 and the ICL3 (263-335) of D2 interacts with the C-terminal of A2A (probably with the sequence 215-224; [31]). However, other possibilities should be considered, such that of TM-interactions between, e.g., CB1 and D2 or between CB1 and A2A. In the latter case, the ‘central position’ in the trimeric RM is occupied by A2A, which interacts through its C-terminal with ICL3 of D2 and by a TM-TM with CB-1.
Fig. (7).
Possible topological arrangement of the trimeric RM formed by CB1, A2A and D2. The hypothetical topology is based on Table 1. and on the plot and the inserted table of Fig. 7. For further details, see text.
Fig. (8).
Per-residue analysis of D2-ICL3 (163 AA). The plot clearly indicates the region where disordered sequences are present. These sequences are precisely given in the inserted table. For further details, see text.
6. A BIOINFORMATICS ANALYSIS OF A PUTATIVE A2A-D2-MGLUR5 TRIMERIC RECEPTOR-MOSAIC
L-glutamate was early on found to reduce the affinity of the D2 agonist binding sites in striatal membranes giving the first indications of the existence of intra-membrane receptor-receptor interactions hence of the possible existence of glutamate receptor/D2 heteromers [63]. Later on by demonstration that group 1 mGluR agonists caused a similar reduction of D2 agonist affinity in striatal membranes indications were obtained that a type 1 mGluR was the glutamate receptor present in this putative heteromer [64]. Subsequently evidence was obtained by using mGluR5 agonists that the mGluR5 was the specific glutamate receptor forming a putative mGluR5-D2 heteromer with the D2 receptor [65]. In this analysis evidence was also obtained that A2A agonists in combination with group 1 mGluR agonists or a mGluR5 agonist could produce an increased reduction of the affinity of the D2 agonist binding sites in striatal membranes which was synergistic in the case of the group 1 mGluR agonist, indicating the existence of a possible trimeric A2A-D2-mGluR5 receptor mosaic [64, 65].
Subsequently coimmunoprecipitation studies were compatible with this hypothesis and strongly indicated the existence of receptor heteromers containing A2A and mGluR5 where the synergism between A2A and mGluR5 may take place [66]. A2A and mGluR5 immunoreactivities were also colocated in striatal neurons of primary cultures [67] as well as in striatal glutamate nerve terminals [68]. Concurrent stimulation of A2A and mGluR5 resulted in synergistic interactions at the level of c-fos expression and phosphorylation of ERK and DARPP-32 [66, 69]. Such combined activation also led to synergistic effects in the ventral striatopallidal GABA neurons as seen from larger than additive effects in increasing GABA release [70]. Such synergism and the synergism of A2A and mGluR5 antagonists to increase locomotion in reserpinized mice [71, 72] can be elegantly explained by the existence of A2A-D2-mGluR5 RM where A2A-mGluR5 synergize to counteract D2 signalling [73]. Taken together, the existence of striatal A2A-D2-mGluR5 receptor mosaics is strongly indicated with multiple receptor-receptor interactions taking place and with A2A-mGluR5 synergistically counteracting D2 recognition and signalling [4, 67, 73, 74].
