Abstract
Realization of the full potential of human pluripotent stem cells (hPSCs) in regenerative medicine requires the development of well-defined culture conditions for their long-term growth and directed differentiation. Current practices for maintaining hPSCs generally utilize empirically determined combinations of feeder cells and other animal-based products, which are expensive, difficult to isolate, subject to batch-to-batch variations, and unsuitable for cell-based therapies. Using a high-throughput screening approach, we identified several polymers that can support self-renewal of hPSCs. While most of these polymers provide support for only a short period of time, we identified a synthetic polymer poly(methyl vinyl ether-alt-maleic anhydride) (PMVE-alt-MA) that supported attachment, proliferation and self-renewal of HUES1, HUES9, and iPSCs over five passages. The hPSCs cultured on PMVE-alt-MA maintained their characteristic morphology, expressed high levels of markers of pluripotency, and retained a normal karyotype. Such cost-effective, polymer-based matrices that support long-term self-renewal and proliferation of hPSCs will not only help to accelerate the translational perspectives of hPSCs, but also provide a platform to elucidate the underlying molecular mechanisms that regulate stem cell proliferation and differentiation.
Keywords: Human embryonic stem cells, human induced pluripotent stem cells, polymer arrays, defined culture conditions, extracellular matrix
INTRODUCTION
In order to use human pluripotent stem cells (hPSCs) for the treatment of a variety of debilitating diseases, it is necessary to harness their ability to differentiate into virtually all mature cell types. Many challenges need be addressed before such therapies can become a reality: (a) lack of well-defined xeno-free conditions for derivation and long-term expansion of hPSCs, (b) insufficient systematic control over conditions that regulate hPSC behavior, and (c) the inability to produce hPSCs on a large-scale in defined conditions. The derivation and proliferation of hPSCs depend on their microenvironment, including the chemical and physical properties of the extracellular matrix (ECM) and the presence of growth factors (GFs). We and others have identified unique factors and signaling pathways that regulate hPSC self-renewal, thus leading to the development of fully defined culture systems, free of animal factors for the long-term self-renewal of hPSCs [1–8]. However, these culture systems rely on recombinant or purified human proteins that are expensive, difficult to isolate, subject to batch-to-batch variations, and hence are not suitable for large-scale expansion of hPSCs.
In contrast, synthetic polymers that are inexpensive and easily fabricated represent a reliable alternative for in vitro hPSC expansion. Polymeric biomaterials have been utilized as substrates for the growth of a variety of cell types [9]. Specifically, they have been used for the expansion of many human adult stem cell and progenitor populations such as hematopoietic [10–13], mesenchymal [14–18], and neural stem cells [19]. Although polymeric biomaterials have been used extensively to support ex vivo expansion and differentiation of adult stem cells, their application as artificial matrices to support self-renewal of hPSCs is only beginning to be explored [20]. The identification of optimal polymer biomaterials for this application is a daunting task with the conventional iterative approaches because of the large number of biomaterials with varying properties to be explored and the lack of identification of a unique set of physicochemical properties that control cell-material interactions.
Recent developments in high-throughput screening of combinatorial libraries of materials and soluble factors offer a cost-effective tool to identify conditions and materials that support self-renewal and differentiation of stem cells [21–28]. In this study, we modified our array-based high-throughput approach[29, 30] for systematic investigation of the effects of polymeric biomaterials on hPSC self-renewal. Using this new technology, polymers with varying chemical compositions, functional groups and molecular weights were screened for their ability to promote hPSC proliferation while supporting pluripotency. These screening experiments led to the identification of a number of candidate polymers that supported short-term hPSC attachment, growth, and maintenance of pluripotency. Furthermore, one of these polymers, poly(methyl vinyl ether-alt-maleic anhydride) (PMVE-alt-MA), supports long-term maintenance of two human embryonic stem cell (hESC) lines, HUES1 and HUES9, and one induced pluripotent stem cell line (iPSC) over 5 passages. This is the first example of long-term maintenance of hPSCs on completely synthetic materials free from exogenous adhesion molecules such as extracellular matrix proteins (ECMPs).
MATERIALS AND METHODS
Polymer Array Fabrication
Glass slides were coated with acrylamide gels as previously described [29]. Briefly, glass slides were cleaned, silanized, and then functionalized with a polyacrylamide gel layer. Polymers were dissolved in the appropriate solvents (PBS, DMSO, DMF, or toluene) at five different concentrations (40, 20, 10, 5, and 1 mg/ml) and placed in polypropylene 384-well plates. Polymers 1–10 were synthesized by free radical polymerization, polymers 11–80 were purchased from Sigma, and polymers 81–91 were purchased from PolySciences. A contact arrayer (SpotArray 24; Perkin Elmer) was used to print the polymers. The printing conditions were a 1000-ms inking time and a 250-ms stamping time. To control for variability, each polymer was printed in replicates of 5 spots. Each spot had a diameter of 150–200 μm, and neighboring microenvironments were separated by a center-to-center distance of 450 μm. A single slide carried 6,400 spots arranged in sixteen 20×20 matrices so that one slide carried 1280 unique biomaterial conditions. Slides were inspected manually under a light microscope for consistent and uniform polymer deposition. Prior to their use, slides were soaked in PBS while being exposed to UVC germicidal radiation in a sterile flow hood for 10 min.
Cell Culture
The following media were used: H1299 (1X high glucose DMEM, 10% fetal bovine serum, 1% (v/v) L-glutamine, 1% (v/v) penicillin/streptomycin); iPSCs (1X DMEM-F12, 20% (v/v) Knockout Serum Replacement, 1% (v/v) non-essential amino acids, 0.5% (v/v) glutamine, 120 μM 2-mercaptoethanol [Sigma]); HUES9 and HUES1 hESCs (1X Knockout DMEM, 10% (v/v) Knockout Serum Replacement, 10% (v/v) human plasmanate (Talecris Biotherapeutics), 1% (v/v) non-essential amino acids, 1% (v/v) penicillin/streptomycin, 1% (v/v) Gluta-MAX, 55 μM 2-mercaptoethanol [Sigma]). All media components are from Life Technologies unless indicated otherwise. HPSC lines were maintained on feeder layers of mitotically inactivated mouse embryonic fibroblasts (2×104/cm2; Chemicon). All hPSC cultures were supplemented with 30 ng/ml bFGF (Life Technologies). MEF-CM was produced by culturing the appropriate hPSC medium on MEFs for 24 hr. StemPro consisted of the StemPro supplement (Invitrogen) diluted in DMEM-F12 with 2% (v/v) BSA (Millipore) and 55 μM 2-mercaptoethanol. HPSCs were routinely passaged as single cells by exposure to Accutase (Millipore) for 5 min, followed by one rinse with media and centrifugation at 200 × g. Cells were then resuspended and plated.
H1299 were passaged (5.0 × 105 cells per slide) directly onto the array slides and allowed to settle on the spots for 18 hr prior to rinsing with H1299 medium 3 times to remove residual cells and debris. Prior to seeding onto the arrays, hPSCs were cultured for two passages on Matrigel (BD Biosciences) with MEF-CM supplemented with 30 ng/ml bFGF to remove residual feeder cells. hPSCs were then Accutase-passaged onto the array slides (1.5 × 106 cells per slide) and allowed to settle on the spots for 18 hr prior to rinsing once with medium to remove unattached cells and debris. Culture medium was replenished daily. Due to the non-fouling nature of the polymerized acrylamide, cells were confined to the printed microenvironment spots.
