Abstract
Proteogenomics has emerged as a field at the junction of genomics and proteomics. It is a loose collection of technologies that allow the search of tandem mass spectra against genomic databases to identify and characterize protein-coding genes. Proteogenomic peptides provide invaluable information for gene annotation, which is difficult or impossible to ascertain using standard annotation methods. Examples include confirmation of translation, reading-frame determination, identification of gene and exon boundaries, evidence for post-translational processing, identification of splice-forms including alternative splicing, and also, prediction of completely novel genes. For proteogenomics to deliver on its promise, however, it must overcome a number of technological hurdles, including speed and accuracy of peptide identification, construction and search of specialized databases, correction of sampling bias, and others. This article reviews the state of the art of the field, focusing on the current successes, and the role of computation in overcoming these challenges. We describe how technological and algorithmic advances have already enabled large-scale proteogenomic studies in many model organisms, including arabidopsis, yeast, fly, and human. We also provide a preview of the field going forward, describing early efforts in tackling the problems of complex gene structures, searching against genomes of related species, and immunoglobulin gene reconstruction.
Keywords: proteogenomics, mass spectrometry, gene annotation
1. What is Proteogenomics?
The term proteogenomics has been used in more than one context, and we must begin by describing the scope of this survey. We focus exclusively on bottom-up tandem mass spectrometry analysis. While there are many different techniques for separating peptides, ionizing fragments, and mass analysis, all producing different results, we consider an end product of this process: the tandem mass spectrum, or MS2 spectrum. The MS2 spectrum is a collection of ionized fragments masses (with intensities) for a peptide that serves as a fingerprint for peptide identification and quantification.
To identify peptides, the standard approach today originated with the seminal paper by Eng and Yates on Sequest [1], and can be abstracted as follows: (1) Consider each peptide from an available database; (2) generate a theoretical spectrum of the peptide by considering the masses of fragments that are most likely to be observed; (3) score, and rank all peptides according to the match between the experimental and theoretical spectrum; and (4) output the peptide with the best match.
If a database of peptides is not available, we can either sequence the peptide through a de novo analysis of the spectrum, or score candidate peptides generated from a genomic (DNA based) template. We call the second approach, in which peptides identified against the genome are used for gene annotation, proteogenomics. Recently, Ansong and colleagues provided an excellent review of the field [2]. Our review shares many of the ideas presented, but has a specific focus on computation.
2. Why Proteogenomics? a primer on gene finding
Scientific progress is often associated with abstraction and compaction of available knowledge, so as to create a foundation on which future discoveries can be made. Our understanding of the gene has unfortunately bucked this trend. The recently concluded ENCODE project resulted in further ambiguity of the concept. The classical definition of the gene being a “unit of heredity” (from Mendel’s work), has now evolved into “… a union of genomic sequences encoding a coherent set of potentially overlapping functional products” [3]. Other examples point to the plasticity of the genome itself, with large genomic rearrangements disrupting genes on the genome [4]. All of this has implications for proteomics.
Historically, the genomics and proteomics communities acted independently. It was the role of the genomics community to identify genes and the corresponding protein sequences. This was often done through large-scale annotation efforts, during and after the sequencing of the genomes (see below). The collection of derived proteins was considered to be a fixed set, although it was recognized that not all proteins are expressed in every cell. It was the role of the proteomics community to understand which proteins are expressed under specific conditions, or tissues, and to identify the various post-translational modifications, and other processing of the proteome. Proteogenomics challenges this perspective: if the definition of the gene itself is not clear, the proteomics (mass spectrometry) and genomics communities should work together from the beginning, to clarify gene structures. Therefore, a good place to start is to look at gene-finding.
2.1. Gene Structure
The central dogma of molecular biology suggests a flow of information from DNA to protein. First, the genic region of the DNA is ‘transcribed’ (copied) into mRNA (also called a transcript, or message). Special ‘promoter’, ‘enhancer’, and ‘repressor’ regions proximal to the gene help switch the transcription on and off, thereby regulating the production of protein. Next, the ribosomal machinery reads the message, and ‘translates’ it into proteins. Note that the beginning and end of the transcript are not translated and are referred to as the 5′ and 3′ untranslated regions (UTRs), respectively. While the process of protein production is common to both prokaryotic and eukaryotic organisms, the structure and organization of genes is quite different between the two, and will be discussed separately.
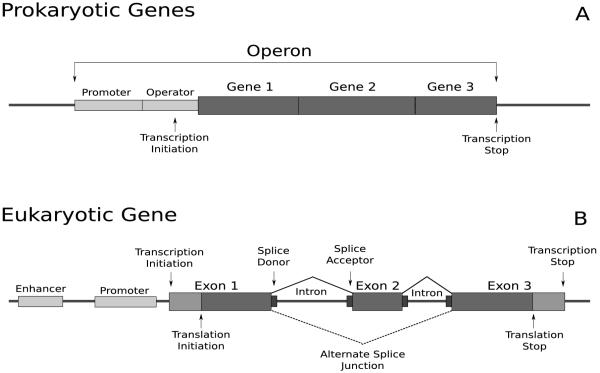
Prokaryotic Genes
In prokaryotes, related genes may be clustered into operons (Figure 1A). All genes in an operon share the same promoter region, and are transcribed as a single mRNA. While the transcript produced by an operon contains mRNA from all the genes, regulation at a higher granularity occurs at translation. Even with this simple structure, there is genic diversity. Programmed frame-shifts can produce alternate or truncated proteins [5], but are nearly impossible to predict from genomic data.
Figure 1.
Prokaryotic and eukaryotic gene structures. A: Prokaryotic genes may be arranged in an operon, sharing the same promoter. B: A eukaryotic gene contains protein-coding regions called exons, separated by non-protein-coding regions called introns. Once transcribed, the introns are spliced out. An alternate splice junction is shown using a dotted line.
Eukaryotic Genes
In eukaryotes, the coding regions of the gene are often present in discontinuous regions called exons. Multiple exons are separated by introns: regions that are transcribed, but not translated (Figure 1B). Introns are spliced out of the mRNA prior to translation by an RNA-protein complex called the spliceosome, producing the mature mRNA. For a given transcript, there may be alternate splice patterns each of which produces a different mature mRNA and may cause the protein sequence to be altered.
2.1.1. Gene Annotation
The goal of gene finding can be roughly stated as the process of identifying the genomic coordinates of exons, and the splicing patterns. Here we focus only on the coding exons. Established methods of gene annotation today combine evidence from multiple orthogonal sources [6]. One form of evidence is from ab initio gene predictors that analyze genomic signals for coding exons and splice junctions. In addition, large-scale transcript sequencing projects (often in the form of expressed sequence tags, or ESTs [7]) yield cDNA sequences that can be mapped against the genome to identify coordinates. Finally, evolutionary conservation with related species is often suggestive of genes, and other functional elements [8]. Even so, gene finding is challenging. The recent release of the Arabidopsis Information resource (TAIR8 to TAIR9) modified over 1,000 genes and added 282 new protein-coding loci [9]. Even with the well-studied human genome, a manual investigation by the ENCODE Consortium [10] resulted in the assignment of additional exons to 80% of studied genes.
