Summary
Glycogen Storage Disease Type II (GSDII) is a recessively inherited disorder due to the deficiency of acid α-glucosidase (GAA) that results in glycogen accumulation in the lysosomes. The molecular analysis of the GAA gene was performed on 45 Italian patients with late onset GSDII. DHPLC analysis revealed 28 polymorphisms spread all over the GAA gene. Direct sequencing identified the 96% of the mutant alleles, 12 of which are novel. Missense mutations were functionally characterized by enzyme activity and protein processing in a human GAA deficient cell line while splicing mutations were studied by RT-PCR and in silico analysis. A complex allele was also identified carrying three different alterations in cis. All the patients studied carried a severe mutation in combination with a milder one, which explains the late onset of the disease. The c.-32-13T > G was the most frequent mutation, present as compound heterozygote in 85% of the patients as described in other late onset GSDII Caucasian populations. Interestingly, 10 of the 45 patients carried the c.-32-13T > G associated to the severe c.2237G > A (p.W746X) mutation. However, despite the common genotype, patients presented with a wide variability in residual enzyme activity, age of appearance of clinical signs and rate of disease progression, suggesting that other genetic/environment factors may modulate clinical presentation.
Introduction
Glycogen Storage Disease Type II (GSDII; Pompe disease, acid maltase deficiency, MIM# 232300) is an autosomal recessive inherited disorder due to the deficiency of acid α-glucosidase (GAA; E.C.3.2.1.20) that results in impaired glycogen degradation which accumulates within the lysosomes.
Clinically, GSDII encompasses a continuous spectrum of phenotypes, from a rapidly progressive infantile form leading to death within the first year of life to a slowly progressive late-onset form of the disease that affects mobility and respiratory function. Classic infantile GSDII manifests soon after birth and is characterized by absent or nearly absent enzyme activity, severe muscle weakness, cardiomegaly/cardiomyopathy and respiratory insufficiency, that typically lead to death within the first year of life (1–4). Some infantile patients have less severe cardiac involvement without left cardiac output obstruction, survive longer and die because of pulmonary infections with secondary ventilatory insufficiency (5, 6). Late onset GSDII comprises all milder subtypes: partial enzyme deficiency manifests in children and adults as slowly progressive skeletal muscle weakness without cardiac involvement. Respiratory muscle weakness, particularly of the diaphragm, is the leading cause of death in the late-onset cases. (1, 2, 4, 7–9).
The GAA gene (MIM# 606800) located in the human chromosome 17q25.2-25.3 produces an inactive 110 kD precursor which is transported to the lysosomal compartment and processed into the 95 kD intermediate and the fully active forms of 76 and 70 kD (1, 10–13). More than 200 mutations in the GAA gene have been described up to date (http://www2.eur.nl/fgg/ch1/pompe).
In an extensive collaborative study we analysed the complete mutational profile in 45 Italian patients affected by the late onset GSDII. We were able to characterize 27 mutant alleles leading to the identification of 12 novel mutations. Missense mutations were functionally characterized in vitro by enzyme activity and protein processing and splicing mutations were studied by RT-PCR or in silico analysis. This work offers a complete picture of the late onset GSDII molecular genetics in Italy which contributes in the understanding of the natural history and in the evaluation of emerging ERT efficacy.
Material and Methods
Patients
We studied 45 Italian patients with late onset GSDII (19 females and 26 males). The diagnosis was based on clinical data and confirmed by reduced GAA activity in lymphocytes or muscle. The age at diagnosis varied from 2 to 68 years. Almost all the patients underwent a program of physiotherapy, high protein diet and respiratory management in their reference centres. Most of the patients have had mild muscular symptoms since childhood. First complaints were mostly related to mobility problems, weakness and fatigue.
GAA mutation analysis
Genomic DNA was extracted with the QIAamp DNA blood Mini Kit (Qiagen GmbH, Hilden, Germany). GAA gene was amplified as described (14). PCR products were screened by denaturing High Performance Liquid Chromatography (dHPLC, Varian, Palo Alto, CA, USA) and in the presence of heteroduplex, sequenced on the ABI PRISM 3700 DNA Analyzer. Mutations were confirmed by sequencing duplicate PCR products and confirmed by the analysis of parental DNA.
