Abstract
Streptococcus pneumoniae D39 AdcR (adhesin competence repressor) is the first metal-sensing member of the MarR (multiple antibiotic resistance repressor) family to be characterized. Expression profiling with a ΔadcR strain grown in liquid culture (brain heart infusion; BHI) under microaerobic conditions reveals upregulation of 13 genes including adcR and adcCBA, encoding a high affinity ABC uptake system for zinc, and genes encoding cell-surface zinc-binding pneumococcal histidine triad (Pht) proteins and AdcAII (Lmb, laminin binding). The ΔadcR, H108Q and H112Q adcR mutant allelic strains grown in 0.2 mM Zn(II) exhibit a slow-growth phenotype and a ≈2-fold increase in cell-associated Zn(II). Apo- and Zn(II)-bound AdcR are homodimers in solution and binding to a 28-mer DNA containing an adc operator is strongly stimulated by Zn(II) with KDNA-Zn = 2.4 ×108 M−1 (pH 6.0, 0.2 M NaCl, 25 °C). AdcR binds two Zn(II) per dimer, with step-wise Zn(II) affinities KZn1 and KZn2 of ≥109 M−1 at pH 6.0 and ≥1012 M−1 at pH 8.0. X-ray absorption spectroscopy (XAS) of the high affinity site reveals a pentacoordinate N/O complex and no cysteine coordination, the latter finding corroborated by wild-type-like functional properties of C30A AdcR. Alanine substitution of conserved residues His42 in the DNA binding domain, and His108 and His112 in the C-terminal regulatory domain, abolish high affinity Zn(II) binding and greatly reduce Zn(II)-activated binding to DNA. NMR studies reveal that these mutants adopt the same folded conformation as dimeric wild-type apo AdcR, but fail to conformationally switch upon Zn(II) binding. These studies clearly identify His42, His108 and H112 as metalloregulatory zinc ligands in S. pneumoniae AdcR.
Introduction
Streptococcus pneumoniae (S. pneumoniae) is a Gram-positive respiratory pathogen that colonizes the upper respiratory tract (the nasopharynx) as a commensal organism. The adcRCBA operon is conserved in all Streptococci and encodes an ABC (ATP-binding cassette) transporter, AdcCBA, that is believed to function as a high affinity importer for Zn(II).1 AdcCBA is a member of the Cluster IX family of metal specific transporters,1–3 and may well mediate uptake of noncognate Mn(II) at high extracellular concentrations.1 AdcR is a proposed Zn(II)-sensing transcriptional regulator4 which sequence analysis suggests is a MarR family repressor. MarR proteins are known to regulate aromatic catabolism, the expression of virulence factors,5,6 and the response to antibiotic and antimicrobial stress and oxidative stress. Multiple crystallographic structures are available that provide insights into MarR-family architecture and in some cases, the mechanism of regulation of operator DNA binding.7–12
The adc operon is reported to be expressed during active S. pneumoniae infection and is essential for genetic transformation, although the molecular basis of this requirement remains unclear.1,3,13 AdcA, the extracellular, metal (solute)-binding lipoprotein component of AdcCAB is strongly immunogenic in mice and is an excellent vaccine candidate for the porcine pathogen Streptococcus suis.14 In S. suis an adcR deletion mutant exhibits attenuated virulence.15 Antibodies raised against a number of solute binding components from ABC transporters found in the Gram-negative pathogen Yersina pestis and other bacterial pathogens are also protective against bacterial infection.16,17 An adcB mutant S. pneumoniae strain shows attenuated binding to human epithelial cells in vitro18 and AdcB is a documented virulence factor in a signature-tagged mutagenesis screen.19 In Streptococcus gordonii, an oral plaque pathogen, AdcR is reported to play an essential role in biofilm formation.20,21
Genomically unlinked AdcR-regulated genes were first predicted on the basis of in silico analysis of the S. pneumoniae genome for what is now known to be the AdcR operator sequence.4 One predicted AdcR-regulated gene in Streptococcus pyogenes is a paralog of 30S ribosomal subunit S14 (rpsN) that has a disrupted C-terminal Zn-ribbon domain similar to those under transcriptional control of the master Zn(II) uptake regulator Zur (Fur family) in Streptomyces coelicolor22 and Bacillus subtilis23 during zinc starvation. Another adcR-regulated locus predicted in Streptococcus pyogenes is pht (pneumococcal histidine triad proteins) which consists of four homologous genes (phtA, phtB, phtD, phtE) which encode highly immunogenic pneumococcal surface proteins24 each of which contains up to five repeats of the sequence Dx(20)HxxHxH now known to coordinate Zn(II) tetrahedrally.25 Pht proteins play a role in immune evasion and may inhibit complement deposition thereby impairing opsonization and phagocytosis, although these effects appear to be strain-specific with the mechanism still under investigation.26 AdcAII, an orphan metal binding receptor of known structure that is homologous to AdcA27 is also regulated transcriptionally by AdcR. In Streptococcus pyogenes, the AdcA homolog Lbp functions in colonization as an adhesin that binds to the extracellular matrix protein laminin, with zinc suggested to play a role in this interaction.28 An Lbp mutant shows attenuated virulence in a murine model of infection and defective growth in zinc depleted media.29
The clear importance of Zn(II) homoeostasis in Streptococcus stands in sharp contrast with the degree to which we understand AdcR structure and function. Zur, the Fur-family30 transcriptional regulator of Zn(II) uptake in Escherichia coli31,32 and Gram-positive organisms Bacillus subtilis and Mycobacterium tuberculosis,33,34 is encoded by some Streptococci with a primary role in regulating Zn(II) homeostasis clearly demonstrated.35 In contrast, Zur is not encoded by any sequenced S. pneumoniae strain, with zinc uptake regulation handled exclusively by AdcR36 which is functionally analogous to Lactobacillus lactis ZitR.37 SczA, a predicted tetracycline repressor (TetR)-family transcriptional regulator controls the expression of a divalent metal ion efflux transporter encoded by czcD38,39 which belongs to the cation diffusion facilitator (CDF) family of transporters.40,41 As such, S. pneumoniae encodes a complete Zn(II) homeostasis system controlled by novel regulators from well-known protein structural families.42 This homeostasis system likely allows S. pneumoniae to effectively respond to changes in external zinc availability during the course of a bacterial infection of the human host, with zinc concentrations reported to vary from the low μM in the nasopharynx, to ≈220 μM in lung tissue to 15 μM in the serum,43 while during inflammation [Zn] is known to increase in the blood and other tissues.44
In this work, we present a detailed biological and structural characterization of the Zn(II) dependent repressor AdcR from S. pneumoniae strain D39. A comparative DNA microarray analysis of isogenic wild-type and ΔadcR strains allows us to clearly define the AdcR regulon in S. pneumoniae strain D39, while revealing that the ΔadcR strain contains ≈2-fold more Zn(II) that the wild-type strain, and exhibits a measurable slower-growth phenotype. We exploit this finding to identify candidate Zn(II)-coordinating residues on AdcR by characterizing strains containing adcR missense alleles, and confirm these results using NMR spectroscopy and quantitative adc operator DNA binding measurements. These studies reveal that Zn(II) strongly activates DNA binding by the AdcR homodimer upon formation of a high affinity, five-coordinate nitrogen/oxygen-rich Zn(II) complex via His108 and H112 in the C-terminal regulatory domain, and His42 in the N-terminal winged helical DNA-binding domain. Other non-cognate metals, Mn(II) and Co(II), also activate DNA binding in vitro, the functional implications of which are discussed.
Results
Microarray determination of the AdcR regulon in Streptococcus pneumoniae
Relative transcript amounts of 15 genes increased or decreased (Fig. 1) with p ≤ 0.001 in D39 ΔadcR (IU2594) compared to adcR+ (IU1781). As anticipated and previously observed using RT-PCR in Streptococcus pyogenes45, each of the genes encoding the Zn(II)-specific ABC uptake system adcCBA (SPD_1999, SPD_1998, SPD_1997, respectively) increased in transcript abundance in the ΔadcR strain. In addition, as was predicted in a genomic analysis4 and shown by RT-PCR analysis,26 transcript amounts of the pneumococcal histidine triad (Pht) proteins also increased in the ΔadcR strain. These Pht genes include phtA (SPD_1038), phpA (SPD_1037), phtD (SPD_0889), phtE (SPD_0890), and a pht truncation (SPD_0892) and frameshift (SPD_0891) gene. In addition, the cell surface proteins adcAII (SPD_0888), which had previously been hypothesized to be regulated by AdcR in S. pneumoniae,27 and pspC (SPD_2017) are increased in transcript abundance in the ΔadcR strain. Interestingly, two putative zinc-containing alcohol dehydrogenases are down-regulated in the ΔadcR strain, SPD_0265 and SPD_1865 (Fig. 1). Although it is unknown if this is a direct or indirect effect of the deletion, the intergenic region upstream of the promoter for SPD_1865 harbors one near-consensus AdcR operator sequence.
Fig. 1.
Microarray analyses of relative transcript amounts in strains IU2594 (ΔadcR) and IU1781 (adcR+) grown exponentially in BHI. Microarray analyses were performed as described in Materials and Methods. A representative log-scale scatter plot of relative transcript amounts is shown, and the fold changes and Bayesian P values for transcripts changing at least 2-fold are listed in Supplementary Table S2.
AdcR residues H112 and H108 are essential for AdcR function in vivo
Growth analysis was done comparing adcR+ parent (IU1781), ΔadcR (IU2594), adcR H112Q (IU2600), adcR C30A (IU2657), adcR H111Q (IU2661), and adcR H108Q (IU4036) strains (Fig. 2a). The strains were grown in brain heart infusion media (BHI) overnight and then diluted to an OD620 of 0.001 in BHI with 200 μM ZnSO4 added. Under this moderate zinc stress a difference in lag time and yield can be observed in the parent adcR+ and ΔadcR strains. The ΔadcR strain reproducibly exhibits a slightly longer lag phase and a lower overall growth yield. Sequence changes in adcR generating amino acid substitutions H112Q and H108Q in AdcR replicate the growth effect observed for ΔadcR, while strains encoding substitutions H111Q and C30A grow similarly to the parent strain adcR+. This implicates H108 and H112 as essential for the function of AdcR in vivo.
Fig 2.
Growth and cellular zinc content of adcR mutants. (a) Growth curves of adcR parent, ΔadcR, adcR C30A, adcR H108Q, adcR H111Q, and adcR H112Q strains of S. pneumoniae in the presence of 200 μM ZnSO4. (b) Cellular zinc concentration in bacteria growing exponentially (OD620 = 0.1 to 0.3). The average of three biological replicates are shown with SEMs (P < 0.05). See Materials and Methods for ICP-MS and calculation of cellular concentration.
ICP-MS analysis (Fig. 2b) shows that the ΔadcR strain has a 2-fold higher cell-associated zinc concentration compared to the adcR+ parent strain (0.4 mM vs. 0.8 mM, respectively). A two-fold elevated zinc concentration is also observed in the adcR H112Q and H108Q mutants, which indicates that these residues are essential for the in vivo functionality of AdcR. The strains with adcR C30A and H111Q mutations have near wild type levels of zinc, which when in combination with the growth data indicate that they are non-essential residues for in vivo AdcR function.