Array Slide Staining, Imaging and Quantification
Because fixing and staining protocols may cause cell detachment and alter the cell counts on each spot, arrays were stained live for DNA with Hoechst 33342 (2 μg/ml; Invitrogen) for 5 min. The arrays were washed 3 times with the medium and then imaged. After live imaging, the arrays were fixed in 4% PFA for 5 min at 4°C, followed by 10 min at room temperature. Live imaging of slides was performed using an automated confocal microscope (Olympus Fluoview 1000 with motorized stage and incubation chamber). Slides were imaged as 3 × 3 arrays at 10× magnification using a focus height that gave the maximum signal in the Z-direction for each channel at the center of the array. Images were then quantified using GenePix software (MDS Analytical Technologies).
OCT4-GFP Reporter Line
The lenti construct that was used to generate the OCT4-GFP reporter line was kindly provided by Dr. Alexey Terskikh. High titer lenti was produced as previously described [53, 54]. HUES9 cells were infected overnight with lenti Oct4-GFP and single clones were isolated and screened for (i) stable GFP expression levels, (ii) low GFP expression levels after EB formation, and (iii) rapid decrease in GFP expression upon removal of MEF-CM.
Polymer-Coated Slide Preparation and Characterization
For studies in which hPSCs were cultured on polymers over multiple passages, acrylamide-coated glass slides were fabricated with polymers having larger defined areas using the contact arrayer. Slides were then dried on a hot plate (60°C) for solvent evaporation. The presence of coated PMVE-alt-MA was verified by FTIR-ATR (Supplementary Figure 4). FTIR spectra were acquired on a Nicolet 6700 with Smart-iTR using a N2 purged sample chamber. The acquisition parameters were: 128 scan and 4cm−1 (spectra resolution).
Characterization of Long-term Culture of HPSCs on PMVE-alt-MA
hPSCs were cultured on Matrigel with MEF-CM prior to culture on PMVE-alt-MA. hPSCs were passaged every 4–6 days depending on colony size and density onto fresh PMVE-alt-MA coated slides. Characterization of hPSCs grown on PMVE-alt-MA was performed after five sequential passages. PMVE-alt-MA acrylamide gel-coated slides were fixed in 4% PFA for 5 min at 4°C, followed by 10 min at room temperature. Immediately before staining, the cells were permeabilized with 0.2% (v/v) Triton-X-100 and blocked with 1% (w/v) BSA and 3% (w/v) nonfat dry milk for 30 min. The slides were stained with the primary antibodies rabbit anti-Oct3/4a or rabbit anti-Nanog (Santa Cruz) diluted 1:200 in 1% BSA overnight at 4°C, washed 3 times with TBS, and incubated with goat anti-rabbit Alexa 647 at 1:400 for 1 hr at 37°C. Nucleic acids were stained for DNA with Hoechst 33342 (2 μg/ml; Invitrogen) for 5 min at room temperature. The slides were then washed 3 times with TBS and air dried immediately before imaging. For the in vitro differentiation studies, hPSCs cultured on PMVE-alt-MA were transferred to Matrigel-coated plates. For ectoderm differentiation, hPSCs were treated with 5 μM of ROCK inhibitor (Y27632, Sigma) 24 hr before embryoid body (EB) formation. Cells were trypsinized, transferred into untreated V-shaped 96-well plate (5 × 103 cell/well) and centrifuged at 950 × g to form compact colonies of cells. After 24 hr, the cell clumps were transferred using a P1000 pipette to ultra-low binding 6-well plate. After 7 days, the EBs were replated onto Matrigel-coated plates for an additional 14 days. During the entire duration, EBs were cultured in 1X DMEM, 20% (v/v) FBS and 1% (v/v) penicillin/streptomycin. Cells were fixed, permeabilized, and stained with mouse anti-beta III tubulin (R&D Systems) at 1:100 and donkey anti-mouse Alexa 546 (Invitrogen) at 1:250. For mesoderm differentiation, hPSCs were cultured on Matrigel in 1X DMEM, 20% (v/v) FBS and 1% (v/v) penicillin/streptomycin for 21 days. Cells were fixed, permeabilized, and stained with mouse anti-smooth muscle actin (R&D Systems) at 1:500 and donkey anti-mouse Alexa 546 (Invitrogen) at 1:250. For endoderm differentiation, hPSCs were cultured on Matrigel until confluency. The medium was then changed to RPMI supplemented with 1% (v/v) Gluta-MAX and 100 ng/ml recombinant human Activin A (R&D Systems). Cells were cultured for 3 days, with FBS concentrations at 0% for the first day and 0.2% for the second and third days. Cultures were supplemented with 30 ng/ml purified mouse Wnt3a for the first day. Cells were fixed, permeabilized, and stained with goat anti-Sox17 (R&D Systems) at 1:200 and donkey anti-goat Alexa 546 (Invitrogen) at 1:250.
RNA Isolation and Quantitative PCR
RNA was isolated from cells using TRIzol (Invitrogen), and treated with DNase I (Invitrogen) to remove traces of DNA. Reverse transcription was performed by means of qScript cDNA Supermix (Quanta Biosciences). Quantitative PCR was carried out using TaqMan probes (Applied Biosystems) and TaqMan Fast Universal PCR Master Mix (Applied Biosystems) on a 7900HT Real Time PCR machine (Applied Biosystems). Taqman gene expression assay primers (Applied Biosystems; Supplementary Table 3) were used. Gene expression was normalized to 18S rRNA levels. Delta Ct values were calculated as Cttarget − Ct18s. All experiments were performed with three technical replicates. Data are presented as the average of the biological replicates ± standard error of the mean.
Karyotype Analysis
For each cell line, cytogenetic analysis was performed on 20 metaphase cells using standard protocols for G-banding (Cell Line Genetics).
Data Analysis
All values were presented as mean ± standard error of the mean unless otherwise noted.
RESULTS
Polymer Microarrays to Study Cell Fate
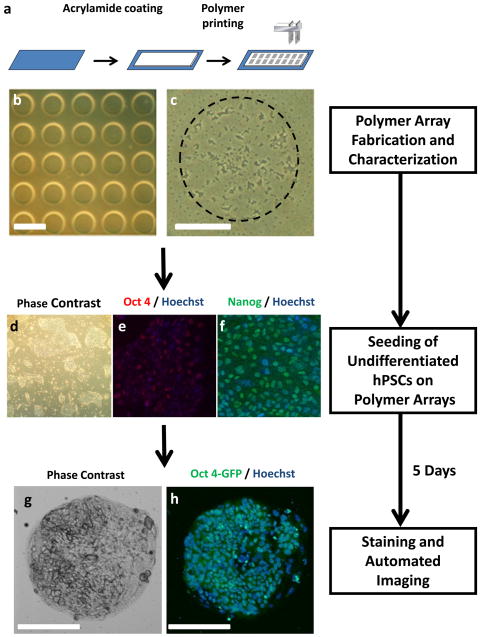
We used a contact DNA microarray spotting instrument to deposit nanoliter amounts of polymer solutions onto glass microscope slides coated with a layer of acrylamide gel (~10 μm thick) (Materials and Methods; Figure 1a). Printing parameters such as inking and printing times were optimized in order to create a uniform distribution of the polymer spots within the array. Polymers were dissolved in the appropriate solvent (PBS, DMSO, DMF, or toluene) and deposited onto the acrylamide gel-coated slides. The spotted polymer chains are mechanically interlocked with the underlying acrylamide gels and anchored in place after solvent evaporation (Figures 1b and 1c). In the spaces devoid of polymer, the acrylamide gel layer inhibited cell growth. A single slide carried 6,400 spots arranged in sixteen 20×20 matrices. Each polymer spot was 150–200 μm in diameter and each polymer solution was spotted in replicates of five so that one slide could carry up to 1280 unique polymeric conditions. Polymer spots showed consistent size between replicate spots and non-replicate spots (Figure 1b).
