Abstract
Multilevel logistic regression models are increasingly being used to analyze clustered data in medical, public health, epidemiological, and educational research. Procedures for estimating the parameters of such models are available in many statistical software packages. There is currently little evidence on the minimum number of clusters necessary to reliably fit multilevel regression models. We conducted a Monte Carlo study to compare the performance of different statistical software procedures for estimating multilevel logistic regression models when the number of clusters was low. We examined procedures available in BUGS, HLM, R, SAS, and Stata. We found that there were qualitative differences in the performance of different software procedures for estimating multilevel logistic models when the number of clusters was low. Among the likelihood-based procedures, estimation methods based on adaptive Gauss-Hermite approximations to the likelihood (glmer in R and xtlogit in Stata) or adaptive Gaussian quadrature (Proc NLMIXED in SAS) tended to have superior performance for estimating variance components when the number of clusters was small, compared to software procedures based on penalized quasi-likelihood. However, only Bayesian estimation with BUGS allowed for accurate estimation of variance components when there were fewer than 10 clusters. For all statistical software procedures, estimation of variance components tended to be poor when there were only five subjects per cluster, regardless of the number of clusters.
Keywords: statistical software, multilevel models, hierarchical models, random effects model, mixed effects model, generalized linear mixed models, Monte Carlo simulations, Bayesian analysis, R, SAS, Stata, BUGS
1. Introduction
Clustered data are frequently encountered in health services, public health, epidemiology, and education research. For instance, data may consist of patients clustered within primary care practices or hospitals, of residents clustered within neighbourhoods, or of students clustered within schools. Subjects nested within the same cluster often exhibit a greater degree of similarity or homogeneity of outcomes compared to randomly selected subjects from different clusters (Snijders and Boskers, 1999; Raudenbush and Bryk, 2002; Kreft and De Leeuw, 1998; Goldstein, 1995; Austin et al., 2001). Due to the possible lack of independence of subjects within the same cluster, traditional statistical methods may not be appropriate for the analysis of clustered data. There is an increasing use of multilevel models for the analysis of clustered data. These models are also referred to as mixed effects models, random effects models, or hierarchical models in the literature (Snijders and Boskers, 1999; Raudenbush and Bryk, 2002; Kreft and De Leeuw, 1998; Goldstein, 1995; Austin et al., 2001). Many popular statistical packages, such as HLM, MLwiN, R, SAS, Stata, and WinBUGS/BUGS have the capacity to fit multilevel models. Some of these are general purpose statistical software packages (R, SAS, Stata, WinBUGS/BUGS), while others were written specifically for fitting mixed effects models (HLM and MLwiN). BUGS/WinBUGS is a statistical programming language for fitting Bayesian models.
In many contexts, the number of clusters may be relatively small. There is a paucity of research examining the effect of a low number of clusters on the estimation of multilevel models. Snijders and Bosker (1999) suggest that multilevel models not be used when there are fewer than 10 clusters. Raudenbush (2008) examined estimation of multilevel models when there are a large number of small clusters.
The goal of our paper is to compare the performance of different statistical software procedures for estimating multilevel models when the number of clusters is low. Since dichotomous outcomes occur frequently in health services, medical, epidemiological, and public health research (Austin et al., 2010), we focus our attention on multilevel logistic regression models. The paper is structured as follows: In Section 2, we describe a series of Monte Carlo simulations that were conducted to examine the performance of different statistical software procedures for estimating multilevel logistic regression models. In Section 3, we describe the different statistical software procedures that were considered. In Section 4, we report the results of these Monte Carlo simulations. Finally, in Section 5, we discuss our findings in the context of prior studies in this area.
2. Design of Monte Carlo simulations
In each simulation there were J clusters with N subjects per cluster. For each subject, an outcome, Yij ∼ Bernoulli(pij), was generated, where the subject-specific probabilities of the outcome was determined using Equation (1):
| (1) |
The fixed effects were β0 = −1.1, β1 = 1, and β2 = 0, while the random effect was β0 j ∼ N(0,1). The covariates values were drawn from a bivariate normal distribution: , for i = 1,…,N and j = 1,…,J. Using β0 = −1.1 implies that the outcome Yij occurs in approximately 25% of subjects in an average cluster.
