Abstract
Properdin, a positive regulator of the complement system, has recently been reported to bind to certain pathogenic microorganisms, to early or late apoptotic and necrotic cells, and to particular live human cell lines, thus providing a platform for de novo convertase assembly and complement activation. These studies, with some contradictory results, have been carried out with purified properdin, which forms a series of oligomers of a ~53,000 Mr subunit, assembling into dimers (P2), trimers (P3), tetramers (P4) and higher forms (Pn). The Pn forms have been shown to likely be an artefact of purification that results from procedures including freeze-thawing of properdin. In this study we isolated the individual natural forms of properdin (P2, P3, and P4) and separated them from the Pn forms present in purified frozen properdin using ion exchange and/or size exclusion chromatography. We analyzed the ability of each form to bind to live or necrotic Jurkat and Raji cells, rabbit erythrocytes (ER), and zymosan by FACS analysis. While the unseparated properdin and the purified Pn forms bound to all the surfaces except ER, the physiological P2-P4 forms specifically bound only to zymosan and to necrotic nucleated cells. Our results indicate that aggregated Pn present in unseparated properdin may bind non-specifically to some surfaces and should be separated before analysis in order to obtain meaningful results. Finally, we have determined for the first time that the physiological forms of human properdin can selectively recognize surfaces and enhance or promote complement activation, which is in agreement with the reported role for properdin as a complement initiator.
Keywords: Alternative pathway, Complement, Human, Properdin, C3
Introduction
Properdin (factor P) is a plasma protein that participates in the alternative pathway of complement within the innate immune system. When it was discovered, more than 50 years ago, it was thought to be an initiator of the alternative pathway, acting in a manner that was analogous to antibodies of the classical pathway (Pillemer et al., 1954). Later, this controversial view was replaced by the widely accepted notion that properdin is a positive regulatory factor that facilitates alternative pathway complement activation by extending the half-life of the nascent C3b,Bb convertase by 5-10 fold (Fearon and Austen, 1975; Schreiber et al., 1975; Medicus et al., 1976) and also by increasing C3 convertase generation (Jelezarova and Lutz, 1999). The C3 convertase (C3b,Bb) then rapidly cleaves more C3 to C3b, which act as an opsonin and can reinitiate the pathway in an amplification loop that proceeds on the pathogenic cell. This cascade does not occur on host cells because of enhanced control by factors H and I and membrane-bound complement regulatory proteins (Pangburn and Muller-Eberhard, 1984).
In contrast to the existence of numerous inhibitory proteins, properdin is the only known physiologic positive regulator of the complement activation cascade. Properdin exists as a set of cyclic polymers, in the ratio 26:54:20 of dimers:trimers:tetramers formed by head-to-tail association of monomers (Pangburn, 1989; Smith et al., 1984). Each monomer is ~ 53 kDa (Nolan and Reid, 1990) and is 26 × 2.5 nm by electron microscopy (Smith et al., 1984) and by neutron and X-ray scattering (Smith et al., 1991). Properdin can by synthesized by monocytes (Whaley, 1980), primary T cells (Schwaeble et al., 1993), T-lymphoblastoid cell lines H-9, HuT78, Jurkat, and T-All (Schwaeble et al., 1993), mast cells (Stover et al., 2008), granulocytes (Wirthmueller et al., 1997), bone marrow progenitor cell line HL-60 (Farries and Atkinson, 1989), the monoblastic cell line U-937 (Minta, 1988), monocytic cell line Mono Mac 6 (Schwaeble et al., 1994), and shear-stressed endothelial cells (Bongrazio et al., 2003). In this respect properdin is unlike other complement proteins, which are produced mainly in the liver. The concentration of properdin has been found to be 4-25 μg/ml in serum (Nolan and Reid, 1993; Fijen et al., 1999; Schwaeble and Reid, 1999; Xu et al., 2008; Pangburn, 1989) and properdin synthesis by monocytic cell lines is upregulated by phorbol esters, bacterial LPS, IL-1β, and TNFα (Schwaeble et al., 1994). TNFα, C5a, IL-8, LPS, and other inflammatory agonists also stimulate release of properdin from stored neutrophil granules (Wirthmueller et al., 1997). Properdin deficiency, a recessive X-linked genetic disorder (Fredrikson et al., 1996; Goundis and Reid, 1988; Westberg et al., 1995), results in defective alternative pathway function, resulting in impaired bactericidal activity. Thus, properdin-deficient patients can be highly susceptible to fulminant meningococcal infections (Sjoholm et al., 1982; Braconier et al., 1983) and have been recently shown to be more susceptible to recurrent pneumonia and otitis media (Schejbel et al., 2009).
Hourcade (Hourcade, 2006) has shown that properdin, non-specifically bound to a biosensor surface, has the ability to bind C3b and the bound C3b,B,P complex can be cleaved by factor D. In addition, various studies, some with contradictory results, have recently shown that properdin can directly bind to non-self surfaces such as zymosan, rabbit erythrocytes (E R), Neisseria gonorrhoeae (Spitzer et al., 2007), and certain E. coli strains (Stover et al., 2008; Spitzer et al., 2007), as well as to “dangerous self” surfaces of early (Kemper et al., 2007; Kemper et al., 2008) or late (Xu et al., 2008) apoptotic and necrotic cells (Xu et al., 2008), as well as to live human leukemia T cell lines (Kemper et al., 2008), and even to normal human proximal tubular epithelial cells (Gaarkeuken et al., 2008) and Chinese hamster ovary cells (Kemper et al., 2008), in the absence of C3b (Xu et al., 2008; Stover et al., 2008). The bound properdin has been further shown to be capable of serving as a platform for de novo C3b,Bb assembly leading to C3 cleavage and complement activation on these surfaces (Spitzer et al., 2007; Gaarkeuken et al., 2008; Kemper et al., 2008; Xu et al., 2008; Kemper et al., 2009). The data from these studies are consistent with the complement initiation function proposed over 50 years ago (Pillemer et al., 1954) and have re-opened the controversy regarding the functions of properdin.
