Abstract
One of the main challenges of Pseudomonas aeruginosa (P. aeruginosa) vaccine development is the design of an antigen that elicits cross-reactive antibodies against multiple virulent strains. Using a rational design approach, we have developed a single 17-residue peptide immunogen that generates antibodies that target the receptor binding domain (RBD) of the type IV pilus of more than one strain of P. aeruginosa. Using the RBD sequence, of native strain PAO as a template, we have systematically changed up to five residues in the PAO sequence of the peptide immunogen, into that of the PAK sequence. We show by indirect and competitive ELISA, that the mutant peptide immunogens elicit the development of polyclonal sera that is cross-reactive to both native strain PAO and PAK pilin. We further show that there are at least two separate antibody populations in the polyclonal sera that possess closely-related epitopes but which are each strain specific. Moreover, part of the epitope for the PAO specific antibodies consists of several residues outside the disulfide loop of the receptor binding domain. This allows us to create two unique epitopes within the same receptor binding domain sequence.
Keywords: anti-adhesin vaccine, cross-reactive antibodies, Pseudomonas aeruginosa, synthetic peptide vaccine
Introduction
Pseudomonas aeruginosa (P. aeruginosa) is a ubiquitous opportunistic pathogen that has become one of the leading causes of hospital acquired infections (1). P. aeruginosa infections are typically treated with antimicrobials which are ineffective against chronic infections. Furthermore, the number of P. aeruginosa virulence factors has made vaccine development difficult. Some vaccines have increased survival rates in animal trials but none have been successful in providing an immunized host with protection against numerous P. aeruginosa strains. P. aeruginosa has one of the largest bacterial genomes sequenced to date (2) and thus is tolerant of a wide variety of physical conditions, which accounts for its pathogenic and ecological success. Since a chronic infection is so difficult to treat, it would be more prudent to target the initial stages of infection.
P. aeruginosa has polar type IV pili that are common among gram-negative bacteria. The pili are made up of thousands of homologous pilin monomers assembled in a helical array. There are 4 or 5 pilin monomers per turn; the exact number is not known (3). The pilus is 5.2 nm in outer diameter and 1.2 nm in central channel diameter and has an average length of 2.5 μm (4). The pili can be extended or retracted through assembly and disassembly of pilin monomers and are used for twitching motility (5).
The pilus extends from the bacterial cell surface and is responsible for the initial contact between the bacterium and the epithelial cell surface (6). Studies have shown that pilus mediated adherence of P. aeruginosa is a tip associated event with the pili adhesin exposed at the pilus tip (7). It is possible that the retractile mechanism of the pili then allows the P. aeruginosa bacterial cell to be drawn in closer to the host cell to allow further binding through other adhesins such as exoenzyme S, thus further mediating a tight attachment to the cell to facilitate infection. Nonretractile mutants were reported to be less adherent than wild type strains (8). P. aeruginosa pili have also been shown to be important in binding to epithelial cell surfaces where non-piliated strains were reported to show a 90% decrease in their ability to bind human A549 pneumocytes (9, 10). Furthermore, another study showed that non-piliated strains caused 28%–96% fewer cases of P. aeruginosa pneumonia as compared to piliated strains in a neonatal mouse model (11).
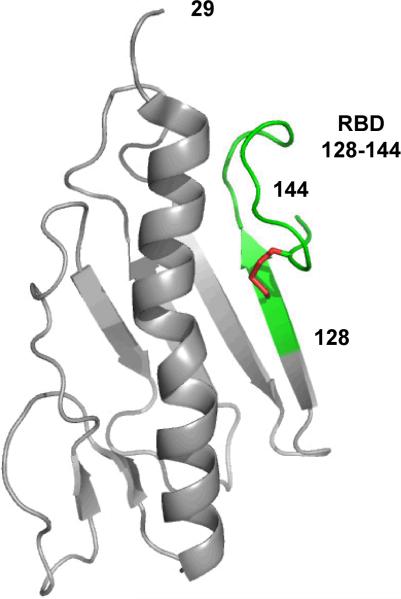
Two native P. aeruginosa strains, PAK and K122-4, have had their pilin monomers structurally characterized (12–16). The pilin monomer can be divided into three regions: a semi-conserved C-terminal region; a hypervariable central region; and a conserved hydrophobic N-terminal region. The receptor binding domain resides in the well structured C-terminal end as a 14-residue disulfide loop (17) (Figure 1). The 14-residue disulfide loop includes the two cysteine residues that form the disulfide bond and the 12 amino acid residues between the two cysteine residues (Figure 2). The 14-residue disulfide loop contains two β-turns, a type I β-turn and a type II β-turn both of which are conserved across known strains of P. aeruginosa. Multimerization of pilin monomers is mediated through a 29-residue portion of the N-terminal helix (12).
Figure 1.
Ribbon diagram showing the structure of PAK monomeric pilin residues 29–144 (PDB ID: 1DZO). The 17-residue receptor binding domain (RBD) 128–144 is highlighted in green and is defined as one residue (128) prior to the 14-residue disulfide loop and the two residues after the disulfide loop (143 and 144). The disulfide bond between residues 129 and 142 is colored red (12).
Figure 2.
Amino acid sequences from the receptor-binding domain (RBD) residues (128–144) of six pilin strains of Pseudomonas aeruginosa. Yellow shaded residues indicate positions of identity or conservative substitutions, grey shaded residues indicate sequence identity between five of the six strains. PAK-K and PAK–R pilin strains are identical in sequence except for the C-terminal residue K or R at position 144. The nine variable positions between the strains are shown in white. Residues in each of these positions vary dramatically in side-chain and type (charged, polar, and non-polar).
The C-terminal receptor binding domain of P. aeruginosa pilin is a good candidate for a peptide vaccine. The 14-residue disulfide loop provides a continuous epitope that can be easily represented by a synthetic peptide. Furthermore, receptor binding domain (RBD) peptides maintain the two β-turns within the loop and are well structured in solution (18, 19). There is structural similarity between the free peptide and the RBD in the full length native pilin protein (16) as well as truncated monomeric pilin proteins (12, 18, 20). Moreover, RBD peptides, from different strains retain receptor-binding activity to host epithelial cells. Disulfide-bridged synthetic peptide derivatives of the pilin RBD competitively inhibit pili binding to cultured epithelial cells showing functional analogy (7). Furthermore, peptide conjugates, when used as immunogens, generate antibodies that can inhibit adhesion of P. aeruginosa to cultured human epithelial cells and have been shown to be protective against bacterial challenge. Antibodies specific for the RBD have been shown to bind only to the tip of the pilus (7). This is consistent with the current model of pilus assembly, where the RBD is partially obscured along the sides of the pilus by the αβ-loop (3). Finally, Type IV pili from all strains of P. aeruginosa share a common receptor; however, the sequence diversity in the epitope presents a significant obstacle to the development of a broadly protective vaccine targeting the RBD of the type IV pilus.