We allowed the number of clusters to range from a low of 5 to a high of 20, in increments of 1. Similarly, the number of subjects per cluster was allowed to range from a low of 5 to a high of 50, in increments of 5. We thus examined 160 scenarios (16 numbers of clusters x 10 sizes of clusters). In each scenario, 1,000 random datasets were generated.
There were several reasons for selecting the model described in Formula (1). First, it involved a non-zero regression parameter (β1), allowing us to examine estimation of non-null regression coefficients. Second, it included a null regression parameter (β2), thereby allowing us to examine empirical type I error rates when testing the null hypothesis that the regression coefficient was not different from zero. Third, having only two covariates, the model was relatively simple, thereby reducing computational demands when Bayesian methods were used to fit the model.
3. Statistical packages and procedures for estimating mixed effects logistic regression models
The variable cluster_id or cluster.id is used to identify subjects who are in the same cluster (the choice of which identifier to use is software dependent – depending on which of “.” or “_” can be used a part of a variable name).
3.1. SAS version 9.2
We examined two different procedures in SAS version 9.2 for estimating two-level multilevel logistic regression models.
3.1.1. Proc NLMIXED
SAS Proc NLMIXED is a procedure for fitting nonlinear mixed models. It fits nonlinear mixed models by maximizing an approximation to the likelihood integrated over the random effects. Different integral approximations are available in NLMIXED. In this study, we used adaptive Gaussian quadrature (Pinheiro and Bates, 1995). Unlike other procedures described below, SAS Proc NLMIXED does not use a fixed number of quadrature points. Rather, Proc NLMIXED selects the number of quadrature points adaptively. Alternatively, one can specify the number of quadrature points. The following SAS code was used to fit the models:
- proc nlmixed data=mc;
- parms a0=−1.1 a1=1 a2=0 s2=1;
- lambda = a0 + a1*x1 + a2*x2 + alpha;
- p = exp(lambda)/(1 + exp(lambda));
- model y ∼ binomial (1,p);
- random alpha ∼ normal (0, s2) subject=cluster_id;
run;
3.1.2. Proc GLIMMIX
Proc GLIMMIX is a SAS procedure that fits generalized linear mixed models (Proc GLIMMIX, which first appeared in SAS 9.2 is not to be confused with the %GLIMMIX macro supplied by SAS that fits generalized linear mixed models using iterative calls to Proc MIXED (Wolfinger and O’Connell, 1993; Breslow and Clayton, 1993)). A variety of estimation methods based on pseudo-likelihood techniques are available in Proc GLIMMIX. The default estimation methods is based on residual pseudo-likelihood methods in which the locus of expansion of the Taylor series expansion of the generalized linear mixed model is the vector of random effects solutions. The following SAS code was used to fit the multilevel logistic regression model:
- proc glimmix method=rspl;
- class cluster_id;
- model y (descending) = x1 x2 /dist=binomial s;
- random intercept /subject=cluster_id;
run;
3.2. HLM 5.00
HLM 5.00 is a stand-alone software package for fitting two and three-level multilevel regression models (Bryk et al., 1996). HLM uses first order penalized quasi-likelihood (PQL-1) for fitting multilevel logistic regression models. HLM/2L is used for fitting 2-level hierarchical models, while HLM/3L is used for fitting 3-level hierarchical models. For a particular analysis, HLM/2L must be used twice. First, it is used to generate a sufficient statistics matrix from the data. Second, it is used for specifying and estimating a multilevel model. The HLM/2L code for specifying and estimating a two-level logistic regression model is given below:
#This command file was run with re.ssm
#This is the sufficient statistics matrix
STOPMICRO : 0.0000010000
STOPMACRO : 0.0001000000
MACROIT : 100, n
MICROIT : 100
NONLIN : BERNOULLI
LAPLACE : n, 50
LAPLACE8 : n, 50
LEVEL1 : OUTCOME=INTRCPT1+X1+X2+RANDOM
LEVEL2 : INTRCPT1=INTRCPT2+RANDOM/
LEVEL2 : X1=INTRCPT2/
LEVEL2 : X2=INTRCPT2/
RESFIL : N
HETEROL1VAR : n
ACCEL : 5
LVR : N
LEV1OLS : 10
HYPOTH : n
FIXSIGMA2 : 1.000000
FIXTAU : 3
CONSTRAIN : N
OUTPUT : hlm.out
TITLE : Random effects model (HLM)
3.3. Stata 9.2 – xtlogit
In Stata 9.2, the xtlogit function can be used for fitting random effects logistic regression models. The default approximation to the likelihood is adaptive Gauss-Hermite approximation (Liu and Pierce, 1994). Optionally, one can specify a nonadaptive Gauss-Hermite approximation. By default, twelve points are used for the Gauss-Hermite quadrature. The following Stata code was used:
xtlogit y x1 x2, i (cluster_id) intmethod (aghermite) intpoints(12)
3.4. R 2.8.0
R is a freely available object-oriented statistical programming language (R Core Development Team, 2005). There are a large number of user-provided packages. We considered two different packages which contain functions for fitting multilevel logistic regression models. The MASS package contains the glmmPQL function, while the lme4 package contains the glmer function.