For RT-PCR analysis the first strand cDNA was synthesized using random hexamer primers and subsequent amplification was done in six overlapping fragments as described by Hermans et al. (15).
All mutations are described according to mutation nomenclature, considering nucleotide + 1 the A of the first ATG translation initiation codon (16, 17, http://www.hgvs.org/mutnomen). Nucleotide numbers are derived from cDNA GAA sequence (RefSeq cDNA Y00839.1).
Site directed mutagenesis
Missense mutations were introduced in the wild type full length cDNA GAA cloned in pcDNA3 (Invitrogen) by site directed mutagenesis using the Quickchange Site-Directed Mutagenesis Kit (Stratagene, Cedar Creek, TX, USA). Each clone was entirely sequenced to confirm that no other mutations were introduced by the PCR-based mutagenesis procedure.
Cell culture and in vitro expression assay
Patient fibroblasts obtained from skin biopsies and the Ad5-SV40 immortalized human GAA-deficient fibroblast cell line were cultured in RPMI 1640 supplemented with 10% fetal calf serum, 2 mM L-glutamine and 50 mg/ml penicillin/streptomycin (Gibco, Paisley, UK).
Cells were transfected with wild type and mutant constructs with a standard calcium/phosphate using 4 µg of total plasmid DNA Endofree purified (Sigma, St. Louis, MO, USA), harvested after 48 h and assayed for GAA activity and Western blot as described elsewhere (18).
Results and discussion
The mutation profile of the GAA gene was analysed in 45 patients with the late onset form of the disease. We identified 27 different alleles corresponding to the 96% of the total alleles: 12 of them are novel including a complex allele that carried three different alterations in cis (Table 1). The GAA profile was characterized by all kind of mutations, including single base changes, both small and large deletions, small insertions and splicing aberrations (19).
Table 1. Mutation profile of the GAA gene in the Italian late onset GSDII population.
| Location | cDNA mutation* | Mutation effect | Allele frequency |
| intron 1 | c.-32-13T > G | leaky splice | 42.0% |
| exon 2 | c.258dupC | p.N87QfsX9 | 1.1% |
| exon 2 | c.525delT | p.E176RfsX45 | 3.4% |
| intron 3 | c.692 + 1G > C | r.0 | 2.2% |
| exon 5 | c.925G > A | p.G309R | 1.1% |
| exon 6 | c.1064 T > C | p.L355P | 3.4% |
| intron 6 | c.1076-1G > C | r.1076-79_1195 + 89ins | 1.1% |
| exon 7 | c.1082C > T | p.P361L | 1.1% |
| intron 7 | c.1194 + 2T > A | r.spl? | 1.1% |
| exon 9 | c.1331C > G | p.P444R | 1.1% |
| exon 9 | c.1333G > C | p.A445P | 1.1% |
| exon 10 | c.1465G > A | p.D489N | 3.4% |
| intron 10 | c.1551 + 1G > C | p.V480_I517del | 1.1% |
| exon 11 | c.1626C > G | r.spl? | 2.2% |
| exon 12 | c.1645G > C | p.G549R | 1.1% |
| exon 12 | c.1655T > C | p.L552P | 2.2% |
| exon 13 | c. 1776del G | p.T593HfsX5 | 1.1% |
| exon 13 | [c.1833_1839del;c.1846G > T; 1847_1848insT] | p.H612_D616delinsRGI | 1.1% |
| exon 13 | c.1836C > G | p.H612Q | 1.1% |
| exon 14 | c.1927G > A | p.G643R | 1.1% |
| exon 14 | c.2014C > T | p.R672W | 3.4% |
| exon 15 | c.2104C > T | p.R702C | 1.1% |
| exon 16 | c.2219_2220delTG | p.V740GfsX55 | 1.1% |
| exon 16 | c.2237G > A | p.W746X | 10.3% |
| exon 16 | c.2242dupG | p.E748GfsX48 | 1.1% |
| intron 17/18 | c.2481 + 102-2646 + 31del | p.G828_N882del | 2.2% |
| exon 18/intron 18 | c.2646_2646 + 1delTG | r.spl? | 1.1% |
* New mutations are indicated in bold; cDNA reference sequence Y00839.1. For cDNA numbering + 1 corresponds to the A of the first ATG translation initiation codon.