Figure 1.
Schematic representation of polymer array assay. (a) Polymers were printed using a contact microarray spotter onto glass slides that have been coated with polyacrylamide gels. Cells were globally seeded onto polymer arrays consisting of spotted polymers arranged in 16 subarrays consisting of a 20 × 20 matrix of spots. (b) Each polymer was printed in five replicates (scale bar = 450 μm). (c) Each polymer spot had a diameter of 150 μm (scale bar = 75 μm). Prior to seeding onto the array, hPSCs were cultured in feeder-free conditions and assessed for their (d) characteristic morphology and (e, f) maintenance of markers of pluripotency. hESCs were grown on the polymer arrays for 5 days and imaged in real time by automated microscopy. hESCs were assessed for their (g) characteristic morphology and proliferation (Hoechst) and (h) maintenance of pluripotency (OCT4).
To determine the efficacy of the polymer array to support cell adhesion and growth, we first tested the polymer array with an immortalized cell line (H1299, non-small-cell lung carcinoma cells), which has been shown to be capable of robust growth on tissue culture treated polystyrene. These cells were seeded onto polymer arrays, cultured for 48 hr after and analyzed for adhesion and growth. Most of the polymers tested were found to support attachment and spreading of H1299.
Polymer Microarray Screens with HESCs
Next, we utilized this array technology to screen for polymers that support hPSC adhesion, growth, and pluripotency (Figure 2). Prior to seeding onto the array, hPSCs were cultured in feeder-free conditions [31] and assessed for their characteristic morphology (Figure 1d), maintenance of markers of pluripotency (Figures 1e and f), and normal karyotype (data not shown). Polymeric biomaterials typically mediate cell adhesion through indirect, integrin-dependent interactions between the cells and extracellular matrix proteins that have been adsorbed from the surrounding media [32]. Recent studies have shown that conditioned media from mouse and human feeder cultures contain several soluble ECMPs, including collagen I, collagen IV, fibronectin, laminin, and heparin sulfate [33]. In order to eliminate undefined adsorption of these soluble ECMPs, we performed all of our experiments in the presence of a defined medium, StemPro™ (Life Technologies), which contains bFGF, IGF1, Heregulin and ActivinA, and no soluble ECMPs. Thus, any observed interactions between polymers and hPSCs would be independent of exogenous soluble ECMPs.
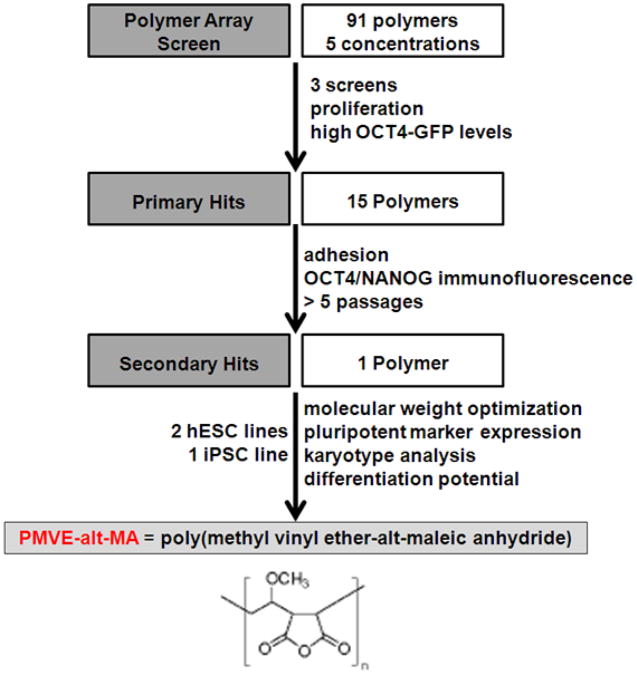
Figure 2.
Screening paradigm for identification of polymers that support hPSC attachment, growth, and maintenance of pluripotency.
To seed the arrays, hPSCs were trypsinized into single cells, and cell suspensions were allowed to settle onto the polymer spots for 18 hr. Thereafter, the medium was replaced to remove non-adhering cells and debris. Seeding the array slides with 1 × 106 cells allowed for the attachment of 10–20 cells per spot and provided sufficient area for subsequent growth. After 5 days, hPSC proliferation and pluripotency were quantified by automated microscopy (Figures 1g and 1h).
In order to have a real-time measure of pluripotency, we generated a hESC line (HUES9) with a GFP gene under transcriptional control of an OCT4 promoter (OCT4-GFP; see Materials and Methods). To validate our assay for detecting small changes in GFP expression, we seeded arrays with defined mixtures of constitutively expressing GFP (CMV-GFP) hESCs and non-fluorescent (NON-GFP) hESCs. After 5 days of culture, the hESC-seeded arrays were imaged. The average GFP signal from an array increased in a linear manner as the proportion of GFP cells in the mixture increased (Supplementary Figure 1). Specifically, we were able to detect ~10% changes in the CMV-GFP: Non-GFP hESC ratio, indicating sufficient sensitivity for screening purposes.
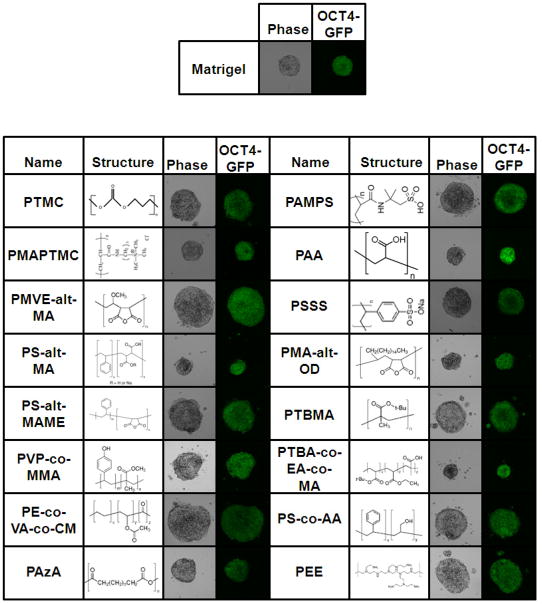
Using this array technology, we next screened a library of diverse polymers (Supplementary Table 1), each at five concentrations (40, 20, 10, 5, and 1 mg/ml). Matrigel was spotted as a positive control. The screening was performed on three independent polymer array slides to ensure reproducibility. The OCT4-GFP values and total cell number (estimated by Hoechst stain) were analyzed and rank-ordered for repeated high hits. Of the 90 polymers tested, 16 polymers demonstrated the ability to support proliferation and maintenance of pluripotency at similar levels as Matrigel (Figure 3 and Supplementary Table 2).
Figure 3.
Hit polymers and their chemical structures.