Wild-type AdcR is a homodimer in solution
Purified wild-type S. pneumoniae AdcR elutes from a gel filtration column (pH 7.0, 0.2 M NaCl) with an elution volume corresponding to a molecular weight of 33 kD or that of a homodimer (monomer MW=16,605.9 D) (Fig. S1). This result is consistent with sedimentation velocity ultracentrifugation experiments (pH 6.0, 0.05 M NaCl, 25.0 °C) which reveal that the predominant apo-AdcR species is a homodimer characterized by s20,w=3.65 S and a frictional coefficient, f/fo, near 1.0, indicative a nearly spherical hydrodynamic particle (Table S3). The same is true for Zn(II)-complexed AdcR. AdcR monomer was not visible within these boundaries, thus placing a lower limit of Kdimer of 106 M−1 under these conditions. A small fraction of apo-AdcR stored at −80 °C becomes resistant to reduction by dithiothreitol or TCEP and migrates on an overloaded denaturing SDS-PAGE gel as a dimer (Fig. S2). Although the chemical nature of this presumed covalent linkage is unknown, crosslinked AdcR dimer can be detected by ESI-MS, the formation of which is lost in the C30A AdcR mutant (Table S4). These data taken collectively are consistent with the interpretation that both wild-type and C30A AdcRs are stable homodimers with and without bound Zn(II).
Zinc binding properties of wild-type AdcR
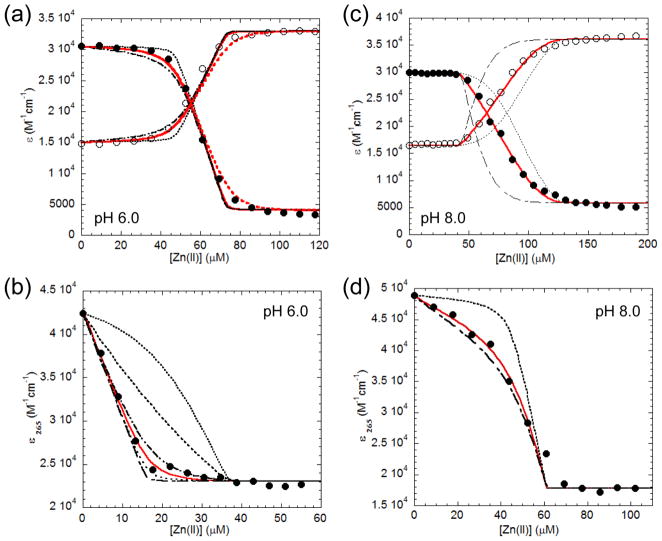
The in vivo experiments above suggest that H108 and H112 play important roles in AdcR function. It was therefore of interest to quantify the Zn(II) binding affinity, stoichiometry and coordination chemistry of Zn(II) in the wild-type and mutant AdcR homodimers. Since the C-terminal region of S. pneumoniae AdcR contains a cluster of five consecutive His residues in an eight amino acid sequence (107EHHHHHEH114) we anticipated a potentially strong pH-dependence on the binding affinity and stoichiometry, and therefore measured these parameters at pH 6.0 and pH 8.0 as representative of conditions below and above the pKa of a typical His residue (neglecting coupling between His side chains).46,47 Zn(II) binding was measured indirectly using a chelator competition experiment with two different chelators, mag-fura-2 (mf2; KZn=5.0×107 M−1)48 and quin-2 (KZn=2.7 × 1011 M−1),49,50 whose affinities are not strongly dependent on pH. Mf2 allows an estimation of the zinc binding affinity, Ki, between ≈106 and ≈109 M−1 while quin-2 effectively brackets ≈1010 and 1013 M−1.48,51
Fig. 3(a)–(b) shows representative zinc binding titrations for wild-type AdcR at pH 6.0, while Fig. 3(c)–(d) shows representative data for wild-type AdcR at the higher pH 8.0, with mf2 competitions shown in panels (a) and (c) and corresponding quin-2 competitions shown in panels (b) and (d). At pH 6.0, analysis of the mf2 titration reveals KZn1 of ≥109 M−1 and KZn2≈109 M−1, since virtually all of the added Zn(II) is bound to AdcR until both protomers are saturated; only then does mf2 coordinate metal giving an absorption change (Fig. 3(a)).52 A third binding event could be also be detected in these experiments (KZn3>106 M−1) (Table 2). In order to better estimate KZn1, Zn(II) was titrated into a solution of AdcR and quin-2 (Fig. 3(b)). These data reveal that KZn1≈1.0 × 1010 M−1. Thus, AdcR binds two mol equiv of Zn(II) per dimer strongly with step-wise affinity constants, KZn1 and KZn2, that differ by no more than a factor of 10, with a third Zn(II) binding much more weakly (Table 2). At pH 8.0, up to five stepwise binding events were required to fit the data, KZn1, KZn2, KZn3, KZn4 and KZn5. KZn1 and KZn2 are presumed to represent binding to one pair of symmetry related sites on the dimer, whereas KZn3 and KZn4 are predicted to represent binding to a second pair of sites on the dimer. The mf2 competition curve (Fig. 3(c) is best fit KZn1 and KZn2≥109 M−1, with KZn3=1.4 (±0.2) × 108 M−1, KZn4=3.0 (±0.3) × 107 M−1 and KZn5=3.8 (±0.8) × 106 M−1. Competition experiments carried out with quin-2 (Fig. 3(d)) allow an estimate of the highest affinity sites with KZn1=KZn2=1.4 (±0.2) × 1012 M−1. All zinc binding affinities of AdcR are larger pH 8.0 relative to pH 6.0, with the highest affinity sites (sites 1 and 2) increasing by 100–1000-fold; in addition, the appearance of several lower affinity sites are detectable at the highest pH.
Fig. 3.
Representative binding isotherms obtained from titrating Zn(II) into a mixture of AdcR wild-type and competitor (mag-fura-2 or quin-2) at pH 6.0 (a–b) or pH 8.0 (c–d). In the mag-fura-2 experiments, filled symbols represent ε366 values which are maximal in the apo-mf2 and open symbols represent ε325 values that are maximal in the Zn(II)-mf2 complex. For quin-2 experiments, filled circles represent ε265 values that are maximal in apo-quin-2. (a) 47.6 μM AdcR and 25 μM mf2. The solid red line represents non-linear least-square fit to a Zn(II):AdcRdimer, 3:1 binding model with the KZn1 stepwise binding affinity fixed to ≥109 M−1 (a lower limit) and optimized to KZn2=1.2 (±0.3) × 109 M−1, KZn3=1.7 (±0.1) × 106 M−1. The black dot and dash lines are simulated data to KZn1 = 1010 and 109 M−1, respectively. (b) 41 μM AdcR and 16 μM quin-2. The solid red line represent non-linear least-square fit to a Zn(II):AdcRdimer, 0.5:1 binding model, where KZn1 = 8.3 (±0.2) × 109 M−1. Simulated data is represented by discontinuous black lines, with KZn1 = 1012, 1011, 1010, 109, 108 M−1, descending from right to left. (c) 42.5 μM AdcR and 17.7 μM mf2. The solid red line represent non-linear least-square fit to a Zn(II):AdcRdimer, 5:1 binding model with KZn1=KZn2 = 1011 M−1, KZn3=1.4 (±0.2) × 108 M−1, KZn4=3.00 (±0.03) × 107, KZn5=3.8 (±0.8) × 106 M−1. The dot and dash black lines are simulated curves one order of magnitude larger and lower, respectively. (d) 44 μM AdcR and 17.1 μM quin-2. The solid red line represent non-linear least-square fit to a Zn(II):AdcRdimer, 2:1 binding model with KZn1=KZn2 = 1.4 (±0.2) × 1012 M−1. The black dot and dashed line are simulated data to KZn1=KZn2=1013 and 1012 M−1, respectively.
Table 2.
Binding affinities of Zn(II) (×109 M−1) and other metal ions for wild-type and selected mutant AdcR homodimersa
| AdcR | pH 6.0b | pH 8.0c | ||||||
|---|---|---|---|---|---|---|---|---|
| KZn1 | KZn2 | KZn3 | KZn1 | KZn2 | KZn3 | KZn4 | KZn5 | |
| WT | 8.3 (±0.5) | 1.2 (±0.3) | 0.0017 (±0.001) | 1400 (±200)d | 1400 (±200)d | 0.14 (±0.02) | 0.030 (±0.003) | 0.0038 (±0.0008) |
| C30A | 1.2 (±0.1) | 0.055 (±0.001) | 0.001 (±0.0001) | 710 (±70) | 710 (±70) | 0.071 (±0.001) | 0.087 (±0.001) | 0.014 (±.001) |
| H42A | 0.012 (±0.004) | –e | – | 7.3 (±1.0) | ≥1.0 | 0.23 (±0.07) | 0.036 (±0.007) | – |
| H108A | – | – | – | 1.2 (±0.1)d | 1.2 (±0.1)d | 0.017 (±0.001) | 0.009 (±0.001) | 0.0020 (±0.0005) |
| H111A | 3.4 (±0.9) | 0.22 (±0.02) | 0.001 (±0.0006) | 903 (±90)d | 903 (±90)d | 0.030 (±0.004) | 0.007 (±0.002) | – |
| H112A | – | – | – | 32 (±10) | 1.2 (±0.5) | 0.090 (±0.001) | 0.009 (±0.001) | 0.0020 (±0.0006) |
| H112Q | 0.010 (±0.004) | – | – | 6.2 (±1.0) | 0.012 (±0.001) | 0.0020 (±0.001) | –e | –e |
| Other metal ions: | ||||||||
| KCo1f | KCo1g | KCo2g | KCo3g | KCo3g | ||||
| C30A | 2.4 (±0.4) × 105 | – | – | 6.7 (±0.7) × 106 | 6.7 (±0.7) × 106 | 3.0 (±0.1) × 104 | 3.0 (±0.1) × 104 | |
| KMn1h | KMn2h | |||||||
| 1.3 (±0.2) × 105 | 1.3 (±0.2) × 105 | 1.0 (±0.1) × 104 | 1.0 (±0.1) × 104 | |||||
| 4.6 (±0.1) × 105,i | 4.6 (±0.1) × 105,i | –e | –e | |||||
25 mM bis-Tris, pH 6.0, 0.2 M NaCl, 25.0 °C or 25 mM Tris-HCl, pH 8.0, 0.5 M NaCl, 25.0 °C. Measured as shown by representative plots in Figs. 3–4 (see text for details) by chelator (mf2 and quin-2) competition assays.
Fit to 2 (KZn1, KZn2) or 3 (KZn1, KZn2, KZn3) step-wise binding events on a nondissociable AdcR dimer ([AdcR monomer]≥25 μM in all cases.
Fit to 4 (KZn1–KZn4) or 5 (KZn1–KZn5) step-wise binding events (see text for details).
Constrained KZn1=KZn2 since goodness-of-fit did justify otherwise.
–, binding event not detected with mf2 (KZn3<5×105 M−1).
Determined by direct titration of wild-type AdcR (Fig. S3)
Determined by isothermal titration calorimetry (Fig. S5(a)) with the constraint that KCo1= KCo2 and KCo3= KCo4.
Determined by ITC (see Fig. S5(b)) with the constraint that KMn1= KMn2 and KMn3= KMn4 (Fig. 5(b)).