Two independent studies (Farries et al., 1987; Pangburn, 1989) from ~ 20 years ago indicated that purified properdin could accumulate in an aggregated form (Pn) during purification and storage. This Pn form, also known as “activated properdin”, has the ability to induce complement activation and consumption in solution, while retaining the ability to stabilize alternative pathway convertases (Pangburn, 1989). Considering the importance of the recent findings that implicate properdin as a complement initiator on surfaces, we sought to separate the physiological forms of properdin from these non-physiological aggregates and determine their ability to bind to promote complement activation on surfaces.
Materials and methods
Buffers and serum
The buffers used were: PBS (10 mM sodium phosphate, 140 mM NaCl, 0.02% NaN3, pH 7.4); veronal buffered saline (VBS, 5 mM veronal, 145 mM NaCl, 0.02% NaN3, pH 7.3); GVB (VBS containing 0.1% gelatin); GVBE (GVB containing 10 mM EDTA); MgEGTA (0.1 M MgCl2, 0.1 M EGTA, pH 7.3); Hank's buffered saline solution (HBSS; Gibco). Normal human serum (NHS) was isolated from human blood collected from two healthy donors by venipuncture. The University of Texas Health Science Center institutional review board approved protocols, and written informed consent was obtained from all human donors. P-depleted serum was generated by immunoadsorption as previously described (Pangburn, 1989).
Proteins
Complement protein properdin was purified from normal human plasma as previously described (Pangburn, 1989), except the plasma was not PEG-precipitated before passing it over the anti-properdin-Sepharose column. Contaminating C1q was removed by passing the sample through an anti-C1q-Sepharose column. The properdin was analyzed by 10% SDS-PAGE, the concentration of the protein was determined spectrophotometrically using an A280nm (1% solution) of 18 (Reid, 1981), and the protein was stored at −80°C.
C3 was purified as described previously (Hammer et al., 1981; Pangburn, 1987) and for some experiments, was labeled with Alexa fluor 488 (Molecular Probes) following manufacturer's instructions.
Separation of native properdin polymeric forms from aggregates
For each set of experiments, pure, frozen properdin was thawed and the physiological P2-P4 forms were separated from the aggregated Pn forms by cation exchange chromatography and/or by gel filtration, as previously described (Pangburn, 1989). Briefly, for separating by charge, 2-5 mg of properdin was diluted with buffer A (50 mM sodium phosphate, pH 6) and loaded onto a 1 ml Mono S column (GE Healthcare). The column was washed with 20% buffer B (50 mM sodium phosphate. 0.5 M NaCl, pH 6) and the properdin eluted with a 20 ml gradient from 20 to 45% buffer B. For separation of the forms of properdin by size, the protein was purified by gel filtration on a BioSep-SEC-S4000 column with guard column (Phenomenex). The thawed properdin sample, in PBS, was loaded onto the 7.8 × 600 mm column and eluted at a flow rate of 0.5 ml/min.
Properdin binding assays
In order to determine if the P2, P3, P4, and Pn forms of properdin bind to alternative pathway activators, zymosan (Sigma) (2×106) or ER (1×106) were incubated with 5 μg properdin (purified by ion exchange chromatography, as described above) in 120 μl GVB with 5 mM MgEGTA for 1 hr at 37°C. The cells were then washed and analyzed by FACS with anti-properdin monoclonal antibody No. 1 or 2 (Quidel) or an IgG1 murine monoclonal antibody isotype control (eBiosciences), followed by an FITC-conjugated, goat anti-mouse IgG antibody (Sigma). Alternatively, the FACs analysis was carried out on the cells treated with goat anti-properdin polyclonal antibody, followed by FITC-donkey-anti goat IgG (Abcam).
The binding of the various forms of properdin (P2-P4 and Pn) to live or necrotic Jurkat (T cell lymphocytic leukemia cell line; ATCC) or Raji (Burkitt's lymphoma B cell lymphoma cell line; ATCC) cells was assessed by incubating 1×106 cells with 4 μg of the different forms of properdin purified by ion exchange chromatography-purified in 120 μl HBSS (with 1.26 mM Ca+2 and 0.49 mM Mg+2; Gibco) for 1 hr at 37°C. The cells were then washed and analyzed by FACS as described above. Necrotic cells were induced by heating the cells for 30 min at 56°C before incubation with the forms of properdin. In assays where P3 forms were used, the P3 form was purified by ion exchange chromatography, followed by size exclusion chromatography, as described above in order to thoroughly eliminate any possible contaminating Pn forms.
Assays to measure properdin-dependent complement activation on surfaces
In order to determine if the physiological forms of properdin, when bound to zymosan, induce complement activation, the P3 form was purified from the unseparated properdin by cation exchange chromatography, followed by size exclusion chromatography, as described above. Zymosan (1×106) was incubated with or without 5 μg P3 in 120 μl GVB with 5 mM MgEGTA for 30 min, at 37°C. The cells were then washed and incubated with 100 μl of 20% P-depleted serum to which 7.5 μg of Alexa fluor 488-labeled C3 was added per reaction, in the presence of GVB with 5mM MgEGTA or with 10 mM EDTA, for 30 min at 37°C. The particles were then washed with cold GVBE and C3b deposition was assessed by FACS. A similar set of experiments to those described above were carried out except that the zymosan was incubated with a pool of P2-P4 or Pn that were separated by size. C3b deposition was measured after properdin binding at different time points after adding the P-depleted serum, using an FITC-labeled goat anti-C3 antibody (Cappel).
To determine if the physiological forms of properdin P2-P4 induce alternative pathway complement activation, when bound to necrotic cells, necrotic Raji and Jurkat cells (1×106) were incubated for 30 min at 37°C with or without 40 μg/ml P2-P4 or Pn forms that were separated by size, in 100 μl HBSS with 5 mM MgEGTA. The cells were then washed and incubated with 100 μl of 20% P-depleted serum in the presence of 5 mM Mg-EGTA or 10 mM EDTA for 15 minutes, at 37°C. Complement activation was stopped by washing with cold HBSS containing 10mM EDTA and deposition of C3b was determined as described above.
