Abstract
Drosophila melanogaster is used as a model system to investigate protein changes associated with the aging process under conditions that alter organism lifespan. Changes in the proteome are assessed at various ages in populations of Oregon-R adult males that have mean lifetimes of 47 and 111 days at 28 and 18 °C, respectively. Peptide hits detected from strong-cation-exchange and reversed-phase liquid chromatography coupled to tandem mass spectrometry analysis are employed to examine patterns in relative protein expression. Thirty-three proteins were identified as having similar patterns of expression at both temperatures investigated when scaling the organism age to lifespan. In addition, the proteins ferritin 2 light chain homologue and larval serum protein 1β were identified in relatively high abundance and displayed distinctly different patterns of expression between the two temperatures. Overall, the results support the notion that aspects of the aging process may be preprogrammed at the protein level.
Keywords: aging, Drosophila melanogaster, proteomics, temperature
Introduction
Generally, chronological age (i.e., calendar time in days, months, or years) is used to characterize deteriorative changes that occur over the course of an organisms’ lifespan.1,2 However, several studies have demonstrated that molecular measures, such as gene expression, can better describe events associated with aging. 3 – 5 For example, physiological (i.e., morphological changes at the organ, tissue, and/or subcellular levels) changes in human muscle tissue were correlated with the expression of 250 age-regulated genes in genome-wide studies.5 Age-regulated transcriptional profiles provided accurate measurements of changes in a middle-aged individual that had undergone considerable aging in muscle tissue (similar to characteristics in individuals 10 to 20 years older).5 These results were also verified in the muscle physiology and thus, genetic measurements may serve as sensitive methods to assess age-related changes.
In Drosophila melanogaster (also known as fruit flies, and hereafter referred to as Drosophila), temporal patterns of gene expression were monitored in the antennae of adult animals using enhancer trap and reporter gene techniques.6 – 8 From these studies, it was demonstrated that the expression of individual genes can scale to lifespan. These results were obtained under ambient temperature conditions whereby animals exhibit a three-fold change in lifespan and in genetic mutants that age at faster rates.6–8 Candidate genes were identified as molecular markers of aging because the change in expression was more indicative of organism age than chronological age alone.6–8 Overall, molecular measurements that examine lifespan effects in response to genetic or environmental perturbations can help to unravel the complexities of the aging process.9
The goal of the present work was to determine if there are differences in protein expression for Drosophila aged under conditions that alter the rate of aging and influence organism lifespan. Drosophila is a poikilothermic organism that has temperature-dependent life spans.10–14 In these studies, the mean lifespan of animals aged at 18 °C is greater than a factor of two in comparison to animals aged at 28 °C; the maximum lifespan is approximately a factor of three times greater. Drosophila were harvested at several ages throughout adult lifespan at both of the temperatures investigated. Proteins extracted from the heads of flies were analyzed using a shotgun proteomics approach that employed strong-cation-exchange (SCX) chromatography coupled to reversed-phase liquid chromatography (LC) and tandem mass spectrometry (MS/MS) techniques. A semi-quantitative measure of relative abundance was used to examine changes in protein expression profiles across adult lifespan.
Experimental Procedures
Drosophila lifespan experiments
Populations of Oregon-R (Bloomington Stock Center, Indiana University) males reared at 24 °C were sorted into cohorts of ~50 flies per vial within 48 hours of eclosure. This allowed the males sufficient time to mate at least once before being segregated from females. Two populations of ~1000 total animals each were transferred to temperature controlled incubators at either 28 or 18 °C and exposed to alternating cycles of 12 h light/12 h darkness. Animals were transferred to fresh cornmeal media every four days at which time deaths were recorded for lifespan experiments.
Drosophila harvesting
Batches of animals grown up at 24 °C were separated into cohorts of ~50 animals per vial within 24 hours post-eclosion. Two populations of ~6000 animals were aged at 28 and 18 °C, respectively. Two hundred heads were harvested from animals as follows: at 28 °C animals were harvested at two, twelve, 22, 32, 36, 42, 46, 50, 52, and, 56 days (after eclosure); and, at 18 °C animals were harvested at two, 22, 42, 62, 82, 102, 112, 122, 132, and, 142 days. Heads were stored at −80 °C until further use.
Sample preparation
Heads were suspended in a phosphate buffer saline solution (containing 4.0 M urea and 0.1 mM α-toluenesulfonyl fluoride) and proteins were extracted using a motorized pestle (KONTES glass company, Vineland, NJ). The Bradford assay indicated protein amounts of ~1–2 mg. Tryptic peptide mixtures were generated as follows. Proteins were reduced with dithiothreitol at a protein:reagent molar ratio of 1:40 at 37 °C for 2 h and alkylated with iodoacetamide at a molar ratio of 1:80 at 0 °C for 2 h in darkness. A 1:40 molar ratio of protein:L-cysteine was used to quench the reaction for 30 min at ambient temperature. The final urea concentration was reduced to 2.0 M with 0.2 M Tris buffer (10 mM CaCl2, pH = 8.0). TPCK-treated trypsin (Sigma-Aldrich, St. Louis, MO) was added at 2% w/w (mass of enzyme to that of protein) to protein solutions and incubated for 24 h at 37 °C. Peptide mixtures were desalted with Oasis HLB cartridges (Waters Inc., Milford, MA), dried with centrifugal evaporation, and stored at −80 °C until further use.