Analogous methods were used to extract Zn(II) binding affinities and stoichiometries for mutant AdcRs (Fig. 4, Table 2). Substitution of the lone Cys in AdcR with Ala (C30A) results in a protein that exhibits properties like wild-type-like AdcR (Table 2); this is consistent with the wild-type like in vivo functional properties of C30A AdcR (Fig. 2; see below). H111A AdcR, like C30A AdcR, is also essentially wild-type in these assays (Table 2). In contrast, H42A, H108A and H112A/Q AdcRs exhibit very weak or no detectable Zn(II) binding at pH 6.0 (KZn1≤107 M−1) with ≈100–1000-fold decreases in zinc affinities of the first two mol equivalents of metal bound at pH 8.0 (Fig. 4, Table 2). These data reveal that introduction of substitutions specifically at conserved residues His42, His108 and His112 leads to a loss of high affinity Zn(II) binding by AdcR.
Fig. 4.
Binding isotherms obtained from titrating Zn(II) into a mixture of AdcR variants and competitor mag-fura-2 at pH 6.0 (a–b) or pH 8.0 (c–d). Closed markers represent ε366 values which are maximal in the apo-mf2 and open markers represent ε325 values that are maximal in the Zn(II)-mf2 complex. (a) 34.4 μM H108A AdcR and 28.6 μM mf2 and (b) 30 μM H112A AdcR and 30 μM mf2. The red solid line represent a non-linear least-square fit to stoichiometric binding of Zn(II) to mf2. (c) 25.3 μM H108A and 22.1 μM mf2. The red solid line represents a non-linear least-square fit to Zn(II):AdcRdimer, 5:1 binding model where KZn1=KZn2=1.17×109 M−1, KZn3=1.77 (±0.1) × 107 M−1, KZn4=9.0 (±0.9) × 106 M−1, KZn5=1.8 (±0.5) × 106 M−1. (d) 41 μM H112A and 27 μM mf2. The red solid line represent a non-linear least-square fit to Zn(II):AdcRdimer, 5:1 binding model where KZn1= 3.5 × 109 M−1 (fixed), KZn2=1.2 (±0.5) × 109 M−1, KZn3=9 (±1) × 107 M−1, KZn4=9 (±1) × 106 M−1, KZn5=2.0 (±0.6) × 106 M−1.
Presumed non-cognate transition metal ions Mn(II) and Co(II) were also tested for their ability to bind to apo-AdcR (Table 2). Titration of Mn(II) into a solution of apo-AdcR and mf2 at pH 8.0 gives two higher affinity sites KMn1,2 of ≈105 M−1 (Fig. S3); by ITC, two pairs of Mn(II) sites are observed with ≈105 M−1 and ≈104 M−1 affinities (Fig. S4(a)). The Mn(II) affinity of AdcR is therefore approximately six orders of magnitude weaker than Zn(II) under the same solution conditions (Table 2). Mn(II) is unlikely to be sensed by AdcR in vivo since stressing the ΔadcR strain with 300 μM Mn(II) has no impact on the growth kinetics of this strain (Fig. S5). The cobalt binding affinity was estimated by direct titration at pH 6.0 (Fig. S6), and gives KCo1 of 2.4 × 105 M−1, with two pairs of Co(II) binding sites clearly observed by ITC at pH 8.0, with step-wise affinities of 6.7 × 106 and 3.0 × 104 M−1 (Fig. S4(b)). Thus, at pH 8.0, metal stoichiometries for both Mn(II) and Co(II) are consistent with that observed for Zn(II) at pH 6.0, and are characterized by relative affinities largely as expected from the trend of stability of small molecule-metal complexes known as the Irving-Williams series.53,54 The molar (monomer) absorptivity of the Co(II) complex in the visible region which reports on d-d electronic transitions of d7 Co(II) ion is weak, ≈60 M monomer−1 cm−1 (Fig. S6) and most consistent with a distorted five-coordinate complex lacking a cysteine ligand, rather than a tetrahedral complex observed in virtually all previously characterized bacterial zinc sensors.33,55–57
X-ray absorption spectroscopy reveals a five-coordinate Zn(II) complex for the high affinity sites
Unlike Co(II), Zn(II) is spectroscopically silent due to a filled d-shell (d10); as a result, we used x-ray absorption spectroscopy55,58 to investigate the coordination geometry of the high affinity site(s) in wild-type AdcR (at pH 6.0 and 8.0) and C30A AdcR (at pH 6.0) that result upon addition of ≈0.8 monomer mol equiv Zn(II). Zn near-edge spectra for all three species are shown in Fig. 5(a), with best-fits of the EXAFS and Fourier transforms (FTs) of the EXAFS data shown in Fig. 5(b) for wild-type AdcR (pH 6.0) or in Fig. S7. Parameters that derive from all three fits are also given (Table 3). The near-edge spectra for all three complexes are very similar, as are the fits to each data set; these data unambiguously reveal that Cys30 does not donate a thiolate ligand to the Zn(II) complex, and the structure of the chelate is not detectably changed as a result of the C30A substitution or a change in pH. All three data sets are best-fit with a five-coordinate (N/O) complex, modeled with three histidine ligands, and two additional N/O ligands. Although we could not obtain a clear determination of the number of coordinated histidine residues, histidine coordination is consistent with significant FT intensity in the 3–4 Å region (Fig. 5(b)).
Fig. 5.
X-ray absorption spectroscopic data of Zn(II)-bound AdcR. (Top) Zn K-edge X-ray absorption spectra of Zn(II)-bound WT AdcR at pH 6 (black), C30A mutant (green), and WT AdcR at pH 8 (blue). (Bottom) EXAFS Fourier transform (k3-weighted, k = 2–13 Å−1 of WT AdcR at pH 6. (Inset) k3-weighted EXAFS spectra for WT AdcR at pH 6. For bottom panel, experimental data are black and best-fit simulations (Table 3, Fit 1) are red.
Table 3.
Curve fitting results for Zn K EXAFSa
| Sample, filename (k range) Δk3 χ |
Fit | Shell | Ras (Å) | σas2 (Å2) | ΔE0 (eV) | f′b |
|---|---|---|---|---|---|---|
| AdcR WT, pH 6.0, ZW01A (k = 2 – 13 Å−1) Δk3 χ = 12.672 |
1 | Zn-N2 | 2.06 | 0.0039 | −0.962 | 0.149 |
| Zn-N3 | 1.98 | 0.0026 | [−0.962] | |||
| Zn-C3 | [2.96] | [0.0039] | [−0.962] | |||
| Zn-C3 | [3.01] | [0.0039] | [−0.962] | |||
| Zn-N3 | [4.14] | [0.0034] | [−0.962] | |||
| Zn-C3 | [4.19] | [0.0034] | [−0.962] | |||
| AdcR C30A, Z301A (k = 2 – 13 Å−1) Δk3 χ = 12.776 |
2 | Zn-N2 | 2.04 | 0.0037 | −0.743 | 0.147 |
| Zn-N3 | 1.98 | 0.0026 | [−0.743] | |||
| Zn-C3 | [2.96] | [0.0039] | [−0.743] | |||
| Zn-C3 | [3.01] | [0.0039] | [−0.743] | |||
| Zn-N3 | [4.14] | [0.0034] | [−0.743] | |||
| Zn-C3 | [4.18] | [0.0034] | [−0.743] | |||
| AdcR WT, pH 8.0, ZW01B (k = 2 – 13 Å−1) Δk3 χ = 12.902 |
3 | Zn-N2 | 2.07 | 0.0021 | 0.361 | 0.154 |
| Zn-N3 | 2.00 | 0.0026 | [0.361] | |||
| Zn-C3 | [3.01] | [0.0039] | [0.361] | |||
| Zn-C3 | [3.06] | [0.0039] | [0.361] | |||
| Zn-N3 | [4.19] | [0.0034] | [0.361] | |||
| Zn-C3 | [4.24] | [0.0034] | [0.361] |
Shell is the chemical unit defined for the multiple scattering calculation. Subscripts denote the number of scatterers per metal. Ras is the metal-scatterer distance. σas2 is a mean square deviation in Ras. ΔE0 is the shift in E0 for the theoretical scattering functions.
Numbers in square brackets were constrained to be either a multiple of the above value (σas2) or to maintain a constant difference from the above value (Ras, ΔE0). Underlined numbers were fixed at the indicated value (not optimized).
NMR studies reveal that Zn(II) coordination by His42, His108 and His112 are required to drive a quaternary structural change in the dimer, while His111 is not
The metal binding experiments presented above reveal H42A, H08A and H112Q/A mutants have greatly perturbed Zn(II) binding equilibria and are therefore strong candidates for metal ligands required to allosterically activate adc operator DNA binding. Uniformly 15N-labeled wild-type AdcR with ≈70% fractional deuteration and selected fully protonated mutant AdcRs were purified and 1H-15N TROSY spectra recorded for each in the presence and absence of Zn(II) at pH 6.0, 35 °C. In all cases, ≈2 mol equiv of Zn(II) per monomer was added to ensure complete saturation of the two highest affinity zinc sites (if present), as well as any lower affinity sites that might be populated under these conditions (see Table 2).
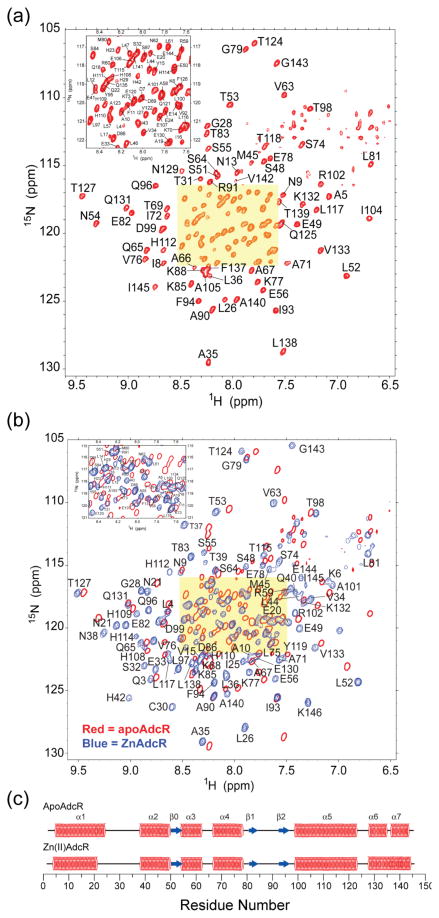
Fig. 6(a) shows a 1H-15N TROSY spectrum of apo-AdcR with sequence specific resonance assignments derived from analysis of triple resonance data indicated for each crosspeak (to be reported elsewhere). All but four (of 143) backbone 1H-15N pairs have been assigned, with the spectrum fully consistent with a highly α-helical two-fold symmetric homodimer. Analysis of backbone 1H, 15N, 13Cα, 13Cβ and 13C′ chemical shifts with TALOS59 gives the secondary structural analysis of the primary structure of apo-AdcR as shown (Fig. 6(c), top). These data reveal seven α-helices (labeled α1-α7) and three short β-strands (labeled β0-β2). A comparison of these features with known structures of MarR family regulators7–12,60 is consistent with an N-terminal winged helical domain consisting of α1–β2, containing an α3-α4 helix-turn-helix and a β1-β2 hairpin wing, and a C-terminal all α–helical regulatory domain linked by a long α5 helix. His42 is in the N-terminal region of the α2 helix in the DNA binding domain, while the poly-His run containing His108 and His112 is positioned in the middle of the α5 helix (Fig. 6(c)).
Fig. 6.