Results
Separation of the polymeric forms of properdin
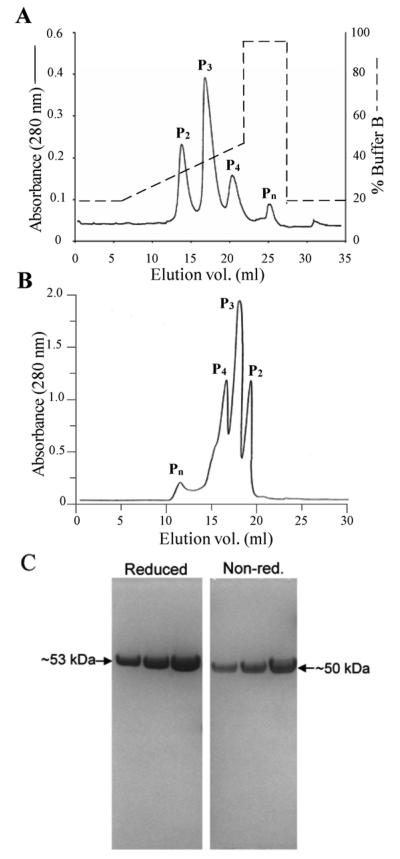
Almost 20 years ago, the Pn forms were shown to likely be an artefact of purification that results from procedures used to purify properdin and from the process of freezing and thawing it (Pangburn, 1989; Farries et al., 1987). The Pn form, also known as “activated properdin”, has the ability to spontaneously activate complement in serum (Farries et al., 1987; Pangburn, 1989). Recent findings implicate properdin as a complement initiator on surfaces (Gaarkeuken et al., 2008; Kemper et al., 2008; Spitzer et al., 2007; Xu et al., 2008), but all this work has been done with frozen and thawed properdin. Considering this and the importance of these findings, we sought to separate the physiological forms of human properdin and determine their ability to bind to and initiate complement activation on different surfaces. For this we isolated the individual natural forms of properdin (P2, P3, and P4) from purified properdin that had been stored at −80°C, by using ion exchange chromatography (Fig. 1A) and/or size exclusion chromatography (Fig. 1B). Both methods separate the natural polymers of properdin found in plasma, the dimers (P2), trimers (P3) and tetramers (P4), from higher polymers (Pn) thought to be artefacts of purification and storage (Pangburn, 1989). The pure unseparated properdin was greater than 99% homogenous by polyacrylamide gel electrophoresis, running as a single band with an apparent m.w. on SDS gel electrophoresis of 53,000 Da in its reduced form, and ~50,000 Da when non-reduced (Fig. 1C), in agreement with previous findings (Farries et al., 1987; Pangburn, 1989). As quality control of the individually isolated forms of properdin, each form was assessed in its ability to act as a stabilizer of the alternative pathway convertases (“native properdin” function) in an alternative pathway-mediated hemolytic assay, as previously described (Pangburn, 1989), but using ER instead of neuraminidase-treated sheep erythrocytes. The ability of P2, P3, or P4 to reconstitute P-depleted serum in complement assays was over 80% that of native properdin found in pooled normal human serum, while Pn conserved less than 14% of this activity (not shown). The low activity of the Pn forms was likely due to Pn-induced complement activation and consumption in the fluid phase, as has been shown previously (Pangburn, 1989; Farries et al., 1987).
Fig. 1.
Separation of the polymeric forms of properdin. The pure, frozen properdin sample was thawed and separated by: (A), Cation exchange chromatography on a Mono S column. The sample was diluted with buffer A (50 mM sodium phosphate, pH 6) and loaded onto a 1 ml Mono S column. The column was washed with 20% buffer B (50 mM sodium phosphate. 0.5 M NaCl, pH 6) and the various forms of properdin were eluted with a 20 ml gradient from 20 to 45% buffer B. (B) Subsequently, or alternatively, the thawed properdin sample, in PBS, was loaded onto a Phenomenex BioSep-SEC-S4000 gel filtration column (7.8 × 600 mm) and was eluted at a flow rate of 0.5 ml/min. (C) 10% SDS-PAGE of 2, 4 or 8 μg of pure properdin under reducing (left) and non-reducing (right) conditions.
Binding of properdin to alternative pathway activators
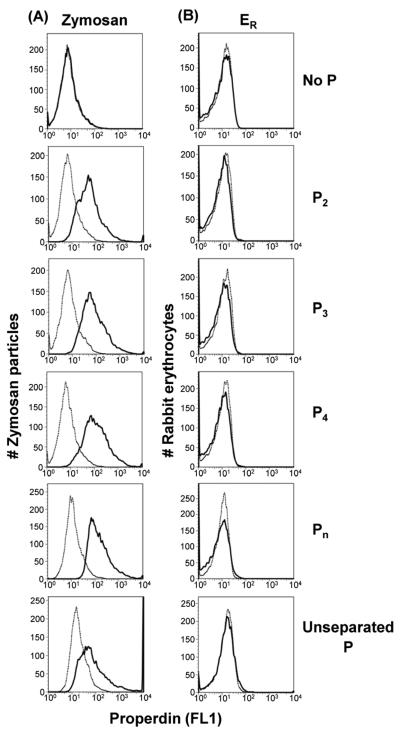
Others have reported that purified properdin binds to and activates complement on both zymosan and ER (Spitzer et al., 2007), two strong alternative pathway activators. We analyzed the ability of each form of properdin to bind to these activators and Figure 2 shows that P2, P3, and P4, the physiologically relevant forms of properdin, as well as Pn, specifically bound to zymosan (Fig. 2A), but not to ER (Fig. 2B). The binding of properdin to ER was assessed using two different anti-properdin monoclonal antibodies from Quidel (anti-P#1 used in Fig. 2B or anti-P#2, not shown) as well as an anti-properdin polyclonal antibody (not shown), and none of these antibodies detected binding of any of the forms of properdin. These results indicate that all forms of properdin show specificity for certain alternative pathway activators and not for others.
Fig. 2.
The P2, P3, P4, and Pn forms of properdin bind to the alternative pathway activator zymosan, but not to ER. (A) Zymosan or (B) ER were incubated with 5 μg properdin (purified by ion exchange chromatography as shown in Fig. 1) in 120 μl GVB with 5 mM MgEGTA for 1 hr at 37°C. The zymosan and ER were then washed and analyzed by FACS with an anti-properdin monoclonal antibody (solid line) or an IgG1 monoclonal antibody isotype control (dotted line), followed by an FITC-conjugated anti-mouse IgG antibody. Results shown are representative of four independent experiments.