Strong-cation-exchange chromatography
A LC system (600 Pump, 2487 Dual Wavelength detector, Waters Inc., Milford, MA) was used for strong-cation-exchange fractionation of tryptic peptide mixtures. Tryptic peptides (~1.2 mg) were injected onto a PolySulfoethyl A column (2.1 mm i.d. × 100 mm, 5 µm, 300 Å; PolyLC, Inc., Columbia, MD). Mobile phases consisted of 5 mM potassium phosphate in 75:25 water:acetonitrile at pH = 3.0 (solvent A) and solvent A with the addition of 350 mM KCl (solvent B). The gradient was delivered at a flow rate of 0.20 mL· min−1 as follows: 0% B for 5 min, 40% B in 40 min, 80% B in 45 min, 100% B in 10 min, and, hold 100% B for 10 min. Peptide absorbance was monitored with UV detection at λ = 214 nm. One minute intervals were collected into 96 well plates and pooled into six fractions. Fractions were cleaned with Oasis HLB cartridges, dried and resuspended in 0.1% formic acid for reversed-phase analysis.
LC-MS/MS
A commercial LCQ Deca XP MS (Thermo Electron Corporation, Waltham, MA) was used to perform triplicate LC-MS/MS experiments of each individual fraction. Samples were injected onto a trapping column (100 µm i.d. × 1.5 cm; New Objective Inc., Woburn, MA) packed with 5 µm, 200 Å Magic C18AQ (Microm BioResources Inc., Auburn, CA) at a flow rate of 4 µL·min−1. Pulled tip fused silica analytical columns were prepared in-house (75 µm i.d. × 150 mm; Polymicro Technology LLC, Phoenix, AZ) and packed with a methanol slurry consisting of 5 µm, 100 Å Magic C18AQ (Microm BioResources Inc., Auburn, CA). Mobile phases, 96.95:2.95:0.1 water:acetonitrile:formic acid (solvent A) and 99.9:0.1 acetonitrile:formic acid (solvent B), were delivered at a flow rate of 250 nL·min−1. Two reversed-phase gradients were used to separate individual SCX fractions that were optimized for the number of components in each fraction. The 1st, 2nd, and 6th fractions of each sample were separated with the following gradient: 0–5% B in 5 min, 5–20% B in 50 min, 20–40% B in 40 min, 40–80% B in 5 min, 80% B for 10 min, 80-0% B in 5 min, and, hold 0% B for 15 min. The 3rd, 4th, and 5th fractions were separated with a longer gradient: 0–5% B in 5 min, 5–20% B in 95 min, 20–40% B in 75 min, 40–80% B in 15 min, 80% B for 10 min, 80-0% B in 5 min, 0% B for 15 min. MS parameters for data dependent acquisition included a mass-to-charge range of 300–1800, 35% normalized collision energy, and, a dynamic exclusion time of one min.
Protein identification
Mass lists of precursor and fragment ions were submitted to MASCOT15 for database searching against the National Center for Biotechnology Information Drosophila protein database.16 Individual gi| accessions were cross correlated with FBgn accession numbers in the Flybase database17 for final protein assignments. The parameters for the MASCOT search allowed two trypsin miscleavages, a fixed carbamidomethyl modification, and parent and fragment mass tolerances of 1.5 and 0.8 Da, respectively. Only peptides having MASCOT scores at or above homology (which indicates less than a 5% chance of occurring at random) were considered for further analysis. Gene ontology (GO)18 information was used to associate individual proteins with biological processes.
Peptide hits analysis
Relative changes in protein abundance were assessed with a peptide hits analysis which considers the total number of observations of peptides matching to particular proteins in the datasets.19–22 Three back-to-back runs of individual SCX fractions were carried out and the total number of peptide hits from individual proteins was combined and treated as a single analysis. In order to account for variations in the total number of hits obtained for a single experiment, peptide hits for individual proteins were normalized to the total number of peptide hits obtained for each individual sample. This approach is analogous to label-free techniques such as spectral counting.