5. FINAL COMMENTS
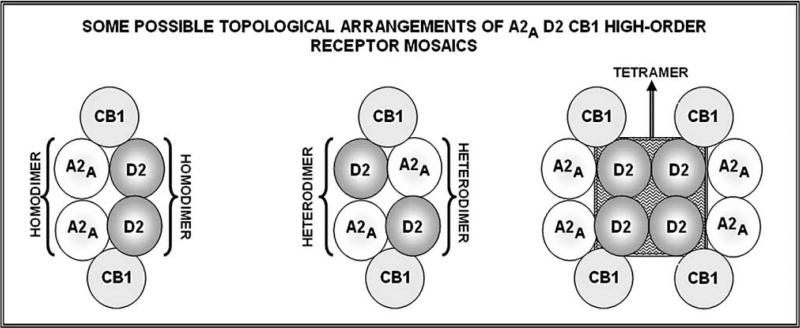
As pointed out in several papers, topology is the most fundamental feature of a RM [12-15, 17], since it opens up an enormous range of possibilities of building up different RMs (hence to get different integrative actions) by using the same set of receptors (tesserae of the mosaic). Thus, topology has the highest interest already from a theoretical standpoint since it underlines the extraordinary plasticity of the system, which can, just by changing the spatial organization of a small number of different receptors, produce an enormous number of different integrated input units to the cell biochemical machinery. In the present paper, this issue has been briefly exemplified by a simple analysis of a hypothetical trimeric RM. Among other potentially relevant implications of topology, the concept of hub receptor has been introduced. The assessment whether in a RM a receptor plays the role of hub node receptor could be of the highest importance since the binding of a transmitter (or a drug) to a hub receptor can have widespread effects on the receptor mosaic and hence on its integrative function [19]. The relevance of the real existence of hetero-trimeric RMs came from the demonstration by the Barcelona's group in a cell model of the CB1-A2A-D2 hetero-trimer. The elegant demonstration is of basic importance but leaves open the question of its functional relevance in vivo (hence the physiological and/or pathological aspects), but also the question of the topology of such a hetero-trimer. It should be noted that topology, as pointed out above, is strictly related to RM function and an altered topology can lead to an altered (pathological) function. Another important aspect that should be investigated is if one or more of the possible hetero-trimers are present as such or can also be building blocks of higher order RMs. As a matter of fact, in view of the experimental evidence that dopamine receptors tend to form dimers and tetramers [75] and of the existence of A2A-D2 heterodimers, several different high order receptor mosaics can be formed from the trimeric organisation for A2A D2 CB1 (some of them are illustrated in the Fig. 9).
Fig. (9).
Some possible topological arrangements of A2A D2 CB1 as building blocks of high-order receptor mosaics. For further details, see text.
The demonstrated possibility of a trimeric RM indicates that high order RMs are possible [15, 76, 77]. Thus, the assemblage of this macro-molecular complex can also occur under the influence of allosteric cooperative interactions as already postulated in a previous paper [37].
ACKNOWLEDGEMENTS
The paper has been supported by a PRIN grant and by the IRCCS (Lido VE, Italy).
The comments to the text of Dr. Benfenati are gratefully acknowledged.
REFERENCES
- 1.Agnati LF, Fuxe K. Subcortical limbic 3H-N-propylnorapomorphine binding sites are markedly modulated by cholecystokinin-8 in vitro. Biosci Rep. 1983;3:1101–1105. doi: 10.1007/BF01120202. [DOI] [PubMed] [Google Scholar]
- 2.Fuxe K, Agnati LF. Receptor-receptor interactions in the central nervous system: a new integrative mechanism in synapses. Med. Res. Rev. 1985;5:441–482. doi: 10.1002/med.2610050404. [DOI] [PubMed] [Google Scholar]