Physiological forms of properdin (P2-P4) bound to zymosan promote complement activation
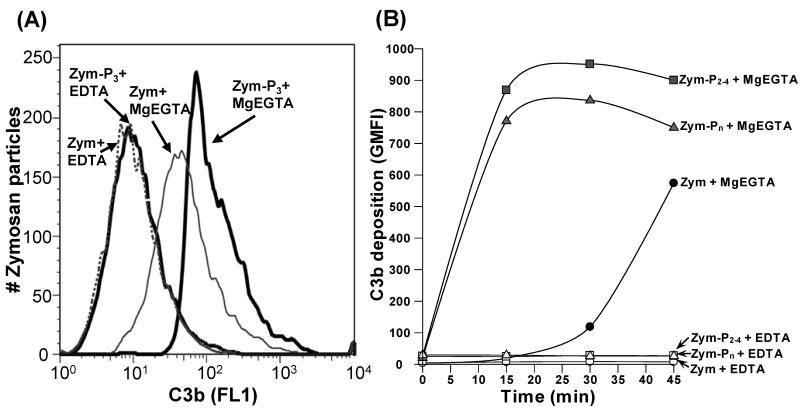
Although others have shown that purified properdin bound to and activated complement on zymosan and ER (Spitzer et al., 2007), this properdin contained Pn which is known to spontaneously activate complement in serum without activating particles (Farries et al., 1987; Pangburn, 1989). Native properdin (P2 - P4) in serum does not activate complement in the absence of an activating particle (Farries et al., 1987; Pangburn, 1989). Thus, we sought to determine if bound native properdin had the ability to activate complement. Zymosan was incubated with or without the highly purified P3 form of properdin. After washing, the particles were exposed to P-depleted serum at 37°C. Figure 3A shows that zymosan bearing purified P3 activated the alternative pathway of complement almost 3-fold more aggressively (measured as C3b deposition) than zymosan lacking bound properdin. A time course of C3b attachment (Fig. 3B) provided more evidence for the ability of bound native forms of properdin to accelerate the activation of complement. The activation of complement on the properdin-coated particles was measured at varying time points (Fig. 3B) by measuring the mean fluorescence intensity of deposited Alexa fluor 488-C3b on the zymosan particles. This graph shows that both native and the aggregated forms of properdin activated complement very rapidly compared to the rate of activation in the absence of properdin. These data are consistent with the possible role of native properdin as a de novo complement activator on certain microorganisms.
Fig. 3.
Physiological forms of properdin (P2, P3, P4), when bound to zymosan, promote complement activation. (A) The P3 form of properdin was separated from the properdin pool by cation exchange chromatography, followed by size exclusion chromatography. Zymosan was incubated with 5 μg P3 (Zym-P3) or without P3 (Zym) in 120 μl GVB-MgEGTA for 30 min, at 37°C. The cells were then washed and incubated with P-depleted serum containing Alexa fluor 488-labeled C3, in the presence of GVB with MgEGTA or with EDTA, for 30 min at 37°C. The particles were then washed and C3b deposition was assessed by FACS. (B) same as (A), except the zymosan was incubated with a pool of P2-P4 or Pn that were purified by size, and C3b deposition was measured at different time points after adding the properdin-depleted serum, using an FITC-labeled anti-C3 antibody. Results shown are representative of three independent experiments.
The physiologic polymeric forms of properdin bind to necrotic cells, but not to live cells
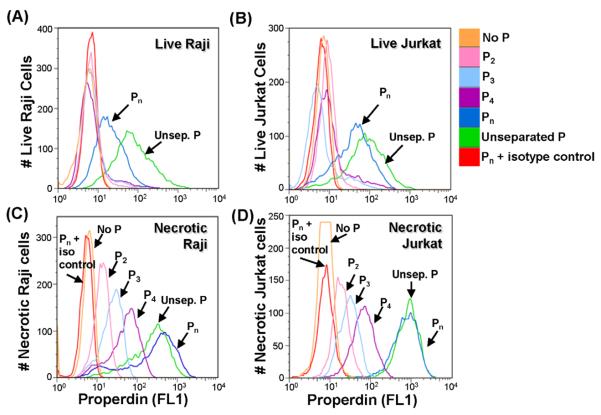
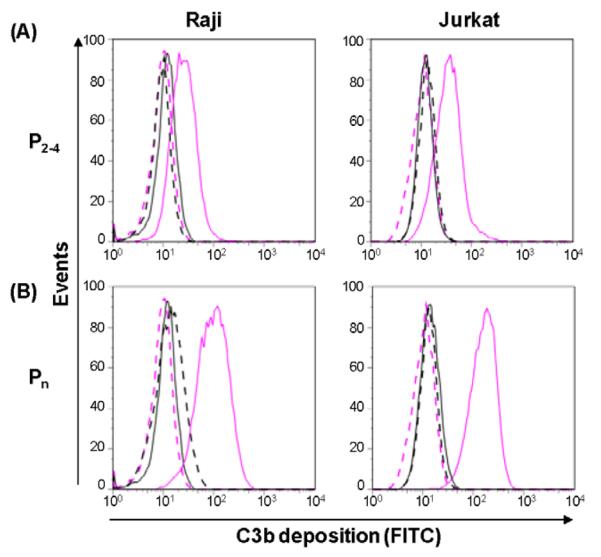
It has been reported that properdin binds to and activates complement on early (Kemper et al., 2008) or late (Xu et al., 2008) apoptotic and necrotic (Xu et al., 2008) cells, as well as on various live cell lines (Gaarkeuken et al., 2008; Kemper et al., 2008) including Jurkat cells (human T cell leukemia line). Concerned that the high molecular weight forms of properdin might be responsible for these effects we tested the ability of each of the purified forms of properdin to bind to live nucleated human Raji B cells and Jurkat T cells. Figures 4A and 4B show that the aggregated Pn form of properdin is the only form of properdin that significantly bound to these live cell lines. The ability of the unseparated properdin to achieve higher levels of binding to live nucleated cells, vs the purified Pn forms, is likely to be due to the presence of higher order oligomers that are not eluted off the ion exchange column at 0.5 M NaCl, or that cannot be washed off the size exclusion matrix. Nevertheless, the results demonstrate that none of the native forms of properdin bound to live cells (Fig. 4A and 4B).
