Summary
Glycosylation is the most common post-translational modification of proteins. The protein sequence data suggested that more than half of all proteins produced in mammalian cells are glycoproteins. Glycans of secreted glycoproteins affect many protein properties such as solubility, stability, protease sensitivity, and polarity, while glycans on cell surface glycoproteins are involved in various cellular functions including cell-cell and cell-matrix interactions during embryogenesis, immune reactions, and tumor development. The past decade of research on glycan function has revealed the etiology of a growing number of human genetic diseases with aberrant glycan formation. This review focuses upon the involvement of O-mannosylglycan in the molecular and cellular mechanisms of muscular dystrophies. Advances in glycobiology are expected to result in a better understanding and in improved treatments of a class of muscular dystrophies called α-dystroglycanopathies.
Keywords: dystroglycan, O-mannosyl glycan, glycosyltransferase, muscular dystrophy
Introduction
Proteins produced by eukaryotic cells are frequently post-translationally modified by the addition of glycans. On the basis of Swiss-Prot data, more than half of proteins are known to undergo glycosylation (1). The glycan moieties of these glycoproteins not only affect their stability and conformation, but also have roles in molecular recognition processes that occur in bacterial and viral infection, cell adhesion in inflammation and metastasis, differentiation, development, and many other events characterized by intercellular communication. The mechanisms underlying the many glycan-mediated recognition processes are not fully understood.
The major glycans of glycoproteins can be classified into two groups according to their glycan-peptide linkage regions. Those that are linked to asparagine (Asn) residues of polypeptides are termed N-glycans, while those that are linked to serine (Ser) or threonine (Thr) residues are called O-glycans. In N-glycans, the reducing terminal N-acetylglucosamine (GlcNAc) is linked to the amide group of Asn, via an aspartylglycosylamine linkage. In O-glycans, the reducing terminal N-acetylgalactosamine (GalNAc) is attached to the hydroxyl groups of Ser and Thr residues of polypeptides. However, in addition to the abundant O-GalNAc forms, several unique types of protein O-glycosylation have been reported, such as O-fucose, O-glucose, O-GlcNAc, O-xylose, O-galactose on hydroxylysine, and O-mannose which will be reviewed here. Recently O-mannosylation of the mammalian glycoprotein dystroglycan has been shown to be important in muscle and brain development.
Dystroglycan
α-Dystroglycan is an extracellular peripheral membrane glycoprotein anchored by binding to a transmembrane glycoprotein, β-dystroglycan. These two dystroglycan subunits were originally identified as members of the sarcolemmal dystrophin-glycoprotein complex. Dystroglycan is thought to act as a transmembrane linker between the extracellular matrix and intracellular cytoskeleton (2). α-Dystroglycan strongly binds to extracellular matrix proteins containing laminin G (LamG) domains, such as laminin, neurexin, and agrin in a calcium-dependent manner. α-Dystroglycan is heavily glycosylated, and its glycans have a role in the binding to these molecules. Previously we reported that the glycans of α-dystroglycan include O-mannosylglycan: Siaα2–3Galβ1–4GlcNAcβ1–2Man (3). α-Dystroglycan has a mucin-type O-glycosylation site in the central region of the molecule. Thr, Ser and proline (Pro) are densely distributed into the predicted mucin domain (amino acid residues 316-489), which is thought to have the form of a rigid rod, since complex secondary and tertiary structures are hindered by heavy glycosylation.
Dystroglycan is encoded by a single gene (DAG1) (2). The function of dystroglycan in the body has been examined by targeting the DAG1 gene in mice. However, disruption of this gene in mice results in embryonic lethality. To allow the embryo to develop, chimeric mice generated from targeted embryonic stem cells have been produced. Dystroglycan-null chimeric mice showed muscular dystrophy, although muscle basement membrane formation was normal (4). The function of dystroglycan in specific tissues was examined with the Cre/LoxP system. Targeting the dystroglycan gene specifically in differentiated skeletal muscle did not affect muscle basement membrane formation but resulted in a mild dystrophic phenotype (5). Targeting the dystroglycan gene in brain resulted in abnormal cerebral cortical layering resembling human cobblestone lissencephaly, and in abnormal cerebellar granule cell migration (6). Targeting the dystroglycan gene in peripheral nerves caused defects in both myelination and nodal architecture (7). Dystroglycan is also required for polarizing epithelial cells and oocytes in Drosophila (8) and removal of dystroglycan causes severe muscular dystrophy in zebrafish embryos (9). These results indicate that dystroglycan is essential for normal development.