1H-15N TROSY spectra of wild-type AdcR in the absence (a) and presence (b) of Zn(II). Sequence-specific resonance assignments are shown for each conformer, with the single-contour red crosspeaks in panel (b) corresponding to those of apo-AdcR. (c) Secondary structural analysis of apo- and Zn(II)-bound AdcR as determined by analysis of the backbone chemical shifts in TALOS.59 Backbone resonances for residues R2, T37, R136 and K146 are unassigned in apo-AdcR. Likewise, backbone resonances for residues L17, L46, P103, Q121, P128, Q135 and K146 are unassigned in Zn(II)-bound AdcR.
Fig. 6(b) shows a 1H-15N TROSY spectrum of Zn(II) complexed AdcR (cyan crosspeaks, labeled) superimposed on the spectrum of apo-AdcR (red crosspeaks, single contour line), with a secondary structural analysis of these data shown in Fig. 6(c) (bottom trace). Zn(II) binding results in large changes in the spectrum that are global in nature extending from the N-terminal α1 to the C-terminal α7 helix (Fig. 7, bottom trace). Interestingly, while the secondary structure of the core of the molecule is unchanged, the N-terminal α1 helix appears to be shortened by three amino acids, with the α6 and α7 helices apparently making a continuous helix. In some other MarR regulators, these two terminal helical regions derived from opposite subunits (e.g., α1 and α6′-α7′) are in close proximity,10 and in AdcR may well reorient as a result of Zn(II) binding. In any case, we reasoned that these spectral changes could be used as a sensitive reporter of quaternary conformational transition in mutant AdcR homodimers upon binding activating metal ions.
Fig. 7.
Chemical shift (1H, 15N) perturbation maps |apo – Zn| of AdcR variants (35 °C, pH 6.0).
Uniformly 15N-labeled H42A, H108A, H111A and H112A AdcRs were purified and a 1H-15N TROSY spectra of apo- and Zn(II) bound forms acquired (spectra not shown). Chemical shift perturbation maps of all four apoproteins relative to the wild-type AdcR (apo-apo) reveals that each mutant is stably folded and adopts a structure very similar to that of apo-wild-type AdcR (Fig. S8). The largest perturbations (Δδ≤0.3 ppm) are obtained with an Ala substitution of H42 in the α2 helix, which influences the chemical shifts of residues not only in the α2 helix, but also in the α5 and α7 helical regions, indicative a long-range perturbations even in the apo-state. Strikingly, addition of Zn(II) to H42A, H108A and H112A results in virtually no spectral changes (Fig. 7), direct evidence that Zn(II) binding (albeit weakly; see Table 2) to these mutants fails to switch the conformation to a high affinity DNA binding state (see below). This is in contrast to H111A AdcR which gives a perturbation map that is nearly identical to that of wild-type AdcR (Fig. 7). These data reveal that loss of a high affinity binding site(s) on the dimer (see Table 2) results in loss of the ability to undergo quaternary structural conformational switching. Thus, these structural data provide further evidence that H42, H108 and H112 are direct ligands to the regulatory pair of Zn(II) binding sites on AdcR.
Zinc binding by AdcR activates adc operator DNA binding
In S. pneumoniae D39, an imperfect 5-2-5 inverted repeat sequence 5′-TTAACTGGTAAA is positioned between the adcR translation start codon and the −10 region of the promoter that conforms to the consensus 5′-TTAACNRGTTAA sequence.4,14 This AdcR operator site is identical to that found in ZitR-regulated genes in Lactobacilis lactis.37 A standard fluorescence anisotropy-based DNA binding experiment61 was used to measure the affinity of apo- and Zn(II)-bound wild-type and mutant AdcRs for a fluorescein-labeled 28-nucleotide duplex DNA harboring a single adc operator (AdcO) derived from the adcR gene. In all cases, the binding was measured in presence of non-specific competitor DNA to ensure specificity in either excess Zn(II) or 0.5 mM EDTA and the resulting data were fit to a non-dissociable AdcR dimer binding model to obtain KDNA-Zn or KDNA-apo. Wild-type AdcR binds with an affinity of 2.4 (±0.3) × 108 M−1 in the presence of Zn(II) (KDNA-Zn), with no detectable binding observed in the presence of EDTA or chelexed buffer (KDNA-apo≤105 M−1) (Fig. 8). These data reveal that AdcR is a Zn(II) activated DNA binding protein, characterized by a lower limit for the allosteric coupling free energy, ΔGc≤–4.6 kcal mol−1 (Table 3).62 Interestingly, non-cognate metal ions Co(II) and Mn(II) also activate AdcR binding to the adc operator, with these metals nearly indistinguishable from that of Zn(II) in this assay (Table 4). This finding suggests that Co(II) and Mn(II) form coordination complexes that are isostructural with that of Zn(II), at least when bound to the operator DNA (see Discussion).
Fig. 8.

Representative isotherms of the binding of Zn(II)-AdcR variants to a fluorescein-labeled 28-mer double stranded DNA containing the AdcO sequence. The anisotropy change is normalized to that observed for wild-type AdcR (circles); for all the other variants, C30A (squares), H42A (diamonds), H108A (triangles) and H112A (gridded square) AdcRs, the fractional change in anisotropy was calculated relative to the total change observed for wild-type AdcR. The Zn(II)-activated binding is reversible at the end of the experiment by addition of 500 μM EDTA (not shown). The red continuous lines represent non-linear least-square fits for the binding of a non-dissociable dimer to DNA with the affinities (KDNA Zn) complied in Table 4.
Table 4.
Equilibrium binding affinities (Ki) resolved from fluorescein anisotropy-based AdcR operator binding isotherms for individual AdcR derivatives in the presence and absence of Zn(II).a
| AdcR variant | KDNA Znb (×108 M−1) | KDNA Meb (×108 M−1) | KDNA apoc (×108 M−1) | ΔGcd (kcal mol−1) |
|---|---|---|---|---|
| Wild-type + Zn | 2.4 (±0.3) | – | ≤ 0.002 | −4.2 |
| Wild-type + Mn | – | 0.82 (±0.05) | ≤ 0.002 | −3.6 |
| Wild-type + Co | – | 1.2 (±0.2) | ≤ 0.002 | −3.8 |
| S-methylated wild-type | 1.1 (±0.1) | – | NDe | ND |
| C30A | 1.1 (±0.1) | – | ≤ 0.002 | −3.7 |
| H42A | 0.047 (±0.001) | – | ≤ 0.002 | −1.9 |
| H108A | 0.020 (±0.0007) | – | ≤ 0.002 | −1.4 |
| H111A | 1.4 (±0.1) | – | ≤ 0.002 | −3.9 |
| H112A | 0.038 (±0.001) | – | ≤ 0.002 | −1.7 |
| H112Q | 0.098 (±0.007) | – | ≤ 0.002 | −2.3 |
Conditions: 25 mM bis-Tris, pH 6.0, 25.0 °C, 0.2 M NaCl, 0.005% Tween 20, 0.8 μg/mL of nonspecific deoxyribonucleic acid from salmon sperm.
For determination of KDNA Zn, 100 μM ZnSO4 was added to the binding reactions.
For determination of KDNA Co and KDNA Mn, 10 μM of metal was added to the binding reactions.
For determinatin of KDNA-apo, 500 μM EDTA was present.
ΔGc = −RT·ln(KDNA-Zn/KDNA-apo) and represents a lower limit given the upper limit on KDNA-apo.
ND, not deternined.
Exactly analogous DNA binding experiments were carried out with each of the missense mutants with the Ki and ΔGc,i compiled in Table 4 with selected DNA binding curves shown in Fig. 8 for the indicated Zn(II) complexes (no binding was observed for any mutant in the presence of 0.5 mM EDTA). As expected, all mutant AdcRs that display high affinity Zn(II) binding (C30A, H111A) and exhibit conformational switching by NMR spectroscopy, e.g. H111A, are strongly activated to bind the AdcO DNA by Zn(II). S-methylation of Cys30 also has no effect on Zn(II)-activated DNA binding, further evidence of the non-essentiality of this cysteine in AdcR function. In contrast, H42A, H108A and H112A mutants are all weakly activated by Zn(II) binding, with affinities 50–200-fold reduced in the presence of Zn(II). Zn(II)-bound H112Q AdcR binds to the operator about three-fold more tightly than the other inactive mutants, with an affinity ≈20-fold reduced relative to wild-type AdcR (Fig. 8), despite the fact that this mutant is inactive in vivo. In fact, all functionally compromised mutants are activated to bind to the DNA operator to a detectable degree in vitro, although clearly not to an extent that yields functional co-repression in vivo (Fig. 2) (see Discussion).
Discussion
Transition metal sensing bacterial repressors comprise a collection of transcriptional regulatory proteins that allow an organism to control the intracellular availability of biological required metal ions.42,63 It is hypothesized that each metalloregulatory protein must be tuned such that it responds only to a single metal ion at a thermodynamic “set-point’ in the context of the intracellular milieu. Such a set-point is proposed to correspond to a concentration of “buffered” or “weakly chelated” metal ion in the intracellular compartment in which metalloregulation or sensing occurs. In bacteria, this compartment corresponds to the cytoplasm. It is therefore important to determine the affinity and stoichiometry of a sensor for cognate and noncognate metal ions, and determine how metal binding allosterically activates or inhibits repressor binding to its DNA operator.50,62,64,65
In this work, we show that S. pneumoniae AdcR is a zinc-sensing metalloregulatory protein that is characterized by a strongly pH-dependent affinity for metal ions, and forms a coordination complex that is thus far unique to our knowledge among zinc-specific bacterial regulators. Although the AdcR homodimer harbors two pairs of the zinc binding sites characterized by KZn ≥ 107 M at pH 8.0 (Table 2), the two highest affinity sites appear necessary and sufficient for metalloregulation of DNA binding in vitro and AdcR function in vivo. This pair of sites likely involves direct coordination of the imidazole side chains of His42 in the α2 helix within the DNA binding domain, and His108 and His112 on opposite ends of a poly-histidine tract in the α5 helix. These residues are absolutely conserved in all known and predicted members of the zinc-sensing AdcR/ZitR subfamily of MarR family regulators, and thus collectively donate three of the five ligand donor atoms to the regulatory Zn(II) ion in each subunit. This follows the predicted trend that two histidine residues separated by four residues apart in a helix (in this case H108 and H112) are used to bind metal ions.66 The affinity of this site is ≈109–1010 M−1 at pH 6.0 and increases by approximately two orders of magnitude, to ≈1012 M−1 at pH 8.0. The origin of the strong pH-dependence is unknown and has not been directly measured for any other bacterial zinc sensor. For S. aureus CzrA, formation of the two regulatory zinc complexes of KZn1=1012 M−1 and KZn2=1011 M−1, each composed of three His ligands and a carboxylate residue,50 occurs with the concomitant release of a net of ≈1 proton per monomer (or ≈2 per dimer) as measured by isothermal titration calorimetry at pH 7.0.67 On the other hand, the instrinsic pKa of transition metal ligands can be quite acidic, and thus would be fully deprotonated at neutral pH. However, neighboring non-liganding histidines and local electrostatics may strongly influence the degree of ligand deprotonation in apo-AdcR68–70 and thus the pH-dependence of the metal-binding affinity.