P. aeruginosa pili can be divided into two groups; those with a 14-residue disulfide loop in the receptor binding domain and those with a 19-residue disulfide loop in the receptor binding domain (21). The amino acid sequence of the receptor binding domain is semi-conserved across known P. aeruginosa strains (Figure 2). Among P. aeruginosa strains with 14-residue receptor binding domains, there are three highly conserved residues (C129, P139 and C142), two residues are conserved in all but one strain (K140 and G141), and three other residues show conservative substitutions (131, 134 and 137). The remaining six positions are variable (130, 132, 133, 135, 136 and 138) (Figure 2).
We have hypothesized that the eight conserved and semi-conserved residues are framework residues that define the peptide backbone of the RBD (Figure 2). The remaining variable positions have an uncertain effect on RBD structure and function. It is likely that the variation of these residues decreases the chances that more than one strain would be recognized by the immune system. Due to complications arising from antigenic competition, a multivalent pilus-based vaccine representing multiple strains of P. aeruginosa is not effective at generating immunity against multiple strains, so we have pursued a consensus sequence approach to vaccine design (21, 22). The feasibility of this approach has been demonstrated, where peptide immunogens with single amino acid substitutions were shown to modulate antibody cross-reactivity in rabbits (22).
A consensus sequence immunogen has been designed in this laboratory based on the hypothesis that making substitutions from the native strain PAO sequence into the native strain PAK sequence would increase cross-reactivity of antibodies raised against the modified sequence (21). A series of peptide immunogens with one, two, or three substitutions of the PAO sequence into the PAK sequence was designed. It was found that certain di-substituted immunogens generated antisera with increased cross-reactivity for other P. aeruginosa strains (21). One promising PAK analog with two substitutions, T130K-E135P, was chosen as a putative consensus sequence and designated Cs1. The Cs1 peptide immunogen has an improved immunological activity in comparison to native-strain immunogens (23). Furthermore, animal protections studies show that Cs1 peptide immunogen can induce an immune response that can be protective against challenge by a number of piliated strains of P. aeruginosa.
To understand better how amino acid substitutions in peptide immunogens, from one native strain to another, affect peptide immunogenicity, we have designed a series of peptide immunogens. Thus, using the native strain PAO RBD sequence as a template, we have substituted two, three, four, or five amino acid residues from the native strain PAK RBD sequence into the PAO peptide immunogen (Figure 3). These synthetic peptides were then conjugated to keyhole limpet hemocyanin (KLH) and used as immunogens in New Zealand White Rabbits. IgG was purified from the sera and then used in indirect and competitive ELISAs to determine antibody cross-reactivity.
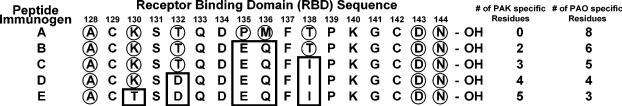
Figure 3.
Peptide immunogens synthesized, using the PAO receptor binding domain (RBD) sequence as a template. Boxed residues indicate substitutions from the native strain PAK RBD sequence, into the native strain PAO RBD sequence of the peptides. Boxed residues also indicate native strain PAK specific residues while the circled residues indicate native strain PAO specific residues; all other residues are identical in PAK and PAO RBDs. Immunogen A (PAO(128–144) native sequence), Immunogen B (PAO(128–144) P135E/M136Q), Immunogen C (PAO(128–144) P135E/M136Q/T138I), Immunogen D (PAO(128–144) T132D/P135E/M136Q/T138I), and Immunogen E (PAO(128–144) K130T/T132D/P135E/M136Q/T138I). −OH denotes a C-terminal carboxyl group. The peptides were covalently attached to keyhole limpet hemocyanin (KLH), via the α-amino group, for immunization as described in the methods.
Materials and Methods
Synthetic Peptides
Synthetic peptide immunogens, based on the RBD sequence (128–144, ACKSTQDPMFTPKGCDN), of the P. aeruginosa strain PAO, were prepared with a three amino acid residue linker (norleucine–Gly–Gly) on the N-terminal to produce the peptide nLGGACKSTQDPMFTPKGCDN for the preparation of peptide-conjugates. Peptides were manually synthesized by solid-phase synthesis chemistry using standard t-BOC chemistry and 4-methyl-benzhydrylamine (MBHA) resin (24). Crude peptides were purified by reversed-phase high-performance liquid chromatography (RP-HPLC) on a semi-preparative Agilent Zorbax 300SB-C8 column (250 mm × 9.4 mm I.D., 5 μm particle size, 300 Å pore size, Agilent Technologies, Palo Alto, CA, USA). Fractions were analysed on a Agilent Zorbax 300SB-C8 (150 mm × 2.1 mm I.D.) narrow bore column. Peptide identity was verified using a Mariner ESITOF mass spectrometer (Applied Biosystems, Foster City, CA, USA). The intrachain disulfide bond was formed by dissolving the synthetic peptide in 0.1 M ammonium bicarbonate, pH 8.0, at a peptide concentration of 0.5 mg / mL and overnight incubation at 25 °C. Disulfide formation was confirmed by reaction with N-ethylmaleimide (NEM) followed by RP-HPLC (25).
Synthetic peptides to be conjugated to a carrier protein keyhole limpet hemocyanin [KLH] were selectively iodoacetylated at the N-terminus (26). Briefly, the peptide was dissolved in 100 mM 2 (N-morpholino) ethanesulfonic acid buffer, pH 6 at 4 °C at 2 mg/mL. Two molar equivalents of iodoacetic anhydride, in dry 1,4-dioxane, were added to the peptide solution with stirring. The peptide solution was kept at a pH of 6 to avoid iodoacetylation of lysine side-chain ε-amino groups. The extent of the reaction was monitored by RP-HPLC. Upon completion, the reaction mixture was acidified with 0.5 mL glacial acetic acid and applied to a Zorbax 300SB-C8 RPHPLC column (9.4 mm × 150 mm) and purified. The identity of the product was confirmed by mass spectrometry. The peptide-KLH conjugate was prepared by first dissolving endotoxin-free KLH (Sigma, St. Louis, MO, USA) at a concentration of 10 mg / mL in freshly prepared 8 M urea. EDTA was added to a concentration of 5 mM. KLH was derivatized with free sulfhydryls groups by adding a 50-fold molar excess of Traut's reagent (2-iminothiolane; Sigma-Aldrich, St. Louis, MO, USA) to the KLH solution. The reaction mixture was protected from light and stirred at 25 °C for 1 h. Unreacted Traut's reagent was removed with a PD10 desalting column (Amersham Biosciences, Piscataway, NJ, USA) using a 50 mM sodium phosphate, 5 mM EDTA eluent. The protein-containing fractions were concentrated to 15–20 mg / mL using an Ultrafree-15 Centrifugal Filter Device (MWCO 10 000, Millipore, Bedford, MA, USA). The iodoacetyl-peptide was added to the mixture in a 10-fold molar excess to KLH. The reaction mixture was protected from light and incubated at 25 °C for 1 h. Unreacted sulfhydryl groups were capped by addition of 20-fold molar excess iodoacetamide (Sigma-Aldrich, St. Louis, MO, USA) and incubated for 30 min at 25 °C. Peptide-conjugates were dialysed extensively against phosphate-buffered saline (PBS) with 5 mM EDTA (pH 7.4) to remove the iodoacetamide. The peptide-KLH conjugate was subjected to amino acid analysis. By calculating the number of moles of norleucine, which was incorporated in the N-terminal linker of the synthetic peptide, and using the known amino acid composition of KLH, the total amount of synthetic peptide that had been conjugated to KLH as well as the synthetic peptide:KLH molar ratio were determined (varied between 1.5 and 8 peptides per KLH monomer).