Fig. 4.
Unseparated properdin and Pn bind to live cells, but physiological forms of properdin (P2, P3, and P4) only bind to necrotic Raji and Jurkat cells. (A) live Raji or (B) live Jurkat (1×106) were incubated with 4 μg of the different forms of properdin (purified by ion exchange chromatography as shown in Fig. 1) in 120 μl HBSS for 1 hr at 37°C. The cells were then washed and analyzed by FACS with an anti-properdin monoclonal antibody or an IgG1 monoclonal antibody isotype control, followed by an FITC-conjugated anti-mouse IgG antibody. (C) and (D), same as (A) and (B), respectively, except the cells underwent necrosis by heating for 30 min at 56°C before incubating them with the different forms of properdin. Results shown are representative of two independent experiments.
We also tested the ability of the physiological forms of properdin to bind to necrotic cells. Fig 4C and 4D show that, in this scenario, the physiological P2-P4 forms of properdin specifically bound to the necrotic cells, even though they did not bind to the live cells (Fig. 4A and 4B).
Bound properdin enhances complement activation on necrotic cells
Figure 5 shows that the bound native forms of properdin were able to promote complement and rapidly deposit C3b on necrotic Raji and Jurkat cells. Figure 5A demonstrates that both cell types with native P2-P4 bound to their surface (solid pink lines), acquired surface-bound C3b when incubated in P-depleted human serum. The cells lacking properdin (Fig. 5A, solid black lines) showed no detectable C3b deposition. The aggregated forms of properdin, which spontaneously activate and consume complement in serum (Farries et al., 1987; Pangburn, 1989), were also capable of binding to and activating complement on necrotic cells in P-depleted serum (Fig. 5B) Nevertheless, these forms cannot be present in blood or they would produce hypocomplementemia due to C3 consumption in serum. Binding of native properdin to necrotic cells may play an important role in necrotic cell clearance which has been shown to involve complement activation (Surh and Sprent, 1994; Munoz et al., 2005).
Fig. 5.
Physiological forms of properdin promote complement activation on necrotic cells. Necrotic Raji (left panel) and Jurkat cells (right panel) (1×106) were incubated for 30 min at 37°C with (pink lines) and without (black lines) P2-4 (A) or Pn (B) forms of properdin in HBSS. The cells were then washed and incubated with P-depleted serum in the presence of 5 mM MgEGTA (solid lines) or EDTA (dashed lines) for 15 minutes at 37°C. Complement activation was stopped by washing with cold HBSS containing EDTA and the deposition of C3b was determined by flow cytometry using an FITC-labeled anti-C3 antibody. Results shown are representative of three independent experiments.
Discussion
The data presented in this study show that the physiological P2-P4 forms of properdin specifically bind to zymosan and to necrotic cells and initiate complement activation on these surfaces, but do not bind to ER, another strong alternative pathway activator, nor to live cells. This conclusion is supported by the observation that non-physiological Pn forms (“activated properdin”) that are commonly present in pure properdin preparations and can induce complement activation and consumption in solution (Farries et al., 1987; Pangburn, 1989), can bind non-specifically to live nucleated cells. This study is the first to show that the purified physiological forms of properdin, in the absence of Pn, can bind to and promote complement activation when directly bound to certain surfaces. Our results provide evidence to support the proposal of properdin as a selective alternative pathway recognition molecule, as proposed by Pillemer over 50 years ago (Pillemer et al., 1954) and more recently by others (Kemper et al., 2007; Xu et al., 2008; Kemper et al., 2008; Gaarkeuken et al., 2008; Kimura et al., 2008).
In our study we have used properdin that was purified by affinity chromatography and anion exchange (Pangburn, 1989) from normal human plasma. We observed a single band under reduced and non-reducing conditions with fresh and freeze-thawed preparations (Fig 1C), which corresponds to a properdin monomer (Nolan and Reid, 1990; Pangburn, 1989; Smith et al., 1984). Immuno-western blot carried out with non-reduced pure properdin, using an anti-properdin monoclonal antibody for detection, also revealed a single band at the same molecular weight (not shown). To our knowledge, no evidence of disulfide-linked or covalently coupled properdin aggregates have been shown for purified properdin. Yet recent reports (Xu et al., 2008; Gaarkeuken et al., 2008), have cited the use of purified properdin that yielded a single 200 kDa band under non-reducing conditions in the presence of SDS. Although the source, form, or purity of the properdin used in their (Xu et al., 2008; Gaarkeuken et al., 2008) and our study must differ, our studies do agree with the ability of properdin to bind to necrotic cells and promote complement activation.
There are conflicting reports in the literature regarding preferential targets for properdin (Kemper et al., 2008; Xu et al., 2008). For example, Kemper et al. (Kemper et al., 2008) determined that properdin binds to apoptotic primary T cells and to live Jurkat (an immortalized cancerous cell line) and CHO cells, but not to necrotic cells or to live primary cells. Our data and that of Xu et al. (Xu et al., 2008), agree with the conclusion that properdin binds to necrotic cells of different origins. It is important to note that Pn (the aggregated form of properdin) contributed significantly to the overall binding detected to both the necrotic and live Raji and Jurkat cells (Fig. 4) and to complement activation on the cells (Fig. 5). Nevertheless, the physiological forms (P2, P3, P4) bind specifically only to the necrotic cells. Some of these discrepancies may be explained at least in part by the source of the properdin used, the level of Pn forms that may or may not have been present in the preparations, as well as the cell type characteristics. For instance, in our study, unseparated properdin achieved even higher levels of binding to live nucleated cells, vs the purified Pn forms (Figs. 4A and 4B), possibly due to the presence of higher order oligomers that could not be eluted from the purification columns.