Samples were first screened by DHPLC and subsequently sequenced, revealing 28 polymorphisms spread all over the GAA gene (Table 2). DHPLC technique allows an accurate and rapid mutation screening which reduces costs and working time but is not useful in the presence of highly polymorphic genes as the GAA.
Table 2. GAA polymorphisms in Italian population.
| Location | Single base change§ | Protein Sequence | Frequency* |
| Exon 2 | c.271GA c.324TC |
p.D91N p.C108C |
0.6% 28% |
| Intron 2 | c.547-4CG c.547-39TG |
Non coding Non coding |
31% 25% |
| Exon 3 | c.596GA c.642CT c.668AG |
p.H199R p.S214S p.H223R |
42% 18% 44% |
| Intron 4 | c.854ins7nt + 7 c.858 + 29TC |
Non coding Non coding |
32% 15% |
| Exon 5 | c.921AT | p.A307A | 5.4% |
| Intron 5 | c.955 + 12 AG | Non coding | 33% |
| Exon 8 | c.1203AG | p.Q401Q | 3.9% |
| Intron 8 | c.1327-18GA | Non coding | 35% |
| Exon 9 | c.1374CT | p.Y458Y | 5.1% |
| Intron 9 | c.1438-19CG | Non coding | 42% |
| Exon 11 | c.1581GA | p.R527R | 32% |
| Intron 12 | c.1754 + 12GA | Non coding | 24% |
| Exon 13 | c.1830CT | p.A610A | 11% |
| Intron 14 | c.2040 + 20AG | Non coding | 36% |
| Exon 15 | c.2133AG c.2154TC |
p.T711T p.V718V |
15% 35% |
| Intron 15 | c.2190-13GT | Non coding | 40% |
| Exon 16 | c.2238GC | p.W746C | 10% |
| Intron 16 | c.2331 + 20GA | Non coding | 38% |
| Exon 17 | c.2338AG | p.I780V | 42% |
| Exon 18 | c.2553GA | p.G851G | 40% |
| Exon 20 | c.3082CT c.3277TC |
Non coding Non coding |
24% 30% |
For cDNA numbering + 1 corresponds to the A of the first ATG translation initiation codon (cDNA reference sequence Y00839.1). § New polymorphisms are indicated in bold. * Frequency is calculated on 100 normal alleles
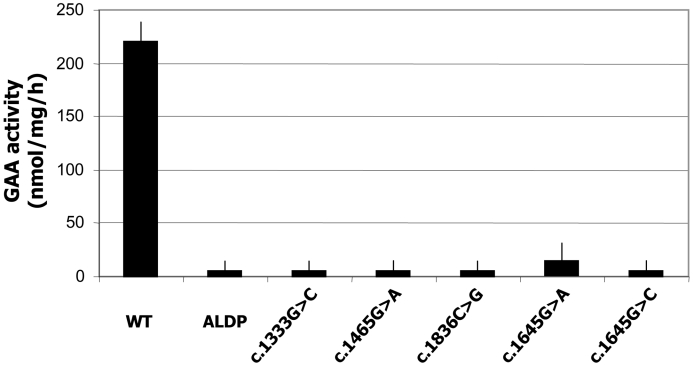
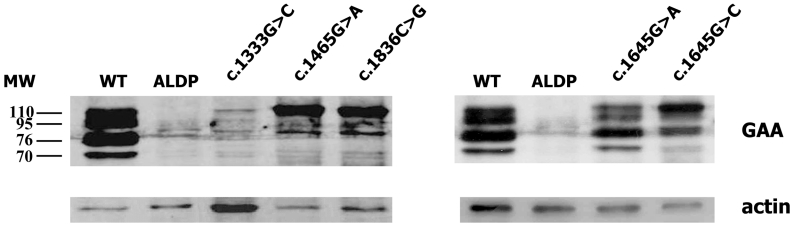
The deleterious effect of the novel missense mutations was confirmed by in vitro expression analysis in human GAA-deficient fibroblasts transiently transfected with the wild type and mutant GAA. As shown in Figure 1A, neither of the mutant proteins expressed residual enzyme activity. Moreover, Western blot analysis demonstrated that the expression pattern of the mutant proteins differed in all cases from the wild type GAA, which confirms they are disease causing mutations (Fig. 1B).