Results and Discussion
Drosophila lifespan as a function of different ambient temperature
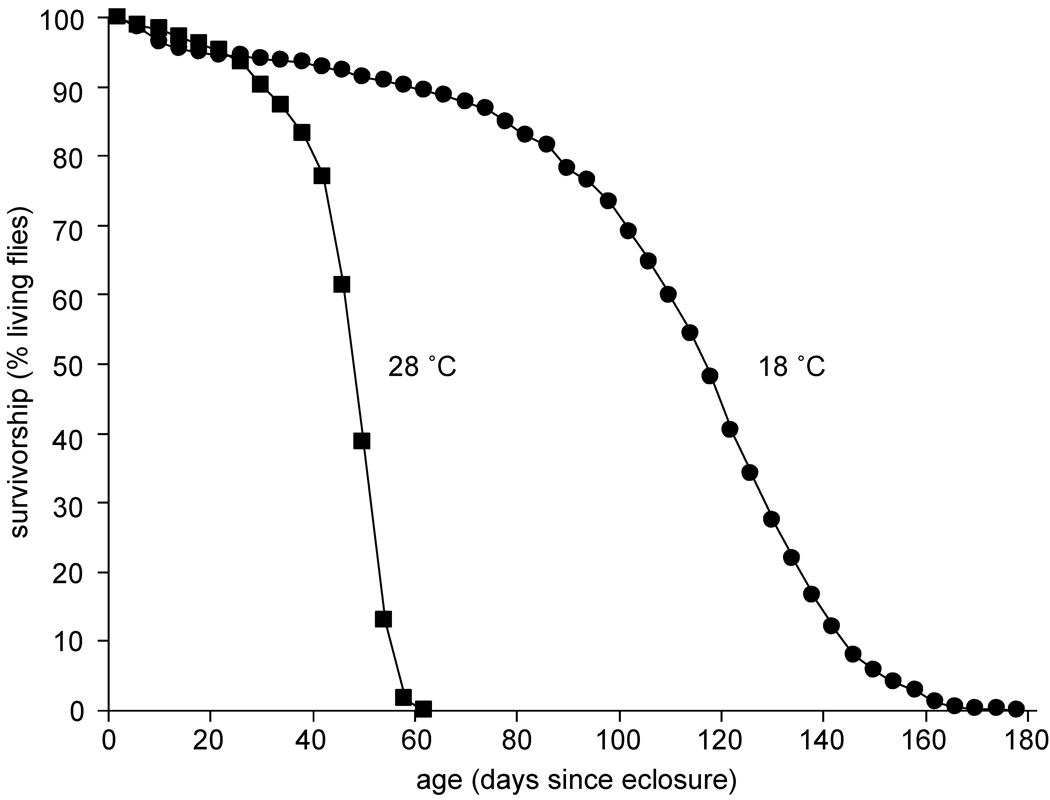
Figure 1 shows the survivorship of adult male Oregon-R that were reared at 24 °C and allowed to age at 28 or 18 °C. From this survival curve, it appears that the maximum lifespan of the population aged at 18 °C is three times greater than the population aged at 28 °C. The results from twenty independent trials of lifespan measurements are listed in Table I. The average lifespans and standard deviations are 47±13 days (N=1003, where N represents the total number of flies) and 111±38 days (N=973) at 28 and 18 °C, respectively. A significant difference in mean lifespan (using a pairwise student’s t-test, P<6×10−16) is observed between the flies aged at 28 and 18 °C. These results are consistent with previously published reports of Drosophila survival at different environmental temperatures.11,14
Figure 1.
Survival curves for Oregon-R male populations at 28 °C (■) and 18 °C (●). The curves shown represent cumulative results from twenty independent trials (see Table I).
Table I.
Mean Lifespan of Oregon R males.
| 28°C | 18°C | |||
|---|---|---|---|---|
| Triala | Nb | x̄ ±σc | Nb | x̄ ±σc |
| 1 | 49 | 47±13 | 49 | 115±40 |
| 2 | 50 | 50±8 | 48 | 86±35 |
| 3 | 50 | 44±11 | 51 | 114±40 |
| 4 | 50 | 46±11 | 50 | 128±46 |
| 5 | 50 | 35±9 | 48 | 132±33 |
| 6 | 50 | 47±11 | 50 | 120±25 |
| 7 | 50 | 53±8 | 51 | 113±33 |
| 8 | 58 | 49±13 | 51 | 117±39 |
| 9 | 50 | 50±8 | 39 | 99±32 |
| 10 | 50 | 50±12 | 50 | 114±31 |
| 11 | 49 | 46±10 | 49 | 122±39 |
| 12 | 50 | 36±11 | 49 | 103±18 |
| 13 | 49 | 51±6 | 49 | 117±37 |
| 14 | 49 | 50±9 | 46 | 89±40 |
| 15 | 50 | 46±12 | 50 | 83±25 |
| 16 | 49 | 50±6 | 50 | 110±29 |
| 17 | 50 | 49±8 | 50 | 118±40 |
| 18 | 50 | 49±10 | 43 | 118±21 |
| 19 | 50 | 50±7 | 50 | 113±23 |
| 20 | 50 | 49±7 | 50 | 98±23 |
| Total | 1003d | 47±13e | 973d | 111±38e |
Each trial is in an independent measure that was initiated at the same time.
The total number of flies (N) measured in each cohort.
The mean lifespan (x̄) ±standard deviation (σ), in days.
The total number of flies measured across all cohorts.
The weighted average ±pooled standard deviation, in days, for each genotype.
Summary of assignments from SCX-LC-MS/MS analyses
It is worthwhile to provide a summary of the overall assignments obtained from these experiments, although the following discussion focuses on only a subset of the proteins identified from these analyses. The total number of peptides identified across all ages was 8,716 and 7,508 leading to the assignment of 760 and 761 proteins at 28 and 18 °C, respectively. Additionally, a total of 17,613 and 13,606 peptide hits (i.e., spectral counts) were detected for all age samples across the 28 and 18 °C datasets, respectively. Cumulative results provide evidence for 1,019 unique protein assignments. We note that 66% (668 proteins) of these identifications were assigned with at least two peptide hits and that caution should be taken with regards to proteins identified with a single peptide as these data are from a low-resolution ion trap MS. A complete listing of peptide and protein assignments is provided in supplementary information (Table SI). Below we focus our attention to example proteins that have distinct patterns of expression as a function of age.
Peptide hits analyses
From the information listed in Table SI, relative abundances of proteins were determined using peptide hits information. Here peptide hits is analogous to spectral counts which refers to the number of MS/MS spectra that lead to confident identification of a peptide. In this manner, there may be multiple peptide hits for a single or several peptide sequences belonging to an individual protein. This approach does not provide absolute quantitative information but rather relative abundance information whereby generally speaking, the more peptide hits a protein has the more abundant it is in these analyses. For each of the time point samples at either 28 or 18 °C, the number of peptide hits for a specific protein is equivalent to the total sum of peptide hits detected for all identified peptide sequences across triplicate RP LC-MS/MS analyses of each the SCX fractions. Because of the sampling issues associated with data-dependent ion trap MS analyses, we report the sum of peptide hits obtained across replicate RP LC-MS/MS analyses. When doing comparative analyses of the data, we normalized peptide hits (e.g., total number of peptide hits for individual protein/total peptides hits across all proteins detected at a particular timepoint) in order to minimize differences that may result due to errors in sample preparation and data dependent analyses. Peptide hits data is provided in Tables S1 and S2.