P2, P3, and P4, the predominant physiological forms of properdin in serum, specifically bind zymosan, but not to another strong alternative pathway activator, ER (Fig. 2). Although others have recently observed binding of properdin to ER (Spitzer et al., 2007), we did not detect binding even of the Pn forms of properdin under similar experimental conditions. This was confirmed with two different anti-properdin monoclonal antibodies and a polyclonal antibody used for detecting purposes. Our data indicate selectivity of properdin for certain AP activators and not others. Other examples of this selectivity include our recent findings that show that physiological P2-P4 forms of human properdin bind to, and promote complement activation on, the surface of Chlamydia pneumoniae (Cortes et al., unpublished data), but do not bind to Neisseria sp, even though the non-physiological Pn forms were found to efficiently bind to this organism (Agarwal et al., 2010).
Kemper, et al (Kemper et al., 2008) have reported substantial evidence to support the binding of properdin to early apoptotic T cells since the properdin that was determined to bind (in the absence C3 binding) was freshly released from co-cultured neutrophils and was thus not likely to have significant amounts of non-native properdin. Thus, properdin binding to apoptotic primary T cells may indeed be a characteristic particular to that cell type. Whether the cells are apoptotic or necrotic, they must be quickly eliminated from tissues in order to prevent further damage. Under physiological conditions, the clearance of dying and dead cells is tightly regulated by a highly redundant system of receptors on phagocytic cells and bridging molecules that detect molecules specific for dying cells. The complement system is known to play an important role in the clearance of dead cells through opsonization and promotion of phagocytosis (Taylor et al., 2000; Flierman and Daha, 2007; Trouw et al., 2008; Lutz et al., 2009). Our results, in agreement with those of others (Kemper et al., 2008; Xu et al., 2008), show that binding of properdin and activation of the alternative pathway of complement on dead or dying cells may also play an important role. The reports indicating that properdin, in the context of complete or C3-deficient serum, is able to bind to the surface of late apoptotic and necrotic cells (Xu et al., 2008) and to E. coli (Stover et al., 2008), support this conclusion.
In our hands, the binding of properdin to zymosan and necrotic cells was inhibited by NHS in a dose-dependent manner (not shown). It is therefore possible that one or more inhibitors of this interaction may exist in serum, such as Amyloid P (Mitchell and Hourcade, 2008) or C3b2-natural IgG complexes that have been shown to stimulate complement amplification in a properdin-dependent manner (Lutz, 2004; Lutz et al., 2009). Nevertheless, properdin is produced by different circulating cell types including peripheral blood monocytes, T cells, and neutrophils (Schwaeble et al., 1993; Whaley, 1980; Wirthmueller et al., 1997). These cells can release properdin upon stimulation, and may significantly increase the local concentration of properdin, especially at sites of inflammation (Schwaeble and Reid, 1999). Native properdin that emerges from neutrophil granules has been shown to be capable of binding specifically to apoptotic T cells in vitro (Kemper et al., 2008). Thus, selected microorganisms, fungi (i.e. zymosan) and necrotic/late apoptotic human cells may acquire properdin directly from the cells that make it provided that interaction occurs close to the site of granule release. The progressive inactivation of this property of nascent properdin may protect blood cells and distant tissues from complement-mediated damage.
Recently, new interactions and possible functions have been proposed for properdin based on its ability to interact with surfaces including binding to certain live cells (Kemper et al., 2008; Gaarkeuken et al., 2008), early (Kemper et al., 2008) or late (Xu et al., 2008) apoptotic, or necrotic (Xu et al., 2008) cells, binding to certain alternative pathway activating particles (ER (Spitzer et al., 2007), zymosan (Spitzer et al., 2007), E. coli (Spitzer et al., 2007; Stover et al., 2008), and Neisseria (Spitzer et al., 2007)), binding to DNA (Xu et al., 2008), glycosaminoglycans (Kemper et al., 2008), bacterial LPS, and lipooligosaccharide (Kimura et al., 2008). In many cases, properdin-initiated complement activation was proposed. In addition, direct opsonization of apoptotic T cells by purified properdin results in ingestion by macrophages in the absence of complement activation (Kemper et al., 2008). Purified properdin has also been recently found to induce the formation of platelet-leukocyte aggregates (Ruef et al., 2008). All these studies, which attribute significant and diverse roles to properdin, were done, at least in part, with purified properdin, which usually contains non-physiological aggregates (Farries et al., 1987; Pangburn, 1989). The data presented herein demonstrate non-specific binding of non-physiological aggregates (Pn and unseparated properdin) to live cells. It is therefore possible that the presence of the Pn forms in purified preparations of properdin could yield misleading binding results and may explain some of the recently reported contradictory observations. It is important to carry out studies with native properdin in order to not overestimate the contribution of surface-bound properdin to complement activation and to effectively determine specific interactions between the physiological forms and as yet to be defined cellular targets for properdin binding.
Acknowledgments
This research was supported by National Institutes of Health Grant DK-35081 (M.K.P.) and American Heart Association National Scientist Development Grant 0735101N (V.P.F.). The authors express their appreciation to Staci Snyder and Connie Elliot for their excellent technical assistance.
Abbreviations used
- Factor P
properdin
- ER
rabbit erythrocytes
- P2, P3, and P4
properdin species composed of two, three, or four 53,000 -Da subunits, respectively
- Pn
high m.w. properdin polymers
- VBS
Veronal-buffered saline
- GVB
VBS containing 0.1% gelatin
- GVBE
GVB containing 10 mM EDTA
- MgEGTA
equimolar MgCl2 and EGTA
- NHS
normal human serum
- HBSS
Hank's buffered saline solution
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure: One of the authors (M.K.P.) is an officer of and has a financial interest in Complement Technology, Inc. (www.ComplementTech.com), a supplier of complement reagents.