Predicting the beginning of translation is a major challenge [11] for current annotation pipelines. Translation start is usually marked by one of a handful of canonical start codons, the most common of which codes for the amino acid methionine. Studies have shown that this is not a hard rule, with many non-standard start sites observed in prokaryotes [12]. Eukaryotic gene annotation is further complicated by the prevalence of alternatively spliced genes, which play a key role in generating proteome diversity. The reliable detection of splice-junctions is difficult, and most ab initio gene finding algorithms only predict a single transcript at a locus, ignoring completely alternate splice isoforms. Several tools have been developed to identify alternative splice variants using ESTs [13, 14], but accurate annotation remains a challenge due to intrinsic problems in EST sequencing including incomplete gene coverage, low sequencing accuracy, and chimerism. The issue of determining whether the alternative transcripts differ in protein-coding regions or UTRs also remains.
While gene annotation efforts for model organisms rely heavily on transcript sequencing, recent studies suggest that evidence of transcription might not be evidence of translation. Clamp et al. [15] suggest that approximately 4,000 genes in human do not code for protein despite cDNA evidence, citing their lack of conservation with primates. Genomic signals, which are the primary component of ab initio gene predictors, may be equally misleading. For example, the codons ‘TGA’, ‘TAA’, and ‘TAG’ are very strong indicators of translation stop. However, in order to accurately determine translation stop, the frame for the terminal exon must be correctly predicted. Coding signals, based on hexamer compositions, are not sufficient to determine frame in short exons [16], and sometimes cause annotators to miss the exon completely. Similar challenges make it hard to identify short genes (< 100 amino acids), which constitute a significant portion of protein-coding genes [17]. Even for longer genes, differing GC composition change coding signals to the point that the tools have to be retrained for each new genome [18].
3. The promise and challenge of proteogenomics
The idea of searching un-interpreted mass spectra against a translated nucleotide database is hardly new. We see an early account in the paper by Yates et al. [19]. However, the true power of the approach comes from a holistic use of these peptides in gene finding. See Figure 2. A proteogenomically identified peptide provides unique information for gene annotation by (a) confirming translation and separating pseudogenes(see below) from coding genes [20]; (b) establishing that a protein is not targeted for degradation (c) automatically determining frame, even multiple overlapping frames; (d) constraining the location of the translation start and end sites, as well as sites of post-translational processing (e.g. signal cleavage); (e) identifying exact splicing boundaries and alternative splice-forms, if the peptide is split across exons; and, finally (f) predicting a completely novel gene, by mapping to an uncharacterized genomic location. One may argue that next-generation sequencing of transcripts is a more promising technology for sampling the translated genome for the purpose of gene annotation. However, recent studies suggest that many transcripts are targeted for nonsense-mediated decay [20], or upon translation are unable to form stable, functional proteins[15]. Indeed, the optimist in us would claim that proteogenomics is the panacea for the ailments that plague gene annotation. Proteomic analysis also carries bene cial side-effects like peptide abundance calculations, and the identification of post-translational modifications.
Figure 2.
Peptide sequences mapped to a genomic region. Several peptides fall within an annotated gene locus, validating translation of two exons and an intron. One peptide indicates a novel splice isoform which skips the internal exon. Additional peptides fall within an annotated pseudogene giving strong indication for its translation in the cell. Peptides which fall within the intergenic region may indicate novel protein-coding loci. Cyan colored peptides would likely not be identified using a standard proteomic database.
However, proteogenomic studies are not without substantial challenges. First, peptide identification is arguably more error-prone than matching cDNA. Incomplete fragmentation, noise, and ‘isometric’ peptides can all lead to erroneous identification. The problem is compounded for proteogenomics as genomic databases are much larger than existing protein databases. A 6-frame translation of the human genome has 6 billion residues in it; contrast that with 180Mb needed for the UniProt database [21] consisting of proteins from over 250 organisms. The number of spectra for single proteogenomic studies is also large, often on the order of tens of millions of spectra.
Second, sampling and dynamic range is a concern in nearly all mass spectrometry studies. Current techniques can reliably sample proteins over 3-4 orders of magnitude [22], which is smaller than the estimated true abundance range (~ 6 orders) [23]. Detectability of peptides is a concern as not all peptides show up in mass spectrometric scans due to chemical attributes unfavorable to ionization or fragmentation.
Peptides which span splice junctions contain a wealth of information useful to gene structure prediction. In the ENSEMBL [24] database of human proteins (assembly GRCh37, release 57), approximately 26% of fully tryptic peptides of length 7 or greater span a splice boundary. These peptides are invaluable, as reliable prediction of splice-junctions is a major challenge for gene finding. However, identifying split peptides using proteogenomics seems to be equally challenging, if not more.
Finally, the output of proteogenomics is peptides, and peptides are not complete genes. Determining the gene structure from incomplete coverage by peptides is a difficult task. While these are all valid criticisms, we assert below that recent developments in technologies and computation are tipping the balance.
4. The proteogenomics solution (eukaryotes)
In the wake of technological advances in DNA sequencing, the number of eukaryotic genomes sequenced has increased dramatically in the past 20 years, with full genomes available for Saccharomyces cerevsiae [25], Caenorhabditis elegans [26], Arabidopsis thaliana [27], Drosophila melanogaster [28], Homo sapiens [29, 30], Anopheles gambiae [31], and recently, Zea mays [32]. As the genome sequences of many model organisms become available, so too are large-scale proteogenomic studies beginning to fill a much needed niche in gene annotation. In the past ten years, proteogenomic studies have confirmed expression of 25% of ORFs in Yeast [17], 73 transcripts in A. gambiae [33], 9,124 transcripts in D. melanogaster [34], 224 hypothetical proteins in Human [35], and over 13,000 transcripts in Arabidopsis [36, 37]. Peptides identified in these studies provide validation of putative genes. These successes are due in part to recent developments described below.