- 3.Fuxe K, Agnati LF. Receptor-Receptor Interactions: a New Intramembrane Integrative Mechanism. MacMillan; London: 1987. pp. 14–18. [Google Scholar]
- 4.Agnati LF, Ferré S, Lluis C, Franco R, Fuxe K. Molecular mechanisms and therapeutical implications of intramembrane receptor / receptor interactions among heptahelical receptors with examples from the striopallidal GABA neurons. Pharmacol. Rev. 2003;55:509–550. doi: 10.1124/pr.55.3.2. [DOI] [PubMed] [Google Scholar]
- 5.Agnati LF, Fuxe K, Zoli M, Ozini I, Toffano G, Ferraguti F. A correlation analysis of the regional distribution of central enkephalin and beta-endorphin immunoreactive terminals and of opiate receptors in adult and old male rats: evidence for the existence of two main types of communication in the central nervous system: the volume transmission and the wiring transmission. Acta Physiol. Scand. 1986;128:201–207. doi: 10.1111/j.1748-1716.1986.tb07967.x. [DOI] [PubMed] [Google Scholar]
- 6.Fuxe K, Agnati LF, Martire M, Neumeyer A, Benfenati F, Frey P. Studies of neurotensin-dopamine receptor interactions in striatal membranes of the male rat. The influence of 6-hydroxydopamine-induced dopamine receptor supersensitivity. Acta Physiol. Scand. 1986;126:147–149. doi: 10.1111/j.1748-1716.1986.tb07798.x. [DOI] [PubMed] [Google Scholar]
- 7.Fuxe K, Agnati LF, Härfstrand A, Zoli M, von Euler G, Grimaldi R, Merlo Pich E, Bjelke B, Eneroth P, Benfenati F. On the role of neuropeptide Y in information handling in the central nervous system in normal and physiopathological states: focus on volume transmission and neuropeptide Y/alpha 2 receptor interactions. Ann. NY Acad. Sci. 1990;579:28–67. doi: 10.1111/j.1749-6632.1990.tb48351.x. [DOI] [PubMed] [Google Scholar]
- 8.Fuxe K, von Euler G, van der Ploeg I, Fredholm BB, Agnati LF. Pertussis toxin treatment counteracts the cardiovascular effects of neuropeptide Y and clonidine in the awake unrestrained rat. Neurosci. Lett. 1989;101:337–341. doi: 10.1016/0304-3940(89)90556-9. [DOI] [PubMed] [Google Scholar]
- 9.Yang SN, Fior DR, Hedlund PB, Agnati LF, Fuxe K. Antagonistic regulation of alpha 2-adrenoceptors by neuropeptide Y receptor subtypes in the nucleus tractus solitarii. Eur. J. Pharmacol. 1994;271:201–212. doi: 10.1016/0014-2999(94)90281-x. [DOI] [PubMed] [Google Scholar]
- 10.Boyd CAR, Noble D. The Logic of Life: The Challenge of Integrative Physiology. Oxford University Press; Oxford: 1993. [Google Scholar]
- 11.Agnati LF, Fuxe K, Zoli M, Rondanini C, Ögren SO. New vistas on synaptic plasticity: the receptor mosaic hypothesis of the engram. Med. Biol. 1982;60:183–190. [PubMed] [Google Scholar]
- 12.Agnati LF, Santarossa L, Benfenati F, Ferri M, Morpurgo A, Apolloni B, Fuxe K. In: From Synapses to Rules. Apolloni B, Kurfes F, editors. Kluwer Academic/Plenum Publishers; New York: 2002. pp. 165–196. [Google Scholar]