Polymer Molecular Weight Influences HPSC Proliferation
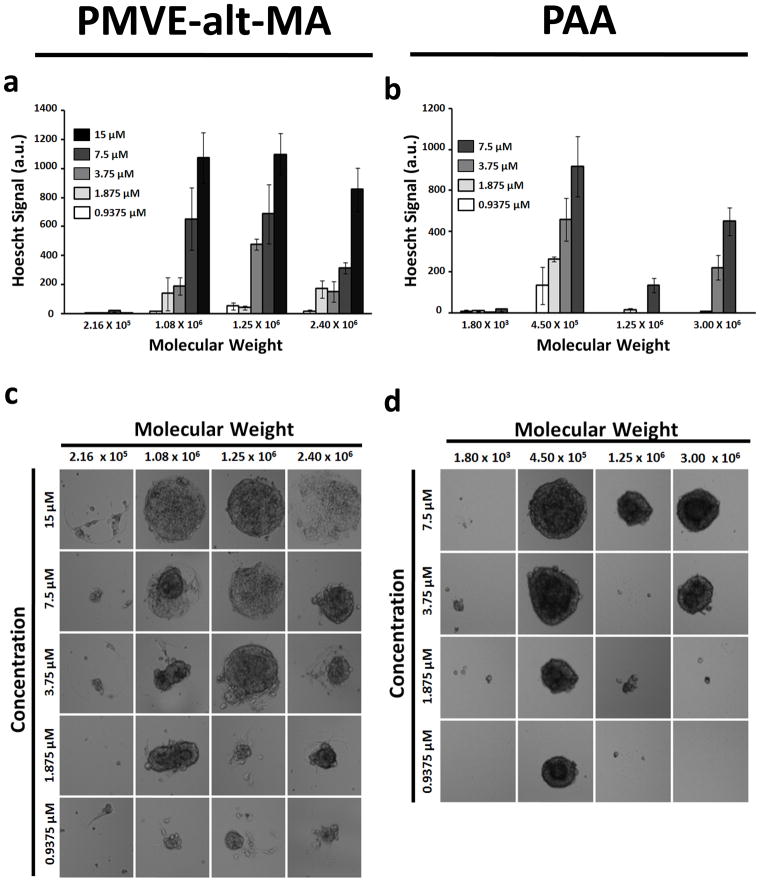
Since polymer molecular weight has been shown to influence cell behavior [34], we next evaluated the effect of molecular weight of two of our hits, poly(methyl vinyl ether-alt-maleic anhydride) (PMVE-alt-MA) and poly(acrylic acid) (PAA), on self renewal of hPSCs. HUES9 cells were cultured on arrays of PMVE-alt-MA and PAA with varying molecular weights and concentrations for 5 days. In general, the PMVE-alt-MA and PAA polymers supported hESC proliferation in a concentration-dependent manner with the highest amount of proliferation typically occurring at the highest concentration of the polymer tested (Figures 4a and 4b). Conversely, hESC proliferation exhibited a non-monotonic dependence on the molecular weight of the polymer. PMVE-alt-MA supported the highest amount of hESC proliferation at a molecular weight of 1.25 × 106 Da (Figure 4c), while PAA supported the highest amount of hESC proliferation at a molecular weight of 4.5 × 105 Da (Figure 4d). Increasing or decreasing the molecular weight of these specific polymers reduced the amount of hESC growth. On the other hand, the molecular weight of the polymers did not elicit a significant effect on proliferation of H1229 cells (Supplementary Figures 2a and 2b). Taken together these results indicate that the polymer molecular weight influences hESC growth and that this effect is unique to hESCs.
Figure 4.
Polymer molecular weight influences hESC proliferation. Hues9 cells were cultured on arrays of (a) PMVE-alt-MA and (b) PAA with varying molecular weights and concentrations for 5 days. Quantitative analysis (relative Hoechst staining) reveals that hESC proliferation on (c) PMVE-alt-MA and (d) PAA is influenced by the polymer’s molecular weight. Data are representative of 3 independent array experiments. All values are presented as mean ± standard error of the mean of 5 replicate spots.
Long-term Culture of HESCS on Defined Polymers
We next tested whether the 16 polymers that were identified to support short-term hPSC self-renewal could support hESC adhesion, growth, and maintenance of pluripotency over multiple passages. Acrylamide gel-coated slides were designed with defined areas fabricated with the hit polymers (see Materials and Methods). HUES9 cells were trypsinized from feeder-free cultures on Matrigel and passaged onto the polymer coated slides. Detachment or spontaneous differentiation, as indicated by changes in morphology and reduced OCT4 and NANOG immunofluorescence, respectively, was observed within the first two passages on all the hit polymers with the exception of poly(methyl vinyl ether-alt-maleic anhydride) (PMVE-alt-MA; Figures 5a and 5b).
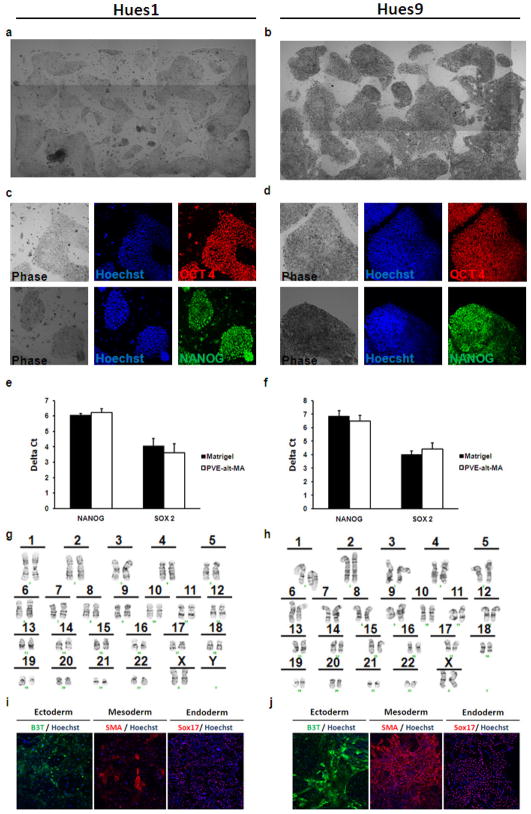
Figure 5.
PMVE-alt-MA supports long-term culture of hESCs in defined medium. (a) Hues1 and (b) Hues9 were cultured on PMVE-alt-MA for 5 passages in StemPro™. Immunofluorescence of (c) Hues1 and (d) Hues9 cultured on PMVE-alt-MA showing expression of markers of pluripotency, NANOG and OCT4. Quantitative RT-PCR of (e) Hues1 and (f) Hues9 cultured on PMVE-alt-MA demonstrates similar expression levels of markers of pluripotency compared to hESCs cultured on Matrigel. Delta Ct values were calculated as Cttarget − Ct18s. Data presented as mean ± standard error of the mean of three independent experiments. Karyotype analysis reveals a normal euploid karyotype of (g) Hues1 and (h) Hues9 cultured on PMVE-alt-MA. Immunofluorescence of in vitro differentiation of (i) Hues1 and (j) Hues9 cultured on PMVE-alt-MA shows expression of ectoderm (beta III tubulin; B3T), mesoderm (smooth muscle actin; SMA), and endoderm (Sox17) cell derivatives.
Next, we tested the acrylamide-covered slides coated with PMVE-alt-MA at the optimum molecular weight (MW= 1.25 × 106 Da) for its ability to support consistent expansion of two hESC lines (HUES1 and HUES9) and one iPSC line over 5 passages. Under these conditions, hESCs (Figures 5a and 5b) and iPSCs (Supplementary Figure 3) exhibited their characteristic morphology and grew as tightly clustered colonies. Maintenance of pluripotency was assessed by immunostaining (Figures 5c and 5d; Supplementary Figure 3) and quantitative RT-PCR (Figures 5e and 5f) for stem cell markers such as OCT4, NANOG, and SOX2. HESCs grown on PMVE-alt-MA maintained expression of these pluripotency markers at levels similar to that of hESCs grown on Matrigel (Figure 5e and 5f). Cells grown on PMVE-alt-MA maintained a normal karotype (Figures 5g and 5h). Finally, hESCs grown on PMVE-alt-MA were differentiated into all three germ layers (ectoderm, mesoderm, and endoderm) (Figures 5i and 5j), thus confirming their pluripotency. Together, these results demonstrated the ability of PMVE-alt-MA to support long-term culture of hPSCs in defined media conditions.