3.4.1. glmmPQL function
The glmmPQL function fits generalized linear mixed models using penalized quasi-likelihood (PQL) (Wolfinger and O’Connell, 1993; Breslow and Clayton, 1993; Schall, 1991). It works by iteratively calling the lme function from the nlme package. The following R code was used:
glmmPQL (y ∼ x1 + x2, random = ∼1|cluster.id,family=binomial)
3.4.2. glmer function
The glmer function fits generalized linear mixed models using the adaptive Gauss-Hermite approximation to the likelihood (Liu and Pierce, 1994). The default number of points per axis for evaluating this approximation is one. In this case, the approximation corresponds to the Laplacian approximation. The following R code was used:
glmer (y ∼ x1 + x2 + (1|cluster.id), family=binomial, nACQ=1)
3.5. BUGS version 0.603
BUGS (Bayesian inference Using Gibbs Sampling) is a software programme for fitting Bayesian models (Gilks et al., 1994). In these simulations, we used the Classic BUGS software programme for a Unix platform. Classic BUGS uses Gibbs Sampling, an implementation of Markov Chain Monte Carlo (MCMC) methods for estimating the posterior distribution of the model parameters (Gilks et al., 1996). The following code was used:
- for (i in 1:N){
- y[i] ∼ dbin (p[i], 1);
- logit (p[i]) <- intercept[clusterid[i]] +
- beta[1]*x1[i] + beta[2]*x2[i];
}
- for (j in 1:M){
- intercept[j] ∼ dnorm(lambda, tau);
}
sigma <- 1/sqrt(tau);
tau ∼ dgamma (0.001,0.001);
lambda ∼ dnorm (0,0.0001);
beta[1] ∼ dnorm (0,0.0001);
beta[2] ∼ dnorm (0,0.0001);
In the above model, diffuse, non-informative priors were assumed for the parameters of the regression model. The random intercepts were assumed to have a normal distribution. The prior distribution of the mean of this normal distribution was assumed to be a normal distribution with mean 0 and variance 10,000. The prior distribution for the precision (the inverse of the variance) of this normal distribution was assumed to be a Gamma distribution with shape and scale parameters both equal to 0.001. The posterior mean of each regression parameter was determined from the monitored samples from the posterior distribution.
When fitting the multilevel model using BUGS in the current simulations, 1,000 burn-in iterations of the Gibbs sampler were performed when there were more than 5 clusters. In scenarios in which there were 5 clusters, then 10,000 burn-in iterations were employed. In all scenarios, the Gibbs sampler was then monitored for an additional 1,000 iterations. The Geweke convergence diagnostic was used to assess the convergence of Gibbs sampler for the parameters of interest (β1, β2, and τ) (Geweke, 1994). If the z-statistic for the Geweke convergence diagnostic exceeded 1.96 in absolute value for any of the three parameters, then that analysis was discarded. For the Bayesian analysis, sufficient iterations of the Monte Carlo simulations were used so that there were 1,000 Bayesian analyses that converged per scenario. Thus, the actual numbers of simulated datasets for the Bayesian analyses varied across the different scenarios.