Not only dystroglycan itself but also the attached glycans are important. O-Mannosylation of proteins has been clearly shown to be vital in unicellular eukaryotic organisms (10), and its absence severely decreases cell wall rigidity and normal cellular morphogenesis. Deficiency in protein O-mannosylation in the fungal pathogen Candida albicans was shown to cause defects in multiple cellular functions including expression of virulence. Protein O-mannosylation has also been suggested to be involved in the ER quality control system. In yeast, protein O-mannosylation is necessary for intracellular protein trafficking. For example, it was found that non-native proteins are O-mannosylated in the endoplasmic reticulum (ER) which causes them to be excreted from the cell without aggregating and without the accumulation of aberrant proteins in the ER (11, 12). These results suggest that yeast O-mannosyltransferases can recognize proteins that have undergone a conformational change. As reviewed here, O-mannosylation of α-dystroglycan is important in muscle and brain development.
Initiation of protein O-mannosylation
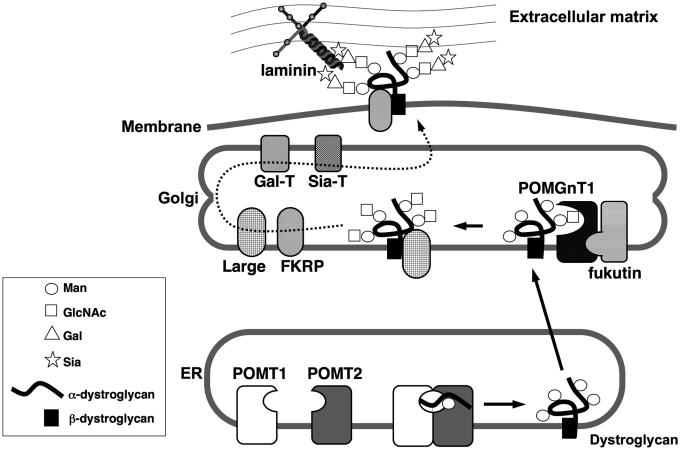
Protein O-mannosyltransferase (PMT) is evolutionarily conserved from prokaryotes, such as Mycobacterium tuberculosis, to eukaryotes, such as yeast, Drosophila, mouse, and human (3). In yeast, Saccharomyces cerevisiae, O-mannosylation is required for the stability, correct localization, and/or function of proteins. Yeast O-mannosylation is initiated in the lumen of the ER by a family of PMTs that catalyze the transfer of a mannosyl residue from dolichyl phosphate mannose (Dol-P-Man) to Ser/Thr residues of proteins (10). S. cerevisiae has seven PMT homologues (Pmt1p-7p) that share almost identical hydropathy profiles. The hydropathy profiles predict that PMTs are integral membrane proteins with multiple trans-membrane domains (10). The PMT family is classified phylogenetically into the PMT1, PMT2, and PMT4 subfamilies. Members of the PMT1 subfamily (Pmt1p and Pmt5p) interact heterophilically with those of the PMT2 subfamily (Pmt2p and Pmt3p), whereas the single member of the PMT4 subfamily (Pmt4p) acts as a homophilic complex (13). Although Pmt1p-4p and Pmt6p have O-mannosyltransferase activity by themselves, complex formation is essential for maximal transferase activity of yeast PMT family members (13). Human have two homologues of yeast PMT (POMT1 and POMT2). We recently demonstrated that formation of a POMT1/POMT2 complex was required for O-mannosyltransferase activity (Fig. 1) (14–16).
Figure 1.