Example data of protein expression as a function of chronological age
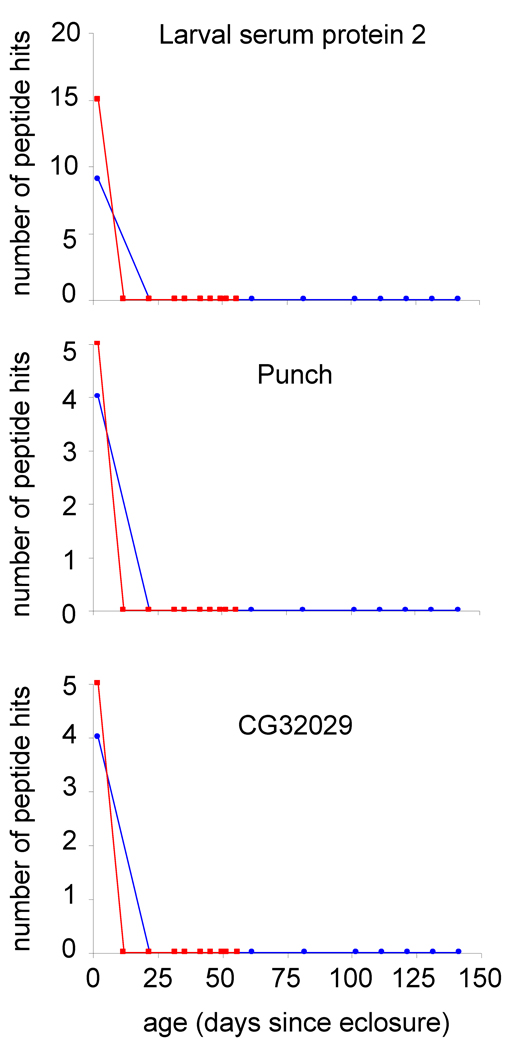
Figure 2 shows the number of peptide hits (without normalization) obtained for larval serum protein 2, punch, and CG32029. Each of these proteins has peptides that are detected in 2-day-old adult Drosophila; however, as the animals grow older no peptide hits are detected at other ages. Moreover, this rapid decrease in expression in early adulthood is observed in both the 28 and 18 °C datasets. Larval serum protein 2 is suggested to be involved in amino acid storage which is necessary for the production of adult cuticle and has previously been shown to be expressed at lower levels in early aged adults relative to the third instar larval stage. 23,24 In general, our findings are consistent with these previous reports although one report detects an additional larval serum protein 2 constituent in gel bands of animals up to 20-days-old.23 The punch gene encodes an enzyme, guanosine triphosphate cyclohydrolase, that functions in the biosynthesis of pteridines which are involved in the synthesis of neurotransmitters and serve as eye pigments in Drosophila.25,26 We have previously observed similar patterns of protein expression for larval serum protein 2 and punch in other proteomics experiments performed by our lab.27,28 CG32029 is a structural constituent of cuticle in the larval stage17 and to-date measurements of protein expression in adults have not been noted.
Figure 2.
Plots of the number of peptide hits (without normalization) as a function of Drosophila age (in days since eclosure) for larval serum protein 2 (FBgn0002565), punch (FBgn0003162), and CG32029 (FBgn0052029 at 28 °C (■) and 18 °C (●).
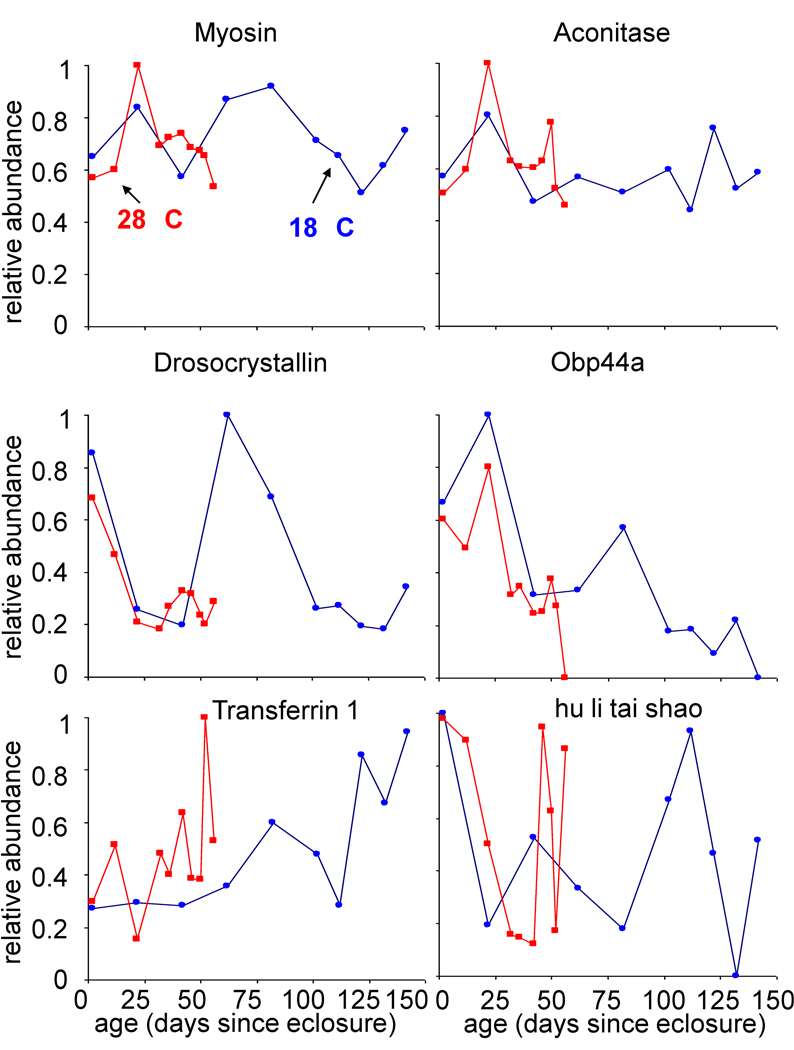
Figure 3a shows the relative abundances of six example proteins as a function of fly age (in days since eclosure) at each temperature. These plots can be examined by considering the overall trend in protein abundance with age. For example, the proteins myosin and aconitase appear to have slight fluctuations in expression over the course of adulthood at each temperature but remain at a relatively constant level with age. On the other hand, the proteins drosocrystallin and odorant binding protein 44a (Obp44a) show a distinct decrease in relative abundance as flies grow older whereas transferrin 1 appears to have a steady increase in relative abundance at each temperature. A rather variable pattern is observed for the hu li tai shao protein which has alternating cycles of decreased and increased expression throughout adulthood.
Figure 3.
Relative protein abundance as a function of a) Drosophila age, defined here as days since eclosure as estimated from Figure 1. The normalized number of peptide hits is plotted on a relative abundance scale of 0 to 1, where 1 represents the maximum number of normalized peptide hits obtained across all ages at both temperatures for individual proteins. Relative abundances at 28 °C (■) and 18 °C (●) are shown for the following proteins: myosin (FBgn0002741), aconitase (FBgn0010100), drosocrystallin (FBgn0005664), Obp44a (FBgn0033268), transferrin 1 (FBgn0022355), and hu li tai shao (FBgn0004873).
Although these proteins display an overall similar change in expression for each temperature, the timing associated with these changes differs as a function of chronological age. For example, at 56 days no peptide hits are detected from Obp44a in the 28 °C population although there is still expression in the 18 °C population. While it would be typical to state that this protein is down regulated in the 28 °C population, the age-related comparison is somewhat skewed. At 56 days, more than 90% of the population is alive at 18 °C whereas less than 2% of the population is alive at 28 °C (Figure 1). Thus, comparisons of protein expression as a function of time (in days) are somewhat misleading and partially limit understanding of the overall aging process in Drosophila. A more reasonable comparison is to assess differences in expression of individual proteins as a function of population lifespan. Such a comparison may provide insight to similar mechanisms of aging at both temperatures.