- 13.Agnati LF, Ferré S, Leo G, Lluis C, Canela EI, Franco R, Fuxe K. On the molecular basis of the receptor mosaic hypothesis of the engram. J. Mol. Cell. Neurobiol. 2004;24:501–516. doi: 10.1023/B:CEMN.0000023626.35717.5d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agnati LF, Santarossa L, Genedani S, Canela EI, Leo G, Franco R, Woods A, Lluis C, Ferrè S, Fuxe K. In: Computational Neuroscience: Cortical Dynamics, Lecture Notes in Computer Sciences. Erdi P, Esposito A, Marinaro M, Scarpetta S, editors. Springer; Berlin: 2004. pp. 24–54. [Google Scholar]
- 15.Agnati LF, Tarakanov AO, Ferré S, Fuxe K, Guidolin D. Receptor-receptor interaction, receptor mosaics and basic principles of molecular network organization. Possible implications for drug development. J. Mol. Neurosci. 2005;26:193–208. doi: 10.1385/JMN:26:2-3:193. [DOI] [PubMed] [Google Scholar]
- 16.Agnati LF, Leo G, Zanardi A, Genedani S, Rivera A, Fuxe K, Guidolin D. Volume transmission and wiring transmission from cellular to molecular networks: history and perspectives. Acta Physiol. 2006;187:329–344. doi: 10.1111/j.1748-1716.2006.01579.x. [DOI] [PubMed] [Google Scholar]
- 17.Agnati LF, Guidolin D, Leo G, Fuxe K. A boolean network modelling of receptor mosaics: relevance of topology and cooperativity. J. Neural. Transm. 2007;114:77–92. doi: 10.1007/s00702-006-0567-6. [DOI] [PubMed] [Google Scholar]
- 18.Agnati LF, Guidolin D, Fuxe K. The brain as a system of nested but partially overlapping networks: heuristic relevance of the model for brain physiology and pathology. J. Neural. Transm. 2007;114:3–19. doi: 10.1007/s00702-006-0563-x. [DOI] [PubMed] [Google Scholar]
- 19.Agnati LF, Guidolin D, Carone C, Dam M, Genedani S, Fuxe K. Understanding neural molecular networks builds on neural network architecture. Brain Res. Rev. 2008;58:379–399. doi: 10.1016/j.brainresrev.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 20.Agnati LF, Guidolin D, Genedani S, Ferré S, Bigiani A, Woods A, Fuxe K. How proteins come together in the plasma membrane and function in macromolecular assemblies: focus on receptor mosaics. J. Mol. Neurosci. 2005;26:133–154. doi: 10.1385/JMN:26:2-3:133. [DOI] [PubMed] [Google Scholar]
- 21.Watts DJ, Strogatz SH. Collective dynamics of small world networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 22.Kenakin T. Ligand-selective receptor conformations revisited: the promise and the problem. Trends Pharmacol. Sci. 2003;24:346–354. doi: 10.1016/S0165-6147(03)00167-6. [DOI] [PubMed] [Google Scholar]
- 23.Park PS, Filipek S, Wells JW, Palczewski K. Oligomerization of G protein-coupled receptors: past, present, and future. Biochemistry. 2004;21:15643–15656. doi: 10.1021/bi047907k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breitwieser GE. G protein-coupled receptor oligomerization: implications for G protein activation and cell signaling. Circ. Res. 2004;94:17–27. doi: 10.1161/01.RES.0000110420.68526.19. [DOI] [PubMed] [Google Scholar]
- 25.Gurevich VV, Gurevich EV. GPCR monomers and oligomers: it takes all kinds. Trends Neurosci. 2008;31:74–81. doi: 10.1016/j.tins.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ofran Y;, Rost B. Protein-protein interaction hotspots carved into sequences. PLoS Comput. Biol. 2007;3:e119. doi: 10.1371/journal.pcbi.0030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agnati LF, Ferré S, Cortelli P, Fuxe K. A brief appraisal on some aspects of the receptor-receptor interaction. Neurochem. Int. 1995;27:139–146. doi: 10.1016/0197-0186(95)00009-w. [DOI] [PubMed] [Google Scholar]