Proposed O-mannosyl glycan processing of dystroglycan. POMT1 and POMT2 form a hetero-complex to express protein O-mannosyltransferase activity in the ER. After O-mannosylated dystroglycan is transported from the ER to the Golgi apparatus, and then POMGnT1 forms a GlcNAcβ1–2Man linkage of O-mannosylglycan. Then galactosyltransferase (Gal-T) and sialyltransferase (Sia-T) work sequentially on O-mannosylglycan. Dystroglycan having mature O-mannosylglycan is located on the cell surface and is known to bind to extracellular matrix proteins such as laminin. Recent studies suggest that fukutin is associated with POMGnT1, and that Large and FKRP are as-sociated with the dystroglycan complex, although the precise associations are unclear.
Individual PMTs have different specificities for protein substrates (10), suggesting the presence of some sequence for recognition by PMTs, but the sequence has not been identified. On the other hand, in mammals, O-mannosylated proteins are rare. In mammals, O-mannosylation may require a specific sequence because we detected O-mannosyltransferase activity when a GST-fused mucin-like domain of α-dystroglycan (amino acid residues 316-489) was used as an acceptor (14). To address the regulation of O-mannosylation, we tried to determine whether substrates require a specific amino acid sequence to be recognized by O-mannosyltransferases. To answer this question, we synthesized a series of peptides that fully covered the mucin-like domain of α-dystroglycan and we examined whether these peptides worked as acceptors for protein O-mannosylation. The results showed that two similar peptide sequences, corresponding to residues 401-420 (IRPTMTIPGYVEPTAVATP) and 336-355 (SRIVPTPTSPAIAPPTETM), respectively, were strongly mannosylated, while other peptides from α-dystroglycan and peptides of various mucin tandem-repeat regions were poorly mannosylated (17). Peptides 401-420 and 336-355 contained four and six Ser/Thr residues, respectively. Substitution of Ala residues for the Ser or Thr residues showed that Thr414 of peptide 401-420 and Thr351 of peptide 336-355 were prominently modified by O-mannosylation. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) and Edman degradation analysis of the mannosylated peptide 401-420 indicated that Thr414 was the Thr residue that was most prominently modified by O-mannosylation and that O-mannosylation occurred sequentially rather than at random. Based on these results, we proposed a preferred amino acid sequence (IXPT(P/X)TXPXXXXPTX(T/X)XX) for mammalian O-mannosylation. A BLAST search for proteins with this sequence turned up only α-dystroglycan, suggesting that the primal O-mannosylated protein is α-dystroglycan (17).
In contrast, O-mannosylation may occur by a different mechanism in S. cerevisiae. Recently, Hutzler et al. reported (18) that Pmt4p mediates O-mannosylation of Ser/Thr-rich membrane-bound proteins, whereas Pmt1/Pmt2 complexes act on both, soluble and membrane-bound proteins. O-Mannosyltransferase activity does not depend on the membrane-anchoring sequence, as long as it is flanked by a Ser/Thr-rich domain facing the ER lumen. In contrast to human O-mannosylation signals, Pmt4 O-mannosylation signals are not just linear sequences of proteins. Thus, it is possible that mammalian O-mannosylation requires a specific amino acid sequence while yeast O-mannosylation does not. If true, this should not be surprising in view of the fact that many proteins are modified by O-mannosylation in yeast while only a few are O-mannosylated in mammals. Further studies are necessary to elucidate the regulatory mechanism of protein O-mannosylation.
Three other types of protein O-glycosylation are initiated in different ways. O-GalNAc glycosylation is the most common protein-modification and is initiated by the action of a family of pp-GalNAc-Ts. So far, no consensus sequence has emerged that is both necessary and sufficient for O-GalNAc glycosylation to occur. Many nuclear and cytosolic proteins are O-GlcNAc glycosylated, but a consensus sequence for O-GlcNAc transferase has not been reported. On the other hand, O-Fuc glycosylation exists in direct O-linkage to Ser or Thr residues in two different types of Cys-knot motifs: epidermal growth factor-like (EGF) repeats and thrombospondin type 1 repeats (TSR). The enzyme responsible for adding O-Fuc to EGF repeats was identified as protein O-fucosyltransferase 1 (POFUT1) and the enzyme for adding to TSR was identified as POFUT2 (19, 20). A consensus sequence for O-Fuc glycosylation in EGF is proposed CysX4–5(Ser/Thr)Cys between the second and third Cys residues and a consensus sequence for O-Fuc in TSR is TrpX5CysX2/3Ser/ThrCysX2G between the first and second Cys residues, respectively. Both POFUT1 and POFUT2 require a specific sequence for O-Fuc glycosylation.