Polyclonal Antibody Production
8 week old New Zealand White Rabbits from Jackson Laboratories were used for polyclonal antibody production. There were three rabbits for each synthetic peptide immunogen with an additional three rabbits receiving the PAO peptide-conjugate as a positive control. Pre-immune blood samples were collected before the first injection. A series of four 0.5 mL 100ug antigen injections were given in four week intervals. Complete Freund's adjuvant was used for the first injection and then incomplete Freund's adjuvant thereafter. Three weeks after the fourth injection the rabbits were exsanguinated.
Polyclonal Antibody and IgG Purification
After the collection of the blood, it was allowed to clot for one hour at 37 °C. The clot was separated from the sides of the 50 mL conical and placed at 4 °C overnight. The blood was then centrifuged at 10,000g for 10 min at 4 °C and the serum supernatant was then removed. IgG was purified by treatment with caprylic acid and ammonium sulfate. Briefly, 2 volumes of 60 mM sodium acetate buffer (pH 4.0) were added to serum and then titrated to a pH of 4.8. Caprylic acid (octanoic acid) was added drop wise to a final volume of 0.75 ml per 10 ml of original volume and then centrifuged at 5,000g for 10 minutes. The supernatant was removed and saturated ammonium sulfate was added slowly, with stirring, to bring the solution to 50% saturation and then stored with stirring at 4 °C overnight. The solution was then centrifuged at 3,000g for 30 minutes. The supernatant was discarded and the pellet was brought up in 0.3 volumes of the starting supernatant volume in PBS. The antibody solution was then dialyzed overnight at 4 °C. The concentration of IgG was determined by UV spectroscopy using an extinction coefficient of 1.35 M−1 cm−1 at 280 nm.
Pilin protein expression
The gene encoding the PAK pilin protein (29–144) was cloned into the pRLDE vector which contains a His tag for purification. Pilin proteins were expressed in BL21 (DE3) E. coli cells and purified from the periplasmic fraction by an osmotic shock protocol. An overnight culture was used to inoculate a larger culture, which was grown at 37 °C until an UV absorbance of 0.4 was reached at 600 nm. The culture was transferred to incubation at 25 °C and allowed to equilibrate for 30 min before inducing protein expression with 0.5 mM isopropyl-β-D-thiogalactopyranoside. Induction proceeded overnight at 25 °C. Protein expression was monitored by SDS-PAGE. Cells were harvested by centrifugation at 3,000g for 10 min at 25 °C. The periplasmic fraction was isolated by an osmotic shock protocol. Briefly, cells were resuspended in TES buffer (100 mM Tris–HCl, 5 mM EDTA, 20% sucrose) at 80 mL / g wet cell mass, shaken at 25 °C for 10 min then centrifuged at 3,500 × g for 20 min. The supernatant was discarded and cells were resuspended in ice cold 5 mM Mg2SO4 at 80 mL / g wet cell mass, shaken on ice for 30 min then centrifuged at 4,000g for 20 min. The supernatant was decanted and passed through a 0.45 μm filter. The sample was then lyophilized prior to purification. Sigma His-Select HF Nickel Affinity Gel resin was packed in a GE XK 16/20 Column (200mm × 16mm I.D.) for protein purification. The lyophilized pilin was dissolved in 50 ml of wash buffer (50 mM sodium phosphate, 300 mM NaCl, 10 mM imidazole, pH 7.2) and loaded on the column. The column was washed with wash buffer until the UV absorbance at 280 nm was close to 0. The pilin protein was eluted with an A–B gradient, where A is the wash buffer and B consists of 50 mM sodium phosphate, 300 mM NaCl, 300 mM imidazole, pH 7.2. The gradient was 1% B/min for 10 min (up to 30 mM imidazole) and then 2% B/min for 45 minutes (up to 300 mM imidazole). The fractions containing the pilin protein were pooled and lyophilized and then dissolved in 100 mM sodium bicarbonate for storage and usage. The pilin protein concentration was determined by UV absorbance at 280 nm using extinction coefficients determined by ExPASy Proteomics Server.
Pilin Mutagenesis
The PAK (ADN) and PAO (KSK) mutant pilin genes were created by site directed mutagenesis. Mutagenesis of the truncated native strain PAK and PAO pilins (Δ 1–28) in the pRLDE plasmids were performed (Quick Change; Strategene, La Jolla, CA). Two oligonucleotides were used to create each mutant (Oligo 1: 5'- GCAGCTGATGGTCTCTGGGCGTGCACCAGTGATCAGG-3'; Oligo 2: 5'- CCGAAAGGTTGCGATAATTAATTTTGTAAGAAGCTTGACC-3') was used to introduce K128A, S143D and R144N into native strain PAK pilin, creating PAK (ADN). To introduce A128K, D143S and R144K into native strain PAO pilin, creating PAO (KSK), the following oligos were used (Oligo 1: 5'-GCGGATGGGGTCTGGAAGTGTAAATCTACCCAGG-3'; Oligo 2: 5'-CACTCCGAAAGGTTGTTCTAAGTAAGGTGATCGAAAGCTTGACC -3').
Amino acid analysis
Peptide concentrations were determined by amino acid analysis by acid hydrolysis of the peptide in 6 N HCl for 24 h at 110 °C and dried under vacuum. Samples were resuspended in sodium diluent, pH 2.2 (Pickering Laboratories, Mountain View, CA, USA) and analysed on a Beckman 6300 amino acid analyzer (Beckman-Coulter, Fullerton, CA, USA).
Indirect ELISA
High-binding polystyrene 96-well immunoassay plates (Costar 3590, Corning, NY, USA) were coated with 100 μL of a 0.1 μM solution of pilin protein in 100 mM sodium carbonate / sodium bicarbonate buffer, pH 9.5, for two hours at 37 °C. Plates were washed three times with phosphate buffered saline with Tween (PBS-T, 50 mM phosphate, 750 mM sodium chloride, 0.05% Tween-20, pH 7.4). Excess binding sites were blocked with 5% bovine serum albumin (BSA) in PBS-T for 1 h at 37 °C. Plates were washed again three times before adding 100 μL of serially diluted rabbit IgG in PBS-T to each well. The starting concentrations ranged from 0.2 μM to 0.4 μM of IgG with a two-fold dilution per well down to approximately 2×10−4 μM. Plates were incubated for 1 h at 37 °C. Plates were washed three times before adding 100 μL / well of a 1 / 5,000 dilution of anti-rabbit IgG horseradish peroxidase (HRP) conjugate (Jackson ImmunoResearch Laboratories, Inc, West Grove, PA, USA). The plates were incubated for 1 h at 37 °C. Plates were washed three times with PBS-T. 100 μL of a 1 mM solution of 2,2'-azinobis-3-ethylbenzthiazoline-6-sulfonic acid (ABTS; Sigma-Alrdich, St. Louis, MO, USA) in 10 mM sodium citrate pH 4.2 with 0.03% H2O2 was incubated for 30 min with shaking. Plates were read on a SpectraMax 386 Plus plate reader (Molecular Devices, Sunnyvale, CA, USA) at 405 nm. Binding curves were fit to a four-parameter logistic curve using SigmaPlot 7.0 (SPSS Inc., Chicago, IL, USA) to determine curve mid-points.