Sampling the proteome
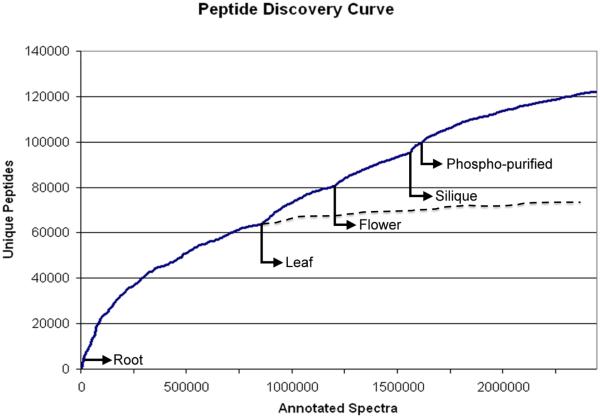
Achieving broad coverage of the proteome is critical to constructing a complete and accurate catalog of genes. A distinct set of proteins is expressed by cells of different tissues or conditions, and sampling each reveals a unique cross-section of the proteome. By acquiring multiple biological replicates of samples from different organs [36, 34, 35, 37] and developmental stages [34] a wider range of proteins can be identified. While broadly sampling the proteome increases the number of proteins detected, absence of peptides from a protein cannot be used as an indicator for absence of the protein in the sample. As Figure 3 shows, the number of unique peptides identified in Arabidopsis nearly doubled with a broader sampling strategy [37]. Coupled with technological advances in the form of high-resolution mass spectrometers [23], spectra can be collected from peptides with a wider dynamic range, while providing accurate spectral information for downstream analysis. Improved protein separation techniques [38] have also enabled the identification of more peptides in a single mass spectrometer run. In addition, fractionation methods can be used to isolate underrepresented subsets of the proteome such as small proteins [17], phosphoproteins [37], and basic proteins [34]. Brunner et al. achieved coverage of 63% of the Drosophila melanogaster proteome by utilizing these techniques as part of an ‘analysis-driven experimentation feedback loop’. They used the analysis of previous data sets to determine categories of the proteome where their sampling was deficient.[Figure 3]
Figure 3.
A graph showing the discovery curve for Arabidopsis peptides [37]. The x-axis in the figure is the number of spectra considered, while the y-axis is the number of distinct peptides recovered from the spectra. As spectra were added to the experiment, the rate of distinct peptide sequences identified slows. The figure suggests that including more spectra from root tissue will not substantially increase the number of peptides identified. However, including spectra from a different tissue type provided additional distinct peptides. Extrapolation suggests that the number of distinct peptides identified is nearly doubled by including additional tissues and additional fractionation.
Error rates
The problem of erroneous identifications is common to all proteomics projects, but is magnified for proteogenomics. Searching large spectral data sets (~ 10M) against large databases (~ 1B AA) translates into a large number of erroneous identifications even at a low error rate. At the same time, the evidence for a new gene (usually low-abundance) might only come from a small number of peptides.
Algorithmically, the identification problem is broken up into two parts: scoring/ranking of each candidate peptide for a spectrum, so that the correct identification gets the top score, and is well separated from the others; and, validation, which provides confidence that the top-scoring peptide is the right identification. Scoring has seen great improvement, based on probabilistic models [39, 40, 41, 42, 43, 44, 45] for peptide fragmentation. Large data sets of annotated peptides allow for a systematic data-mining of fragmentation patterns, which are then encoded into scoring models.
To understand why a secondary validation step is needed, consider the case when the correct peptide is not in the database, and never considered. Even if the ranking of candidates is perfect, the validation part is needed to reject the top scoring peptide. To assess the likelihood of the top-scoring peptide being the correct identification, parametric and non-parametric approaches have been tried. In the model-based approach, it is suggested that the correct and incorrect peptides follow a distinct distribution of scores. By modeling the two distributions, once can use a statistical test to identify the correct peptides [46]. Gygi and colleagues suggest a model-free approach based on constructing a decoy database [47]. The spectra are simultaneously searched against the standard and the decoy database (typically a scrambled version of the standard database). Peptides identified using the decoy are all spurious and can be used to estimate the false discovery rate (FDR).
An issue with FDR is that all peptides that exceed the score cutoff are treated equally (have the same FDR). However, we know that higher scoring peptides are more likely to be correct. Additionally, our confidence in peptide identification also depends upon its size, charge, and physico-chemical characteristics. One direction to improve FDR is to bin peptides that are similar (by score, size, charge, etc.), and compute FDR separately for each bin [37]. This local FDR (ℓ-FDR) [48] computation is possible in proteogenomics, where the large number of peptides allow each bin to be populated. Second, the goal of proteogenomics is to find protein-coding regions, not just peptides. If 2-3 peptides support the same genic locus, or gene model refinement event, then the error occurs only if all of the identifications are wrong. In this case, the ℓ-FDR values of these peptides (under the assumption that the identifications are independent) can be multiplied to give an event level FDR (eFDR). The generic approach is as follows: a list of proteogenomic events is created, such as ‘spliced exons’, ‘translated ORF’. Each event is supported by a collection of peptides and their associated ℓ-FDR values. A Bayesian approach is used to compute eFDR values for the event being incorrect [37]. For example, a spliced peptide may have a high probability of being correct, but makes a small contribution to a splicing event because of a small overlap with the second exon. On the other hand, a small collection of peptides that hit two exons, with a few spliced-peptides is strong evidence for a splicing event, even when each of the peptides has a poor ℓ-FDR score. In the set of 591 gene models submitted by Castellana et al. to TAIR, a lower eFDR correlated with manual curation and acceptance into TAIR9 resource.
The decoy database approach, and its variations, have been widely adopted. However, critics point out that including the decoy database doubles the search time and, more importantly, the FDR values greatly depend on the size of the database and the distribution of peptides in it. The most obvious difficulty is in the construction of a decoy database. One desirable attribute of the decoy database is that it does not share peptide sequences with the target database. This becomes a difficult task when the target database exceeds 6 billion amino acids.
Another difficulty stems from the construction of the target database. In proteogenomic studies, a major goal in constructing a database from genomic data is to include as many putative protein sequences as possible. This often is done by performing a translation of the genome in all six-frames. In addition to containing all putative proteins sequences, the resulting database also contains spurious sequences, often at a much higher rate than standard proteomic databases. This implicit addition of decoy peptides in the database results in diminished sensitivity at the same FDR. To combat this, an additional goal of constructing proteogenomic databases is compactness. For example, the six frame translation can be restricted to sequences exceeding the average size of an exon in the organism of interest, or to regions which receive high scores by ab initio gene predictors.
One might argue that instead of attempting to construct a database of putative protein sequences, interpreting the peptide sequence de novo will guarantee that any possible peptide sequence is considered. Several groups have proposed database-independent p-value computation methods [49, 50]. These methods rate peptide-spectrum matches using spectrum-specific score distributions, but make the assumption that all peptides are equally likely a priori.
This argument illustrates a philosophical difference regarding the importance of prior information (the database) in peptide identification, and is analogous to the debate between Bayesian and frequentist inferences. De novo approaches to peptide identification seek to distinguish the correct peptide among all possible peptides, and consequently are highly error-prone. In a database search, the space of candidate peptides is greatly reduced. This automatically increases the confidence in identification, but only if we agree that the database is complete. In proteogenomic studies, the search is on ever larger genomic databases, and the differences between de novo and database searches diminishes, particularly when modifications and mutations are permitted. If we consider only the set of peptides of length 9, de novo algorithms must consider 209 < 1012 candidate peptides. The six-frame translation of the human genome contains about 109 peptides. However, if we allow a single mutation per peptide (which would have no effect on de novo algorithms), the size of the genomic database increases 200 times, to 1011. For large databases typical of proteogenomics, the boundary between de novo and database search is blurred. Confident assessment of a genomic region being translated must depend upon the discovery of multiple, large peptides with accurate fragmentation patterns.