Glycosylation is basically controlled by the combined action of each glycosyltransferase. However, recent studies indicate that glycosylation is regulated in a complicated manner. Glycosyltransferase activities are regulated by other factors or by complex formation. For example, human core 1 β3-galactosyltransferase activity requires the expression of Cosmc (21). Cosmc is a molecular chaperone that specifically assists the folding/stability of core 1 β3-galactosyltransferase and is required for a glycosyltransferase expression. Mutations of COSMC were recently found in patients with Tn syndrome who could not produce core 1 structure (Galβ1-3GalNAc) (22). Another glycosyltransferase with complex regulation is human chondroitin synthase, which cannot polymerize chondroitin sulfate in vitro; rather its activity requires the coexpression of chondroitin polymerizing factor (23). As a third example, the bifunctional glycosyltransferases EXT1 and EXT2, which polymerize heparan sulfate, need to form a hetero-oligomeric complex to exert their optimal catalytic activities and to exist in the appropriate intracellular locations (24). We observed that protein O-mannosylation can be initiated by direct complex-formation of POMT1 and POMT2, but not by either enzyme by itself. POMT1 or POMT2 are thus different from EXT1 and EXT2 because the latter enzymes are active by themselves. One possibility is that formation of the POMT1-POMT2 complex creates a new catalytic domain (15). Further studies are needed to elucidate the mechanism of complex formation between POMT1 and POMT2, and the regulation of O-mannosyltransferase activity.
Glycosyltransferase activity in lymphocytes of patients with muscular dystrophies
Muscular dystrophies are genetic diseases that cause progressive muscle weakness and wasting. Recent data suggest that aberrant glycosylation of α-dystroglycan is the primary cause of some forms of congenital muscular dystrophy and neuronal migration disorder called α-dystroglycanopathies (25). At least six genes are known to cause α-dystroglycanopathies. Previously we reported that defects in O-mannosyl glycan cause a type of muscular dystrophy (25).
Protein O-linked mannose β1,2-N-acetylglucosaminyltransferase 1 (POMGnT1) forms a GlcNAcβ1–2Man linkage of O-mannosyl glycans on α-dystroglycan (Fig. 1) (26). We have demonstrated that POMGnT1 is responsible for muscle-eye-brain disease (MEB: OMIM 253280), whose symptoms are severe cerebral and ocular anomalies. We previously demonstrated that all known mutations in the POMGnT1 gene in MEB patients caused loss of enzymatic activity (25). These findings indicate that MEB is inherited as a loss-of-function of the POMGnT1 gene.
POMT1 and POMT2 are responsible for the catalysis of the first step in O-mannosyl glycan synthesis as described above (14). Mutations in the POMT1 and POMT2 genes are the cause of Walker-Warburg syndrome (WWS: OMIM 236670), an autosomal recessive developmental disorder associated with congenital muscular dystrophy, neuronal migration defects and ocular abnormalities (27, 28). We have demonstrated that the mutations of POMT1 protein found in WWS patients do not prevent complex formation with POMT2 but they do abolish O-mannosyltransferase activity of the complex (15). Thus, O-mannosylation is indispensable for normal structure and function of α-dystroglycan in muscle and brain.