4. Results of the Monte Carlo simulations
SAS Proc NLMIXED uses an adaptive algorithm to determine the number of quadrature points used for the adaptive Gaussian quadrature. Across all analyses in all the simulated datasets, the selected number of quadrature points ranged from a low of 1 to a high of 5, with a mean of 4.1. When the number of clusters was fixed, the mean number of quadrature points across the simulated datasets tended to decrease as the number of subjects per cluster increased. In general, when the number of subjects per cluster was fixed, the effect of the number of clusters on the mean number of quadrature points was negligible.
4.1. Estimation of β1
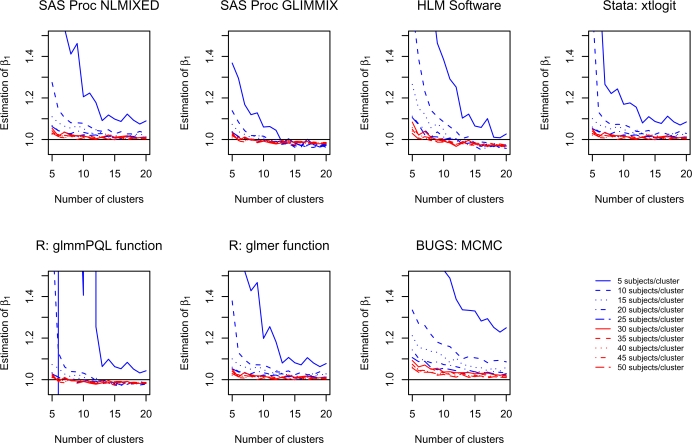
For each scenario and for each software procedure, the mean estimated regression coefficient for β1 was determined across the 1,000 simulated datasets. The influence of the number of clusters and the number of subjects per cluster on estimating β1 is reported graphically in Figure 1. There is one panel for each of the seven software procedures (one in BUGS, one in HLM, two in R, two in SAS and one in Stata). Due to the large biases that occurred when there were 5 clusters and 5 subjects per cluster, we have fixed the range of the vertical axes so that these values are not visible in the plots. Had the vertical axes been extended to include the entire range of the estimated values of β1, then most of the patterns would not have been discernable. When there were five subjects per cluster and fewer than 10–15 clusters, then each software procedure performed poorly for estimating β1. In some scenarios, the bias was substantial, particularly when there were only five clusters. In many scenarios, the use of BUGS tended to result in estimation that was modestly more biased compared to most of the other software procedures. In general, estimation improved as the number of subjects per cluster increased. When using Proc GLIMMIX in SAS or the HLM software, there was a trend towards increasing bias as the number of clusters increased from 10 to 20. A similar, though attenuated, pattern was observed when using the glmmPQL function in R. For the remaining three likelihood-based software procedures, one observes that biases in estimating β1 were less than approximately 5% once there were at least 20 subjects per cluster, regardless of the number of clusters. Thus, even with 5 clusters, once there were at least 20 subjects per cluster, bias tended to be less than 5%. These findings suggest that, from an estimation perspective, previous guidelines suggesting the need for at least 10 clusters may have been overly conservative. The above results suggest that, in our setting, estimation is minimally biased when there are a small number of clusters, provided the number of subjects per cluster is large enough.
Figure 1.