Another aspect of large genomic databases is the non-random distribution of nucleotides. In fact, segmental duplications, and retrotransposon mediated elements, often create multiple copies of the same gene [30, 29], leading to identical peptides at multiple locations in the genome (shared peptides). Sometimes, only one of the gene copies is active. The inactive genes (possibly transcribed into RNA, but not translated), are called pseudogenes, and will cause problems for proteogenomic identification. Identifying pseudogenes is one of the major challenges for gene annotation.
One approach to handling genomic redundancy is to consider all locations of the shared peptide [33]. However, this may lead to the false reporting of proteins. A stricter approach is to ignore the shared peptides [51, 36], significantly reducing the number of protein identifications. Grobei et al., developed a classification method of peptides based on their occurrence in the database. Peptides which could uniquely identify a protein sequence were labeled Class 1. Peptides which mapped to multiple locations were classified depending on whether their matches were to isoforms of the same gene (Class 2), members of the same gene family (Class 3a), or from multiple gene families (Class 3b). In the study by Grobei et al [52], all Class 3b peptides were discarded. Other groups [37] have used peptide locality to decide whether to keep or discard shared peptides. If a shared peptide appears in close proximity to a uniquely-located peptide, the additional identification boosts the confidence in the shared peptide location.
Spliced peptides
In humans, approximately one quarter of peptides cross a splice junction. These peptides are especially informative for gene annotation, giving boundary and frame information for two exons and a splice junction. The identification of these spliced peptides is a challenge unique to eukaryotic proteogenomics.
Historically, peptides identified against predicted or known proteins were mapped back to the genome to validate splicing events [53, 54, 35]. The detection of new splice-junctions, however, calls for a special database that encodes putative splice-forms. Such a database can be constructed using ESTs which are produced from mature mRNA and have the introns already spliced out, thus enabling the identification of peptides which span the intron boundaries [51, 55]. However, ESTs are error-prone, and highly redundant. Edwards proposed a compression scheme for reducing EST database size, based on a de Bruijn graph representation of cDNA fragments [55, 56], while retaining all potential peptide sequences.
Even with large sampling efforts, ESTs do not adequately cover all splice-junctions since many ESTs are sequenced from the 3′ end which provides unique tags for identification, but only limited representation of the coding sequence. A second source of putative spliced sequences is ab initio gene prediction tools, such as GeneID [57], Fgenesh [58], Augustus [59], and GeneMark [60]. Kuster et al. [61] used a two-pass system to identify spliced peptides by first identifying likely novel coding regions using unspliced peptides, then predicting a new model for that region and searching the spectra against the new model.
Tanner et al. [35] proposed a spliced-exon graph to compactly represent all gene structures and splice-junctions generated by gene prediction tools and EST mappings. In the graph, each exon is a node and each edge between exons represents a putative splice junction. While the graph provides a compact encoding of all splice-forms, the MS2 identification tools need to be modified to search the specialized database. In recent studies, this approach confirmed over 15, 000 spliced peptides in human, including over 40 instances of alternative splicing, where peptides confirm the splicing of one exon with multiple partner exons [35]. In arabidopsis, 4, 018 novel spliced peptides were identified when compared to the TAIR7 annotations [37]. By structuring proteogenomic databases as spliced-exon graphs and de Bruijn graphs, the sequence redundancy that is inherently present in the proteomes of higher organisms is reduced. This is of particular importance in proteogenomics where database size has significant impact on error rates and search time.
The general issue of identifying ‘discontinuous’ peptides (of which spliced-peptides are a special case) is likely to persist. The genomes of individual humans are now being sequenced [62], and show remarkable plasticity, with large rearrangements leading to gene disruptions, fusions, and trans-splicing [4]. Additionally, the rearrangements often characterize the transition of a normal genome to a tumor genome [63, 64]. Identification of discontinuous peptides confirming fusion events is likely to expand the role of proteogenomics in cancer detection and therapy. It is important to note that in the case of diseases which result from genome rearrangements, such as cancer, that the reference genome should not be limited to the wild type individual. Proteogenomic studies, coupled with deep genomic and transcript sequencing, can provide valuable information on aberrant protein expression.
Search speed
In 2001, Choudhary et al. [65] constructed the 6-frame translations of the human draft genome sequence. On a single processor, searching 169 spectra required 10 hours of compute time. Since then search algorithms and computing resources have improved greatly, while the framework of proteogenomic studies has remained largely unchanged. Filtering spectra for quality [66, 67], or clustering them to increase the signal to noise ratio of each spectrum [68] are techniques employed to improve both the quality of identifications, as well as the speed of the search.
A second advance is in ‘database filtering’, where a two-pass search is employed for MS2 identifications. The goal of the first search (the filter step) is simply to discard most of the database, while retaining the correct peptides, using minimal computation. The more expensive scoring is relegated to a second stage, and is fast because only the filtered peptides are scored. Novel strategies for filtering are under active development, including tagbased filtering, and peak-based filtering resulting in two orders of magnitude speedup, with little loss of sensitivity [69, 70, 42]. The database size can be reduced, resulting in faster searches, by applying outside knowledge of regions which are unlikely to be coding for proteins, such as repeat regions or open reading frames of insufficient length to contain an exon. Structuring the database as a graph, as described above, can also reduce the amount of redundant sequence.
Today, many database search engines can be run on multiple cores, or in parallel on large compute clusters. Proteogenomic studies are able to bene t from the enormous advances in processor speed and parallelism. A typical search of 1M spectra against the 6-frame translation of the maize genome containing over 1B amino acids takes on the order of days using a compute cluster of 100 nodes, while also identifying difficult peptides with unexpected modifications and mutations [70, 71].
4.1. Improving gene annotation
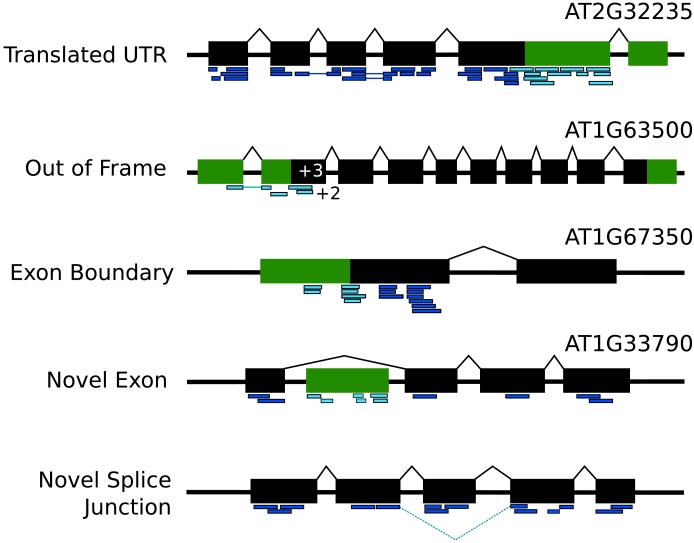
The proteogenomic identification of a peptide might come from a region of the genome not previously known to code for protein. We refer to these peptides as ‘novel’. Novel peptides might be intragenic (fall within the locus of a known gene structure), or intergenic (fall outside the locus of a known gene model), and suggest different categories of genome annotation. A set of possible events with supporting peptides is shown in Figure 4.