Figure 1.
A) Enzyme activity in GAA deficient fibroblasts transiently transfected with the wild type pCDNA3-GAA and mutant constructs, measured using the fluorogenic substrate 4-MU-a-D-glucopyranoside. B) Western blot analysis of the cellular extracts described above All GAA forms are detected in the wild type transfected cells (WT) while no GAA is detected in the mock transfected cells (ALDP). Almost all the protein obtained from the c.1465G > A and c.1836C > G constructs remained as the GAA inactive precursor of 110kD. The c.1645G > A mutant was correctly processed while c.1645G > C accumulates predominantly as the 110kD and 76kD forms. For the c.1333G > C construct, a faint band of 110 kD was detected when a higher amount of protein was loaded onto the gel, suggesting that the protein is highly unstable and it is rapidly degraded.
Only four patients had one of the two still unknown alleles. However, in one of these patients, the paternally inherited mutation (c.-32-13T > G) was observed as compound heterozygosity in genomic DNA and in apparent homozygosity in cDNA. Based on these findings we assumed that the unknown allele may harbor an unidentified mutation in the non coding regions of the GAA gene that prevents the formation of a stable mRNA.
The mutation profile of the GAA gene in Italian late onset GSDII patients was quite heterogeneous, similar to what has been previously described in the French late onset GSDII population (19). As described in the Caucasian late onset GSDII population the c.-32-13T > G resulted the most frequent mutation (allele frequency 42%) (2). In all cases studied, the combination of known severe mutations with milder mutations explained the late onset of the disease. Interestingly, the c.-32-13T > G was associated to the severe c.2237G > A (p.W746X) in 10 of the 45 patients studied. Despite the common genotype, patients presented with a wide variability in residual enzyme activity, age of appearance of clinical signs and rate of disease progression.
This work represents the largest study of GSDII conducted in Italy to date. It should be pointed out that almost half of the mutant alleles found are due to novel mutations. Therefore, in vitro analysis resulted an useful tool in discriminating disease-causing mutations and evaluating their effect on the normal enzyme function.
Increasing knowledge on the mutant protein structure may be potentially used in the development of novel therapeutic strategies (Parenti, et al., in press). However, in vivo enzyme function determination is still preferable for genotype/phenotype correlation (20, 21).
Our data confirmed the wide spectrum of clinical manifestations observed in GSDII and the phenotypic variability among patients, even carrying the same genotype. Moreover, continued mutational analysis contributes in the understanding of genotype/phenotype correlations and this may be useful in the evaluation of emerging ERT efficacy.