Competitive ELISA
High-binding polystyrene 96-well immunoassay plates (Costar 3590, Corning, Lowell, MA, USA ) were coated with 100 μL / well of a 0.1 μM solution of pilin protein in 100 mM sodium carbonate / sodium bicarbonate buffer, pH 9.5, at 37° C for 2h. Plates were washed three times with PBS-T (50 mM phosphate, 750 mM sodium chloride, 0.05% Tween-20, pH 7.4). Excess binding sites were blocked with 5% BSA in PBS-T for 1 h at 37 °C. Based on the indirect ELISA results, three to six times the IC50 IgG concentrations were used in the competition ELISAs. For the competition experiments, the primary antibody at the dilution determined was preincubated with serial dilutions of the competitor (native strain PAO or PAK pilin proteins or mutant PAO (KSK) or PAK (ADN) pilin proteins) in PBS-T with 0.5% BSA for 2 h at 37 °C. Then, 50 μL of each antibody / competitor solution was added to the pilin-coated ELISA plates and incubated for 1 h at 37 °C. Plates were washed three times before adding 100 μL / well of a 1 / 10,000 dilution of anti-rabbit IgG (HRP) conjugate. After thorough washing, 50 μL of Ultra-TMB (3,3',5,5'-tetramethylbenzidine, Pierce, Rockford, IL, USA) was added to each plate for quantitation and developed according to the manufacturer's instructions. The ELISA plates were read on a plate reader at 450 nm. Data were normalized by dividing the raw absorbance data by the absorbance of wells without competitors. The resulting curves were fit to a modified four parameter logistic binding model using GraphPad Prism 5 to determine IC50 values (pilin concentration at required for 50% inhibition of antibody binding).
Results
Strain-specific Antibodies
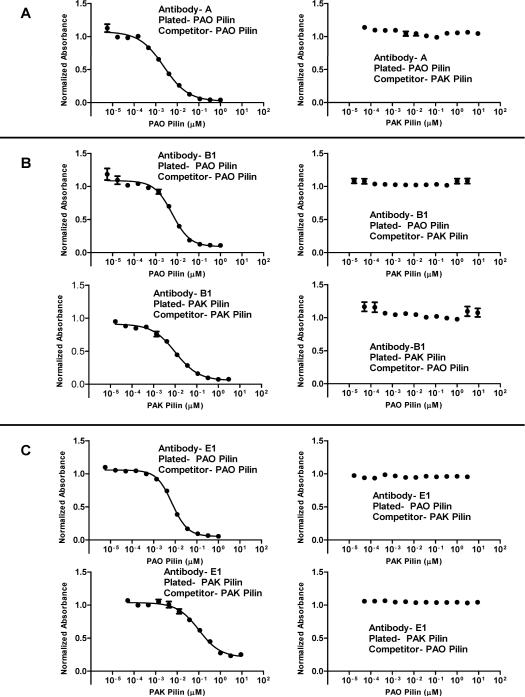
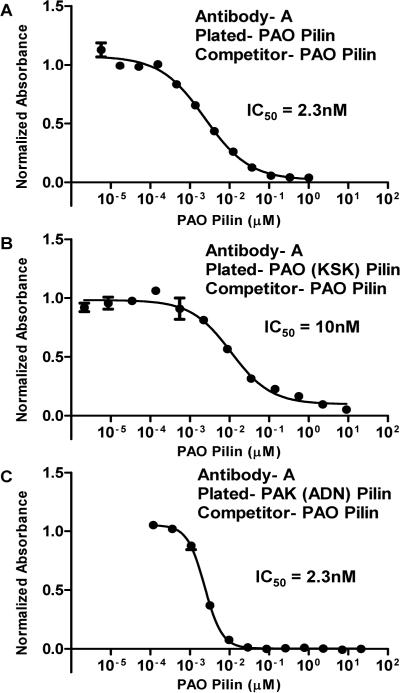
Strain-specific antibodies are generated from native sequence peptide immunogens of the receptor binding domain, which is the major problem for anti-adhesion vaccine development. This is shown for the native PAO sequence immunogen in (Figure 3). The polyclonal antibodies generated to immunogen A are strain-specific in that they bind PAO pilin and do not bind to PAK pilin by indirect ELISA. Moreover, when PAO pilin is plated in competitive ELISAs, only PAO pilin can compete any of the A antibodies off plated PAO pilin, while PAK pilin cannot (Figure 4A). This result is understandable since 8 of the 17 residues in the receptor binding domain sequence (128–144) are PAO specific residues with 9 residues common to both PAO and PAK sequences (Figure 3).
Figure 4.
Competitive ELISAs using three representative groups of antibodies; A, PAO pilin specific antibodies, B1, PAK/PAO cross-reactive antibodies, and E1, PAK/PAO cross-reactive antibodies. The antibodies were purified as described in the methods. Panel A shows antibody A in a PAO-PAO pilin and PAO-PAK pilin competition ELISA. The results, coupled with indirect ELISA results, show that antibody A is PAO pilin specific. Panel B shows antibody B1 in PAO-PAO pilin, PAO-PAK pilin, PAK-PAK pilin, and PAK-PAO pilin competition ELISAs. As shown, PAK pilin cannot compete B1 antibodies off PAO pilin and vice versa; PAO pilin cannot compete B1 antibodies off PAK pilin. This indicates that there are at least two distinct antibodies in B1 with different epitopes, one PAO pilin specific and one PAK pilin specific. Panel C shows antibody E1 in PAO-PAO pilin, PAO-PAK pilin, PAK-PAK pilin, and PAK-PAO pilin competition ELISAs. The results are similar to the B1 antibody results and indicate that there are at least two distinct antibodies in E1, one PAO pilin specific and one PAK pilin specific. Antibodies are designated by letter based on the peptide immunogens shown in Figure 2.
Peptide Immunogen Design
The goal of this project was to design a 17-residue peptide immunogen of the receptor binding domain that contained two distinct epitopes, each of which would generate antibodies that were strain specific, one to PAO pilin and one to PAK pilin of P. aeruginosa. To design such an immunogen, we used the native PAO sequence shown in Figure 3, known to generate strain-specific antibodies and systematically substituted two, three, four and five amino acid residues from the PAK sequence into the PAO sequence. With five amino acid substitutions, from the PAK sequence into the peptide immunogen, we have a native strain PAK epitope within the disulfide loop region (Figure 3) while maintaining a native strain PAO specific epitope outside the disulfide loop. In other words, we hypothesized that antibodies derived from this immunogen would bind to both native PAK and PAO strain pilin receptor binding domains.