- 28.Agnati LF, Leo G, Genedani S, Andreoli N, Marcellino D, Woods A, Piron L, Guidolin D, Fuxe K. Structural plasticity in G-protein coupled receptors as demonstrated by the allosteric actions of homocysteine and computer-assisted analysis of disordered domains. Brain Res. Rev. 2008;58:459–474. doi: 10.1016/j.brainresrev.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 29.Thummer RP, Campbell MP, Dean MK, Frusher MJ, Scott PD, Reynolds CA. Entropy and oligomerization in GPCRs. J. Mol. Neurosci. 2005;26:113–122. doi: 10.1385/JMN:26:2-3:113. [DOI] [PubMed] [Google Scholar]
- 30.Milligan G. Opioid receptors and their interacting proteins. Neuromol. Med. 2005;7:51–59. doi: 10.1385/NMM:7:1-2:051. [DOI] [PubMed] [Google Scholar]
- 31.Ciruela F, Burgueño J, Casadó V, Canals M, Marcellino D, Goldberg SR, Bader M, Fuxe K, Agnati LF, Lluis C, Franco R, Ferré S, Woods AS. Combining mass spectrometry and pull-down techniques for the study of receptor heteromerization: direct epitope-epitope electrostatic interactions between adenosine A2A and dopamine D2 receptors. Anal. Chem. 2004;76:5354–5363. doi: 10.1021/ac049295f. [DOI] [PubMed] [Google Scholar]
- 32.Agnati LF, Ferre S, Genedani S, Leo G, Guidolin D, Filaferro M, Carriba P, Casado V, Lluis C, Franco R, Woods AS, Fuxe K. Allosteric modultion of dopamine D2 receptors by homocysteine. J. Proteome Res. 2006;5:3077–3083. doi: 10.1021/pr0601382. [DOI] [PubMed] [Google Scholar]
- 33.Karnik SS, Gogonea C, Patil S, Saad Y, Takezako T. Activation of G-protein-coupled receptors: a common molecular mechanism. Trends Endocrinol. Metab. 2003;14:431–437. doi: 10.1016/j.tem.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 34.Stanfield P. Voltage sparks a GPCR. Nat. Cell Biol. 2006;8:1323–1325. doi: 10.1038/ncb1206-1323. [DOI] [PubMed] [Google Scholar]
- 35.Gouldson PR, Higgs C, Smith RE, Dean MK, Gkoutos GV, Reynolds CA. Dimerization and domain swapping in G-protein-coupled receptors: a computational study. Neuropsychopharmacology. 2000;23:S60–S77. doi: 10.1016/S0893-133X(00)00153-6. [DOI] [PubMed] [Google Scholar]
- 36.Thévenin D, Roberts MF, Lazarova T, Robinson CR. Identifying interactions between transmembrane helices from the adeno-sine A2A receptor. Biochemistry. 2005;44:16239–16245. doi: 10.1021/bi051422u. [DOI] [PubMed] [Google Scholar]
- 37.Agnati LF, Fuxe K, Ferre S. How receptor mosaics decode transmitter signals. Possible relevance of cooperativity. Trends Biochem. Sci. 2005;30:188–193. doi: 10.1016/j.tibs.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 38.Jaffe EK. Morpheeins--a new structural paradigm for allosteric regulation. Trends Biochem. Sci. 2005;30:490–497. doi: 10.1016/j.tibs.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 39.Fuxe K, Agnati LF, Franco R, Roberts DC, Tanganelli S, Bader M, Huebner N, Schiffmann S, Scheel-Kruger J, Stromberg I, Marsden C, Filip M, Jaber M, Bergquist J. EU DDTOMICS. proposal N 004863, LifeSciHealth-I, 2003, OJ 2003/C164,FP6-2003.