References
- Agarwal S, Ferreira V, Cortes C, Pangburn MK, Rice PA, Ram S. An evaluation of the role of properdin in alternative pathway activation on Neisseria meningitidis and Neisseria gonorrhoeae. J. Immunol. 2010 doi: 10.4049/jimmunol.0903598. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongrazio M, Pries AR, Zakrzewicz A. The endothelium as physiological source of properdin: role of wall shear stress. Mol. Immunol. 2003;39:669–675. doi: 10.1016/s0161-5890(02)00215-8. [DOI] [PubMed] [Google Scholar]
- Braconier JH, Sjoholm AG, Soderstrom C. Fulminant meningococcal infections in a family with inherited deficiency of properdin. Scand. J. Infect. Dis. 1983;15:339–345. doi: 10.3109/inf.1983.15.issue-4.04. [DOI] [PubMed] [Google Scholar]
- Farries TC, Atkinson JP. Biosynthesis of properdin. J. Immunol. 1989;142:842–847. [PubMed] [Google Scholar]
- Farries TC, Finch JT, Lachmann PJ, Harrison RA. Resolution and analysis of ‘native’ and ‘activated’ properdin. Biochem. J. 1987;243:507–517. doi: 10.1042/bj2430507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon DT, Austen KF. Properdin: Binding to C3b and stabilization of the C3bdependent C3 convertase. J. Exp. Med. 1975;142:856–863. doi: 10.1084/jem.142.4.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fijen CA, van den BR, Schipper M, Mannens M, Schlesinger M, Nordin FG, Dankert J, Daha MR, Sjoholm AG, Truedsson L, Kuijper EJ. Properdin deficiency: molecular basis and disease association. Mol. Immunol. 1999;36:863–867. doi: 10.1016/s0161-5890(99)00107-8. [DOI] [PubMed] [Google Scholar]
- Flierman R, Daha MR. The clearance of apoptotic cells by complement. Immunobiology. 2007;212:363–370. doi: 10.1016/j.imbio.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Fredrikson GN, Westberg J, Kuijper EJ, Tijssen CC, Sjoholm AG, Uhlen M, Truedsson L. Molecular characterization of properdin deficiency type III: dysfunction produced by a single point mutation in exon 9 of the structural gene causing a tyrosine to aspartic acid interchange. J. Immunol. 1996;157:3666–3671. [PubMed] [Google Scholar]
- Gaarkeuken H, Siezenga MA, Zuidwijk K, van KC, Rabelink TJ, Daha MR, Berger SP. Complement activation by tubular cells is mediated by properdin binding. Am. J. Physiol Renal Physiol. 2008;295:F1397–F1403. doi: 10.1152/ajprenal.90313.2008. [DOI] [PubMed] [Google Scholar]
- Goundis D, Reid KBM. Properdin, the terminal complement components, thrombospondin and the circumsporozoite protein of malaria parasites contain similar sequence motifs. Nature. 1988;335:82–85. doi: 10.1038/335082a0. [DOI] [PubMed] [Google Scholar]
- Hammer CH, Wirtz GH, Renfer L, Gresham HD, Tack BF. Large scale isolation of functionally active components of the human complement system. J. Biol. Chem. 1981;256:3995–4006. [PubMed] [Google Scholar]
- Hourcade DE. The role of properdin in the assembly of the alternative pathway C3 convertases of complement. J. Biol. Chem. 2006;281:2128–2132. doi: 10.1074/jbc.M508928200. [DOI] [PubMed] [Google Scholar]
- Jelezarova E, Lutz HU. Assembly and regulation of the complement amplification loop in blood: the role of C3b-C3b-IgG complexes. Mol. Immunol. 1999;36:837–842. doi: 10.1016/s0161-5890(99)00104-2. [DOI] [PubMed] [Google Scholar]
- Kemper C, Atkinson JP, Hourcade DE. Properdin: Emerging Roles of a Pattern-Recognition Molecule. Annu. Rev. Immunol. 2009 doi: 10.1146/annurev-immunol-030409-101250. doi:10.1146/annurev-immunol-030409-101250. [DOI] [PubMed] [Google Scholar]
- Kemper C, Mitchell LM, Hourcade DE. Properdin: Cell Death's Little Helper? J. Immunol. 2007;178:53.3. Abstract. [Google Scholar]
- Kemper C, Mitchell LM, Zhang L, Hourcade DE. The complement protein properdin binds apoptotic T cells and promotes complement activation and phagocytosis. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9023–9028. doi: 10.1073/pnas.0801015105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Miwa T, Zhou L, Song WC. Activator-specific requirement of properdin in the initiation and amplification of the alternative pathway complement. Blood. 2008;111:732–740. doi: 10.1182/blood-2007-05-089821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz HU. Innate immune and non-immune mediators of erythrocyte clearance. Cell Mol. Biol. (Noisy. -le-grand) 2004;50:107–116. [PubMed] [Google Scholar]
- Lutz HU, Binder CJ, Kaveri S. Naturally occurring auto-antibodies in homeostasis and disease. Trends Immunol. 2009;30:43–51. doi: 10.1016/j.it.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Medicus RG, Schreiber RD, Gotze O, Muller-Eberhard HJ. A molecular concept of the properdin pathway. Proc. Natl. Acad. Sci. USA. 1976;73:612–616. doi: 10.1073/pnas.73.2.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minta JO. Biosynthesis of complement factor P (properdin) by the human pre-monocyte cell line (U-937) Mol. Immunol. 1988;25:1363–1370. doi: 10.1016/0161-5890(88)90052-1. [DOI] [PubMed] [Google Scholar]
- Mitchell L, Hourcade D. Inhibition of properdin-directed complement activation by serum amyloid P component. Mol Immunol. 2008;45:4103. Mol. Immunol. 45, 4103. [Google Scholar]
- Munoz LE, Gaipl US, Franz S, Sheriff A, Voll RE, Kalden JR, Herrmann M. SLE--a disease of clearance deficiency? Rheumatology. (Oxford) 2005;44:1101–1107. doi: 10.1093/rheumatology/keh693. [DOI] [PubMed] [Google Scholar]
- Nolan KF, Reid KB. Complete primary structure of human properdin: a positive regulator of the alternative pathway of the serum complement system. Biochem. Soc. Trans. 1990;18:1161–1162. doi: 10.1042/bst0181161. [DOI] [PubMed] [Google Scholar]
- Nolan KF, Reid KB. Properdin. Methods Enzymol. 1993;223:35–46. doi: 10.1016/0076-6879(93)23036-m. [DOI] [PubMed] [Google Scholar]