In addition to MEB and WWS, four other muscular dystrophies have been reported to be associated with an abnormal glycosylation of α-dystroglycan: Fukuyama-type congenital muscular dystrophy (FCMD: OMIM 253800), CMD type 1C (MDC1C: OMIM 606612), limbgirdle muscular dystrophy type 2I (LGMD2I: OMIM 607155), and CMD type 1D (MDC1D: OMIM 608840). FCMD, which is due to mutations in fukutin, is an autosomal recessive disorder that is characterized by congenital muscular dystrophy, lissencephaly, and eye anomalies. It is relatively common in the Japanese population. A sequence analysis of fukutin predicts it to be an enzyme that glycosylates proteins or lipids. MDC1C is caused by a defect of fukutin-related protein (FKRP), a homologue of fukutin and is characterized by severe muscle weakness and degeneration, and cardiomyopathy. Mental retardation and cerebellar cysts have been observed in some cases. Allelic mutations in the FKRP gene also cause a milder and more common form of muscular dystrophy called LGMD2I, which is frequently associated with cardiomyopathy and shows variable onsets ranging from adolescence to adult-hood. Patients with mutations in the FKRP gene invariably exhibit a reduced expression of glycosylated α-dystroglycan, which is strongly correlated with disease severity. Although the function of FKRP is unknown, FKRP has been suggested to be a Golgiresident protein and to be involved in the glycosylation of α-dystroglycan as a glycosyltransferase or a kind of modulator. A recent study described a patient with congenital muscular dystrophy, profound mental retardation, white matter changes, and subtle structural abnormalities in the brain and a reduction of immunologically detectable α-dystroglycan. The patient was found to have mutations in the LARGE gene. This type of muscular dystrophy was named MDC1D.
Since multiple genes are known to cause α-dystroglycanopathies, with an extremely broad clinical spectrum and relatively poor phenotype-genotype correlation, at present molecular diagnosis of α-dystroglycanopathy patients is difficult and requires searching for mutations gene by gene. These methods are expensive and time-consuming. At present, of the six known α-dystroglycanopathy genes, the function of the protein product is clear only for POMT1, POMT2 and POMGnT1 (25). Vajsar et al. and we have developed assay methods for lymphoblast POMGnT1 (29, 30) and POMT activity (29, 30), respectively, for patients with confirmed α-dystroglycanopathy. To screen patients with suspected forms of α-dystroglycanopathy, we measured the activities of both POMT and POMGnT1 in lymphoblasts from a series of patients (29, 30). We observed reductions in POMGnT1 or POMT activity in several uncharacterized patients, in whom secondary targeted sequencing led to the identification of mutations in POMT1, POMT2 or POMGnT1. This lymphoblast-based assay was proposed as a rapid and relatively simple diagnostic test for MEB and WWS patients, and may bypass the need for invasive muscle biopsies when clinical findings are highly suggestive of an α-dystroglycanopathy.
It is noteworthy that patients with FKRP mutations did not show reduced activity for POMT and POMGnT1, suggesting that FKRP is not associated with POMT1 or POMT2, or with POMGnT1 (30). Recently, FKRP was reported to be associated with the sarcolemmal dystrophin-glycoprotein complex and may influence the glycosylation of α-dystroglycan, although the precise function of FKRP remains unknown (31). On the other hand, fukutin was reported to be associated with POMGnT1 in the Golgi compartment (Fig. 1) (32). Although fukutin has no proven glycosyltransferase activity, transgenic knock-in mouse carrying a retrotransposon insertion in the fukutin gene showed a 30% reduction of POMGnT1 activity (32), suggesting that fukutin modulates POMGnT1 activity in muscle. It will be interesting to test POMGnT1 activity in muscle, lymphoblasts, and fibroblasts from FCMD patients to determine whether mutations in the fukutin gene could modulate POMGnT1 activity.
Perspectives
In summary, O-mannosylation is important in muscle and brain development. Although highly glycosylated α-dystroglycan has been found to be selectively deficient in the skeletal muscle of α-dystroglycanopathies and the products of the mutated genes are putative glycosyltransferases, the functions of these gene products are still unclear except for POMT1/2 and POMGnT1 (25). Identification of the functions of the other mutated genes associated with α-dystroglycanopathies will make it possible to diagnose patients with an α-dystroglycanopathy with an assay for glycosyltransferase activity. Future studies may reveal how α-dystroglycan glycosylation contributes to muscular dystrophy and neuronal migration disorder and how normal glycosylation restores functions of dystroglycan. Such studies may lead to therapies for incomplete glycosylation-induced dystroglycanopathies.