Of the five consecutive His residues in the α5 helix in AdcR, only His108, His111 and His112 are invariant and thus were strong candidates for direct coordination to the regulatory zinc ion. We show here that His108 and His112, predicted to be on the same side of the α5 helix, are required for AdcR function in vivo and in vitro. X-ray absorption spectroscopy, as well as spectroscopic features of the Co(II) complex, are most consistent with a coordination number (n) of five in this chelate; however, n=6 can not be rigorously excluded from these data. The identity of the additional ligands to the regulatory site, as well as any of the metal ligands to the second pair of lower affinity sites on the dimer are currently unknown. Zn(II) is most often coordinated by carboxylate, imidazole and/or thiolate ligands and typically adopts a tetrahedral (n=4) coordination geometry.33,50,56 On the basis of sequence conservation, strong candidates for other metal ligands are Glu20 and Glu24 in the α1 helical region, C30 in the α1-α2 loop, Glu41 and Glu107, each of which are immediately adjacent to ligands His42 and His108 (Fig. 6). The backbone 1H-15N TROSY crosspeak positions of Glu20 and Glu24 are not strongly perturbed on Zn(II) binding, evidence that this region is unlikely to donate ligands to the regulatory metal. Cys30 also does not donate a ligand to the regulatory pair of sites, since we could not obtain spectroscopic evidence for thiolate coordination in the regulatory site, nor is Cys30 essential for AdcR function. This finding does not rule out an additional regulatory role of Cys30 in an oxidative stress response superimposed on the primary zinc-sensing role of AdcR as recently proposed in S. suis.14,15 The physiological significance, if any, of redox chemistry occurring on Cys30 in S. pneumoniae AdcR remains unknown and contrasts sharply with bona fide MarR family members that sense peroxide or other ROS stress via formation of higher oxidation states of cysteine.11,71,72
Glu41 and/or Glu107 are the strongest candidates for donating a carboxylate ligand to the regulatory site in AdcR. Carboxylate side chains can assume monodentate or bidentate coordination, and thus one or both may be involved in Zn(II) coordination. It is interesting to note that efforts to create S. pneumoniae D39 strains harboring E41A and H42Q alleles yielded either wild-type revertants or suppressor mutants (data not shown). High resolution structural studies will be required to obtain a comprehensive picture of the (N/O)5 regulatory coordination complex in AdcR. The pentavalent coordination structure of the activating metal site in AdcR is unusual, as an increased coordination number is often associated with transition metal ions Fe(II) and Mn(II), with biological Zn(II) complexes overwhelmingly tetrahedral (n=4),73,74 despite the fact that there is no fundamental a priori reason why this should be the case.75 In fact, this feature alone might be expected to decrease the specificity of this sensor for Zn(II) in the cytoplasm. Other transition metals are capable of activating DNA binding to an extent quantitatively similar to that of Zn(II) (Table 4), a finding that contrasts sharply with non-cognate metal complexes formed by well characterized Ni(II) sensor NikR,76 and ArsR and CsoR/RcnR family sensors.77–79 In these cases, non-cognate metal ions nearly always adopt coordination complexes that differ structurally from that formed by the cognate metal ion.80 In AdcR, a fundamentally distinct mechanism of specificity is likely operative, with the relative metal binding affinities (Table 2) likely controlling AdcR function in the cell, i.e., bioavailable concentrations of Co(II) and Mn(II) never rise to the degree necessary to activate DNA binding in the cell. This ensures that only Zn(II), which may well be buffered to very low (femtomolar) “free” or weakly chelated intracellular concentrations81 far below that of Mn(II), Fe(II), Co(II) and Ni(II), will strongly activate DNA binding in the cytoplasm of S. pneumoniae.82 For AdcR, KZn≈1012 M−1 which is two-three orders of magnitude lower than that reported for zinc uptake and efflux regulators E. coli Zur and ZntR, respectively,81 but comparable to that of S. aureus efflux regulator CzrA.67 The functional significance of this discrepancy is currently unknown.
This model for the zinc specificity of AdcR also partly explains the apparent discrepancy between the ability of Zn(II) to activate AdcO binding in vitro vs. that which occurs in vivo. All AdcR mutants in which the high affinity Zn(II) sites are destroyed, are activated to small but significant degrees to bind to the operator under standard conditions; furthermore, this binding is sequence-specific since it is measured in the presence of excess non-specific DNA (Fig. 8). Thus, substitution of single metal ligands in the regulatory site does not abolish metalloregulation of DNA binding, although the magnitude of the allosteric coupling free energy (ΔGc) is significantly decreased, indicative of a perturbation of the communication between metal and DNA binding sites (Table 4). However, a regulatory KZn reduced by ≈200–1000 fold renders these AdcR mutants capable of sensing free Zn(II) concentrations only if they rise to nM range, thus making them non-functional in vivo (Fig. 2). Elucidation of the structural and dynamical mechanism of allosteric activation of DNA binding by Zn(II) by this novel MarR regulator, particularly in the context of well-studied allosterically inhibited zinc sensors, promises to reveal novel insights into MarR family function5 and zinc homeostasis in S. pneumoniae.38
Materials and Methods
Chemicals and reagents
All water used in these experiments was Milli-Q deionized (>18MΩ) and the buffers were obtained from Sigma. All glassware was acid washed before use and rinsed exhaustively with metal-free water. Indicator dyes mag-fura-2 and quin-2 were obtained from Invitrogen and Sigma, respectively. Metal stocks made with Ultra Pure Alfa Aesar metals. All other reagents are as indicated in the text. For all the chromatography steps an Ätka 10 purifier (GE) was used. All metal binding experiments were performed under anaerobic conditions. Dialysis into experimental buffer was under an Argon saturated atmosphere of no more than 8 ppm of oxygen.
Bacterial strains and growth conditions
Strains used in this study are listed in Table 1. Bacteria were grown on plates containing trypticase soy agar II (modified; Becton-Dickinson) and 5% (vol/vol) defibrinated sheep blood (TSAII BA) and incubated at 37 °C in an atmosphere of 5% CO2. Strains were also cultured statically in Becton-Dickinson brain heart infusion (BHI) broth at 37°C in an atmosphere of 5% CO2, and growth was monitored by optical density at 620 nm (OD620) using a Spectronic 20 spectrophotometer fitted for measurement of capped tubes (outer diameter, 16 mm). Bacteria were inoculated into BHI broth from frozen cultures, serially diluted into the same medium, and propagated overnight. Overnight cultures that were still in exponential phase (OD620, 0.1 to 0.3) were diluted back to an OD620 of ≈0.001 to start final cultures. Final cultures were grown in either BHI [containing ≈20 μM Zn(II) by ICP-MS) or BHI with 200 μM ZnSO4 added (for ≈220 μM total Zn(II)].
Table 1.
Strains used in the studya
| Strain | Genotype (derivation) | Antibiotic resistanceb | Reference or source |
|---|---|---|---|
| IU1781 | D39 rpsL1 | Strr | {Ramos-Montañez, 2008 #1} |
| IU2571 | D39 ΔadcR::kan-rpsL | Kanr | This paper |
| IU2594 | D39 rpsL1 ΔadcR | Strr | This paper |
| IU2600 | D39 rpsL1 AdcR H112Qc | Strr | This paper |
| IU2657 | D39 rpsL1 AdcR C30Ac | Strr | This paper |
| IU2661 | D39 rpsL1 AdcR H111Qc | Strr | This paper |
| IU4036 | D39 rpsL1 AdcR H108Qc | Strr | This paper |
Strains were constructed by transformation of indicated recipients with PCR fragments synthesized by fusion PCR (see Materials and Methods).
Antibiotic resistance markers: Kanr, kanamycin; Strr, streptomycin.
Codon changes: H112Q CAT to CAA; H111Q & H108Q CAC to CAA; C30A TGC to GCT
Microarray analysis
Bacteria were grown in BHI broth as described83 but were diluted 100-fold into BHI to start final cultures, which were harvested at an OD620 of ~0.2. RNA extraction and purification, cDNA synthesis, labeling and hybridization to S. pneumoniae R6 microarrays (Ocimum Biosolutions), array washing and data collection were performed as described previously.83 Data were collected from three independent biological replicates, including one dye swap, and analyzed using software from the TM4 Microarray Software Suite (www.tm4.org). Results files generated by GenePix Pro 6.0 software (Molecular Devices) were converted to TIGR MultiExperiment Viewer file format using ExpressConverter 2.1 software. Lowess (block) data normalization was performed using TIGR MIDAS 2.21 software. Spots with background signals higher than foreground signals and spots that were flagged as bad by the investigator or MIDAS software were removed from the analysis. Expression ratios and Bayesian P values were calculated as described previously.83 The cutoff for significant changes in relative transcript amounts was set at positive or negative 2-fold with a Bayesian P value of <0.001. Intensity and expression ratio data have been deposited in the GEO database (www.ncbi.nlm.nih.gov/geo/; accession no. GSE21506).
ICP-MS analysis
1 mL aliquots of S. pneumoniae cells in exponential phase were centrifuged and washed twice with BHI containing 5 mM nitrilotriacetic acid (Aldrich), then once with PBS that had been treated overnight with chelex (Biorad) according to the manufactures’ protocol. The cell pellets were dried overnight in a rotary evaporator. 400 μL of 2.5% v/v nitric acid (Ultrapure, Sigma-Aldrich) with 0.1% v/v Triton-X 100 were added to the cell pellets which were then lysed for 10 min at 95 °C with shaking at 500 rpm followed by vigorous vortexing for 20 s. 200 μL of the lysed cell solution was added to 1.3 mL of 2.5% v/v nitric acid for ICP-MS analysis. Analyses were performed using a Perkin Elmer ELAN DRCII ICP-MS. The instrument was equipped with a Microflow PFA-ST concentric nebulizer with a 100 μL/min self-aspiration capillary, a cyclonic spray chamber, a quartz torch and platinum sampler/skimmer cones. Germanium at 50 ppb was added as an internal standard using an EzyFit glass mixing chamber. Metal content was determined per cell using the OD620 to obtain the number of cells analyzed. Molar metal content was determined assuming a cell volume of 0.644 × 10−15 L84 and 10 cells per chain.
Cloning and mutations
The AdcR gene was PCR-amplified from S. pneumoniae D39 genomic DNA using the cloning oligonucleotides (Integrated DNA Technologies) 5′-CCGGATCCTCTACCGTAATATATCTCATTATTTGATTTCTC and 5′-CCCATATGAGACAGCTAGCAAAGGATATCAATGCTTTTTTG (BamHI and NdeI restriction sites are underlined, respectively) and cloned into the expression vector pET3a. The final construction has the entire native sequence of the AdcR gene under the control of a T7 promoter. AdcR single residue mutants were obtain using the protocol supplied by QuikChange from Stratagene using pET-AdcR as the template for PCR and appropriate mutagenic primers. All expression constructs were sequenced to verify the integrity of the plasmids on an ABI 3730 DNA Analyzer (Applied Biosystems) located in the Indiana Molecular Biology Institute. The plasmids were transformed into either BL21(DE3) or BL21 pLysS cells for protein expression.