Culture of hESCS on PMVE-alt-MA Results in Increased Expression of Endogenous ECMPs and Integrins
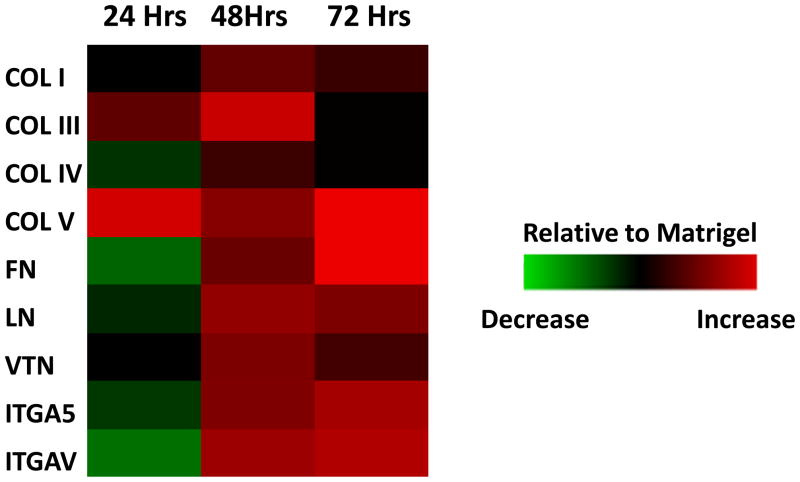
To determine whether culture of hPSCs on PMVE-alt-MA modulated endogenous ECMP and integrin levels, we analyzed the expression levels of genes encoding ECMPs, including collagen I (COL I), collagen III (COL III), collagen IV (COL IV), collagen V (COL V), fibronectin (FN), laminin (LN), and vitronectin (VTN), and cell adhesion molecules, including integrin α5 (ITGA5) and integrin αv (ITGAV), by quantitative RT-PCR. ECMPs have been previously shown to be secreted by both hPSCs and supportive feeder cells and to play a critical role in attachment and maintenance of pluripotency of hPSCs [35]. Similarly, it has been demonstrated that ITGA5 and ITGAV are the primary integrins that mediate hPSC attachment and interaction with these ECMPs and with Matrigel [35]. Expression levels of endogenous ECMPs and integrins in hESCs grown for 48 hr of culture on PMVE-alt-MA are significantly higher in comparison to hESCs grown on Matrigel for the same duration of time (Figure 6). These data indicate that hPSCs cultured free of exogenous ECMPs on PMVE-alt-MA increased the production and secretion of ECMPs to assemble a microenvironment that supports their attachment, proliferation, and pluripotency.
Figure 6.
Culture of hESCs on PMVE-alt-MA leads to increased expression of endogenous ECMPs and integrins. Hues9 cells were cultured on Matrigel and PMVE-alt-MA for 24, 48, and 72 hr. Quantitative RT-PCR of expression levels of ECMPs (collagen I:COL I, collagen III: COL III, collagen IV: COL IV, collagen V:COL V, fibronectin: FN, laminin: LN, and vitronectin: VTN) and integrins (integrin alpha 5: ITGA5, integrin alpha V: ITGAV) was performed. Data are displayed in a heat map where each block represents (Delta CtPMVE-alt-MA − Delta CtMatrigel) at the same time point.
DISCUSSION
Realization of the multitude of potential hPSC applications is dependent upon our ability to design novel culture conditions which are chemically defined, robust, cost-effective, and devoid of animal-derived components. Much of the research carried out on hPSCs thus far has focused on soluble signals and their effects on hPSC self renewal [3, 5, 36], thus providing a well-defined alternative combination of soluble factors to replace conditioned medium for hPSC expansion [1–8]. Since signaling molecule responses are affected by interactions between the cell and its surrounding matrix, we [29] and others [37–40] have investigated the effect of the extracellular matrix components on influencing hPSC fate. Subsequently, various ECMP combinations have been developed for the expansion of hPSCs [33, 41–43]. However, these approaches utilize purified or recombinant proteins, which are expensive, thus making the use of these systems for large-scale expansion of hPSCs cost-prohibitive. Compared to ECMPs and synthetic peptides, polymer biomaterials are inexpensive and provide a robust platform to produce large numbers of hPSCs.
In this study, we employed an unbiased high-throughput screening approach to systematically screen in a concentration-varying manner, a library of synthetic polymers to develop a chemically defined, cost-effective, robust culture system for in vitro expansion of hPSCs. From our initial screens, we identified several polymers that were able to support short-term hPSC proliferation and maintenance of pluripotency. Although most of these polymers could not support long-term self-renewal of hPSCs, we identified one polymer, PMVE-alt-MA, that was able to sustain the long-term culture of two hESC lines (HUES1 and HUES9) and one hiPSC line, while maintaining their morphology, expression of stem cell markers, genetic integrity, and in vitro pluripotency. Furthermore, we verified that this polymer substrate was compatible with the defined medium StemPro™ (Life Technologies). Our results also show a molecular weight-dependent effect of the polymer on hPSC growth supporting the notion that the physicochemical properties of polymers play an important role in modulating cell-matrix interactions. There could also be topographical changes associated with the molecular weight-dependent entanglement of polymer chains and/or changes in interaction between the polymer-coated matrix and its surrounding medium.
The specific mechanism by which PMVE-alt-MA supports self-renewal of hPSCs is not entirely clear. Cell surface integrins of the hPSCs play an important role in their interactions with the surrounding matrix and thus playing a role in their self-renewal [35, 44]. The hPSCs cultured on PMVE-alt-MA exhibited higher expression levels of integrin α5 and αv. These integrins have been shown to mediate adhesion of hPSCs to various ECMPs and Matrigel [35, 44]. It has also been shown that synthetic peptides designed to engage these integrins and cell surface heparan sulfates support self-renewal of hESCs in vitro [45].
We also found that hPSCs cultured free of exogenous ECMPs on PMVE-alt-MA show increased expression of several endogenous ECMPs. This suggests that although PMVE-alt-MA fosters initial hPSC adhesion independent of non-specific adsorption of proteins from the surrounding medium, there are additional contributions of proteins secreted by the growing hPSCs. Even though a defined medium free of exogenous ECMPs was used in these experiments, growth factors in the medium, such as bFGF can bind to the polymer. In an aqueous environment, PMVE-alt-MA undergoes hydrolysis and forms poly[(methyl vinyl ether)-alt-(maleic acid)] [46]. Such anionic polymers bearing carboxyl and sulfonyl groups have been shown to mimic functional features of heparin [47–49]. Heparin-mimicking polymers have been shown to bind to various growth factors, such as bFGF, mainly through favorable energetic interactions [49, 50]. In fact, maleic acid-based heparin-mimetic polymers have been used in vivo for regulating various heparin binding growth factors [51]. Furthermore, heparin is an important component of the microenvironment, which has recently been recognized for its role in regulating self-renewal of hESCs [52]. Taken together, these findings suggest a possible mechanism by which PMVE-alt-MA promotes proliferation of hPSCs while maintaining their undifferentiated state in a manner similar to heparin in MEF-coculture or MEF-conditioned medium.