Antibody Cross-reactivity
The key result of this study is that all peptide immunogens (B, C, D and E shown in Figure 3, that contain two to five PAK residue substitutions into the PAO sequence) generate antibodies that bind to both native strain PAK and PAO pilins. That is, the polyclonal antibodies are cross-reactive (Figure 4B and 4C).
For example, both indirect and competitive ELISAs, show that the B1 antibodies generated to peptide immunogen B (two PAK residues substituted for two PAO residues: Figure 3) bind PAO pilin and can be competed by PAO pilin and bind to PAK pilin and can be competed by PAK pilin. However, the antibodies that bind to PAO pilin cannot be competed by PAK pilin and the antibodies that bind to PAK pilin cannot be competed by PAO pilin. These results show that there are two different antibodies in the polyclonal sera B1, with at least two different epitopes. One population of antibodies recognizes PAO pilin and the other population of antibodies recognizes PAK pilin. By our definition, this makes the polyclonal antibodies cross-reactive.
In a similar fashion, both indirect and competitive ELISAs (Figure 4C) show that E1 antibodies generated to peptide immunogen E (five PAK residue substituted for five PAO residues, Figure 3) bind to PAO pilin and can be competed by PAO pilin and bind to PAK pilin and can be competed by PAK pilin. Moreover, the results are similar to those for immunogen B, i.e., the antibodies that bind to PAO pilin cannot be competed by PAK pilin and the antibodies that bind to PAK pilin cannot be competed by PAO pilin. The results show again that there are two different antibodies in the polyclonal sera E1, with at least two different epitopes. One population of antibodies recognizes PAO pilin and the other population of antibodies recognizes PAK pilin.
In all cases, the other immunogens, C (with three PAK residue substitutions) and D (with four PAK residue substitutions) generated cross-reactive antibodies with similar properties to antibodies generated from immunogens B and E (discussed above and shown in Figure 4, panels B and C). Furthermore, as shown in Table 1 the affinities of antibodies for PAO pilin as determined by competitive ELISA using IC50 values were similar for antibodies isolated from different rabbits using the same immunogen. For example, immunogen B gave IC50 values for antibodies between different rabbits of 5.8 and 7.8 nM, while immunogen E gave IC50 values of 7.1 and 7.6 nM. The largest difference of antibody IC50 values was for immunogen C, with IC50 values of 1.3 and 25 nM. Overall, very similar antibody affinities for PAO pilin were generated for immunogens B, C, D and E (Table 1). As shown in Table 1, the affinities of different antibodies for PAK pilin, as shown by competitive ELISA using IC50 values, were similar to PAO IC50 values. For example, B1 antibodies give values of 10 nM for PAK pilin and 5.8 nM for PAO pilin. C1 antibodies give values of 34 nM for PAK and 1.3 nM for PAO pilin and the reverse situation for C2 antibodies with values of 3.3 nM for PAK pilin and 25 nM for PAO pilin (Table 1).
Table 1.
Direct and competitive ELISA results for polyclonal antibodies A (PAO pilin specific), B1, B2, C1, C2, D1, E1, and E2 (PAO pilin and PAK pilin cross-reactive antibodies).
| Pilin Competitor |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Aba | Plated Pilin | Direct ELISA | PAK | IC50a,b (nM) | PAO | IC50a,b (nM) | PAK (ADN)c | IC50 (nM) | PAO (KSK)d | IC50 (nM) |
| A | PAK | nbe | ncf | nag | nc | na | nc | na | nc | na |
| PAO | + | nc | na | + | 2.3 | nc | na | nc | na | |
|
| ||||||||||
| B1 | PAK | + | + | 10 | nc | na | + | 15 | nc | na |
| PAO | + | nc | na | + | 5.8 | + | 20 | nc | na | |
|
| ||||||||||
| B2 | PAK | + | + | 92 | nc | na | + | 64 | nc | na |
| PAO | + | nc | na | + | 7.8 | + | 565 | nc | na | |
|
| ||||||||||
| C1 | PAK | + | + | 34 | nc | na | + | 32 | nc | na |
| PAO | + | nc | na | + | 1.3 | + | 9.6 | nc | na | |
|
| ||||||||||
| C2 | PAK | + | + | 3.3 | nc | na | + | 37 | nc | na |
| PAO | + | nc | na | + | 25 | + | 61 | nc | na | |
|
| ||||||||||
| D1 | PAK | + | + | 110 | nc | na | + | 57 | nc | na |
| PAO | + | nc | na | + | 9.0 | + | 8.8 | nc | na | |
|
| ||||||||||
| E1 | PAK | + | + | 100 | nc | na | + | 71 | nc | na |
| PAO | + | nc | na | + | 7.1 | + | 42 | nc | na | |
|
| ||||||||||
| E2 | PAK | + | + | 31 | nc | na | + | 11 | nc | na |
| PAO | + | nc | na | + | 7.6 | + | 160 | nc | na | |
IC50, The pilin competitor concentration at which 50% of the antibodies (Ab) are competed off the plated pilin. Antibodies designated A, B, C, D, and E are obtained from immunogens shown in Fig. 3. The number following the letter denotes antibodies from different rabbits. Antibodies from different rabbits that were similar to the reported antibodies are not shown. For example, all antibodies to immunogen A from three different rabbits were similar and PAO specific.
Geometric mean of IC5D values for cross reactive antibodies B1, B2, C1, C2, D1 , E1, and E2 for PAK (32 nM) and PAO (7.7 nM).
PAK (ADN) is a PAK pilin mutant where ADN denotes residue substitutions, K128A/S143D/K144N, frorn PAO pilin
PAO (KSK) is a PAO pilin mutant where KSK denotes residue substitutions, A128K/D143S/N144K, from PAK pilin.
nb, no binding of the antibody to the pilin by direct ELISA
nc, no competition from the pilin competitor for the antibodies binding to the plated pilin.
na, not applicable.
The geometric mean of the IC50 values, for the antibody affinities for PAO pilin from the seven examples shown in Table 1 is 7.7 nM and for PAK pilin the geometric mean is 32 nM. Thus, the affinity for the two different pilins is within a factor of four. This suggests that the immunogens are generating antibodies with excellent cross-reactivity between PAO and PAK pilins. Clearly antibody cross-reactivity is not enhanced by increasing the number of PAK residues substituted into the PAO immunogen.
Compare the results in Table 1 for immunogen B (two PAK residues E135 and Q136 substituted in the PAO immunogen) to immunogen E (five PAK residues T130, D132 and I138 in addition to E135 and Q136 substituted in the PAO immunogen). These results suggest that antibodies that bind PAK pilin can be generated by two substitutions only, i.e., E135 and Q136 into the PAO sequence.