- Pangburn MK. A fluorimetric assay for native C3. The hemolytically active form of the third component of human complement. J. Immunol. Methods. 1987;102:7–14. doi: 10.1016/s0022-1759(87)80003-0. [DOI] [PubMed] [Google Scholar]
- Pangburn MK. Analysis of the natural polymeric forms of human properdin and their functions in complement activation. J. Immunol. 1989;142:202–207. [PubMed] [Google Scholar]
- Pangburn MK, Muller-Eberhard HJ. The alternative pathway of complement. Springer Seminars in Immunopathology. 1984;7:163–192. doi: 10.1007/BF01893019. [DOI] [PubMed] [Google Scholar]
- Pillemer L, Blum L, Lepow IH, Ross OA, Todd EW, Wardlaw AC. The properdin system and immunity. I. Demonstration and isolation of a new serum protein, properdin, and its role in immune phenomena. Science. 1954;120:279–285. doi: 10.1126/science.120.3112.279. [DOI] [PubMed] [Google Scholar]
- Reid KBM. Preparation of human properdin. Methods Enzymol. 1981;80:143–150. [Google Scholar]
- Ruef J, Kuehnl P, Meinertz T, Merten M. The complement factor properdin induces formation of platelet-leukocyte aggregates via leukocyte activation. Platelets. 2008;19:359–364. doi: 10.1080/09537100802105040. [DOI] [PubMed] [Google Scholar]
- Schejbel L, Rosenfeldt V, Marquart H, Valerius NH, Garred P. Properdin deficiency associated with recurrent otitis media and pneumonia, and identification of male carrier with Klinefelter syndrome. Clin. Immunol. 2009;131:456–462. doi: 10.1016/j.clim.2009.02.008. [DOI] [PubMed] [Google Scholar]
- Schreiber RD, Medicus RG, Gotze O, Muller-Eberhard HJ. Properdin- and nephritic factor-dependent C3 convertases: Requirement of native C3 for enzyme formation and the function of bound C3b as properdin receptor. J. Exp. Med. 1975;142:760–772. doi: 10.1084/jem.142.3.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaeble W, Dippold WG, Schafer MK, Pohla H, Jonas D, Luttig B, Weihe E, Huemer HP, Dierich MP, Reid KB. Properdin, a positive regulator of complement activation, is expressed in human T cell lines and peripheral blood T cells. J. Immunol. 1993;151:2521–2528. [PubMed] [Google Scholar]
- Schwaeble W, Huemer HP, Most J, Dierich MP, Strobel M, Claus C, Reid KB, Ziegler-Heitbrock HW. Expression of properdin in human monocytes. Eur. J. Biochem. 1994;219:759–764. doi: 10.1111/j.1432-1033.1994.tb18555.x. [DOI] [PubMed] [Google Scholar]
- Schwaeble WJ, Reid KB. Does properdin crosslink the cellular and the humoral immune response? Immunol. Today. 1999;20:17–21. doi: 10.1016/s0167-5699(98)01376-0. [DOI] [PubMed] [Google Scholar]
- Sjoholm AG, Braconier J, Soderstrom C. Properdin deficiency in a family with fulminant meningococcal infections. Clin. Exp. Immunol. 1982;50:291–297. [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Pangburn MK, Vogel C-W, Muller-Eberhard HJ. Molecular architecture of human properdin, a positive regulator of the alternative pathway of complement. J. Biol. Chem. 1984;259:4582–4588. [PubMed] [Google Scholar]
- Smith KF, Nolan KF, Reid KB, Perkins SJ. Neutron and X-ray scattering studies on the human complement protein properdin provide an analysis of the thrombospondin repeat. Biochemistry. 1991;30:8000–8008. doi: 10.1021/bi00246a018. [DOI] [PubMed] [Google Scholar]
- Spitzer D, Mitchell LM, Atkinson JP, Hourcade DE. Properdin can initiate complement activation by binding specific target surfaces and providing a platform for de novo convertase assembly. J. Immunol. 2007;179:2600–2608. doi: 10.4049/jimmunol.179.4.2600. [DOI] [PubMed] [Google Scholar]
- Stover CM, Luckett JC, Echtenacher B, Dupont A, Figgitt SE, Brown J, Mannel DN, Schwaeble WJ. Properdin plays a protective role in polymicrobial septic peritonitis. J. Immunol. 2008;180:3313–3318. doi: 10.4049/jimmunol.180.5.3313. [DOI] [PubMed] [Google Scholar]
- Surh CD, Sprent J. T-cell apoptosis detected in situ during positive and negative selection in the thymus. Nature. 1994;372:100–103. [PubMed] [Google Scholar]
- Taylor PR, Carugati A, Fadok VA, Cook HT, Andrews M, Carroll MC, Savill JS, Henson PM, Botto M, Walport MJ. A hierarchical role for classical pathway complement proteins in the clearance of apoptotic cells in vivo. J. Exp. Med. 2000;192:359–366. doi: 10.1084/jem.192.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouw LA, Blom AM, Gasque P. Role of complement and complement regulators in the removal of apoptotic cells. Mol. Immunol. 2008;45:1199–1207. doi: 10.1016/j.molimm.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Westberg J, Fredrikson GN, Truedsson L, Sjoholm AG, Uhlen M. Sequence-based analysis of properdin deficiency: identification of point mutations in two phenotypic forms of an X-linked immunodeficiency. Genomics. 1995;29:1–8. doi: 10.1006/geno.1995.1208. [DOI] [PubMed] [Google Scholar]
- Whaley K. Biosynthesis of the complement components and the regulatory proteins of the alternative complement pathway by human peripheral blood monocytes. J. Exp. Med. 1980;151:501–516. doi: 10.1084/jem.151.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirthmueller U, Dewald B, Thelen M, Schafer MK, Stover C, Whaley K, North J, Eggleton P, Reid KB, Schwaeble WJ. Properdin, a positive regulator of complement activation, is released from secondary granules of stimulated peripheral blood neutrophils. J. Immunol. 1997;158:4444–4451. [PubMed] [Google Scholar]
- Xu W, Berger SP, Trouw LA, de Boer HC, Schlagwein N, Mutsaers C, Daha MR, van KC. Properdin binds to late apoptotic and necrotic cells independently of c3b and regulates alternative pathway complement activation. J. Immunol. 2008;180:7613–7621. doi: 10.4049/jimmunol.180.11.7613. [DOI] [PubMed] [Google Scholar]