A recent report by Villa-Diaz et al. demonstrated the use of synthetic polymer coatings for the growth of hESCs [20]. The authors observed five polymer coatings that were able to support short term self-renewal of hESCs while only one polymer, poly[2-(methacryloyloxy)ethyl dimethyl-(3-sulfopropyl)ammonium hydroxide] (PMEDSAH), was able to sustain long-term growth of hESCs without inducing differentiation. Although we did not test PMEDSAH in our screen, we did test several chemically similar polymers containing sulfonyl groups, and found that two of these polymers, poly(acrylamido-methyl-propane sulfonate) (PAMPS) and poly(sodium 4-styrenesulfonate) (PSSS), were able to support short-term self-renewal of hPSCs. However, unlike PMVE-alt-MA, PSSS and PAMPS were unable to support long-term culture of hPSCs. Nonetheless, like PMVE-alt-MA, PAMPS, PSSS, and PMEDSAH are anionic polymers which may assist in hPSC self renewal by mimicking heparin activities, such as bFGF binding. Our study distinguishes itself from that by Villa-Diaz et al. in several ways: 1) PMVE-alt-MA was identified from an unbiased screening approach in which we analyzed a large number of polymers at varying concentrations. 2) PMVE-alt-MA is available off the shelf with controlled molecular weights. As seen from our findings, polymer molecular weight plays a significant role in hPSC attachment and growth. 3) PMVE-alt-MA supports the long-term maintenance of hiPSCs in addition to hESCs.
CONCLUSION
In summary, we demonstrated that polymeric biomaterials can be used as an artificial matrix to support ex vivo expansion of hPSCs while maintaining their undifferentiated state. Specifically, we found that one inexpensive polymer, PMVE-alt-MA, can support long-term self-renewal and proliferation of hPSCs. PMVE-alt-MA is available off the shelf, thus avoiding complications associated with synthesis and purifications. PMVE-alt-MA supports the long-term culture of both hESCs and hiPSCs despite the reported differences between the two cell types, suggesting that this polymer could be widely used for expansion of hPSCs. Furthermore, the described array-based high-throughput screening technology can be used for investigating and characterizing large numbers of polymer biomaterials and culture conditions for proliferation and/or directed differentiation. The use of polymeric biomaterials along with defined medium conditions to support in vitro expansion of hPSCs not only reduces the risk of contamination with animal derived pathogens and transmission of cell secreted components from the feeder cells but also offers a cost-effective, scalable, robust platform to generate large number of hPSCs. The completely synthetic nature of polymers avoids batch-to-batch variations, and the physical and chemical properties of polymers can be precisely controlled and tuned.
Supplementary Material
Supplementary Figure 1: Validation of GFP detection. To validate the array assay for detecting small changes in GFP expression, polymer arrays were seeded with defined mixtures of hESCs constitutively expressing GFP and non-fluorescent hESCs. After 5 days of culture, the arrays were imaged. The average GFP signal from an array increased in a linear manner as the proportion of GFP cells in the mixture increased.
Supplementary Figure 2: Influence of molecular weight of polymers is specific to hPSCs. H1299s were seeded onto arrays consisting of varying molecular weights and concentrations of (a) PMVE-alt-MA and (b) PAA. Neither polymer molecular weight nor concentration had an effect on H1299 proliferation.
Supplementary Figure 3: PMVE-alt-MA supports growth of hiPSCs. HiPSCs were cultured on PMVE-alt-MA for 5 passages. Cells retained their characteristic morphology and high expression levels of markers of pluripotency such as OCT4.
Supplementary Figure 4: Characterization of PMVE-alt-MA coated acrylamide slides. FITR spectrum of PMVE-alt-MA polyacrylamide surface. Peaks from both polyacrylamide (1647 & 3334 cm−1 (N-H stretching); 3185 (C-H stretching); 2934 cm−1 (aliphatic asymmetric C-H stretching)) and polyanhydride (1848 cm−1 [symmetric C=O stretching]; 1776 cm−1 [asymmetric C=O stretching]) were identified.
Supplementary Table 1: Polymers used for microarray synthesis.
Supplementary Table 2: Hit polymers and abbreviations.
Supplementary Table 3: Taqman primers used for quantitative RT-PCR.
Acknowledgments
D.A.B. was supported by funding from the University of California Biotechnology Research and Education Program (2007-006), the National Science Foundation Graduate Research Fellowship Program, and the UCSD California Institute of Regenerative Postdoctoral Fellowship Program. This research was supported in part by the California Institute of Regenerative Medicine (RS1-00172-1) to S.C., and NHLBI Research Grant HL080518 to S.C. We would also like to thank the UCSD Human Embryonic Core Facility for generation and characterization of the hiPSC lines.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
DA Brafman, Departments of Bioengineering, Cellular and Molecular Medicine, and Institute of Engineering in Medicine, University of California-San Diego, 9500 Gilman Dr., La Jolla, CA 92093-0695.
CW Chang, Department of Bioengineering, University of California-San Diego, 9500 Gilman Dr., La Jolla, CA 92093-0412.
A Fernandez, Department of Cellular and Molecular Medicine, University of California-San Diego, 9500 Gilman Dr., La Jolla, CA 92093-0695.
K Willert, Department of Cellular and Molecular Medicine, University of California-San Diego, 9500 Gilman Dr., La Jolla, CA 92093-0695.
S Varghese, Department of Bioengineering, University of California-San Diego, 9500 Gilman Dr., La Jolla, CA 92093-0412.
S Chien, Departments of Bioengineering, Medicine, and Institute of Engineering in Medicine, University of California-San Diego, 9500 Gilman Dr., La Jolla, CA 92093-0412.
References
- 1.Xu RH, Peck RM, Li DS, Feng X, Ludwig T, Thomson JA. Basic FGF and suppression of BMP signaling sustain undifferentiated proliferation of human ES cells. Nat Methods. 2005;2(3):185–190. doi: 10.1038/nmeth744. [DOI] [PubMed] [Google Scholar]
- 2.Xu C, Rosler E, Jiang J, Lebkowski JS, Gold JD, O’Sullivan C, et al. Basic fibroblast growth factor supports undifferentiated human embryonic stem cell growth without conditioned medium. Stem Cells. 2005;23(3):315–323. doi: 10.1634/stemcells.2004-0211. [DOI] [PubMed] [Google Scholar]
- 3.Wang L, Schulz TC, Sherrer ES, Dauphin DS, Shin S, Nelson AM, et al. Self-renewal of human embryonic stem cells requires insulin-like growth factor-1 receptor and ERBB2 receptor signaling. Blood. 2007;110(12):4111–4119. doi: 10.1182/blood-2007-03-082586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10(1):55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 5.Ludwig TE, Levenstein ME, Jones JM, Berggren WT, Mitchen ER, Frane JL, et al. Derivation of human embryonic stem cells in defined conditions. Nat Biotechnol. 2006;24(2):185–187. doi: 10.1038/nbt1177. [DOI] [PubMed] [Google Scholar]