Example data of protein expression as a function of population lifespan
The relative abundances for the six aforementioned example proteins can also be considered as a function of lifespan [(i.e., the fraction of animals no longer living as estimated from Figure 1), data not shown]. In order to account for differences that may arise due to errors introduced during sample preparation or during MS/MS sampling, the peptide hits data were normalized for comparisons across timepoints and temperatures. The data provide evidence that these proteins have overall similar patterns in expression at each temperature. For example, drosocrystallin has an initial sharp decrease in relative abundance at both temperatures and then displays an increase when ~11% of the population is dead. A more substantial increase is observed at 18 °C than at 28 °C before expression declines again and levels off at each temperature. In the case of the hu li tai shao protein, the overlap in expression between the two temperatures is rather remarkable. An initial decline in abundance is observed in early adulthood, and then an increase occurs until a peak in expression is obtained when ~55–60% of the population is still alive. This protein then has another sharp decrease and increase in expression towards the latest survival times. Overall the profile patterns of the proteins shown in Figure 3 are very similar at each temperature, suggesting that mechanisms of aging are similar at each of the temperatures
Biological processes of proteins with similar expression patterns at each temperature
Overall, the expression profiles of proteins in these studies are highly dynamic in nature; however, many proteins exhibit similar temporal patterns of expression at 28 and 18 °C. Because we are most interested in the age-related expression on an individual protein basis, we manually inspected proteins that were detected with high confidence (MASCOT scores with p<0.05) and relatively high abundance (i.e., at least ten peptide hits detected at any single age across both temperatures) for similarities at 28 and 18 °C. We note that while clustering of the data may provide general information about families of proteins, delineating the mechanisms of aging can best be gained by studies that provide leads to individual molecules that require further investigation. In addition, the data are provided as supplemental information (Table SII) so that protein changes may be assessed by others as desired.
Table II lists 33 proteins that have overall superimposable patterns in expression at both 28 and 18 °C when plotted as a function of population lifespan. We note that this small number of proteins in relation to the 668 total proteins identified, were extracted based on confidence in identification resulting from being considered as relatively abundant proteins (i.e., at least 10 peptide hits at any one time point). It is apparent that these proteins are associated with a variety of biological processes utilizing GO information. Many of the proteins have multiple associations, thus their expression profiles may be reflective of the sum of the different biological functions that are carried out during adulthood. In assessing the trends for the 21 proteins that have metabolic functions it appears that the expression patterns primarily are constant or variable as the animals age (Table SII). Thus, no distinct decrease or increase in relative abundance for the entire group of metabolic proteins is evident.