Location of the Antibody Epitopes on the PAK and PAO pilins
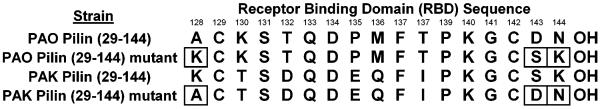
To help determine the location in the peptide sequence of the two epitopes that explain the antibody cross-reactivity to PAK and PAO pilins, we prepared two different pilin mutants in addition to the two native strain pilins, PAK and PAO pilins. The two native strain pilin sequences and the two mutant pilins are shown in Figure 5. The PAO mutant involves three PAK residue substitutions into the PAO pilin sequence outside of the RBD disulfide loop. PAK residues K128, S143 and K144 were substituted into the PAO sequence to generate a PAO pilin mutant denoted PAO (KSK). Similarly, the PAK mutant involved three substitutions of PAO residues into the PAK pilin sequence outside of the RBD disulfide loop. PAO residues A128, D143 and N144 were substituted into the PAK sequence to generate a PAK pilin mutant denoted PAK (ADN). The idea was to determine the key residues required for antibody binding. That is, to determine if the key residues responsible for antibody binding to PAO pilin were within the disulfide loop or involved residues outside the loop.
Figure 5.
Shown are the native strain PAO and PAK pilin sequences of the receptor binding domain, residues 128–144, and two mutants. The mutated residues in PAO and PAK pilins (29–144) are boxed. The PAO pilin (29–144) mutant contains three PAK pilin residues, A128K, D143S, and N144K and is denoted PAO (KSK). The PAK pilin (29–144) mutant contains three PAO pilin residues, K128A, S143D, and K144N and is denoted PAK (ADN).
The results of PAO strain specific A antibodies, binding to PAO pilin, PAO (KSK) pilin and PAK (ADN) pilin are shown in Figure 6. As stated previously, the antibodies generated to the PAO peptide immunogen A (Figure 3) recognize PAO pilin in both indirect and competitive ELISAs and A antibodies do not bind PAK pilin. The IC50 value for A antibodies binding to PAO pilin, with PAO pilin as a competitor was found to be 2.3 nM (Figure 6, Panel A). Antibody A binding to the PAO pilin mutant PAO (KSK) with PAO pilin as a competitor is shown in Figure 6B, with an IC50 value of 10 nM. This result shows that native strain PAO specific A antibodies recognize an epitope within the disulfide loop. This is because mutating the residues at positions A128K, D143S, and N144K, which are outside of the disulfide loop region to PAK residues, does not alter the ability of PAO pilin to compete A antibodies off PAO (KSK). Antibody A binding to the PAK pilin mutant PAK (ADN) with PAO pilin as a competitor is shown in Figure 6C. Keep in mind that the PAO specific A antibodies do not bind to PAK pilin (Figure 4A).
Figure 6.
Competitive ELISAs using antibody A, which is specific for PAO pilin. In Panels A, B, and C, PAO pilin is the competitor. In Panel A, PAO pilin is plated; in Panel B, PAO (KSK) mutant pilin is plated and in Panel C PAK (ADN) mutant pilin is plated. IC50, the concentration of pilin protein resulting in 50% inhibition of antibody binding in the competitive ELISA.
Since the A antibodies bind to PAK (ADN) pilin (three PAO residues A128, D143 and N144 substituted into the PAK pilin sequence outside the disulfide loop) this shows that there is another epitope for PAO specific antibodies. This epitope is outside of the RBD disulfide loop and involves the three substituted PAO pilin residues A128, D143 and N144. Thus, PAO specific polyclonal antibodies must contain antibodies that recognize at least two epitopes, one epitope within the disulfide loop and one epitope outside the disulfide loop involving residues A128, D143 and N144. Furthermore, the antibody affinities for these two epitopes are similar, with IC50 values of 10 nM and 2.3 nM respectively (Figure 6B and 6C). This result strongly supports the concept of designing a single peptide immunogen with two closely related epitopes, where one of the epitopes generates antibodies that recognize one particular P. aeruginosa strain, while the other epitope generates antibodies that recognize another strain.
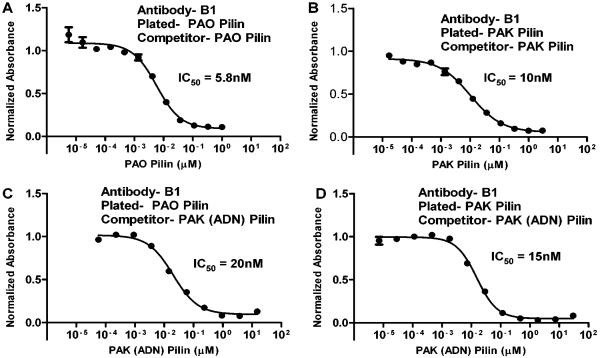
The results of cross-reactive B1 antibodies in competitive ELISAs, with native strain PAO and PAK pilins and the mutant pilin PAK (ADN) are shown in Figure 7. The B1 antibodies recognize both PAO and PAK pilins with IC50 values of 5.8 nM and 10 nM, respectively (Figure 7A and 7B). The antibodies that bind PAO pilin can be competed by PAK (ADN) pilin with an IC50 value of 20 nM (Figure 7C) which is similar to the IC50 value of 5.8 nM for PAO pilin. However, these antibodies do not bind to PAO (KSK) pilin (Table 1). This result shows that the key residues responsible for generating antibodies that bind PAO pilin are the three residues outside the disulfide loop, A128, D143 and N144 (Figure 8). The antibodies that bind PAK pilin can be competed by PAK (ADN) pilin with an IC50 of 15 nM which is similar to the IC50 value for PAK pilin of 10 nM. Thus, the key residues for generating antibodies that bind PAK in the PAO immunogen B are the two PAK residues within the disulfide loop, E135 and Q136 (Figure 8). Although we have shown the results for the B1 antibodies only, the results for the other cross-reactive antibodies derived from immunogens, C, D and E, were all similar (Table 1).
Figure 7.
Competitive ELISAs using antibody B1, which is cross-reactive to PAO pilin (panel A) and PAK pilin (Panel B). Antibodies which bind to PAO pilin (Panel C) or PAK pilin (Panel D) can be readily competed by PAK mutated pilin, PAK (ADN). IC50, the concentration of pilin protein resulting in 50% inhibition of antibody binding in the competitive ELISA.
Figure 8.
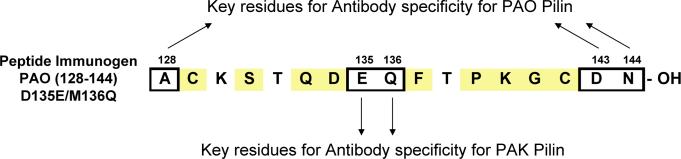
Shown is the amino acid sequence for one peptide immunogen, PAO (128–144) (D135E/M136Q). The residues shaded yellow are identical between native strains PAO and PAK receptor binding domains. The boxed residues are the suggested key residues, (A128, D143, N144) and (E135, Q136), responsible for generating antibodies that are PAO or PAK specific, respectively.