Estimation of β1
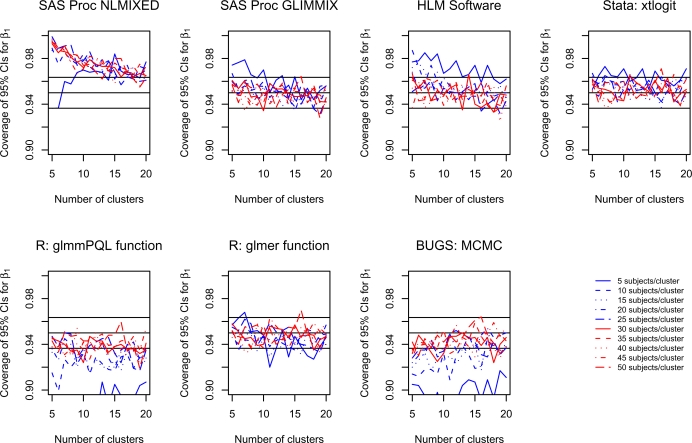
4.2. Coverage of the 95% confidence intervals for β1
For each scenario and for each software procedure, the proportion of estimated 95% confidence intervals for β1 that contained the true value of β1 was determined across the 1,000 simulated datasets. These proportions are reported in Figure 2. Due to the use of 1,000 iterations for each scenario, any coverage rate that exceeds 0.9635 or that is less than 0.9365 is statistically significantly different from 0.95 at a significance level less than 0.05 using a standard test based on the normal approximation due the binomial distribution. On each panel we have superimposed three horizontal denoting coverage rates of 0.9365, 0.95, and 0.9635. For Bayesian estimation using the BUGS software, we are reporting coverage of the 95% credible intervals, thus we are evaluating the frequentist performance of a Bayesian interval.
Figure 2.
Coverage of 95% confidence intervals for β1
Using Proc NLMIXED in SAS, estimated confidence intervals tended to be conservative, with empirical coverage rates that exceeded 95%. Coverage rates approached 95% as the number of clusters increased. However, the number of subjects per cluster did not appear to have an impact upon the empirical coverage rate of the 95% confidence intervals. It was not until the number of clusters was close to 20 that coverage rates were approximately equal to the advertised rates. In contrast, Proc GLIMMIX in SAS and xtlogit in Stata tended to produce confidence intervals with approximately correct coverage rates, except when there were only five subjects per cluster. Even when there were very few clusters, Proc GLIMMIX and xtlogit produced confidence intervals with approximately correct coverage rates when there were at least 10 subjects per cluster. Both the HLM software package and the glmer function in R produced confidence intervals with approximately correct coverage rates, except in some instances when there were only 5 or 10 subjects per cluster. The glmmPQL function in R tended to result in confidence intervals whose coverage rates were lower than the advertised level. However, except when the number of subjects per cluster was very low, the empirical coverage rates tended to not be significantly different from 0.95. In most instances, the number of clusters had, at most, a negligible impact on the coverage rates of confidence intervals.
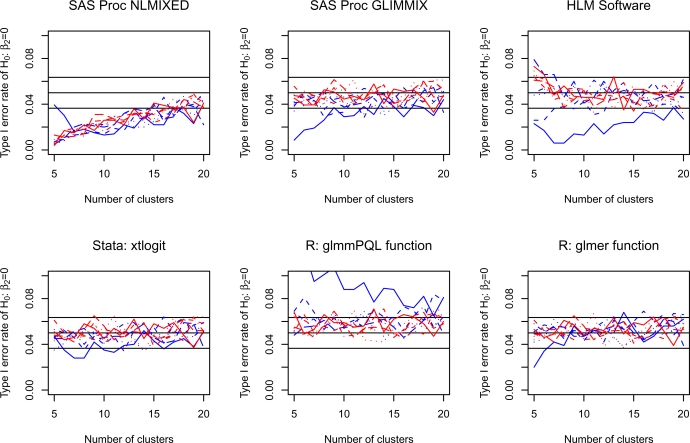
4.3. Type I error rate for H0 : β2 =0
For each scenario and for each software procedure, the proportion of simulated datasets in which the null hypothesis H0 : β2 =0 was rejected was determined across the 1,000 simulated datasets. The empirical type I error rates for testing the hypothesis that β2 was equal to zero are reported in Figure 3. Due to the use of 1,000 iterations of the Monte Carlo simulations for each scenario, empirical type I error rates that are less than 0.0365 or that exceed 0.0635 are statistically significantly different than 0.05, using a test based on the normal approximation to the binomial distribution. Three lines are superimposed on each panel denoting empirical type I error rates of 0.0365, 0.05, and 0.0635.
Figure 3.