Overexpression and purification
Growth was performed as previously described.85 Overexpression of AdcR was accomplished by induction of 1 L mid-log LB cultures with IPTG (Inalco) to a final concentration of 0.4 mM for 4 h at 37 °C or 30 °C overnight. The cells were harvested and resuspended in 25 mM MES, pH 6.0, 0.5 M NaCl, 5 mM EDTA, and 5 mM DTT. The cell suspension was lysed by sonication. After centrifugation, 10% PEI (polyethylenimine) pH 6 was added to the supernatant to a final concentration of 0.015% (v/v) and stirred for 2 h at 4°C. The supernatant was fractionated with two (NH4)2SO4 cuts (35% and 70%) and the pellet was resuspended and equilibrated in 25 mM MES pH 6, 0.1 M NaCl, 5 mM EDTA and 2 mM TCEP (GoldBio). Dialysis to remove excess (NH4)2SO4 was performed with at least four 1 L buffer exchanges over 16 h. This fraction was loaded on an SP-Sepharose Fast Flow column and fractions eluted in a salt gradient from 0.1–1 M NaCl. The cleanest fractions as determined by SDS-PAGE gels, were pooled and equilibrated in 25 mM Tris, pH 8.0, 50 mM NaCl, and 2 mM TCEP and loaded into a Q-Sepharose column. The cleanest fractions were then pooled, concentrated using centrifugal filter units (Millipore) and loaded on a Superdex 75 gel preparative grade column as the last step in the purification. The identity of AdcR wild-type and variant proteins was confirmed by LC-ESI/MS mass spectrometry at the Indiana University Department of Chemistry Mass Spectrometry Facility. Samples for LC-ESI-MS were prepared in the glove box under anaerobic conditions and passed through a Micro Bio Spin P6 desalting column (BioRad). The concentration of the AdcR monomer was determined for all variants using an ε280 of 2980 M−1 cm−1.
Zinc-binding experiments
All zinc binding experiments were carried out by using a Hewlett-Packard model 8452A spectrophotometer. Two different zinc chelator indicator dyes were used as apo-AdcR competitors under anaerobic conditions, mag-fura-2 (mf2) and quin-2. Protein was diluted in 800 μL of buffer (25 mM Bis-Tris, pH 6, 0.2 M NaCl or 25 mM Tris, pH 8, 0.5 M NaCl) that was passed through a Chelex (Bio-Rad) column to remove contaminating metals. These chelator competition experiments were carried out and the data corrected as previously described. 55,61,86 The mf2 isotherm was fit to the model indicated in the text using a KD,Zn = 0.022 μM for mf2.87 To corroborate the affinity of quin-2 for Zn(II) at pH 8.0 and 0.5 M NaCl, a series of measurements were done using 10, 20 and 28 μM EDTA (KZn = 2.7×1014 M−1, pH 8.0, 25.0 °C)88 competing for Zn(II) in presence of 20 μM quin-2. The affinity was fit to a KZn = 2.9 × 1011 M−1, which is in agreement to the previously reported value of 2.7 × 1011 M−1.49 All data was fit to a simple competition model using Dyna-Fit. 89
Fluorescence anisotropy experiments
A 28 mer double stranded 3′-fluorescein labeled DNA (5′-TGATATAATTAACTGGTAAACAAAATGT) and its complementary strand containing the native operator-promotor sequence found upstream AdcR were synthesized using a 3′-fluorescein CPG column (Glen Research) on a MerMade 4 oligonucleotide synthesizer (Bio Automation Corporation), and purified and annealed as described before.48,67 All anisotropy experiments were carried out with an ISS PC1 spectrofluorometer operating in steady-state mode fitted with Glan-Thompson polarizers in the L format (excitation wavelength set at 495 nm with 1 mm slit and the emission collected through a 515 nm filter). The G-factor was set to 0.99 and the average light intensity was 1×105 (a.u) with a decrease of no more than 5% at the end of the titration with all the components in solution. The average of 10 iterations of 40 s each for each ith addition of protein measured with a corresponding standard deviation that was ≤ 0.001. All experiments were carried out with 10 nM DNA in 50 mM Bis-Tris, pH 6.0, 0.2 M NaCl, 0.005% Tween-20, 1 mM TCEP and 80 μg/mL DNA sodium salt from salmon testes (Sigma) as an unspecific competitor in a volume of 1.8 mL. A solution containing 1–10 μM of AdcR and AdcR variants was titrated into the cuvette containing the DNA and allowed to mix for 3 min. To measure the allosteric effect of metals ions, one molar equivalent per monomer of the indicated metal was preincubated with the protein titrant. The binding reaction contained either 100 μM of Zn(II) or 10 μM Mn(II) or Co(II). The anisotropy of the apo-protein was measured in presence of 0.5 mM EDTA and was under the limit of detection. The buffer used to measured the anisotropy of Co(II)/Mn(II)-AdcR was passed through a Chelex (Bio-Rad) column and no anisotropy change was detected in absence of metal, assuring that no metal contaminants were present in the buffer or protein. The data, normalized as described previously,62,65 were fit to a binding model as indicated in the text. The maximal change in anisotropy for wild-type AdcR of 0.023, is consistent with that expected for one dimer bound per DNA duplex when taking into account the effect of the DNA length on Δrobs.61,65,86
Atomic absorption spectroscopy
All the Zn(II) and Co(II) metal stocks were measured as before using metal specific hollow cathode lamps61 and Mn(II) was detected at 279.5 nm (slit-width of 0.2 nm) in a Perkin-Elmer AAnalyst spectrometer. Zn(II) contents of the wild-type and AdcR apoprotein variants ranged from 0.01–0.08 monomer mol equiv. Mn(II) and Co(II) concentrations were negligible.
Quantitation of free thiol
DTNB was used to determine the total concentration of free thiol as previously described.55 The total concentration of free thiols ranged from 80–90% in all the AdcR and AdcR variants as purified. Glu-C in-gel digestion and LC-MS/MS analysis of the sample was done as previously described62 with the exception that 25 μL of a 20 ng/μL GluC was used to digested the samples. The LC-MS/MS was performed in Indiana University, Chemistry Department at the National Center for Glycomics and Glycoproteomics.
Derivatization of thiols
MMTS (methyl methanethiosulfonate) converts free sulfhydryl groups of cysteines side chains into –S-SCH3.90 Wild-type AdcR (100 μL, 690 μM) was modified by incubation with 100 fold molar excess of MMTS for 1 h. The protein was then passed through a Bio-Spin P6 (Bio-Rad) gel filtration column, concentrated using centrifugal filter membranes (Millipore) and buffer exchanged by dialysis to remove any unreacted MMTS. The modified protein was verified by LC-ESI-MS where AdcR (16,649 amu, MW= 16,605 + 2Na) forms a product after MMTS reaction of 16,697 (AdcR+ 2Na + 46 amu, the latter consistent with a single –S-SCH3 derivatization on Cys30).
NMR Experiments
NMR spectra were acquired on a Varian DDR 800 MHz spectrometer equipped with a cryogenic probe at the Indiana University MetaCyt Biomolecular NMR laboratory. NMR spectra were processed and analyzed using NMRPipe91 and SPARKY†. Typical solution conditions were 250–600 μM 15N-labeled or 15N/13C-labeled protein, pH 6.0, MES-d13 (Sigma), 50 mM NaCl, 1 mM tris(2-carboxyethyl)phosphine (TCEP; Gold Biotechnology), 0.02% NaN3 (Fisher) and 10% (v/v) 2H2O (CIL) at a level of 70% random deuteration. Zn(II)-AdcR samples contained 2 monomer mol equiv of Zn(II) and 1 mM imidazole. All spectra were acquired at 35 °C. Chemical shift referencing is relative to 2,2-dimethyl-2-silapentene-5-sulfonic acid (DSS; Sigma).92 Sequential resonance assignments (to be reported elsewhere) for the apo and Zn(II) forms were obtained using 1H-15N HSQC, and triple resonance HNCA, HN(CO)CA, HN(CA)CB, HN(COCA)CB and HNCO spectra.93–95
Zn(II) X-ray Absorption Spectroscopy
Samples were loaded into a polycarbonate XAS cuvette containing five 10-μL wells covered with a Mylar-tape front window and immediately frozen in liquid nitrogen. Zinc K-edge XAS data were collected at Stanford Synchrotron Radiation Lightsource (SSRL) on beam line 9-3 with the SPEAR storage ring operating at 3.0 GeV, 60–100 mA. X-ray absorption spectra were recorded with the sample at 10 K using a 1×1 mm beam incident on a fully tuned Si[220] double-crystal monochromator, downstream of a harmonic rejection mirror set for a 15-keV cutoff. XAS data were collected using a 30-element intrinsic Ge detector windowed to Zn Kα emission with a 6-absorption length Cu fluorescence filter backed by Soller slits. Energies were calibrated using an internal zinc standard, with the first inflection point assigned at 9660.7 eV. k values were calculated using a threshold (k = 0) energy of 9670 eV. The averaged XAS data for a sample represent 6–8 scans, each of 21 min duration, collected on several replicate sample wells. Data reduction and analysis were performed with EXAFSPAK software (www-ssrl.slac.stanford.edu/exafspak.html) according to standard procedures as described previously.96 Fourier transforms of the EXAFS spectra were generated using sulfur-based phase correction. The multiple-scattering model, based on zinc-bound imidazole, was calculated using FEFF version 8.0.97,98 Debye-Waller factors for the multiple-scattering legs were optimized by fitting a structural model of Zn(imid)42+ and were fixed during curve-fitting. Optimal imidazole coordination numbers were chosen based on the size of the 3- and 4-Å FT peaks arising from outer-shell C,N atoms of the imidazole ring. Zn-(N,O) distances and Debye-Waller factors of additional first-shell ligands were then optimized for the final fits.
Supplementary Material
Acknowledgments
We thank Yue Fu for his help in protein purification and Dhriti Mukherjee and Kyle J. Wayne for their assistance with S. pneumoniae D39 strain construction. This work was supported by grants from the NIH GM042569 (to D.P.G.), F32 AI084445 (to F.E.J.), GM042025 (to R.A.S.) and AI060744 (to M.E.W).