References
- 1.Hirschhorn R, Reuser AJJ. Glycogen storage disease type II: acid α-glucosidase (acid maltase) deficiency, In: Scriver CR, Beaudet AL, Sly WS, Valle D, eds. The metabolic and molecular basis of inherited disease. Vol 3, 8th edn. New York: McGraw-Hill; 2001. p. 3389-420. [Google Scholar]
- 2.Raben N, Plotz P, Byrne BJ. Acid alpha-glucosidase deficiency (glycogenosis type II, Pompe disease). Curr Mol Med 2002;2:145-66. [DOI] [PubMed] [Google Scholar]
- 3.Van den Hout HM, Hop W, van Diggelen OP. The natural course of infantile Pompe’s disease: 20 original cases compared with 133 cases from the literature. Pediatrics 2003;112:332-40. [DOI] [PubMed] [Google Scholar]
- 4.Kishnani PS, Howell RR. Pompe disease in infants and children. J Pediatr 2004;144:35-43. [DOI] [PubMed] [Google Scholar]
- 5.Slonim AE, Bulone L, Ritz S, Goldberg T, Chen A, Martiniuk F. Identification of two subtypes of infantile acid maltase deficiency. J Pediatr 2000;137:283-5. [DOI] [PubMed] [Google Scholar]
- 6.Winkel LP, Hagemans ML, van Doorn PA, et al. The natural course of non-classic Pompe’s disease; a review of 225 published cases. J Neurol 2005;252:875-84. [DOI] [PubMed] [Google Scholar]
- 7.Hagemans ML, Winkel LP, Hop WC, et al. Disease severity in children and adults with Pompe disease related to age and disease duration. Neurology 2005a;64:2139-41. [DOI] [PubMed] [Google Scholar]
- 8.Hagemans ML, Winkel LP, Van Doorn PA, et al. Clinical manifestation and natural course of late-onset Pompe’s disease in 54 Dutch patients. Brain 2005b;128:671-7. [DOI] [PubMed] [Google Scholar]
- 9.Hagemans ML, Janssens AC, Winkel LP, et al. Late-onset Pompe disease primarily affects quality of life in physical health domains. Neurology 2004;63:1688-92. [DOI] [PubMed] [Google Scholar]
- 10.Hoefsloot LH, Hoogeveen-Westerveld M, Reuser AJ, Oostra BA. Characterization of the human lysosomal alpha-glucosidase gene. Biochem J 1990a;272:493-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoefsloot LH, Willemsen R, Kroos MA, et al. Expression and routeing of human lysosomal alpha-glucosidase in transiently transfected mammalian cells. Biochem J 1990b;272:485-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martiniuk F, Bodkin M, Tzall S, Hirschhorn R. Isolation and partial characterization of the structural gene for human acid alpha glucosidase. DNA Cell Biol 1991;10:283-92. [DOI] [PubMed] [Google Scholar]
- 13.Moreland RJ, Jin X, Zhang XK, et al. Lysosomal acid alpha-glucosidase consists of four different peptides processed from a single chain precursor. J Biol Chem 2005;280:6780-91. [DOI] [PubMed] [Google Scholar]
- 14.Hermans MM, van Leenen D, Kroos MA, Reuser AJ. Mutation detection in glycogen storage-disease type II by RT-PCR and automated sequencing. Biochem Bioph Res Commun 1997;241:414-8. [DOI] [PubMed] [Google Scholar]
- 15.Ko TM, Hwu WL, Lin YW, et al. Molecular genetic study of Pompe disease in Chinese patients in Taiwan. Hum Mutat 1999;13:380-4. [DOI] [PubMed] [Google Scholar]
- 16.den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat 2000;15:7-12. [DOI] [PubMed] [Google Scholar]
- 17.den Dunnen JT, Paalman MH. Standardizing mutation nomenclature: why bother? Hum Mutat 2003;22:181-2. [DOI] [PubMed] [Google Scholar]
- 18.Montalvo ALE, Bembi B, Donnarumma M, et al. Mutation profile of the GAA gene in 40 Italian patients with late onset Glycogen Storage Disease type II. Hum Mutat 2006;27:999-1006. [DOI] [PubMed] [Google Scholar]
- 19.Laforet P, Nicolino M, Eymard PB, et al. Juvenile and adult-onset acid maltase deficiency in France: genotype-phenotype correlation. Neurology 2000;55:1122-8. [DOI] [PubMed] [Google Scholar]
- 20.Huie ML, Tsujino S, Sklower Brooks S, et al. Glycogen storage disease type II: identification of four novel missense mutations (D645N, G648S, R672W, R672Q) and two insertions/deletions in the acid alpha-glucosidase locus of patients of differing phenotype. Biochem Biophys Res Commun 1998;244:921-7. [DOI] [PubMed] [Google Scholar]
- 21.Montalvo ALE, Cariati R, Deganuto M, et al. Glycogenosis type II: identification and expression of three novel mutations in the acid alpha-glucosidase gene causing the infantile form of the disease. Mol Genet Metab 2004;81:203-8. [DOI] [PubMed] [Google Scholar]