Figure 4.
Five different refinement events which may be suggested by intragenic peptides. Exons are shown in black boxes while novel coding regions suggested by the peptides are shown in green. Peptides (dark blue and cyan) are shown aligned to the gene models. Examples drawn of refined genes in TAIR7 from Castellana et al. [37] are annotated with the updated gene model. Several of these genes have been updated in subsequent gene annotation releases to include the peptide evidence.
Refining gene models
For intragenic novel peptides, it is difficult to distinguish if the gene structure needs to be corrected, or if it can be explained by a novel splice-form. The sampling of the proteome is not dense enough to observe peptides from multiple isoforms. Therefore, extrinsic data, such transcript sequences, or homology to genomic regions, is used to distinguish the two cases [36, 37].
Reconstructing gene models using mapped peptides is non-trivial, mostly because the peptide information is not sufficient to completely determine the structure due to limited coverage. While spliced-peptides provide information on which exons might splice together, they are not informative about distal events (isoforms with multiple alternative splicing patterns). Top-down proteomics, in which intact proteins are analyzed, might help in this case, but has not been used for proteogenomics due to the complexity of the samples.
The peptides can be used to increase the likelihood of a gene model being correct. New gene finding tools such as Augustus [59], are able to combine ab initio signals with external hints, including homology with related species, ESTs, annotated gene models, and now, proteogenomic peptides. Recent proteogenomic studies have proposed automated prediction of the updated gene model including the peptides as hints [61, 35, 37]. A total of 339 arabidopsis gene models predicted in this way were incorporated into the most recent gene annotation release for Arabidopsis, TAIR9 [9].
Gene discovery
Intergenic peptides which are not proximal to a known gene may indicate a novel coding region. To reduce errors, eFDR or Protein-Prophet can be used to combine the evidence from multiple peptides in support of the novel gene [72]. Validation of the novel genes remains a challenge, but supporting evidence is obtained from expressed transcript sequence, RT-PCR validation [73, 74] or homology searches of newly predicted gene models [37, 75]. The homology searches can also be used for functional annotation of the corresponding protein sequence [53, 76].
5. Proteogenomics in prokaryotes
Bacterial genomes are being sequenced at an astonishing rate, and as a consequence that gene annotations are primarily computational predictions. Prokaryotic genomes tend to be smaller and less genetically complex than eukaryotes. As prokaryotic genes do not undergo splicing, all proteins can be captured by translating the genome in all six frames.
Several studies on prokaryotic genomes have shown that ab initio tools alone are insufficient, particularly for identifying gene boundaries, and for short ORFs [77, 53, 12, 75]. Proteogenomic validation of predictions is a pragmatic compromise between computational prediction, and full-experimental validation. Jaffe et al. [53] validated 81% of predicted ORFs in Mycoplasma pneumoniae and Gupta et al. [12] validated 40% of genes in Shewanella oneidensis. Wang et al. [76] constructed a database of gene predictions, and validated 901 proteins in Mycobacterium smegmatis.
The first study to search tandem mass spectra against the 6-frame translation of a fully-sequenced bacterium (Haemophilus influenzae) identified 263 proteins and 2 genomic loci which were not previously believed to be translated. Since then, several high-throughput studies have identified novel translated loci in Mycoplasma pneumoniae (16 ORFs) [53], Rhodopseudomonas palustris (85 ORFs) [73], Shewanella oneidensis (8 ORFs) [12], and Deinococcus deserti (15 ORFs) [78].
Peptides which map in close proximity to annotated genes may suggest changes to the gene model, rather than separate novel loci. While determining the translation end site is simply the location of the first in-frame, down-stream stop codon, determining translation start is much trickier. Proteogenomic mapping of peptides to regions proximal to the N-terminus of the annotation gene may correct these errors. In Shewanella oneidensis, 30 genes appeared to have incorrect 5′ boundaries based on peptides mapping upstream of the annotated start site as well as alignment with proteins in related species [12]. Peptides which are mapped near an annotated gene, but are in a different frame, may indicate the rare event of programmed frame shift [53], which is nearly impossible to predict by other automated methods. Baudet et al [75] derived protein N-termini using a labeling reagent, TMPP, to correct the translation start sites of 60 genes in Deinococcus radiodurans.
With the dramatic increase of sequenced genomes of related prokaryotic organisms, proteogenomics is now being performed on multiple sequences in tandem. Gallien et al. [79] combined comparative genomics and N-terminal protein labeling to correct 19% of translation start sites in M. smegmatis and 601 start sites in 16 other Mycobacterium species. As an extension to previous work in Shewanella oneidensis [12], Gupta et al. [80] sampled the proteomes of three Shewanella species to simultaneous annotate their genomes. Due to the high level of sequence similarity between the species, 2,590 orthologous ORFs were defined as ‘shared genes’. By allowing peptides identified on an orthologous protein to contribute evidence for expression of a protein, Gupta et al. are able to rescue over 140 proteins which would have been excluded from a proteomic experiment using the ‘two peptide per protein’ inference rule. While using comparative proteogenomics represents a new frontier for annotating genomes, methods for determining statistical significance of these inferences have yet to be developed.
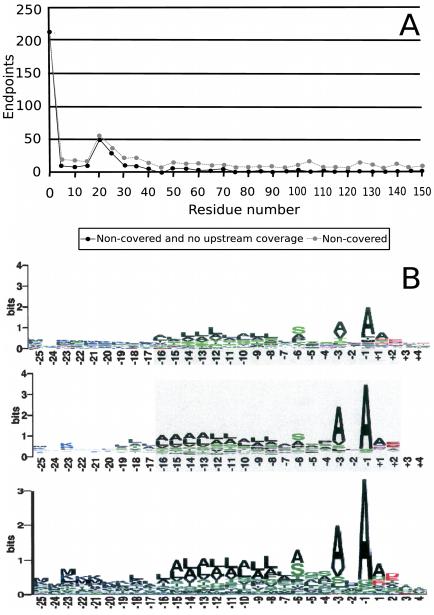
In addition to gene annotation, a study in the bacteria Shewanella onei-densis discovered over 10,000 sites of chemical modification [12]. The diversity of modifications identified is beyond what can be specified by popular database search tools, underscoring one of the main challenges to proteomics and proteogenomics. Gupta et al. also discovered non-chemical protein modifications which reveal the dynamic nature of the proteome. By considering the positions of the most N-terminal peptides observed with relation to the predicted translation start site (Figure 5A), Gupta et al. [12] observed possible instances of signal peptides and N-terminal methionine cleavages. The study was able to distinguish potential signal peptides from post-source decay by identifying non-tryptic peptides contained in tryptic peptides. The peptide compositions also allowed them to determine motifs for signal peptide cleavage sites that closely agree with motifs used by computational predictors (Figure 5B). A comparative analysis reveals a functional role for N-terminal methionine excision [81]. Jaffe et al. [53] showed post-processing of a gene by identifying two halves of the resulting protein appearing separately in the same mass spectrometry run.