Abbreviations
- AdcR
adhesin-competence repressor
- MarR
multiple antibiotic resistance repressor
- TROSY
transverse relaxation optimized spectroscopy
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dintilhac A, Alloing G, Granadel C, Claverys JP. Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and Mn resulting from inactivation of putative ABC metal permeases. Mol Microbiol. 1997;25:727–739. doi: 10.1046/j.1365-2958.1997.5111879.x. [DOI] [PubMed] [Google Scholar]
- 2.Claverys JP, Dintilhac A, Mortier-Barriere I, Martin B, Alloing G. Regulation of competence for genetic transformation in Streptococcus pneumoniae. Soc Appl Bacteriol Symp Ser. 1997;26:32S–41S. [PubMed] [Google Scholar]
- 3.Dintilhac A, Claverys JP. The adc locus, which affects competence for genetic transformation in Streptococcus pneumoniae, encodes an ABC transporter with a putative lipoprotein homologous to a family of streptococcal adhesins. Res Microbiol. 1997;148:119–131. doi: 10.1016/S0923-2508(97)87643-7. [DOI] [PubMed] [Google Scholar]
- 4.Panina EM, Mironov AA, Gelfand MS. Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc Natl Acad Sci U S A. 2003;100:9912–9917. doi: 10.1073/pnas.1733691100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilkinson SP, Grove A. Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins. Curr Issues Mol Biol. 2006;8:51–62. [PubMed] [Google Scholar]
- 6.Ellison DW, Miller VL. Regulation of virulence by members of the MarR/SlyA family. Curr Opin Microbiol. 2006;9:153–159. doi: 10.1016/j.mib.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Alekshun MN, Levy SB, Mealy TR, Seaton BA, Head JF. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 A resolution. Nat Struct Biol. 2001;8:710–714. doi: 10.1038/90429. [DOI] [PubMed] [Google Scholar]
- 8.Lim D, Poole K, Strynadka NC. Crystal structure of the MexR repressor of the mexRAB-oprM multidrug efflux operon of Pseudomonas aeruginosa. J Biol Chem. 2002;277:29253–29259. doi: 10.1074/jbc.M111381200. [DOI] [PubMed] [Google Scholar]
- 9.Wu RY, Zhang RG, Zagnitko O, Dementieva I, Maltzev N, Watson JD, et al. Crystal structure of Enterococcus faecalis SlyA-like transcriptional factor. J Biol Chem. 2003;278:20240–20244. doi: 10.1074/jbc.M300292200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong M, Fuangthong M, Helmann JD, Brennan RG. Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol Cell. 2005;20:131–141. doi: 10.1016/j.molcel.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 11.Chen PR, Bae T, Williams WA, Duguid EM, Rice PA, Schneewind O, et al. An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat Chem Biol. 2006;2:591–595. doi: 10.1038/nchembio820. [DOI] [PubMed] [Google Scholar]
- 12.Bordelon T, Wilkinson SP, Grove A, Newcomer ME. The crystal structure of the transcriptional regulator HucR from Deinococcus radiodurans reveals a repressor preconfigured for DNA binding. J Mol Biol. 2006;360:168–177. doi: 10.1016/j.jmb.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 13.Mahdi LK, Ogunniyi AD, LeMessurier KS, Paton JC. Pneumococcal virulence gene expression and host cytokine profiles during pathogenesis of invasive disease. Infect Immun. 2008;76:646–657. doi: 10.1128/IAI.01161-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aranda J, Garrido ME, Cortes P, Llagostera M, Barbe J. Analysis of the protective capacity of three Streptococcus suis proteins induced under divalent-cation-limited conditions. Infect Immun. 2008;76:1590–1598. doi: 10.1128/IAI.00987-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aranda J, Garrido ME, Fittipaldi N, Cortes P, Llagostera M, Gottschalk M, et al. Protective capacities of cell surface-associated proteins of Streptococcus suis mutants deficient in divalent cation-uptake regulators. Microbiology. 2009;155:1580–1587. doi: 10.1099/mic.0.026278-0. [DOI] [PubMed] [Google Scholar]
- 16.Garmory HS, Titball RW. ATP-binding cassette transporters are targets for the development of antibacterial vaccines and therapies. Infect Immun. 2004;72:6757–6763. doi: 10.1128/IAI.72.12.6757-6763.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanabe M, Atkins HS, Harland DN, Elvin SJ, Stagg AJ, Mirza O, et al. The ABC transporter protein OppA provides protection against experimental Yersinia pestis infection. Infect Immun. 2006;74:3687–3691. doi: 10.1128/IAI.01837-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song XM, Connor W, Jalal S, Hokamp K, Potter AA. Microarray analysis of Streptococcus pneumoniae gene expression changes to human lung epithelial cells. Can J Microbiol. 2008;54:189–200. doi: 10.1139/w07-133. [DOI] [PubMed] [Google Scholar]
- 19.Hava DL, Camilli A. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol. 2002;45:1389–1406. [PMC free article] [PubMed] [Google Scholar]
- 20.Loo CY, Mitrakul K, Voss IB, Hughes CV, Ganeshkumar N. Involvement of the adc operon and manganese homeostasis in Streptococcus gordonii biofilm formation. J Bacteriol. 2003;185:2887–2900. doi: 10.1128/JB.185.9.2887-2900.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitrakul K, Loo CY, Gyurko C, Hughes CV, Ganeshkumar N. Mutational analysis of the adcCBA genes in Streptococcus gordonii biofilm formation. Oral Microbiol Immunol. 2005;20:122–127. doi: 10.1111/j.1399-302X.2004.00205.x. [DOI] [PubMed] [Google Scholar]
- 22.Owen GA, Pascoe B, Kallifidas D, Paget MSB. Zinc-responsive regulation of alternative ribosomal protein genes in Streptomyces coelicolor involves Zur and sigmaR. J Bacteriol. 2007;189:4078–4086. doi: 10.1128/JB.01901-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gabriel SE, Helmann JD. Contributions of Zur-controlled ribosomal proteins to growth under zinc starvation conditions. J Bacteriol. 2009;191:6116–6122. doi: 10.1128/JB.00802-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adamou JE, Heinrichs JH, Erwin AL, Walsh W, Gayle T, Dormitzer M, et al. Identification and characterization of a novel family of pneumococcal proteins that are protective against sepsis. Infect Immun. 2001;69:949–958. doi: 10.1128/IAI.69.2.949-958.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riboldi-Tunnicliffe A, Isaacs NW, Mitchell TJ. 1.2 A crystal structure of the S. pneumoniae PhtA histidine triad domain a novel zinc binding fold. FEBS Lett. 2005;579:5353–5360. doi: 10.1016/j.febslet.2005.08.066. [DOI] [PubMed] [Google Scholar]
- 26.Ogunniyi AD, Grabowicz M, Mahdi LK, Cook J, Gordon DL, Sadlon TA, et al. Pneumococcal histidine triad proteins are regulated by the Zn2+-dependent repressor AdcR and inhibit complement deposition through the recruitment of complement factor H. FASEB J. 2008;23:731–738. doi: 10.1096/fj.08-119537. [DOI] [PubMed] [Google Scholar]
- 27.Loisel E, Jacquamet L, Serre L, Bauvois C, Ferrer JL, Vernet T, et al. AdcAII, a new pneumococcal Zn-binding protein homologous with ABC transporters: Biochemical and structural analysis. J Mol Biol. 2008;381:594–606. doi: 10.1016/j.jmb.2008.05.068. [DOI] [PubMed] [Google Scholar]
- 28.Linke C, Caradoc-Davies TT, Young PG, Proft T, Baker EN. The laminin-binding protein Lbp from Streptococcus pyogenes is a zinc receptor. J Bacteriol. 2009;191:5814–5823. doi: 10.1128/JB.00485-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weston BF, Brenot A, Caparon MG. The metal homeostasis protein, Lsp, of Streptococcus pyogenes is necessary for acquisition of zinc and virulence. Infect Immun. 2009;77:2840–2848. doi: 10.1128/IAI.01299-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JW, Helmann JD. Functional specialization within the Fur family of metalloregulators. Biometals. 2007;20:485–499. doi: 10.1007/s10534-006-9070-7. [DOI] [PubMed] [Google Scholar]
- 31.Patzer SI, Hantke K. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol Microbiol. 1998;28:1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- 32.Patzer SI, Hantke K. The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J Biol Chem. 2000;275:24321–24332. doi: 10.1074/jbc.M001775200. [DOI] [PubMed] [Google Scholar]
- 33.Lucarelli D, Russo S, Garman E, Milano A, Meyer-Klaucke W, Pohl E. Crystal structure and function of the zinc uptake regulator FurB from Mycobacterium tuberculosis. J Biol Chem. 2007;282:9914–9922. doi: 10.1074/jbc.M609974200. [DOI] [PubMed] [Google Scholar]
- 34.Maciag A, Dainese E, Rodriguez GM, Milano A, Provvedi R, Pasca MR, et al. Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J Bacteriol. 2007;189:730–740. doi: 10.1128/JB.01190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng Y, Li M, Zhang H, Zheng B, Han H, Wang C, et al. Functional definition and global regulation of Zur, a zinc uptake regulator in a Streptococcus suis serotype 2 strain causing streptococcal toxic shock syndrome. J Bacteriol. 2008;190:7567–7578. doi: 10.1128/JB.01532-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogunniyi AD, Grabowicz M, Mahdi LK, Cook J, Gordon DL, Sadlon TA, et al. Pneumococcal histidine triad proteins are regulated by the Zn2+-dependent repressor AdcR and inhibit complement deposition through the recruitment of complement factor H. FASEB J. 2009;23:731–738. doi: 10.1096/fj.08-119537. [DOI] [PubMed] [Google Scholar]
- 37.Llull D, Poquet I. New expression system tightly controlled by zinc availability in Lactococcus lactis. Appl Environ Microbiol. 2004;70:5398–5406. doi: 10.1128/AEM.70.9.5398-5406.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kloosterman TG, van der Kooi-Pol MM, Bijlsma JJ, Kuipers OP. The novel transcriptional regulator SczA mediates protection against Zn2+ stress by activation of the Zn2+-resistance gene czcD in Streptococcus pneumoniae. Mol Microbiol. 2007;65:1049–1063. doi: 10.1111/j.1365-2958.2007.05849.x. [DOI] [PubMed] [Google Scholar]
- 39.Bijlsma JJ, Burghout P, Kloosterman TG, Bootsma HJ, de Jong A, Hermans PW, et al. Development of genomic array footprinting for identification of conditionally essential genes in Streptococcus pneumoniae. Appl Environ Microbiol. 2007;73:1514–1524. doi: 10.1128/AEM.01900-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grass G, Fan B, Rosen BP, Franke S, Nies DH, Rensing C. ZitB (YbgR), a member of the cation diffusion facilitator family, is an additional zinc transporter in Escherichia coli. J Bacteriol. 2001;183:4664–4667. doi: 10.1128/JB.183.15.4664-4667.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu M, Fu D. Structure of the zinc transporter YiiP. Science. 2007;317:1746–1748. doi: 10.1126/science.1143748. [DOI] [PubMed] [Google Scholar]
- 42.Ma Z, Jacobsen FE, Giedroc DP. Coordination chemistry of bacterial metal transport and sensing. Chem Rev. 2009;109:4644–4681. doi: 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Versieck J. Trace elements in human body fluids and tissues. Crit Rev Clin Lab Sci. 1985;22:97–184. doi: 10.3109/10408368509165788. [DOI] [PubMed] [Google Scholar]
- 44.Thurnham DI, Mburu AS, Mwaniki DL, De Wagt A. Micronutrients in childhood and the influence of subclinical inflammation. Proc Nutr Soc. 2005;64:502–509. doi: 10.1079/pns2005468. [DOI] [PubMed] [Google Scholar]
- 45.Brenot A, Weston BF, Caparon MG. A PerR-regulated metal transporter (PmtA) is an interface between oxidative stress and metal homeostasis in Streptococcus pyogenes. Mol Microbiol. 2007;63:1185–1196. doi: 10.1111/j.1365-2958.2006.05577.x. [DOI] [PubMed] [Google Scholar]