Table II.
List of proteins that have similar expression patterns when scaled to lifespan.a
| Descriptionb | FBgn no.c | Biological processd |
|---|---|---|
| alcohol dehydrogenase | FBgn0000055 | defense response, localization, metabolism |
| aldolase | FBgn0000064 | development, metabolism |
| arginine kinase | FBgn0000116 | metabolism |
| abnormal wings disc | FBgn0000150 | development, localization, metabolism |
| elongation factor 2b | FBgn0000559 | metabolism |
| enolase | FBgn0000579 | metabolism |
| bangles and beads | FBgn0001090 | development |
| glutamate dehydrogenase | FBgn0001098 | metabolism, reproduction |
| heat shock protein 70 | FBgn0001219 | cell communication, defense response, development, localization metabolism |
| heat shock protein 83 | FBgn0001233 | cell communication, defense response, development, metabolism reproduction |
| larval serum protein 2 | FBgn0002565 | localization |
| malic enzyme | FBgn0002719 | metabolism |
| myosin heavy chain | FBgn0002741 | development |
| Na,K ATPase α subunit | FBgn0002921 | development, localization, metabolism |
| neither inactivation nor afterpotential C | FBgn0002938 | cell communication, defense response, localization, metabolism |
| paramyosin | FBgn0003149 | development |
| α-spectrin | FBgn0003470 | cell communication, development, metabolism, reproduction |
| α-Tubulin at 84B | FBgn0003884 | defense response, localization, metabolism |
| porin | FBgn0004363 | localization |
| hu li tai shao | FBgn0004873 | development, localization, reproduction |
| drosocrystallin | FBgn0005664 | NS |
| aconitase | FBgn0010100 | metabolism |
| bellwether | FBgn0011211 | development, localization, metabolism |
| aldehyde dehydrogenase | FBgn0012036 | metabolism |
| 14-3-3ε | FBgn0020238 | cell communication, defense response, development |
| transferrin 1 | FBgn0022355 | defense response, localization |
| odorant-binding protein 44a | FBgn0033268 | localization |
| CG4784 | FBgn0036619 | NS |
| CG4169 | FBgn0036642 | localization, metabolism |
| CG7433 | FBgn0036927 | metabolism |
| CG11963 | FBgn0037643 | metabolism |
| CG7217 | FBgn0038570 | NS |
| CG7998 | FBgn0038587 | metabolism |
Proteins in this list have similar expression patterns as a function of population survivorship at both 18 and 28 °C.
The name of the protein, gene, or computed gene (CG) number provided is given as a description of the identified protein.
The Flybase gene number (FBgn no.) was obtained from the Flybase database.
The biological processes associated with proteins utilizing gene ontology (GO) information. Proteins were grouped into broad categories. NS represents those proteins for which a process was not specified.
In several cases, proteins listed in Table II, display a distinct trend in protein expression with age (Table SII). For example, two proteins involved in localization, porin and odorant-binding protein 44a (Figure 3), appear to continually decrease in expression with increasing organism age. In addition, transferrin 1 (Figure 3), which is involved in both defense response and localization, has an overall increase in relative abundance over the course of adulthood. The two heat shock proteins (hsps), hsp70 and hsp83 are associated with categories such as cell communication, defense response, development, localization, metabolism and reproduction. Interestingly, hsp70 increases with organism age whereas hsp83 has an overall decrease in expression with age. These patterns are similar for both of the temperatures investigated. It has been previously reported that hsp70 protein expression increases in the thoraces of old Drosophila at both 25 and 29 °C; although no increase in expression was detected in the heads of animals.29,30 This could be because the expression of the protein was in too low of an abundance to detect by Western analysis; no spots were visible on the gels from young or old animals.29
Evidence for temperature-dependent expression patterns
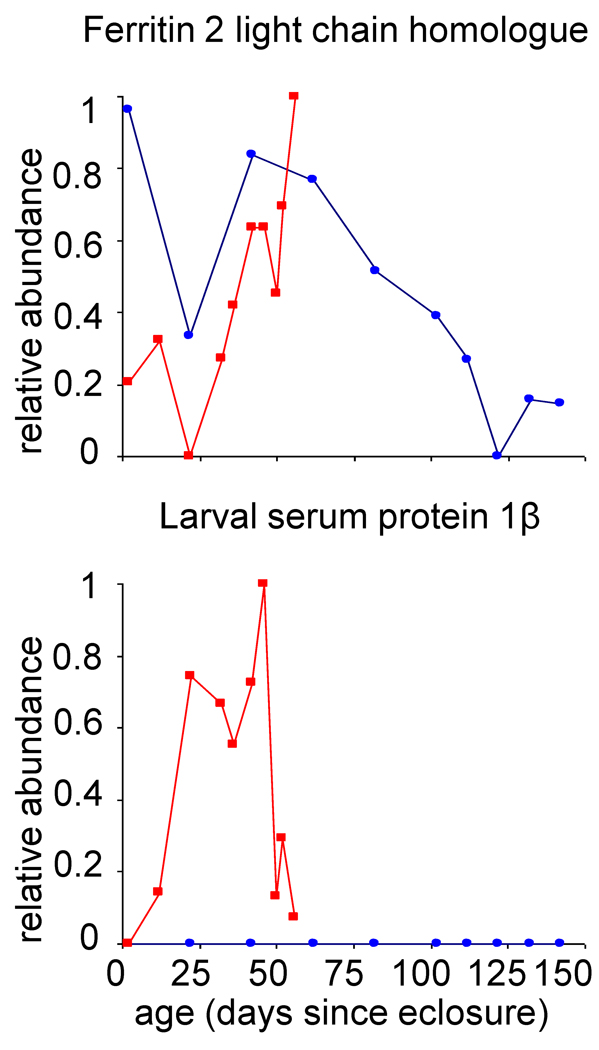
In assessing the temporal patterns of expression for proteins identified in these studies, two proteins displayed drastically different expression patterns at the two temperatures investigated. Figure 4 shows that the protein ferritin 2 light chain homologue decreases in expression in animals by day 22. However, the relative abundance continues to increase in the 28 °C population while it decreases in expression in the 18 °C population. This protein is suggested to be involved in iron ion transport (as inferred from gene ontology information).17 While ferritin 2 light chain homologue protein expression has previously been observed in all stages of development,31 changes associated with age in adult Drosophila have not yet been reported.