Type I error rate of H0: β2=0
When using Proc NLMIXED in SAS, empirical type I error rates were less than 0.05; however, they approached 0.05 as the number of clusters increased. In most instances, the number of subjects per cluster did not have a large effect on the observed type I error rate. In Stata, the use of the xtlogit function tended to result in type I error rates that were not significantly different from 0.05, regardless of the number of clusters or the number of subjects per cluster. Similarly, the glmer function in R tended to have acceptable empirical type I error rates, except when both the number of clusters and the number of subjects per cluster were equal to five. The HLM software, Proc GLIMMIX in SAS, and the glmmPQL function in R tended to produce satisfactory type I error rates, except when there were only five subjects per cluster.
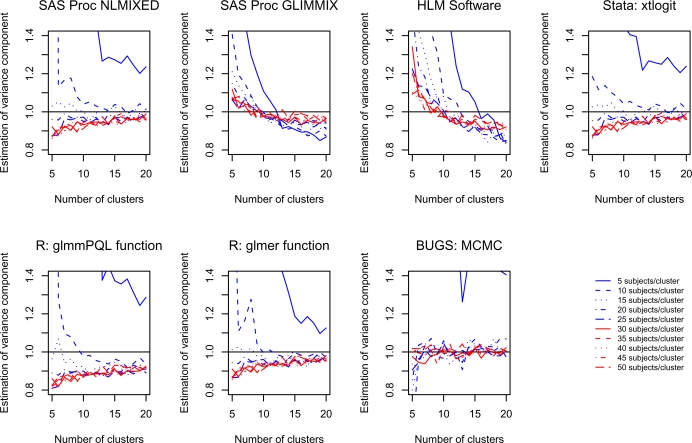
4.4. Estimation of the variance component
For each scenario and for each software procedure, the mean estimated variance of the random effects was determined across the 1,000 simulated datasets. The estimation of the variance of the random effect is reported in Figure 4 for each of the statistical software procedures. For most of the statistical software procedures, estimation of the variance component tended to be poor when there were only 5 or 10 subjects per cluster, regardless of the number of clusters. When using Proc NLMIXED in SAS, estimation of the variance components improved as the number of clusters increased. With the exception of when there were 5 or 10 subjects per cluster, estimates of the variance component tended to be biased downward. When the number of clusters exceeded 10, then the relative bias was less than approximately 10%. When using Proc GLIMMIX in SAS, estimates of the variance component tended to be biased downward when the number of clusters exceeded approximately 10. However, the bias tended to be attenuated with an increasing number of subjects per cluster. When using the xtlogit function in Stata, estimation improved as the number of clusters increased. With the exception of when there were only five subjects per cluster, the relative bias tended to be less than 10% when there were at least 10 clusters. When using R, estimation of the variance component tended to be superior when using the glmer function compared to when the glmmPQL function was used. When using BUGS, estimation of the variance component was minimally biased once there were at least 30 subjects per cluster, regardless of the number of clusters. If there were 10 to 25 subjects per cluster, then estimation of the variance component was minimally biased once there were at least seven clusters.
Figure 4.
Estimation of the variance of the random effects
5. Discussion
Our findings complement and corroborate those of previous studies. Rodriquez and Goldman (1995) conducted a series of Monte Carlo simulations to compare the performance of two software packages (VARCL and ML3) for estimating multilevel logistic regression models (both VARCL and ML3 use an estimation method that is equivalent to marginal quasi-likelihood (MQL) for estimating nonlinear regression models (Rodriquez and Goldman, 1995)). They found substantial biases in the estimation of fixed effects and/or variance components when the random effects were large. Similar to our findings, they also observed that biased estimation occurred when the number of subjects per cluster was small. Breslow and Clayton (1993), in a series of simulations, found that PQL resulted in estimates of variance components that were biased downwards, when estimating multilevel logistic regression models. This was similar to our finding that glmmPQL in R (which employs PQL for estimation) tended to produce estimates of the variance component that were biased downwards. Finally, Browne and Draper (2006) used simulations to compare the performance of MCMC methods for estimating random-effects logistic regression models with that of quasi-likelihood based methods (PQL or MQL). They found that the use of PQL resulted in estimates of variance components that were biased downwards, whereas the use of Bayesian estimation, when done with Gamma prior distributions for the variance components, resulted in negligible bias. These results are similar to ours (the HLM software and the glmmPQL function in R use PQL estimation methods). Furthermore, they found that estimation of the fixed effects was less biased when Bayesian estimation was used compared to when either PQL or MQL was used.