Discussion
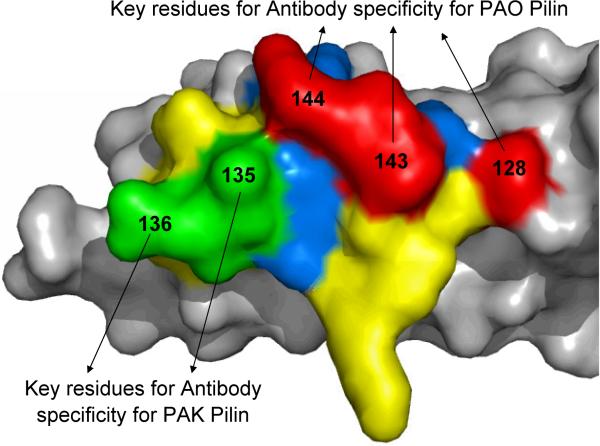
In this study, we have demonstrated that we can design a single peptide immunogen of the 17-residue RBD that modulates the immune response to induce cross-reactive polyclonal sera. This polyclonal sera contains antibodies that bind to the RBDs of two native P. aeruginosa strains, PAK and PAO, through two closely-related epitopes (Figure 9). One epitope is PAK specific and is within the RBD disulfide loop; the other epitope is PAO specific and includes residues 128A, 143D and 144N which are all outside the RBD disulfide loop (Figure 8). Furthermore, this study has shown that peptide immunogens using the PAO sequence as a template can elicit the development of PAK specific antibodies with only two substitutions inside the disulfide loop, P135E and M136Q (Figure 8). Other peptide immunogens had more PAK residue substitutions (Figure 3) but did not elicit antibodies with improved cross-reactivity or affinities. Using the PAK pilin crystal structure as a model, one can visualize the two distinct epitopes for antibody binding on the RBD surface (Figure 9). Residues 135E and 136Q in the peptide immunogen are principally responsible for developing PAK specific antibodies and are positioned prominently to act as an epitope (colored green in Figure 9). Similarly, residues 128A, 143D and 144N, which are responsible for developing PAO specific antibodies, are also positioned prominently to act as an epitope (colored red in Figure 9).
Figure 9.
Molecular surface of the PAK monomeric pilin showing the different residues of the Receptor Binding Domain (RBD). Residues 128, 143, and 144 are shown in red and are the key residues for binding to PAO pilin specific antibodies. Residues 135 and 136 are show in green and are the key residues for binding to PAK pilin specific antibodies. Identical residues between the PAK and PAO strain RBDs are shown in yellow (positions C129, S131, Q133, D134, F137, P139, K140, G141, and C142, Figure 1). Other residues different between the two strains, in addition to those shown in red and green, are shown in blue (positions 130, 132, and 138, Figure 1) and the rest of the pilin protein is shown in gray (12).
The indirect and Competitive ELISAs support these conclusions. Note that, among the different immunogens that generate cross-reactive antibodies (immunogens B, C, D, and E) all of them bound to native strain PAK and PAO pilin by indirect ELISA. Furthermore, these antibodies bound PAK (ADN) by indirect ELISA and none of them bound to PAO (KSK). This alone indicates that residues 128A, 143D and 144N are an important part of the peptide immunogen epitope for generating antibodies to PAO pilin. Once these three residues were substituted with PAK residues (K128, S143 and K144) in the PAO pilin to make the PAO (KSK) pilin mutant, the antibodies did not bind to PAO (KSK). The competitive ELISAs further support these conclusions, for example, in the competitive ELISAs using polyclonal antibody B1 with PAO pilin plated, PAK (ADN) competes the antibodies off PAO pilin with an IC50 of 20nM (Figure 7). This is in contrast to native strain PAK pilin, which will not compete the antibodies off plated native strain PAO pilin. Moreover, PAK (ADN) competes antibodies off PAO pilin because both pilins have residues A128, D143 and N144. These residues are outside the RBD and are part of the epitope for the PAO specific antibodies in the cross-reactive antibody B1. Conversely, B1 antibodies could not bind to PAO (KSK), which lacks the ADN residues, by indirect ELISA. This indicates that the ADN residues are an important part of the PAO pilin epitope. It should be pointed out, that prior published experiments have indicated that antibodies that bind to the RBD have had epitopes that were inside the disulfide loop (17, 27). In particular, in one study using a monoclonal PAK specific antibody, PK99H, generated from native strain PAK pilin, the RBD epitope of PK99H was mapped and was determined to consist of residues 134–140 (D-E-Q-F-I-P-K), which are all within the disulfide loop (27).
It is well established that antibodies produced to a protein immunogen can recognize even smaller epitopes on the surface of a protein. For example, antibovine rhodopsin monoclonal antibodies were shown to recognize a linear determinant at the C-terminus as small as four residues. Furthermore, only three of the four amino acid side-chains were important for antibody binding (28). This epitope is analogous to the PAO epitope derived from the 17-residue RBD immunogen B used in this study, where the polyclonal antibodies that specifically recognize PAO pilin require the three PAO residues, A128, D143 and N144 for binding with nanomolar affinity. These three residues are clustered together in the pilin protein (Figure 9). Similarly, the PAK specific epitope within the RBD disulfide loop has two key residues, E135 and Q136. The polyclonal antibodies that are specific to PAK pilin do not recognize PAO pilin even though nine residues are identical between the PAK and PAO sequences, within the RBD disulfide loop.
Conclusions and Future Directions
In this study, we have shown that residues immediately outside the pilin RBD disulfide loop, are important for P. aeruginosa peptide immunogen design and can be used to generate more cross-reactive antisera to multiple P. aeruginosa stains. Furthermore, we have shown that only two substitutions from the PAK sequence into the PAO peptide immunogen sequence are required for the peptide immunogen to elicit an immune response to native strain PAK pilin. Both of these features help elucidate what amino acid sequences enhance cross-reactivity of antibodies generated from a peptide immunogen. These features shall be used in future plans for our rational design approach to develop a synthetic peptide vaccine that generates antibodies that are effective against multiple strains of P. aeruginosa pili.
Acknowledgments
Financial support for this project was provided by the National Institute of Health; grant number R01-AI 048717 and the John Stewart Chair in Peptide Chemistry to R.S.H. We also thank Eunice York for synthesizing and purifying the peptide immunogens.