- 6.James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132(6):1273–1282. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- 7.Heins N, Englund MC, Sjoblom C, Dahl U, Tonning A, Bergh C, et al. Derivation, characterization, and differentiation of human embryonic stem cells. Stem Cells. 2004;22(3):367–376. doi: 10.1634/stemcells.22-3-367. [DOI] [PubMed] [Google Scholar]
- 8.Amit M, Shariki C, Margulets V, Itskovitz-Eldor J. Feeder layer- and serum-free culture of human embryonic stem cells. Biol Reprod. 2004;70(3):837–845. doi: 10.1095/biolreprod.103.021147. [DOI] [PubMed] [Google Scholar]
- 9.Hubbell JA. Bioactive biomaterials. Curr Opin Biotechnol. 1999;10(2):123–129. doi: 10.1016/s0958-1669(99)80021-4. [DOI] [PubMed] [Google Scholar]
- 10.Bagley J, Rosenzweig M, Marks DF, Pykett MJ. Extended culture of multipotent hematopoietic progenitors without cytokine augmentation in a novel three-dimensional device. Exp Hematol. 1999;27(3):496–504. doi: 10.1016/s0301-472x(98)00053-8. [DOI] [PubMed] [Google Scholar]
- 11.Banu N, Rosenzweig M, Kim H, Bagley J, Pykett M. Cytokine-augmented culture of haematopoietic progenitor cells in a novel three-dimensional cell growth matrix. Cytokine. 2001;13(6):349–358. doi: 10.1006/cyto.2001.0836. [DOI] [PubMed] [Google Scholar]
- 12.Berrios VM, Dooner GJ, Nowakowski G, Frimberger A, Valinski H, Quesenberry PJ, et al. The molecular basis for the cytokine-induced defect in homing and engraftment of hematopoietic stem cells. Exp Hematol. 2001;29(11):1326–1335. doi: 10.1016/s0301-472x(01)00734-2. [DOI] [PubMed] [Google Scholar]
- 13.Ehring B, Biber K, Upton TM, Plosky D, Pykett M, Rosenzweig M. Expansion of HPCs from cord blood in a novel 3D matrix. Cytotherapy. 2003;5(6):490–499. doi: 10.1080/14653240310003585. [DOI] [PubMed] [Google Scholar]
- 14.Curran JM, Chen R, Hunt JA. The guidance of human mesenchymal stem cell differentiation in vitro by controlled modifications to the cell substrate. Biomaterials. 2006;27(27):4783–4793. doi: 10.1016/j.biomaterials.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Kotobuki N, Katsube Y, Katou Y, Tadokoro M, Hirose M, Ohgushi H. In vivo survival and osteogenic differentiation of allogeneic rat bone marrow mesenchymal stem cells (MSCs) Cell Transplant. 2008;17(6):705–712. doi: 10.3727/096368908786092793. [DOI] [PubMed] [Google Scholar]
- 16.Zhao F, Grayson WL, Ma T, Bunnell B, Lu WW. Effects of hydroxyapatite in 3-D chitosan-gelatin polymer network on human mesenchymal stem cell construct development. Biomaterials. 2006;27(9):1859–1867. doi: 10.1016/j.biomaterials.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 17.Fan H, Hu Y, Zhang C, Li X, Lv R, Qin L, et al. Cartilage regeneration using mesenchymal stem cells and a PLGA-gelatin/chondroitin/hyaluronate hybrid scaffold. Biomaterials. 2006;27(26):4573–4580. doi: 10.1016/j.biomaterials.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 18.Richardson SM, Hughes N, Hunt JA, Freemont AJ, Hoyland JA. Human mesenchymal stem cell differentiation to NP-like cells in chitosan-glycerophosphate hydrogels. Biomaterials. 2008;29(1):85–93. doi: 10.1016/j.biomaterials.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 19.Hayman MW, Smith KH, Cameron NR, Przyborski SA. Growth of human stem cell-derived neurons on solid three-dimensional polymers. J Biochem Biophys Methods. 2005;62(3):231–240. doi: 10.1016/j.jbbm.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 20.Villa-Diaz LG, Nandivada H, Ding J, Nogueira-de-Souza NC, Krebsbach PH, O’Shea KS, et al. Synthetic polymer coatings for long-term growth of human embryonic stem cells. Nat Biotechnol. 2010;28(6):581–583. doi: 10.1038/nbt.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tare RS, Khan F, Tourniaire G, Morgan SM, Bradley M, Oreffo RO. A microarray approach to the identification of polyurethanes for the isolation of human skeletal progenitor cells and augmentation of skeletal cell growth. Biomaterials. 2009;30(6):1045–1055. doi: 10.1016/j.biomaterials.2008.10.038. [DOI] [PubMed] [Google Scholar]
- 22.Tourniaire G, Diaz-Mochon JJ, Bradley M. Fingerprinting polymer microarray. Comb Chem High Throughput Screen. 2009;12(7):690–696. doi: 10.2174/138620709788923692. [DOI] [PubMed] [Google Scholar]
- 23.Pernagallo S, Diaz-Mochon JJ, Bradley M. A cooperative polymer-DNA microarray approach to biomaterial investigation. Lab Chip. 2009;9(3):397–403. doi: 10.1039/b808363k. [DOI] [PubMed] [Google Scholar]
- 24.Diaz-Mochon JJ, Tourniaire G, Bradley M. Microarray platforms for enzymatic and cell-based assays. Chem Soc Rev. 2007;36(3):449–457. doi: 10.1039/b511848b. [DOI] [PubMed] [Google Scholar]
- 25.Mant A, Tourniaire G, Diaz-Mochon JJ, Elliott TJ, Williams AP, Bradley M. Polymer microarrays: identification of substrates for phagocytosis assays. Biomaterials. 2006;27(30):5299–5306. doi: 10.1016/j.biomaterials.2006.04.040. [DOI] [PubMed] [Google Scholar]
- 26.Tourniaire G, Collins J, Campbell S, Mizomoto H, Ogawa S, Thaburet JF, et al. Polymer microarrays for cellular adhesion. Chem Commun (Camb) 2006;28(20):2118–2120. doi: 10.1039/b602009g. [DOI] [PubMed] [Google Scholar]
- 27.Anderson DG, Levenberg S, Langer R. Nanoliter-scale synthesis of arrayed biomaterials and application to human embryonic stem cells. Nat Biotechnol. 2004;22(7):863–866. doi: 10.1038/nbt981. [DOI] [PubMed] [Google Scholar]
- 28.Anderson DG, Putnam D, Lavik EB, Mahmood TA, Langer R. Biomaterial microarrays: rapid, microscale screening of polymer-cell interaction. Biomaterials. 2005;26(23):4892–4897. doi: 10.1016/j.biomaterials.2004.11.052. [DOI] [PubMed] [Google Scholar]
- 29.Brafman DA, Shah KD, Fellner T, Chien S, Willert K. Defining long-term maintenance conditions of human embryonic stem cells with arrayed cellular microenvironment technology. Stem Cells Dev. 2009;18(8):1141–1154. doi: 10.1089/scd.2008.0410. [DOI] [PubMed] [Google Scholar]
- 30.Brafman DA, de Minicis S, Seki E, Shah KD, Teng D, Brenner D, et al. Investigating the role of the extracellular environment in modulating hepatic stellate cell biology with arrayed combinatorial microenvironments. Integr Biol (Camb) 2009;1(8–9):513–524. doi: 10.1039/b912926j. [DOI] [PubMed] [Google Scholar]
- 31.Xu C, Inokuma MS, Denham J, Golds K, Kundu P, Gold JD, et al. Feeder-free growth of undifferentiated human embryonic stem cells. Nat Biotechnol. 2001;19(10):971–974. doi: 10.1038/nbt1001-971. [DOI] [PubMed] [Google Scholar]
- 32.Shin H, Jo S, Mikos AG. Biomimetic materials for tissue engineering. Biomaterials. 2003;24(24):4353–4364. doi: 10.1016/s0142-9612(03)00339-9. [DOI] [PubMed] [Google Scholar]
- 33.Braam SR, Zeinstra L, Litjens S, Ward-van Oostwaard D, van den Brink S, van Laake L, et al. Recombinant vitronectin is a functionally defined substrate that supports human embryonic stem cell self-renewal via alphavbeta5 integrin. Stem Cells. 2008;26(9):2257–2265. doi: 10.1634/stemcells.2008-0291. [DOI] [PubMed] [Google Scholar]