Acknowledgements
The author gratefully acknowledges the financial support of Research Grants for Nervous and Mental Disorders (17A-10) and Core-to-Core Program from JSPS throughout this project.
References
- 1.Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta 1999;1473:4-8. [DOI] [PubMed] [Google Scholar]
- 2.Barresi R, Campbell KP. Dystroglycan: from biosynthesis to patho-genesis of human disease. J Cell Sci 2006;119:199-207. [DOI] [PubMed] [Google Scholar]
- 3.Endo T. Structure, function and pathology of O-mannosyl glycans. Glycoconj J 2004;21:3-7. [DOI] [PubMed] [Google Scholar]
- 4.Cote PD, Moukhles H, Lindenbaum M, et al. Chimaeric mice defi-cient in dystroglycans develop muscular dystrophy and have disrupted myoneural synapses. Nat Genet 1999;23:338-42. [DOI] [PubMed] [Google Scholar]
- 5.Cohn RD, Henry MD, Michele DE, et al. Disruption of DAG1 in differ-entiated skeletal muscle reveals a role for dystroglycan in muscle regeneration. Cell 2002;110:639-48. [DOI] [PubMed] [Google Scholar]
- 6.Moore SA, Saito F, Chen J, et al. Deletion of brain dystroglycan re-capitulates aspects of congenital muscular dystrophy. Nature 2002;418:422-5. [DOI] [PubMed] [Google Scholar]
- 7.Saito F, Moore SA, Barresi R, et al. Unique role of dystroglycan in peripheral nerve myelination, nodal structure, and sodium channel stabilization. Neuron 2003;38:747-58. [DOI] [PubMed] [Google Scholar]
- 8.Deng WM, Schneider M, Frock R, et al. Dystroglycan is required for polarizing the epithelial cells and the oocyte in Drosophila. Development 2003;130:173-84. [DOI] [PubMed] [Google Scholar]
- 9.Parsons MJ, Campos I, Hirst EM, et al. Removal of dystroglycan causes severe muscular dystrophy in zebrafish embryos. Development 2002;129:3505-12. [DOI] [PubMed] [Google Scholar]
- 10.Lehle L, Strahl S, Tanner W. Protein glycosylation, conserved from yeast to man: a model organism helps elucidate congenital human diseases. Angew Chem Int Ed Engl 2006;45:6802-18. [DOI] [PubMed] [Google Scholar]
- 11.Harty C, Strahl S, Romisch K. O-mannosylation protects mutant alpha-factor precursor from endoplasmic reticulum-associated degradation. Mol Biol Cell 2001;12:1093-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakatsukasa K, Okada S, Umebayashi K, et al. Roles of O-mannosylation of aberrant proteins in reduction of the load for endoplasmic reticulum chaperones in yeast. J Biol Chem 2004;279:49762-72. [DOI] [PubMed] [Google Scholar]
- 13.Girrbach V, Strahl S. Members of the Evolutionarily Conserved PMT Family of Protein O-Mannosyltransferases Form Distinct Protein Complexes among Themselves. J Biol Chem 2003;278:12554-62. [DOI] [PubMed] [Google Scholar]
- 14.Manya H, Chiba A, Yoshida A, et al. Demonstration of mammalian protein O-mannosyltransferase activity: coexpression of POMT1 and POMT2 required for enzymatic activity. Proc Natl Acad Sci USA 2004;101:500-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akasaka-Manya K, Manya H, Nakajima A, et al. Physical and functional association of human protein O-mannosyltransferases 1 and 2. J Biol Chem 2006;281:19339-45. [DOI] [PubMed] [Google Scholar]
- 16.Manya H, Chiba A, Margolis RU, et al. Molecular cloning and char-acterization of rat Pomt1 and Pomt2. Glycobiology 2006;16:863-73. [DOI] [PubMed] [Google Scholar]