Abbreviations
- RBD
Receptor binding domain
- KLH
keyhole limpet hemocyanin
- BSA
Bovine serum albumin
References
- 1.Gaynes R, Edwards JR. Overview of nosocomial infections caused by gram-negative bacilli. Clin Infect Dis. 2005;41:848–854. doi: 10.1086/432803. [DOI] [PubMed] [Google Scholar]
- 2.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 3.Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol. 2004;2:363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- 4.Paranchych W, Frost LS. The physiology and biochemistry of pili. Adv Microb Physiol. 1988;29:53–113. doi: 10.1016/s0065-2911(08)60346-x. [DOI] [PubMed] [Google Scholar]
- 5.Mattick J. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 6.Irvin RT, Doig PC, Sastry PA, Heller B, Paranchych W. Usefulness of equilibrium parameters of adhesion in predicting the outcome of competition for bacterial receptor sites on respiratory epithelial cells by Pseudomonas aeruginosa strains of heterologous pilus type. Microb Ecol Health Dis. 1990;3:39–47. [Google Scholar]
- 7.Lee KK, Sheth HB, Wong WY, Sherburne R, Paranchych W, Hodges RS, Lingwood CA, Krivan H, Irvin RT. The binding of Pseudomonas aeruginosa pili to glycosphingolipids is a tip-associated event involving the C-terminal region of the structural pilin subunit. Mol Microbiol. 1994;11:705–713. doi: 10.1111/j.1365-2958.1994.tb00348.x. [DOI] [PubMed] [Google Scholar]
- 8.Baker NR. Mucosal adherence of Pseudomonas aeruginosa. In: Fick RB Jr., editor. Pseudomonas aeruginosa the Opportunist: Pathogenesis and Disease. CRC Press, Inc.; Florida: 1993. pp. 7–23. [Google Scholar]
- 9.Chi E, Mehl T, Nunn D, Lory S. Interaction of Pseudomonas aeruginosa with A549 pneumocyte cells. Infection and Immunity. 1991;59:822–828. doi: 10.1128/iai.59.3.822-828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farinha MA, Conway BD, Glasier LM, Ellert NW, Irvin RT, Sherburne R, Paranchych W. Alteration of the pilin adhesin of Pseudomonas aeruginosa PAO results in normal pilus biogenesis but a loss of adherence to human pneumocyte cells and decreased virulence in mice. Infect Immun. 1994;62:4118–4123. doi: 10.1128/iai.62.10.4118-4123.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang H, Kays M, Prince A. Role of Pseudomonas aeruginosa pili in acute pulmonary infection. Infect Immun. 1995;63:1278–1285. doi: 10.1128/iai.63.4.1278-1285.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hazes B, Sastry PA, Hayakawa K, Read RJ, Irvin RT. Crystal structure of Pseudomonas aeruginosa PAK pilin suggests a main- chain-dominated mode of receptor binding. J Mol Biol. 2000;299:1005–1017. doi: 10.1006/jmbi.2000.3801. [DOI] [PubMed] [Google Scholar]
- 13.Keizer DW, Kalisiak M, Crump MP, Suh JY, Irvin RT, Sykes BD. Assignments of 1H and 15N resonances of the Pseudomonas aeruginosa K122- 4 pilin monomer. J Biomol NMR. 2001;19:385–396. doi: 10.1023/a:1011285803129. [DOI] [PubMed] [Google Scholar]
- 14.Suh JY, Spyracopoulos L, Keizer DW, Irvin RT, Sykes BD. Backbone dynamics of receptor binding and antigenic regions of a Pseudomonas aeruginosa pilin monomer. Biochemistry. 2001;40:3985–3995. doi: 10.1021/bi002524h. [DOI] [PubMed] [Google Scholar]
- 15.Craig L, Taylor RK, Pique ME, Adair BD, Arvai AS, Singh M, Lloyd SJ, Shin DS, Getzoff ED, Yeager M, Forest KT, Tainer JA. Type IV Pilin Structure and Assembly. X-Ray and EM Analyses of Vibrio cholerae Toxin-Coregulated Pilus and Pseudomonas aeruginosa PAK Pilin. Mol Cell. 2003;11:1139–1150. doi: 10.1016/s1097-2765(03)00170-9. [DOI] [PubMed] [Google Scholar]
- 16.Audette GF, Irvin RT, Hazes B. Crystallographic analysis of the Pseudomonas aeruginosa strain K122-4 monomeric pilin reveals a conserved receptor-binding architecture. Biochemistry. 2004;43:11427–11435. doi: 10.1021/bi048957s. [DOI] [PubMed] [Google Scholar]
- 17.Lee KK, Wong WY, Sheth HB, Hodges RS, Paranchych W, Irvin RT. Use of synthetic peptides in characterization of microbial adhesins. Methods Enzymol. 1995;253:115–131. doi: 10.1016/s0076-6879(95)53013-4. [DOI] [PubMed] [Google Scholar]
- 18.Campbell AP, McInnes C, Hodges RS, Sykes BD. Comparison of NMR solution structures of the receptor binding domains of Pseudomonas aeruginosa pili strains PAO, KB7, and PAK: implications for receptor binding and synthetic vaccine design. Biochemistry. 1995;34:16255–16268. doi: 10.1021/bi00050a005. [DOI] [PubMed] [Google Scholar]
- 19.McInnes C, Sonnichsen FD, Kay CM, Hodges RS, Sykes BD. NMR solution structure and flexibility of a peptide antigen representing the receptor binding domain of Pseudomonas aeruginosa. Biochemistry. 1993;32:13432–13440. doi: 10.1021/bi00212a008. [DOI] [PubMed] [Google Scholar]
- 20.Keizer DW, Slupsky CM, Kalisiak M, Campbell AP, Crump MP, Sastry PA, Hazes B, Irvin RT, Sykes BD. Structure of a pilin monomer from Pseudomonas aeruginosa: implications for the assembly of pili. J Biol Chem. 2001;276:24186–24193. doi: 10.1074/jbc.M100659200. [DOI] [PubMed] [Google Scholar]
- 21.Cachia PJ, Hodges RS. Synthetic peptide vaccine and antibody therapeutic development: Prevention and treatment of Pseudomonas aeruginosa. Biopolymers. 2003;71:141–168. doi: 10.1002/bip.10395. [DOI] [PubMed] [Google Scholar]
- 22.Cachia PJ, Glasier LM, Hodgins RR, Wong WY, Irvin RT, Hodges RS. The use of synthetic peptides in the design of a consensus sequence vaccine for Pseudomonas aeruginosa. J Pept Res. 1998;52:289–299. doi: 10.1111/j.1399-3011.1998.tb01243.x. [DOI] [PubMed] [Google Scholar]
- 23.Kao DJ, Churchill ME, Irvin RT, Hodges RS. Animal Protection and Structural Studies of a Consensus Sequence Vaccine Targeting the Receptor Binding Domain of the Type IV Pilus of Pseudomonas aeruginosa. J Mol Biol. 2007;374:426–442. doi: 10.1016/j.jmb.2007.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erickson BW, Merrifield RB. Solid-phase peptide synthesis. In: Neurath H, Hill RL, editors. The Proteins. Academic Press; New York, USA: 1976. pp. 255–527. [Google Scholar]
- 25.Lee KK, James AB, Hodges RS. Separation of Intrachain Disulfide Bridged Peptides from their Reduced forms By Reversed-Phase Chromotography. In: Mant CT, Hodges RS, editors. HPLC of Peptides and Proteins: Separation, Analysis, and Conformation. CRC Press; Boca Raton: 1991. [Google Scholar]
- 26.Wetzel R, Halualani R, Stults JT, Quan C. A general method for highly selective cross-linking of unprotected polypeptides via pH-controlled modification of N-terminal alpha-amino groups. Bioconjug Chem. 1990;1:114–122. doi: 10.1021/bc00002a005. [DOI] [PubMed] [Google Scholar]
- 27.Wong WY, Irvin RT, Paranchych W, Hodges RS. Antigen-antibody interactions: Elucidation of the epitope and strain-specificity of a monoclonal antibody directed against the pilin protein adherence binding domain of Pseudomonas aeruginosa strain K. Protein Sci. 1992;1:1308–1318. doi: 10.1002/pro.5560011010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hodges RS, Heaton RJ, Parker JMR, Molday L, Molday RS. Antigen-antibody interaction: Synthetic peptides define linear antigenic determinants recognized by monoclonal antibodies directed to the cytoplasmic carboxyl terminus of rhodopsin. J Biol Chem. 1988;263:11768–11775. [PubMed] [Google Scholar]