- 34.Saltzman WM. Cell interactions with polymers. In: Lanza R, Langer R, Vacanti JP, editors. Principles of tissue engineering. 2. San Diego: Academic Press; 2000. pp. 221–235. [Google Scholar]
- 35.Kohen NT, Little LE, Healy KE. Characterization of matrigel interfaces during defined human embryonic stem cell culture. Biointerphases. 2009;4(4):69–79. doi: 10.1116/1.3274061. [DOI] [PubMed] [Google Scholar]
- 36.Yao S, Chen S, Clark J, Hao E, Beattie GM, Hayek A, et al. Long-term self-renewal and directed differentiation of human embryonic stem cells in chemically defined conditions. Proc Natl Acad Sci U S A. 2006;103(18):6907–6912. doi: 10.1073/pnas.0602280103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma W, Tavakoli T, Derby E, Serebryakova Y, Rao MS, Mattson MP. Cell-extracellular matrix interactions regulate neural differentiation of human embryonic stem cells. BMC Dev Biol. 2008;8:90. doi: 10.1186/1471-213X-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gong J, Sagiv O, Cai H, Tsang SH, Del Priore LV. Effects of extracellular matrix and neighboring cells on induction of human embryonic stem cells into retinal or retinal pigment epithelial progenitors. Exp Eye Res. 2008;86(6):957–965. doi: 10.1016/j.exer.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toh WS, Guo XM, Choo AB, Lu K, Lee EH, Cao T. Differentiation and enrichment of expandable chondrogenic cells from human embryonic stem cells in vitro. J Cell Mol Med. 2009;13(9B):3570–3590. doi: 10.1111/j.1582-4934.2009.00762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gil JE, Woo DH, Shim JH, Kim SE, You HJ, Park SH, et al. Vitronectin promotes oligodendrocyte differentiation during neurogenesis of human embryonic stem cells. FEBS Lett. 2009;583(3):561–567. doi: 10.1016/j.febslet.2008.12.061. [DOI] [PubMed] [Google Scholar]
- 41.Noaksson K, Zoric N, Zeng X, Rao MS, Hyllner J, Semb H, et al. Monitoring differentiation of human embryonic stem cells using real-time PCR. Stem Cells. 2005;23(10):1460–1467. doi: 10.1634/stemcells.2005-0093. [DOI] [PubMed] [Google Scholar]
- 42.Genbacev O, Krtolica A, Zdravkovic T, Brunette E, Powell S, Nath A, et al. Serum-free derivation of human embryonic stem cell lines on human placental fibroblast feeders. Fertil Steril. 2005;83(5):1517–1529. doi: 10.1016/j.fertnstert.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 43.Beattie GM, Lopez AD, Bucay N, Hinton A, Firpo MT, King CC, et al. Activin A maintains pluripotency of human embryonic stem cells in the absence of feeder layers. Stem Cells. 2005;23(4):489–495. doi: 10.1634/stemcells.2004-0279. [DOI] [PubMed] [Google Scholar]
- 44.Lee ST, Yun JI, Jo YS, Mochizuki M, van der Vlies AJ, Kontos S, et al. Engineering integrin signaling for promoting embryonic stem cell self-renewal in a precisely defined niche. Biomaterials. 2010;31(6):1219–1226. doi: 10.1016/j.biomaterials.2009.10.054. [DOI] [PubMed] [Google Scholar]
- 45.Kato H. Chemical analysis of blood coagulation factors--with special reference to the test using synthetic peptide substrates. Rinsho Byori. 1987;70:43–57. [PubMed] [Google Scholar]
- 46.Volet G, Auvray L, Amiel C. Monoalkyl poly(2-methyl-2-oxazoline) micelles. a small-angle neutron scattering study. J Phys Chem B. 2009;113(41):13536–13544. doi: 10.1021/jp9029634. [DOI] [PubMed] [Google Scholar]
- 47.Liekens S, Leali D, Neyts J, Esnouf R, Rusnati M, Dell’Era P, et al. Modulation of fibroblast growth factor-2 receptor binding, signaling, and mitogenic activity by heparin-mimicking polysulfonated compounds. Mol Pharmacol. 1999;56(1):204–213. doi: 10.1124/mol.56.1.204. [DOI] [PubMed] [Google Scholar]
- 48.Christman KL, Vazquez-Dorbatt V, Schopf E, Kolodziej CM, Li RC, Broyer RM, et al. Nanoscale growth factor patterns by immobilization on a heparin-mimicking polymer. J Am Chem Soc. 2008;130(49):16585–16591. doi: 10.1021/ja803676r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miao HQ, Ornitz DM, Aingorn E, Ben-Sasson SA, Vlodavsky I. Modulation of fibroblast growth factor-2 receptor binding, dimerization, signaling, and angiogenic activity by a synthetic heparinmimicking polyanionic compound. J Clin Invest. 1997;99(7):1565–1575. doi: 10.1172/JCI119319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Medalion B, Merin G, Aingorn H, Miao HQ, Nagler A, Elami A, et al. Endogenous basic fibroblast growth factor displaced by heparin from the lumenal surface of human blood vessels is preferentially sequestered by injured regions of the vessel wall. Circulation. 1997;95(7):1853–1862. doi: 10.1161/01.cir.95.7.1853. [DOI] [PubMed] [Google Scholar]
- 51.Jeanbat-Mimaud V, Barbaud C, Caruelle JP, Barritault D, Cammas-Marion S, Langlois V, et al. Bioactive functionalized polymer of malic acid for bone repair and muscle regeneration. J Biomater Sci Polym Ed. 2000;11(9):979–991. doi: 10.1163/156856200744147. [DOI] [PubMed] [Google Scholar]
- 52.Levenstein ME, Berggren WT, Lee JE, Conard KR, Llanas RA, Wagner RJ, et al. Secreted proteoglycans directly mediate human embryonic stem cell-basic fibroblast growth factor 2 interactions critical for proliferation. Stem Cells. 2008;26(12):3099–3107. doi: 10.1634/stemcells.2007-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miyoshi H, Blomer U, Takahashi M, Gage FH, Verma IM. Development of a self-inactivating lentivirus vector. J Virol. 1998;72(10):8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zufferey R, Dull T, Mandel RJ, Bukovsky A, Quiroz D, Naldini L, et al. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol. 1998;72(12):9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Validation of GFP detection. To validate the array assay for detecting small changes in GFP expression, polymer arrays were seeded with defined mixtures of hESCs constitutively expressing GFP and non-fluorescent hESCs. After 5 days of culture, the arrays were imaged. The average GFP signal from an array increased in a linear manner as the proportion of GFP cells in the mixture increased.
Supplementary Figure 2: Influence of molecular weight of polymers is specific to hPSCs. H1299s were seeded onto arrays consisting of varying molecular weights and concentrations of (a) PMVE-alt-MA and (b) PAA. Neither polymer molecular weight nor concentration had an effect on H1299 proliferation.
Supplementary Figure 3: PMVE-alt-MA supports growth of hiPSCs. HiPSCs were cultured on PMVE-alt-MA for 5 passages. Cells retained their characteristic morphology and high expression levels of markers of pluripotency such as OCT4.
Supplementary Figure 4: Characterization of PMVE-alt-MA coated acrylamide slides. FITR spectrum of PMVE-alt-MA polyacrylamide surface. Peaks from both polyacrylamide (1647 & 3334 cm−1 (N-H stretching); 3185 (C-H stretching); 2934 cm−1 (aliphatic asymmetric C-H stretching)) and polyanhydride (1848 cm−1 [symmetric C=O stretching]; 1776 cm−1 [asymmetric C=O stretching]) were identified.
Supplementary Table 1: Polymers used for microarray synthesis.
Supplementary Table 2: Hit polymers and abbreviations.
Supplementary Table 3: Taqman primers used for quantitative RT-PCR.