There are certain limitations to the current paper. First, we only examined procedures in BUGS, HLM, R, SAS, and Stata for fitting multilevel logistic regression models. We did not consider other software packages such as MLwiN. Our simulations were conducted in a Unix environment; MLwiN is available only on a PC environment under a Windows-based operating system. Second, we restricted our focus on the estimation of multilevel logistic regression models. We did not consider multilevel linear models or multilevel Poisson regression models. The reason for this restriction was due to the frequency with which dichotomous outcomes occur in health research. Third, we examined a limited set of scenarios. We considered a bivariate regression model, in which one regression coefficient was zero. Thus, we could examine estimation of non-null regression coefficients, coverage of confidence intervals, empirical type I error rates, and estimation of the variance component. Due to the computationally intensive nature of the simulations when Bayesian methods were used, we were unable to examine more complex regression models, with a larger number of covariates or variance components. Fourth, we only considered models with a random intercept and did not consider models with random slopes. One justification for this decision was that random intercept models may be used more frequently in the medical literature than models that incorporate random slopes. Due to the simplicity of the regression model that we considered, our findings may not be generalizable to other regression models. Fifth, three of the software procedures used adaptive Gauss-Hermite to estimate the likelihood function. However, the default number of quadrature points varied across the procedures. In xtlogit in Stata, the default number was 12, while in the glmer function in R, the default was 1. The default in Proc NLMIXED in SAS is to use an adaptive algorithm to determine the number of quadrature points. We used the default setting in each procedure, to reflect how that procedure would be used by a typical user. Examining a variety quadrature points for each software procedure was beyond the scope of the current simulations. The sixth limitation was that we assumed diffuse, non-informative priors for the model parameters in the Bayesian analysis conducted in BUGS. With small to moderate sized datasets, the posterior distribution may be more influenced by the choice of prior distributions than when the number of clusters and number of subjects per cluster are large. However, due to the time-intensive nature of Monte Carlo simulations of MCMC analyses, it was not feasible to examine the performance of Bayesian modeling under a variety of prior distributions.
It has previously been suggested that one should not fit multilevel models to data consisting of fewer than 10 clusters (Snijders and Boskers, 1999). Our findings, in the context of a bivariate regression model, would suggest that, in some settings, one may take a more nuanced approach to the minimum sample sizes for fitting multilevel models. For estimation and inference concerning regression coefficients, the findings of our Monte Carlo study suggest that one can consider settings in which there are as few as five clusters, as long as the number of subjects per cluster exceeds approximately 30. As noted above, estimation and inference may be superior using one software procedure compared to another software procedure. Conversely, when there were more than 10 clusters, estimation and inference was poor for some software procedures if the number of subjects per cluster was too low. Finally, if the focus is on accurate estimation of the variance component, then the minimum number of clusters depends on the acceptable degree of bias and the software procedure used. When using the BUGS software, then accurate estimation of variance components was achieved regardless of the number of clusters, provided that the number of subjects per cluster was at least 30. For several of the other software procedures, one would suggest that there should be at least 10 to 15 clusters. Among these competing likelihood-based software procedures, estimation of variance components tended to be better when procedures based on adaptive Gaussian quadrature were implemented (Proc NLMIXED in SAS, xtlogit in Stata, and glmer in R).
There are two primary conclusions to our study. First, in the specific regression model that we considered, there were qualitative differences in the performance of different statistical software procedures for estimating multilevel logistic models. Second, depending on the software procedure used, the bivariate logistic regression models that we considered can be reliably fit when the number of clusters is small, provided that there are a sufficient number of subjects per cluster.
Footnotes
This study was supported by the Institute for Clinical Evaluative Sciences (ICES), which is funded by an annual grant from the Ontario Ministry of Health and Long-Term Care (MOHLTC). The opinions, results and conclusions reported in this paper are those of the authors and are independent from the funding sources. No endorsement by ICES or the Ontario MOHLTC is intended or should be inferred. This research was supported by an operating grant from the Canadian Institutes of Health Research (CIHR) (MOP 86508). Dr. Austin is supported in part by a Career Investigator Award from the Heart and Stroke Foundation of Ontario.