- 17.Manya H, Suzuki T, Akasaka-Manya K, et al. Regulation of mam-malian protein O-mannosylation: preferential amino acid sequence for O-mannose modification. J Biol Chem 2007;282:20200-6. [DOI] [PubMed] [Google Scholar]
- 18.Hutzler J, Schmid M, Bernard T, et al. Membrane association is a determinant for substrate recognition by PMT4 protein O-mannosyltransferases. Proc Natl Acad Sci USA 2007;104:7827-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Shao L, Shi S, et al. Modification of epidermal growth factor-like repeats with O-fucose. Molecular cloning and expression of a novel GDP-fucose protein O-fucosyltransferase. J Biol Chem 2001;276:40338-45. [DOI] [PubMed] [Google Scholar]
- 20.Luo Y, Koles K, Vorndam W, et al. Protein O-fucosyltransferase 2 adds O-fucose to thrombospondin type 1 repeats. J Biol Chem 2006;281:9393-9. [DOI] [PubMed] [Google Scholar]
- 21.Ju T, Cummings RD. A unique molecular chaperone Cosmc re-quired for activity of the mammalian core 1 beta 3-galactosyltransferase. Proc Natl Acad Sci USA 2002;99:16613-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ju T, Cummings RD. Protein glycosylation: chaperone mutation in Tn syndrome. Nature 2005;437:1252. [DOI] [PubMed] [Google Scholar]
- 23.Kitagawa H, Izumikawa T, Uyama T, et al. Molecular cloning of a chondroitin polymerizing factor that cooperates with chondroitin synthase for chondroitin polymerization. J Biol Chem 2003;278:23666-71. [DOI] [PubMed] [Google Scholar]
- 24.Kim BT, Kitagawa H, Tanaka J, et al. In vitro heparan sulfate po-lymerization: crucial roles of core protein moieties of primer substrates in addition to the EXT1-EXT2 interaction. J Biol Chem 2003;278:41618-23. [DOI] [PubMed] [Google Scholar]
- 25.Endo T. Aberrant glycosylation of α-dystroglycan and con-genital muscular dystrophies. Acta Myol 2005;24:64-9. [PubMed] [Google Scholar]
- 26.Yoshida A, Kobayashi K, Manya H, et al. Muscular dystrophy and neuronal migration disorder caused by mutations in a glycosyltransferase, POMGnT1. Dev Cell 2001;1:717-24. [DOI] [PubMed] [Google Scholar]
- 27.Beltran-Valero De Bernabe D, Currier S, Steinbrecher A, et al. Mutations in the O-mannosyltransferase gene POMT1 give rise to the severe neuronal migration disorder Walker-Warburg syn-drome. Am J Hum Genet 2002;71:1033-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Reeuwijk J, Janssen M, van den Elzen C, et al. POMT2 muta-tions cause alpha-dystroglycan hypoglycosylation and Walker-Warburg syndrome. J Med Genet 2005;42:907-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vajsar J, Zhang W, Dobyns WB, et al. Carriers and patients with muscle-eye-brain disease can be rapidly diagnosed by enzymatic analysis of fibroblasts and lymphoblasts. Neuromuscul Disord 2006;16:132-6. [DOI] [PubMed] [Google Scholar]
- 30.Manya H, Bouchet C, Yanagisawa A, et al. Protein O-mannosyltransferase activities in lymphoblasts from patients with α-dystroglycanopathies. Neuromuscul Disord 2008;18:45-51. [DOI] [PubMed] [Google Scholar]
- 31.Beedle AM, Nienaber PM, Campbell KP. Fukutin-related protein associates with the sarcolemmal dystrophin-glycoprotein complex. J Biol Chem 2007;282:16713-7. [DOI] [PubMed] [Google Scholar]
- 32.Xiong H, Kobayashi K, Tachikawa M, et al. Molecular interaction between fukutin and POMGnT1 in the glycosylation pathway of α-dystroglycan. Biochem Biophys Res Commun 2006;350:935-41. [DOI] [PubMed] [Google Scholar]