- 40.Fuxe K, Ferre S, Woods A, Rivera A, Hoistad M, Franco R, Kehr J, Agnati LF. Monitoring Molecules in Neuroscience. Karolinska University Press; Stockholm: 2003. pp. 199–202. [Google Scholar]
- 41.Kearn CS, Blake-Palmer K, Daniel E, Mackie K, Glass M. Concurrent stimulation of cannabinoid CB1 and dopamine D2 receptors enhances heterodimer formation: a mechanism for receptor cross-talk? Mol. Pharmacol. 2005;67:1697–1704. doi: 10.1124/mol.104.006882. [DOI] [PubMed] [Google Scholar]
- 42.Marcellino D, Carriba P, Filip M, Borgkvist A, Frankowska M, Bellido I, Tanganelli S, Muller CE, Roberts DC, Fisone G, Lluis C, Agnati L, Franco R, Fuxe K. Antagonistic cannabinoid CB1/dopamine D2 receptor interactions in striatal CB1/D2 heteromers: a combined neurochemical and behavioral analysis. Neuropharmacology. 2008;54:815–823. doi: 10.1016/j.neuropharm.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 43.Carriba P, Ortiz O, Patkar K, Justinova Z, Stroik J, Themann A, Muller C, Woods AS, Hope BT, Ciruela F, Casado V, Canela EI, Lluis C, Goldberg SR, Moratalla R, Franco R, Ferre S. Striatal adenosine A2A and cannabinoid CB1 receptors form functional heteromeric complexes that mediate the motor effects of cannabinoids. Neuropsychopharmacology. 2007;32:2249–2259. doi: 10.1038/sj.npp.1301375. [DOI] [PubMed] [Google Scholar]
- 44.Andersson M, Usiello A, Borgkvist A, Pozzi L, Dominguez C, Fienberg AA, Svenningsson P, Fredholm BB, Borrelli E, Greengard P, Fisone G. Cannabinoid action depends on phosphorylation of dopamine- and cAMP-regulated phosphoprotein of 32 kDa at the protein kinase A site in striatal projection neurons. J. Neurosci. 2005;25:8432–8438. doi: 10.1523/JNEUROSCI.1289-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borgkvist A, Marcellino D, Fuxe K, Greengard P, Fisone G. Regulation of DARPP-32 phosphorylation by Delta9-tetrahydrocannabinol. Neuropharmacology. 2008;54:31–35. doi: 10.1016/j.neuropharm.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 46.Fuxe K, Marcellino D, Rivera A, Diaz-Cabiale Z, Filip M, Gago B, Roberts DC, Langel U, Genedani S, Ferraro L, de la Calle A, Narvaez J, Tanganelli S, Woods A, Agnati LF. Receptor-receptor interactions within receptor mosaics. Impact on neuropsychopharmacology. Brain Res. Rev. 2008;58:415–452. doi: 10.1016/j.brainresrev.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 47.Carriba P, Navarro G, Ciruela F, Ferré S, Casadó V, Agnati L, Cortés A, Mallol J, Fuxe K, Canela EI, Lluís C, Franco R. Detection of heteromerization of more than two proteins by sequential BRET-FRET. Nat. Methods. 2008;5:727–733. doi: 10.1038/nmeth.1229. [DOI] [PubMed] [Google Scholar]
- 48.Kitano H. Computational systems biology. Nature. 2002;420:206–210. doi: 10.1038/nature01254. [DOI] [PubMed] [Google Scholar]
- 49.Dunker AK, Brown CJ, Obradovic Z. Identification and functions of usefully disordered proteins. Adv. Protein Chem. 2002;62:25–49. doi: 10.1016/s0065-3233(02)62004-2. [DOI] [PubMed] [Google Scholar]
- 50.Romero P, Obradovic Z, Dunker AK. Sequence data analysis for long disordered regions prediction in the calcineurin family. Genome Inform. 1997;8:110–124. [PubMed] [Google Scholar]
- 51.Radivojac P, Obradovic Z, Brown CJ, Bunker AK. Prediction of boundaries between intrinsically ordered and disordered protein regions. Pac. Symp. Biocomput. 2003;8:216–227. [PubMed] [Google Scholar]