Figure 4.
Relative protein abundance as a function of a) Drosophila age, defined here as days since eclosure. The normalized number of peptide hits is plotted on a relative abundance scale of 0 to 1, where 1 represents the maximum number of normalized peptide hits obtained across all ages at both temperatures for individual proteins. Relative abundances at 28 °C (■) and 18 °C (●) are shown for ferritin 2 light chain homologue (FBgn0015221) and larval serum protein 1β (FBgn0002563).
The larval serum protein 1β is detected at multiple ages in the population of animals that were aged at 28 °C ; no expression is detected in animals aged at 18 °C. We note that this protein was detected with a total of 59 unique peptide hits (there were also an additional six nonunique hits detected that overlap with larval serum protein 1γ) across all ages at 28 °C. Thus, it is a rather abundant protein in these experiments. This protein is suggested to be involved in nutrient storage and the formation of adult cuticle, and is known to be expressed only in the third instar larval stage.24,32,33 Aging studies of larval serum protein 1β in adult animals have not previously been reported. That larval serum protein 1β is detected in animals over a range of ages suggests that this protein may have differential regulation in adult Drosophila. In addition, that we detect expression only in the 28 °C animals and not in the 18 °C animals, may suggest that there is a temperature-dependent regulation of this protein. While we speculate that this may be associated with elevated stress levels in 28 °C populations, further studies are necessary in order to validate this hypothesis.
Conclusions
Lifespan measurements for Oregon-R males were consistent with previous reports in the literature such that Drosophila has a longer lifespan at 18 °C than at 28 °C. To our knowledge, this is the first report that has assessed proteome profiles in the heads of Drosophila as a function of age and temperature. Many proteins involved in metabolism, defense response, localization, development, and cell communication displayed similar patterns of expression at both temperatures. These results corroborate the idea that some biological pathways are conserved with aging in adult Drosophila and provide evidence for preprogrammed expression for certain proteins with aging. Two proteins (i.e, ferritin 2 light chain homologue and larval serum protein 1β) had distinct temperature-dependent differences in expression. This insight is useful for determining mechanisms that increase the rate of aging and that influence organism lifespan.
Supplementary Material
Acknowledgement
The authors acknowledge the National Institutes of Health (AG-024547-04) for financial support of this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Finch CE. Longevity, senescence, and the genome. Chicago, IL and London, UK: The University of Chicago Press; 1990. pp. 1–42. [Google Scholar]
- 2.Arking R. Biology of Aging. 2nd Ed. Sunderland, MA: Sinauer Associates, Inc; 1998. pp. 3–26. [Google Scholar]
- 3.Rodwell GEJ, Sonu R, Zahn JM, Lund J, Wilhelmy J, Wang L, Xiao W, Mindrinos M, Crane E, Segal E, Myers BD, Brooks JD, Davis RW, Higgins J, Owen AB, Kim SK. A transcriptional profile of aging in the human kidney. PLoS Biol. 2004;2:e427. doi: 10.1371/journal.pbio.0020427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L, Sharpless NE. Ink4a/Arf expression is a biomarker of aging. J. Clin. Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zahn J, Sonu R, Vogel H, Crane E, Mazan-Mamczara K, Rabkin R, Davis RW, Becker KG, Owen AB, Kim SK. Transcriptional profiling of aging in human muscle reveals a common aging signature. PLoS Biol. 2006;2:e115. doi: 10.1371/journal.pgen.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helfand SL, Blake KJ, Rogina B, Stracks MD, Centurion A, Naprta B. Temporal Pattern of Gene Expression in the Antenna of the Adult Drosophila melanogaster. Genetics. 1995;140:549–555. doi: 10.1093/genetics/140.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogina B, Helfand SL. Regulation of gene expression is linked to life span in adult Drosophila. Genetics. 1995;141:1043–1048. doi: 10.1093/genetics/141.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogina B, Benzer S, Helfand SL. Drosophila drop-dead mutations accelerate the time course of age-related markers. Proc. Natl. Acad. Sci., USA. 1997;94:6303–6306. doi: 10.1073/pnas.94.12.6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guarente L, Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408:255–262. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- 10.Loeb J, Northrop JH. Is There a Temperature Coefficient for the Duration of Life? Proc. Natl. Acad. Sci., USA. 1916;2:456–457. doi: 10.1073/pnas.2.8.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loeb J, Northrop JH. On the influence of food and temperature upon the duration of life. J. Biol. Chem. 1917;32:103–121. [Google Scholar]
- 12.Alpatov WW, Pearl R. Influence of different feeding during larval and imaginal stages on duration of life and imago of Drosophila melanogaster. Am. Naturalist. 1929;63:37–67. [Google Scholar]
- 13.Cohet Y. Epigenetic influences on the lifespan of the Drosophila: existence of an optimal growth temperature for adult longevity. Exp. Geront. 1975;10:181–184. doi: 10.1016/0531-5565(75)90029-7. [DOI] [PubMed] [Google Scholar]
- 14.Miquel J, Lundgren PR, Bensch KG, Atlan H. Effects of temperature on the life span, vitality and fine structure of Drosophila melanogaster. Mech. Ageing Dev. 1976;5:347–370. doi: 10.1016/0047-6374(76)90034-8. [DOI] [PubMed] [Google Scholar]
- 15.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 16. http://www.ncbi.nlm.nih.gov/
- 17. http://www.flybase.org.