Figure 5.
Proteomic annotation results courtesy of Gupta et al. [12]. A: A histogram of the position of non-tryptic N-terminal peptides reveals two protein processing events; cleavage of N-terminal methionine and cleavage of signal peptides. B: The signal peptide motif recovered by MS/MS analysis compared to the same motif determined by two computational predictors [95, 96].
6. Conclusion: New directions for proteogenomics
The discussion above assumes that the peptide encoded by the spectrum can be found in the genomic database. This may not always be the case. However, the peptide may be inferred by comparing the spectrum against a related genomic template. We refer to this as comparative proteogenomics. An exciting, if somewhat controversial, recent example is the sequencing of T. rex and mastodon peptides [82, 83, 84, 85].
MS-Blast [86] is often cited as an early tool for comparative proteogenomics. It relies on a de novo analysis to establish tags. A collection of tags is then searched using Blast to identify homologous sequences. Likewise the search for mutated and modified peptides also implies an imperfect genomic template. Tools such as MSAlignment [71], Modi [87], SPIDER [88], and TagRecon [89] perform a search of a homologous database with an unrestricted set of modifications or mutations.
When the genomic templates are very different (a different species), a new set of tools are required. Comparative shotgun protein sequencing [90] uses clustering, spectrum alignment, and de novo sequencing techniques to create sequence contigs of the target protein. Champs [91] identifies the most similar protein to the target protein in the database, and uses SPIDER to correct de novo sequenced peptides against the protein. GenoMS [92] resembles both de novo and database search techniques. It first identifies one or more templates from a database of homologous proteins or a related genome. Mutated or missing portions of the target proteins or proteins are then sequenced using model-based spectral alignment and de novo sequencing.
The gene annotation for an organism is not a once and done enterprise, but relies on a feedback loop involving the genomic and proteomic communities. Proteogenomics has developed beyond the proof-of-principle level, and is becoming an integral part of the annotation pipeline for model organisms. The realization of the method, in studies to date, has only been as a downstream analysis tool, for improvement of a first pass annotation. However, the high-throughput nature and the ability to directly ascertain the elements of the genome which are translated, highly recommend proteogenomics as a method to be used on the front-line of gene annotation.
The need for proteogenomics is highlighted by the exponential rate of growth of genomic databases, not only across species, but of individuals within species. For eukaryotes, the notion of gene is evolving to diverse trans-splicing and rearrangement induced splicing events. For prokaryotes, a vast majority of the genomes will never be sampled due to the difficulty in culturing. Instead, metagenomic studies sample genomic sequence from a community of genetically diverse organisms, which makes even species identification difficult. The development of sequencing technologies is allowing for the sequencing of genomes and meta-genomes at an unprecedented rate [93, 94]. At the same time, advances in instrumentation, MS2 identification algorithms, specialized database construction, and comparative tools suggest that the future is bright for proteogenomics.
Acknowledgements
V. Bafna was supported by 1-P41-RR024851-01. N.E. Castellana was supported by National Science Foundation IGERT Plant Systems Biology training grant # DGE-0504645.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Eng J, McCormack A, Yates J., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- [2].Ansong C, Purvine SO, Adkins JN, Lipton MS, Smith RD. Proteogenomics: needs and roles to be filled by proteomics in genome annotation. Brief Funct Genomic Proteomic. 2008;7:50–62. doi: 10.1093/bfgp/eln010. [DOI] [PubMed] [Google Scholar]
- [3].Gerstein MB, Bruce C, Rozowsky JS, Zheng D, Du J, Korbel JO, Emanuelsson O, Zhang ZD, Weissman S, Snyder M. What is a gene, post-ENCODE? History and updated definition. Genome Res. 2007;17:669–681. doi: 10.1101/gr.6339607. [DOI] [PubMed] [Google Scholar]
- [4].Campbell PJ, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat. Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Farabaugh PJ. Programmed translational frameshifting. Annu. Rev. Genet. 1996;30:507–528. doi: 10.1146/annurev.genet.30.1.507. [DOI] [PubMed] [Google Scholar]
- [6].Curwen V, Eyras E, Andrews TD, Clarke L, Mongin E, Searle SM, Clamp M. The Ensembl automatic gene annotation system. Genome Res. 2004;14:942–950. doi: 10.1101/gr.1858004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Adams M, et al. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- [8].Birney E, et al. Identification and analysis of functional elements in 1of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Huala E, et al. The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 2001;29:102–105. doi: 10.1093/nar/29.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Pennisi E. Genomics. DNA study forces rethink of what it means to be a gene. Science. 2007;316:1556–1557. doi: 10.1126/science.316.5831.1556. [DOI] [PubMed] [Google Scholar]
- [11].Brent MR. Steady progress and recent breakthroughs in the accuracy of automated genome annotation. Nat. Rev. Genet. 2008;9:62–73. doi: 10.1038/nrg2220. [DOI] [PubMed] [Google Scholar]
- [12].Gupta N, et al. Whole proteome analysis of post-translational modifications: applications of mass-spectrometry for proteogenomic annotation. Genome Res. 2007;17:1362–1377. doi: 10.1101/gr.6427907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kan Z, Rouchka EC, Gish WR, States DJ. Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res. 2001;11:889–900. doi: 10.1101/gr.155001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Florea L, et al. Gene and alternative splicing annotation with AIR. Genome Res. 2005;15:54–66. doi: 10.1101/gr.2889405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Clamp M, Fry B, Kamal M, Xie X, Cuff J, Lin MF, Kellis M, Lindblad-Toh K, Lander ES. Distinguishing protein-coding and non-coding genes in the human genome. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19428–19433. doi: 10.1073/pnas.0709013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mathe C, Sagot MF, Schiex T, Rouze P. Current methods of gene prediction, their strengths and weaknesses. Nucleic Acids Res. 2002;30:4103–4117. doi: 10.1093/nar/gkf543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Oshiro G, Wodicka LM, Washburn MP, Yates JR, Lockhart DJ, Winzeler EA. Parallel identification of new genes in Saccharomyces cerevisiae. Genome Res. 2002;12:1210–1220. doi: 10.1101/gr.226802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- [19].Yates JR, Eng JK, McCormack AL. Mining genomes: correlating tandem mass spectra of modified and unmodified peptides to sequences in nucleotide databases. Anal. Chem. 1995;67:3202–3210. doi: 10.1021/ac00114a016. [DOI] [PubMed] [Google Scholar]