- 52.Linding R, Jensen F, Diella P, Bork T, Gibson J, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 53.Vullo A, Bortolami O, Pollastri G, Tosatto SCE. Spritz: a server for the prediction of intrinsically disordered regions in protein sequences using kernel machines. Nucleic Acids Res. 2006;34:W164–W168. doi: 10.1093/nar/gkl166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shimizu K, Muraoka Y, Hirose S, Tomii K, Noguchi T. Predicting mostly disordered proteins by using structure-unknown protein data. BMC Bioinformatics. 2007;8:78. doi: 10.1186/1471-2105-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pawar AP, Dubay KF, Zurdo J, Chiti F, Vendruscolo M, Dobson CM. Prediction of “aggregation-prone” and “aggregation-susceptible” regions in proteins associated with neurodegenerative diseases. J. Mol. Biol. 2005;350:379–392. doi: 10.1016/j.jmb.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 56.Sánchez de Groot N, Pallarés I, Avilés FX, Vendrell J, Ventura S. Prediction of “hot spots” of aggregation in disease-linked polypeptides. BMC Struct. Biol. 2005;5:18. doi: 10.1186/1472-6807-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Conchillo-Solé O, de Groot NS, Avilés FX, Vendrell J, Daura X, Ventura S. AGGRESCAN: a server for the prediction and evaluation of “hot spots” of aggregation in polypeptides. BMC Bioinformatics. 2007;8:65. doi: 10.1186/1471-2105-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dunker AK, Cortese MS, Romero P, Iakoucheva LM. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 59.González-Maeso J, Meana JJ. Heterotrimeric g proteins: insights into the neurobiology of mood disorders. Curr. Neuropharmacol. 2006;4:127–138. doi: 10.2174/157015906776359586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo T, Hobbs DW. Privileged structure-based combinatorial libraries targeting G protein-coupled receptors. Assay Drug Dev. Technol. 2003;1:579–592. doi: 10.1089/154065803322302835. [DOI] [PubMed] [Google Scholar]
- 61.Milligan G, Wilson S, López-Gimenez JF. The specificity and molecular basis of alpha1-adrenoceptor and CXCR chemokine receptor dimerization. J. Mol. Neurosci. 2005;26:161–168. doi: 10.1385/JMN:26:2-3:161. [DOI] [PubMed] [Google Scholar]
- 62.Fuxe K, Marcellino D, Woods AS, Leo G, Antonelli T, Ferraro L, Tanganelli S, Agnati LF. Integrated signaling in heterodimers and receptor mosaics of different types of GPCRs of the forebrain: relevance for schizophrenia. J. Neural Transm. 2009;116:923–939. doi: 10.1007/s00702-008-0174-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fuxe K, Celani MF, Martire M, Zini I, Zoli M, Agnati LF. l-Glutamate reduces the affinity of [3H]N-propylnorapomorphine binding sites in striatal membranes. Eur. J. Pharmacol. 1984;100:127–130. doi: 10.1016/0014-2999(84)90326-1. [DOI] [PubMed] [Google Scholar]
- 64.Ferré S, Popoli P, Rimondini R, Reggio R, Kehr J, Fuxe K. Adenosine A2A and group I metabotropic glutamate receptors synergistically modulate the binding characteristics of dopamine D2 receptors in the rat striatum. Neuropharmacology. 1999;38:129–140. doi: 10.1016/s0028-3908(98)00154-3. [DOI] [PubMed] [Google Scholar]
- 65.Popoli P, Pezzola A, Torvinen M, Reggio R, Pintor A, Scarchilli L, Fuxe K, Ferre S. The selective mGlu(5) receptor agonist CHPG inhibits quinpirole-induced turning in 6-hydroxydopamine-lesioned rats and modulates the binding characteristics of dopamine D(2) receptors in the rat striatum: interactions with adenosine A(2a) receptors. Neuropsychopharmacology. 2001;25:505–513. doi: 10.1016/S0893-133X(01)00256-1. [DOI] [PubMed] [Google Scholar]