- 46.Grimsley GR, Scholtz JM, Pace CN. A summary of the measured pK values of the ionizable groups in folded proteins. Protein Sci. 2009;18:247–251. doi: 10.1002/pro.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thurlkill RL, Grimsley GR, Scholtz JM, Pace CN. pK values of the ionizable groups of proteins. Protein Sci. 2006;15:1214–1218. doi: 10.1110/ps.051840806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.VanZile ML, Chen X, Giedroc DP. Structural characterization of distinct α3N and α5 metal sites in the cyanobacterial zinc sensor SmtB. Biochemistry. 2002;41:9765–9775. doi: 10.1021/bi0201771. [DOI] [PubMed] [Google Scholar]
- 49.Jefferson JR, Hunt JB, Ginsburg A. Characterization of indo-1 and quin-2 as spectroscopic probes for Zn2(+)-protein interactions. Anal Biochem. 1990;187:328–336. doi: 10.1016/0003-2697(90)90465-l. [DOI] [PubMed] [Google Scholar]
- 50.Eicken C, Pennella MA, Chen X, Koshlap KM, VanZile ML, Sacchettini JC, et al. A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins. J Mol Biol. 2003;333:683–695. doi: 10.1016/j.jmb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 51.Zimmermann M, Clarke O, Gulbis JM, Keizer DW, Jarvis RS, Cobbett CS, et al. Metal binding affinities of arabidopsis zinc and copper transporters: Selectivities match the relative, but not the absolute, affinities of their amino-terminal domains. Biochemistry. 2009;48:11640–11654. doi: 10.1021/bi901573b. [DOI] [PubMed] [Google Scholar]
- 52.Liu T, Chen X, Ma Z, Shokes J, Hemmingsen L, Scott RA, et al. A Cu(I)-sensing ArsR family metal sensor protein with a relaxed metal selectivity profile. Biochemistry. 2008;47:10564–10575. doi: 10.1021/bi801313y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frausto da Silva J, Williams R. The Biological Chemistry of Elements: The Inorganic Chemistry of Life. 2. Oxford University Press; Oxford: 2001. [Google Scholar]
- 54.Golynskiy MV, Gunderson WA, Hendrich MP, Cohen SM. Metal binding studies and EPR spectroscopy of the manganese transport regulator MntR. Biochemistry. 2006;45:15359–15372. doi: 10.1021/bi0607406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.VanZile ML, Cosper NJ, Scott RA, Giedroc DP. The zinc metalloregulatory protein Synechococcus PCC7942 SmtB binds a single zinc ion per monomer with high affinity in a tetrahedral coordination geometry. Biochemistry. 2000;39:11818–11829. doi: 10.1021/bi001140o. [DOI] [PubMed] [Google Scholar]
- 56.Outten CE, Tobin DA, Penner-Hahn JE, O’Halloran TV. Characterization of the metal receptor sites in Escherichia coli Zur, an ultrasensitive zinc(II) metalloregulatory protein. Biochemistry. 2001;40:10417–10423. doi: 10.1021/bi0155448. [DOI] [PubMed] [Google Scholar]
- 57.Changela A, Chen K, Xue Y, Holschen J, Outten CE, O’Halloran TV, et al. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science. 2003;301:1383–1387. doi: 10.1126/science.1085950. [DOI] [PubMed] [Google Scholar]
- 58.Guo J, Wang S, Dong J, Qiu H, Scott RA, Giedroc DP. X-ray and visible absorption spectroscopy of wild-type and mutant T4 gene 32 proteins: His64, not His81, is the non-thiolate zinc ligand. J Am Chem Soc. 1995;117:9437–9440. [Google Scholar]
- 59.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 60.Sieminska EA, Xu X, Savchenko A, Sanders DA. The X-ray crystal structure of PA1607 from Pseudomonas aureginosa at 1.9 A resolution--a putative transcription factor. Protein Sci. 2007;16:543–549. doi: 10.1110/ps.062668207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pennella MA, Arunkumar AI, Giedroc DP. Individual metal ligands play distinct functional roles in the zinc sensor Staphylococcus aureus CzrA. J Mol Biol. 2006;356:1124–1136. doi: 10.1016/j.jmb.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 62.Ma Z, Cowart DM, Ward BP, Arnold RJ, DiMarchi RD, Zhang L, et al. Unnatural amino acid substitution as a probe of the allosteric coupling pathway in a mycobacterial Cu(I) sensor. J Am Chem Soc. 2009;131:18044–18045. doi: 10.1021/ja908372b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Giedroc DP, Arunkumar AI. Metal sensor proteins: nature’s metalloregulated allosteric switches. Dalton Trans. 2007;29:3107–3120. doi: 10.1039/b706769k. [DOI] [PubMed] [Google Scholar]
- 64.Golynskiy M, Li S, Woods VL, Jr, Cohen SM. Conformational studies of the manganese transport regulator (MntR) from Bacillus subtilis using deuterium exchange mass spectrometry. J Biol Inorg Chem. 2007;12:699–709. doi: 10.1007/s00775-007-0216-z. [DOI] [PubMed] [Google Scholar]
- 65.Arunkumar AI, Campanello GC, Giedroc DP. Solution structure of a paradigm ArsR family zinc sensor in the DNA-bound state. Proc Natl Acad Sci U S A. 2009;106:18177–18182. doi: 10.1073/pnas.0905558106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chakrabarti P. Geometry of interaction of metal-ions with histidine-residues in protein structures. Protein Eng. 1990;4:57–63. doi: 10.1093/protein/4.1.57. [DOI] [PubMed] [Google Scholar]
- 67.Grossoehme NE, Giedroc DP. Energetics of allosteric negative coupling in the zinc sensor S. aureus CzrA. J Am Chem Soc. 2009;131:17860–17870. doi: 10.1021/ja906131b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Edgcomb SP, Murphy KP. Variability in the pKa of histidine side-chains correlates with burial within proteins. Proteins: Struct, Funct, Genet. 2002;49:1–6. doi: 10.1002/prot.10177. [DOI] [PubMed] [Google Scholar]
- 69.Gunner MR, Nicholls A, Honig B. Electrostatic potentials in Rhodopseudomonas viridis reaction centers: Implications for the driving force and directionality of electron transfer. J Phys Chem. 1996;100:4277–4291. [Google Scholar]
- 70.Becher D, Hempel K, Sievers S, Zuhlke D, Pane-Farre J, Otto A, et al. A proteomic view of an important human pathogen--towards the quantification of the entire Staphylococcus aureus proteome. PLoS One. 2009;4:e8176. doi: 10.1371/journal.pone.0008176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee JW, Soonsanga S, Helmann JD. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc Natl Acad Sci U S A. 2007;104:8743–8748. doi: 10.1073/pnas.0702081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen H, Hu J, Chen PR, Lan L, Li Z, Hicks LM, et al. The Pseudomonas aeruginosa multidrug efflux regulator MexR uses an oxidation-sensing mechanism. Proc Natl Acad Sci U S A. 2008;105:13586–13591. doi: 10.1073/pnas.0803391105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maret W, Li Y. Coordination dynamics of zinc in proteins. Chem Rev. 2009;109:4682–4707. doi: 10.1021/cr800556u. [DOI] [PubMed] [Google Scholar]
- 74.Patel K, Kumar A, Durani S. Analysis of the structural consensus of the zinc coordination centers of metalloprotein structures. Biochim Biophys Acta, Proteins Proteomics. 2007;1774:1247–1253. doi: 10.1016/j.bbapap.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 75.Cotton FA, Hanson HP. Soft X-ray absorption edges of metal ions in complexes. 3 Zinc(II) complexes. J Chem Phys. 1958;28:83–87. [Google Scholar]
- 76.Leitch S, Bradley MJ, Rowe JL, Chivers PT, Maroney MJ. Nickel-specific response in the transcriptional regulator, Escherichia coli NikR. J Am Chem Soc. 2007;129:5085–5095. doi: 10.1021/ja068505y. [DOI] [PubMed] [Google Scholar]
- 77.Pennella MA, Shokes JE, Cosper NJ, Scott RA, Giedroc DP. Structural elements of metal selectivity in metal sensor proteins. Proc Natl Acad Sci U S A. 2003;100:3713–3718. doi: 10.1073/pnas.0636943100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Iwig JS, Leitch S, Herbst RW, Maroney MJ, Chivers PT. Ni(II) and Co(II) sensing by Escherichia coli RcnR. J Am Chem Soc. 2008;130:7592–7606. doi: 10.1021/ja710067d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ma Z, Cowart DM, Scott RA, Giedroc DP. Molecular insights into the metal selectivity of the copper(I)-sensing repressor CsoR from Bacillus subtilis. Biochemistry. 2009;48:3325–3334. doi: 10.1021/bi900115w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pennella MA, Giedroc DP. Structural determinants of metal selectivity in prokaryotic metal-responsive transcriptional regulators. Biometals. 2005;18:413–428. doi: 10.1007/s10534-005-3716-8. [DOI] [PubMed] [Google Scholar]
- 81.Outten CE, O’Halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 82.Waldron KJ, Rutherford JC, Ford D, Robinson NJ. Metalloproteins and metal sensing. Nature. 2009;460:823–830. doi: 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- 83.Lanie JA, Ng WL, Kazmierczak KM, Andrzejewski TM, Davidsen TM, Wayne KJ, et al. Genome sequence of Avery’s virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J Bacteriol. 2007;189:38–51. doi: 10.1128/JB.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Barendt SM, Land AD, Sham LT, Ng WL, Tsui HCT, Arnold RJ, et al. Influences of capsule on cell shape and chain formation of wild-type and pcsB mutants of serotype 2 Streptococcus pneumoniae. J Bacteriol. 2009;191:3024–3040. doi: 10.1128/JB.01505-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Busenlehner LS, Giedroc DP. Kinetics of metal binding by the toxic metal-sensing transcriptional repressor Staphylococcus aureus pI258 CadC. J Inorg Biochem. 2006;100:1024–1034. doi: 10.1016/j.jinorgbio.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 86.VanZile ML, Chen X, Giedroc DP. Allosteric negative regulation of smt O/P binding of the zinc sensor, SmtB, by metal ions: a coupled equilibrium analysis. Biochemistry. 2002;41:9776–9786. doi: 10.1021/bi020178t. [DOI] [PubMed] [Google Scholar]
- 87.Walkup GK, Imperiali B. Fluorescent chemosensors for divalent zinc based on zinc finger domains. Enhanced oxidative stability, metal binding affinity, and structural and functional characterization. J Am Chem Soc. 1997;119:3443–3450. [Google Scholar]
- 88.Perrin DD, Dempsey B. Buffers for pH and Metal Ion Control. 2. Chapman and Hall; New York: 1979. [Google Scholar]
- 89.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 90.Hansen RE, Winther JR. An introduction to methods for analyzing thiols and disulfides: Reactions, reagents, and practical considerations. Anal Biochem. 2009;394:147–158. doi: 10.1016/j.ab.2009.07.051. [DOI] [PubMed] [Google Scholar]
- 91.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 92.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, et al. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 93.Kay LE, Keifer P, Saarinen T. Pure absorption gradient-enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J Am Chem Soc. 1992;114:10663–10665. [Google Scholar]
- 94.Yamazaki T, Lee W, Arrowsmith CH, Muhandiram DR, Kay LE. A suite of triple resonance NMR experiments for the backbone assignment of 15N, 13C, 2H labeled proteins with high sensitivity. J Am Chem Soc. 1994;116:11655–11666. [Google Scholar]
- 95.Yamazaki T, Lee W, Revington M, Mattiello DL, Dahlquist FW, Arrowsmith CH, et al. An HNCA pulse scheme for the backbone assignment of 15N,13C,2H-labeled proteins: Application to a 37-kDa Trp repressor-DNA complex. J Am Chem Soc. 1994;116:6464–6465. [Google Scholar]
- 96.Scott RA. X-ray Absorption Spectroscopy. In: Que L, editor. Physical Methods in Bioinorganic Chemistry: Spectroscopy and Magnetism. University Science Books; Sausalito, CA: 2000. pp. 465–503. [Google Scholar]
- 97.Ankudinov AL, Ravel B, Rehr JJ, Conradson SD. Real-space multiple-scattering calculation and interpretation of X-ray-absorption near-edge structure. Phys Rev B. 1998;58:7565–7576. [Google Scholar]
- 98.Poiarkova AV, Rehr JJ. Multiple-scattering X-ray absorption fine-structure Debye–Waller factor calculations. Phys Rev B. 1999;59:948–957. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.