- [20].Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. U.S.A. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Apweiler R, et al. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- [23].Yates JR, Ruse CI, Nakorchevsky A. Proteomics by mass spectrometry: approaches, advances, and applications. Annu Rev Biomed Eng. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- [24].Flicek P, et al. Ensembl’s 10th year. Nucleic Acids Res. 2010;38:D557–562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Goffeau A, et al. Life with 6000 genes. Science. 1996;274:563–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- [26].C. elegans Sequencing Consortium Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- [27].Initiative TAG. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- [28].Adams MD, et al. The Genome Sequence of Drosophila melanogaster. Science. 2000;287(5461):2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- [29].Venter JC, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- [30].Lander E, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- [31].Holt RA, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- [32].Schnable PS, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- [33].Kalume DE, Peri S, Reddy R, Zhong J, Okulate M, Kumar N, Pandey A. Genome annotation of Anopheles gambiae using mass spectrometry-derived data. BMC Genomics. 2005;6:128. doi: 10.1186/1471-2164-6-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Brunner E, et al. A high-quality catalog of the Drosophila melanogaster proteome. Nat. Biotechnol. 2007;25:576–583. doi: 10.1038/nbt1300. [DOI] [PubMed] [Google Scholar]
- [35].Tanner S, Shen Z, Ng J, Florea L, Guigo R, Briggs SP, Bafna V. Improving gene annotation using peptide mass spectrometry. Genome Res. 2007;17:231–239. doi: 10.1101/gr.5646507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Baerenfaller K, et al. Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science. 2008;320:938–941. doi: 10.1126/science.1157956. [DOI] [PubMed] [Google Scholar]
- [37].Castellana NE, Payne SH, Shen Z, Stanke M, Bafna V, Briggs SP. Discovery and revision of Arabidopsis genes by proteogenomics. Proc. Natl. Acad. Sci. U.S.A. 2008;105:21034–21038. doi: 10.1073/pnas.0811066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Washburn MP, Wolters D, Yates JR. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- [39].Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- [40].Elias JE, Gibbons FD, King OD, Roth FP, Gygi SP. Intensity-based protein identification by machine learning from a library of tandem mass spectra. Nat. Biotechnol. 2004;22:214–219. doi: 10.1038/nbt930. [DOI] [PubMed] [Google Scholar]
- [41].Wan Y, Yang A, Chen T. PepHMM: a hidden Markov model based scoring function for mass spectrometry database search. Anal. Chem. 2006;78:432–437. doi: 10.1021/ac051319a. [DOI] [PubMed] [Google Scholar]
- [42].Bern M, Cai Y, Goldberg D. Lookup peaks: a hybrid of de novo sequencing and database search for protein identification by tandem mass spectrometry. Anal. Chem. 2007;79:1393–1400. doi: 10.1021/ac0617013. [DOI] [PubMed] [Google Scholar]
- [43].Klammer AA, Reynolds SM, Bilmes JA, MacCoss MJ, Noble WS. Modeling peptide fragmentation with dynamic Bayesian networks for peptide identification. Bioinformatics. 2008;24:i348–356. doi: 10.1093/bioinformatics/btn189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Frank AM. A ranking-based scoring function for peptide-spectrum matches. J. Proteome Res. 2009;8:2241–2252. doi: 10.1021/pr800678b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Frank AM. Predicting intensity ranks of peptide fragment ions. J. Proteome Res. 2009;8:2226–2240. doi: 10.1021/pr800677f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- [47].Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- [48].Efron B, Tibshirani R, Storey JD, Tusher V. Empirical Bayes analysis of a microarray experiment. J. Am. Stat. Assoc. 2001;96:1151–1160. [Google Scholar]
- [49].Kim S, Gupta N, Pevzner PA. Spectral probabilities and generating functions of tandem mass spectra: a strike against decoy databases. J. Proteome Res. 2008;7:3354–3363. doi: 10.1021/pr8001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Alves G, Yu YK. Statistical Characterization of a 1D Random Potential Problem - with applications in score statistics of MS-based peptide sequencing. Physica A. 2008;387:6538–6544. doi: 10.1016/j.physa.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Fermin D, Allen BB, Blackwell TW, Menon R, Adamski M, Xu Y, Ulintz P, Omenn GS, States DJ. Novel gene and gene model detection using a whole genome open reading frame analysis in proteomics. Genome Biol. 2006;7:R35. doi: 10.1186/gb-2006-7-4-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Grobei MA, Qeli E, Brunner E, Rehrauer H, Zhang R, Roschitzki B, Basler K, Ahrens CH, Grossniklaus U. Deterministic protein inference for shotgun proteomics data provides new insights into Arabidopsis pollen development and function. Genome Res. 2009;19:1786–1800. doi: 10.1101/gr.089060.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Jaffe JD, Berg HC, Church GM. Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics. 2004;4:59–77. doi: 10.1002/pmic.200300511. [DOI] [PubMed] [Google Scholar]
- [54].Desiere F, et al. Integration with the human genome of peptide sequences obtained by high-throughput mass spectrometry. Genome Biol. 2005;6:R9. doi: 10.1186/gb-2004-6-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Edwards NJ. Novel peptide identification from tandem mass spectra using ESTs and sequence database compression. Mol. Syst. Biol. 2007;3:102. doi: 10.1038/msb4100142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].de Bruijn N. A combinatorial problem. Proc. Kon. Ned. Akad. Wetensch. 1946;49:758–764. [Google Scholar]
- [57].Blanco E, Parra G, Guigo R. Using geneid to identify genes. Curr Protoc Bioinformatics Chapter. 2007;4 doi: 10.1002/0471250953.bi0403s18. Unit 4.3. [DOI] [PubMed] [Google Scholar]
- [58].Salamov AA, Solovyev VV. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Stanke M, Schoffmann O, Morgenstern B, Waack S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics. 2006;7:62. doi: 10.1186/1471-2105-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lomsadze A, Ter-Hovhannisyan V, Chernoff YO, Borodovsky M. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 2005;33:6494–6506. doi: 10.1093/nar/gki937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kuster B, Mortensen P, Andersen JS, Mann M. Mass spectrometry allows direct identification of proteins in large genomes. Proteomics. 2001;1:641–650. doi: 10.1002/1615-9861(200104)1:5<641::AID-PROT641>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- [62].The 1000 genome project.