- 18.The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pang JX, Ginanni N, Dongre AR, Hefta SA, Opiteck GJ. Biomarker Discovery in Urine by Proteomics. J. Proteome Res. 2002;1:161–169. doi: 10.1021/pr015518w. [DOI] [PubMed] [Google Scholar]
- 20.Qian W-J, Jacobs JM, Camp DG, II, Monroe ME, Moore RJ, Gritsenko MA, Calvano SE, Lowry SF, Xiao W, Moldawer LL, Davis RW, Tompkins RG, Smith RD. Comparative proteome analyses of human plasma following in vivo lipopolysaccharide administration using multidimensional separations coupled with tandem mass spectrometry. Proteomics. 2005;5:572–584. doi: 10.1002/pmic.200400942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao J, Opiteck GJ, Friedrichs M, Dongre AR, Hefta SA. Changes in the protein expression of yeast as a function of carbon source. J. Proteome Res. 2003;2:643–649. doi: 10.1021/pr034038x. [DOI] [PubMed] [Google Scholar]
- 22.Gao J, Friedrichs MS, Dongre AR, Opiteck GJ. Guidelines for the Routine Application of the Peptide Hits Technique. J. Am. Soc. Mass. Spectrom. 2005;16:1231–1238. doi: 10.1016/j.jasms.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Beneš H, Edmondson RG, Fink P, Kejzlarová-Lepesant J, Lepesant J-A, Miles JP, Spivey DW. Adult expression of the Drosophila Lsp-2 gene. Dev. Biol. 1990;142:138–146. doi: 10.1016/0012-1606(90)90157-e. [DOI] [PubMed] [Google Scholar]
- 24.Roberts DB, Wolfe J, Akam ME. The developmental profiles of two major haemolymph proteins from Drosophila melanogaster. J. Insect Physiol. 1977;23:871–878. doi: 10.1016/0022-1910(77)90013-0. [DOI] [PubMed] [Google Scholar]
- 25.MacKay WJ, O’Donnel JM. A Genetic Analysis of the Pteridine Biosynthetic Enzyme, Guanosine Triphosphate Cyclohydrolase, in Drosophila Melanogaster. Genetics. 1983;105:35–53. doi: 10.1093/genetics/105.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLean JR, Krishnakumar S, O’Donnell JM. Multiple mRNAs from the Punch locus of Drosophila melanogaster encode isoforms of GTP cyclohydrolase I with distinct Nterminal domains. J. Biol. Chem. 1993;268:27191–27197. [PubMed] [Google Scholar]
- 27.Xun Z, Sowell RA, Kaufman TC, Clemmer DE. Protein Expression in a Drosophila Model of Parkinson’s Disease. J. Proteome Res. 2007;6:348–357. doi: 10.1021/pr060488o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sowell RA, Hersberger KE, Kaufman TC, Clemmer DE. Examining the Proteome of Drosophila Across Organism Lifespan. J. Proteome Res. 2007;6:3637–3647. doi: 10.1021/pr070224h. [DOI] [PubMed] [Google Scholar]
- 29.Wheeler JC, Bieschke ET, Tower J. Muscle-specific expression of Drosophila hsp70 in response to aging and oxidative stress. Proc. Nat’l. Acad. Sci. USA. 1995;92:10408–10412. doi: 10.1073/pnas.92.22.10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wheeler JC, King V, Tower J. Sequence requirements for upregulated expression of Drosophila hsp70 transgenes during aging. Neurobiol. Aging. 1999;20:545–553. doi: 10.1016/s0197-4580(99)00088-3. [DOI] [PubMed] [Google Scholar]
- 31.Georgieva T, Dunkov BC, Dimov S, Ralchev K, Law JH. Drosophila melanogaster ferritin: cDNA encoding a light chain homologue, temporal and tissue specific expression of both subunit types. Insect Biochem. Mol. Biol. 2002;32:295–302. doi: 10.1016/s0965-1748(01)00090-x. [DOI] [PubMed] [Google Scholar]
- 32.Roberts D. The functions of the major serum proteins of Drosophila larvae. Biol. Chem. Hoppe-Seyler. 1987;368:572. [Google Scholar]
- 33.Roberts DB, Jowett T, Hughes J, Smith DF, Glover DM. The major serum protein of Drosophila larvae, larval serum protein 1, is dispensable. Eur. J. Biochem. 1991;195:195–201. doi: 10.1111/j.1432-1033.1991.tb15695.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.