References
- Austin PC, Goel V, van Walraven C. An Introduction to Multilevel Regression Models. Canadian Journal of Public Health. 2001;92:150–154. doi: 10.1007/BF03404950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin PC, Manca A, Zwarenstein M, Juurlink DN, Stanbrook MB. A substantial and confusing variation exists in handling of baseline covariates in randomized controlled trials: a review of trials published in leading medical journals. Journal of Clinical Epidemiology. 2010;63:142–153. doi: 10.1016/j.jclinepi.2009.06.002. DOI:10.1016/j.jclinepi.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Breslow NE, Clayton DG. Approximate inference in generalized linear mixed models. Journal of the American Statistical Association. 1993;88:9–25. doi: 10.2307/2290687. [DOI] [Google Scholar]
- Browne WJ, Draper D. A comparison of Bayesian and likelihood-based methods for fitting multilevel models. Bayesian Analysis. 2006;1:473–514. doi: 10.1214/06-BA117. [DOI] [Google Scholar]
- Bryk AS, Raudenbush SW, Congdon RT., Jr . HLM: Hierarchical Linear and Nonlinear Modeling with the HLM/2L and HLM/3L Programs. Chicago, IL: Scientific Software International Inc; 1996. [Google Scholar]
- Geweke J. Evaluating the accuracy of sampling-based approaches to calculating posterior moments. In: Bernardo JM, Berger JO, Dawid AP, Smith AFM, editors. Bayesian Statistics. Vol. 4. Oxford: Clarendon Press; 1994. pp. 169–193. [Google Scholar]
- Gilks WR, Thomas A, Spiegelhalter DJ. A language and program for complex Bayesian modelling. The Statistician. 1994;43:169–78. doi: 10.2307/2348941. [DOI] [Google Scholar]
- Gilks WR, Richardson S, Spiegelhalter DJ. Introducing Markov chain Monte Carlo. In: Gilks WR, Richardson S, Spiegelhalter DJ, editors. Markov chain Monte Carlo in practice. London: Chapman & Hall; 1996. pp. 1–19. [Google Scholar]
- Goldstein H. Multilevel Statistical Models, second edition. London: Edward Arnold; 1995. [Google Scholar]
- Kreft I, De Leeuw J. Introduction to Multilevel Modeling. Thousand Oaks, CA: Sage Publications Inc; 1998. [Google Scholar]
- Liu Q, Pierce DA. A note on Gauss-Hermite Quadrature. Biometrika. 1994;81:624–629. doi: 10.2307/2337136. [DOI] [Google Scholar]
- Pinheiro JC, Bates DM. Approximations to the log-likelihood function in the nonlinear mixed-effects model. Journal of Computational and Graphical Statistics. 1995;4:12–35. doi: 10.2307/1390625. [DOI] [Google Scholar]
- Schall R. Estimation in generalized linear models with random effects. Biometrika. 1991;78:719–727. doi: 10.1093/biomet/78.4.719. [DOI] [Google Scholar]
- R Core Development Team . R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2005. [Google Scholar]
- Raudenbush SW. Many small groups. In: de Leeuw J, Meijer E, editors. Handbook of Multilevel Analysis. New York, NY: Springer; 2008. pp. 207–236. [Google Scholar]
- Raudenbush SW, Bryk AS. Hierarchical Linear Models: Applications and Data Analysis Methods, second edition. Thousand Oaks, CA: Sage Publications Inc; 2002. [Google Scholar]
- Rodriquez G, Goldman N. An assessment of estimation procedures for multilevel models with binary responses. Journal of the Royal Statistical Society, Series A. 1995;158:73–89. [Google Scholar]
- Snijders T, Boskers R. Multilevel Analysis: An introduction to basic and advanced multilevel modeling. Thousand Oaks, CA: Sage Publications Inc; 1999. [Google Scholar]
- Wolfinger R, O’Connell M. Generalized linear mixed models: A pseudo-likelihood approach. Journal of Statistical Computation and Simulation. 1993;48:223–243. doi: 10.1080/00949659308811554. [DOI] [Google Scholar]