- [63].Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat. Rev. Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Mani RS, Tomlins SA, Callahan K, Ghosh A, Nyati MK, Varambally S, Palanisamy N, Chinnaiyan AM. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Choudhary JS, Blackstock WP, Creasy DM, Cottrell JS. Interrogating the human genome using uninterpreted mass spectrometry data. Proteomics. 2001;1:651–667. doi: 10.1002/1615-9861(200104)1:5<651::AID-PROT651>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- [66].Bern M, Goldberg D, McDonald WH, Yates JR. Automatic quality assessment of peptide tandem mass spectra. Bioinformatics. 2004;20(Suppl 1):49–54. doi: 10.1093/bioinformatics/bth947. [DOI] [PubMed] [Google Scholar]
- [67].Nesvizhskii AI, Roos FF, Grossmann J, Vogelzang M, Eddes JS, Gruissem W, Baginsky S, Aebersold R. Dynamic spectrum quality assessment and iterative computational analysis of shotgun proteomic data: toward more efficient identification of post-translational modifications, sequence polymorphisms, and novel peptides. Mol. Cell Proteomics. 2006;5:652–670. doi: 10.1074/mcp.M500319-MCP200. [DOI] [PubMed] [Google Scholar]
- [68].Frank AM, Bandeira N, Shen Z, Tanner S, Briggs SP, Smith RD, Pevzner PA. Clustering millions of tandem mass spectra. J. Proteome Res. 2008;7:113–122. doi: 10.1021/pr070361e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- [70].Tanner S, Shu H, Frank A, Wang LC, Zandi E, Mumby M, Pevzner PA, Bafna V. InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal. Chem. 2005;77:4626–4639. doi: 10.1021/ac050102d. [DOI] [PubMed] [Google Scholar]
- [71].Tsur D, Tanner S, Zandi E, Bafna V, Pevzner PA. Identification of post-translational modifications by blind search of mass spectra. Nat. Biotechnol. 2005;23:1562–1567. doi: 10.1038/nbt1168. [DOI] [PubMed] [Google Scholar]
- [72].Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- [73].Savidor A, Donahoo RS, Hurtado-Gonzales O, Verberkmoes NC, Shah MB, Lamour KH, McDonald WH. Expressed peptide tags: an additional layer of data for genome annotation. J. Proteome Res. 2006;5:3048–3058. doi: 10.1021/pr060134x. [DOI] [PubMed] [Google Scholar]
- [74].Merrihew GE, Davis C, Ewing B, Williams G, Kall L, Frewen BE, Noble WS, Green P, Thomas JH, MacCoss MJ. Use of shotgun proteomics for the identification, confirmation, and correction of C. elegans gene annotations. Genome Res. 2008;18:1660–1669. doi: 10.1101/gr.077644.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Baudet M, et al. Proteomic-based refinement of Deinococcus deserti genome annotation reveals an unwonted use of non-canonical translation initiation codons. Mol. Cell Proteomics. doi: 10.1074/mcp.M900359-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Wang R, Prince JT, Marcotte EM. Mass spectrometry of the M. smegmatis proteome: protein expression levels correlate with function, operons, and codon bias. Genome Res. 2005;15:1118–1126. doi: 10.1101/gr.3994105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Lasonder E, et al. Analysis of the Plasmodium falciparum proteome by high-accuracy mass spectrometry. Nature. 2002;419:537–542. doi: 10.1038/nature01111. [DOI] [PubMed] [Google Scholar]
- [78].de Groot A, et al. Alliance of proteomics and genomics to unravel the specificities of Sahara bacterium Deinococcus deserti. PLoS Genet. 2009;5:e1000434. doi: 10.1371/journal.pgen.1000434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Gallien S, Perrodou E, Carapito C, Deshayes C, Reyrat JM, Van Dorsselaer A, Poch O, Schaeffer C, Lecompte O. Orthoproteogenomics: multiple proteomes investigation through orthology and a new MS-based protocol. Genome Res. 2009;19:128–135. doi: 10.1101/gr.081901.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Gupta N, et al. Comparative proteogenomics: combining mass spectrometry and comparative genomics to analyze multiple genomes. Genome Res. 2008;18:1133–1142. doi: 10.1101/gr.074344.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Bonissone S, Gupta N, Romine M, Pevzner P. Comparative proteogenomics reveals a possible functional role of N-terminal methionine excision. submitted. [Google Scholar]
- [82].Asara JM, Schweitzer MH, Freimark LM, Phillips M, Cantley LC. Protein sequences from mastodon and Tyrannosaurus rex revealed by mass spectrometry. Science. 2007;316:280–285. doi: 10.1126/science.1137614. [DOI] [PubMed] [Google Scholar]
- [83].Asara JM, Garavelli JS, Slatter DA, Schweitzer MH, Freimark LM, Phillips M, Cantley LC. Interpreting sequences from mastodon and T. rex. Science. 2007;317:1324–1325. doi: 10.1126/science.317.5843.1324. [DOI] [PubMed] [Google Scholar]
- [84].Buckley M, et al. Comment on ”Protein sequences from mastodon and Tyrannosaurus rex revealed by mass spectrometry”. Science. 2008;319:33. doi: 10.1126/science.1147046. author reply 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Pevzner PA, Kim S, Ng J. Comment on ”Protein sequences from mastodon and Tyrannosaurus rex revealed by mass spectrometry”. Science. 2008;321:1040. doi: 10.1126/science.1155006. author reply 1040. [DOI] [PubMed] [Google Scholar]
- [86].Shevchenko A, Sunyaev S, Loboda A, Shevchenko A, Bork P, Ens W, Standing KG. Charting the proteomes of organisms with unsequenced genomes by MALDI-quadrupole time-of-flight mass spectrometry and BLAST homology searching. Anal. Chem. 2001;73:1917–1926. doi: 10.1021/ac0013709. [DOI] [PubMed] [Google Scholar]
- [87].Na S, Jeong J, Park H, Lee KJ, Paek E. Unrestrictive identification of multiple post-translational modifications from tandem mass spectrometry using an error-tolerant algorithm based on an extended sequence tag approach. Mol. Cell Proteomics. 2008;7:2452–2463. doi: 10.1074/mcp.M800101-MCP200. [DOI] [PubMed] [Google Scholar]
- [88].Han Y, Ma B, Zhang K. SPIDER: software for protein identification from sequence tags with de novo sequencing error. J Bioinform Comput Biol. 2005;3:697–716. doi: 10.1142/s0219720005001247. [DOI] [PubMed] [Google Scholar]
- [89].Dasari S, Chambers MC, Slebos RJ, Zimmerman L, Ham AJ, Tabb DL. TagRecon: high-throughput mutation identification through sequence tagging. J Proteome Res. doi: 10.1021/pr900850m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Bandeira N, Pham V, Pevzner P, Arnott D, Lill JR. Automated de novo protein sequencing of monoclonal antibodies. Nat. Biotechnol. 2008;26:1336–1338. doi: 10.1038/nbt1208-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Liu X, Han Y, Yuen D, Ma B. Automated protein (re)sequencing with MS/MS and a homologous database yields almost full coverage and accuracy. Bioinformatics. 2009;25:2174–2180. doi: 10.1093/bioinformatics/btp366. [DOI] [PubMed] [Google Scholar]
- [92].Castellana NE, Pham V, Arnott D, Lill JR, Bafna V. Template proteogenomics: sequencing whole proteins using an imperfect database. Mol Cell Proteomics. doi: 10.1074/mcp.M900504-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Ram RJ, Verberkmoes NC, Thelen MP, Tyson GW, Baker BJ, Blake RC, Shah M, Hettich RL, Banfield JF. Community proteomics of a natural microbial biofilm. Science. 2005;308:1915–1920. [PubMed] [Google Scholar]
- [94].Klaassens ES, de Vos WM, Vaughan EE. Metaproteomics approach to study the functionality of the microbiota in the human infant gastrointestinal tract. Appl. Environ. Microbiol. 2007;73:1388–1392. doi: 10.1128/AEM.01921-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- [96].Hiller K, Grote A, Scheer M, Munch R, Jahn D. PrediSi: prediction of signal peptides and their cleavage positions. Nucleic Acids Res. 2004;32:W375–379. doi: 10.1093/nar/gkh378. [DOI] [PMC free article] [PubMed] [Google Scholar]