- 66.Ferre S, Karcz-Kubicha M, Hope BT, Popoli P, Burgueno J, Gutierrez MA, Casado V, Fuxe K, Goldberg SR, Lluis C, Franco R, Ciruela F. Synergistic interaction between adenosine A2A and glutamate mGlu5 receptors: implications for striatal neuronal function. Proc. Natl. Acad. Sci. USA. 2002;99:11940–11945. doi: 10.1073/pnas.172393799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fuxe K, Agnati LF, Jacobsen K, Hillion J, Canals M, Torvinen M, Tinner-Staines B, Staines W, Rosin D, Terasmaa A, Popoli P, Leo G, Vergoni V, Lluis C, Ciruela F, Franco R, Ferre S. Receptor heteromerization in adenosine A2A receptor signaling: relevance for striatal function and Parkinson's disease. Neurology. 2003;61:S19–S23. doi: 10.1212/01.wnl.0000095206.44418.5c. [DOI] [PubMed] [Google Scholar]
- 68.Rodrigues RJ, Alfaro TM, Rebola N, Oliveira CR, Cunha RA. Co-localization and functional interaction between adenosine A(2A) and metabotropic group 5 receptors in glutamatergic nerve terminals of the rat striatum. J. Neurochem. 2005;92:433–441. doi: 10.1111/j.1471-4159.2004.02887.x. [DOI] [PubMed] [Google Scholar]
- 69.Nishi A, Liu F, Matsuyama S, Hamada M, Higashi H, Nairn AC, Greengard P. Metabotropic mGlu5 receptors regulate adeno-sine A2A receptor signaling. Proc. Natl. Acad. Sci. USA. 2003;100:1322–1327. doi: 10.1073/pnas.0237126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Diaz-Cabiale Z, Vivo M, Del Arco A, O'Connor WT, Harte MK, Muller CE, Martinez E, Popoli P, Fuxe K, Ferre S. Metabotropic glutamate mGlu5 receptor-mediated modulation of the ventral striopallidal GABA pathway in rats. Interactions with adenosine A(2A) and dopamine D(2) receptors. Neurosci. Lett. 2002;324:154–158. doi: 10.1016/s0304-3940(02)00179-9. [DOI] [PubMed] [Google Scholar]
- 71.Coccurello R, Breysse N, Amalric M. Simultaneous blockade of adenosine A2A and metabotropic glutamate mGlu5 receptors increase their efficacy in reversing Parkinsonian deficits in rats. Neuropsychopharmacology. 2004;29:1451–1461. doi: 10.1038/sj.npp.1300444. [DOI] [PubMed] [Google Scholar]
- 72.Kachroo A, Orlando LR, Grandy DK, Chen JF, Young AB, Schwarzschild MA. Interactions between metabotropic glutamate 5 and adenosine A2A receptors in normal and parkinsonian mice. J. Neurosci. 2005;25:10414–10419. doi: 10.1523/JNEUROSCI.3660-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schwarzschild MA, Agnati L, Fuxe K, Chen JF, Morelli M. Targeting adenosine A2A receptors in Parkinson's disease. Trends Neurosci. 2006;29:647–654. doi: 10.1016/j.tins.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 74.Ferre S, Agnati LF, Ciruela F, Lluis C, Woods AS, Fuxe K, Franco R. Neurotransmitter receptor heteromers and their integrative role in ‘local modules’: the striatal spine module. Brain Res. Rev. 2007;55:55–67. doi: 10.1016/j.brainresrev.2007.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nimchinsky EA, Hof PR, Janssen WG, Morrison JH, Schmauss C. Expression of dopamine D3 receptor dimers and tetramers in brain and in transfected cells. J. Biol. Chem. 1997;272:29229–29237. doi: 10.1074/jbc.272.46.29229. [DOI] [PubMed] [Google Scholar]
- 76.Agnati LF, Guidolin D, Leo G, Carone C, Genedani S, Fuxe K. Receptor-receptor interactions: history, concepts and implications of a membrane molecular integrative mechanism. Prog. Neurobiol. 2009 In press. [Google Scholar]
- 77.Agnati LF, Guidolin D, Leo G, Genedani S, Arhem P, Forni A, Andreoli N, Fuxe K. Role of cooperativity in protein folding and protein mosaic assemblage relevance for protein conformational diseases. Curr. Protein Pept. Sci. 2007;8:460–470. doi: 10.2174/138920307782411419. [DOI] [PubMed] [Google Scholar]