Abstract
Genome-wide association studies have underscored the importance of the clustered neuronal nicotinic acetylcholine receptor subunit genes with respect to nicotine dependence as well as lung cancer susceptibility. CHRNB4, which encodes the nicotinic acetylcholine receptor β4 subunit, plays a major role in the molecular mechanisms that govern nicotine withdrawal. Thus, elucidating how expression of the β4 gene is regulated is critical for understanding the pathophysiology of nicotine addiction. We previously identified a CA box regulatory element, (5′ – CCACCCCT –3′) critical for β4 promoter activity in vitro. We further demonstrated that a 2.3-kb fragment of the β4 promoter region containing the CA box is capable of directing cell-type specific expression of a reporter gene to a myriad of brain regions that endogenously express the β4 gene. To test the hypothesis that the CA box is critical for β4 promoter activity in vivo, transgenic animals expressing a mutant form of the β4 promoter were generated. Reporter gene expression was not detected in any tissue or cell type at embryonic day 18.5. Similarly, we observed drastically reduced reporter gene expression at postnatal day 30 when compared to wild type transgenic animals. Finally, we demonstrated that CA box mutation results in decreased interaction of the transcription factor Sp1 with the mutant β4 promoter. Taken together these results demonstrate that the CA box is critical for β4 promoter activity in vivo.
Keywords: Nicotinic Acetylcholine Receptors, CHRNB4, Sp1, CACCC, Transgenic Mice
During the development of the mammalian nervous system, transcriptional regulatory cascades act to define the properties of individual nerve cells by controlling gene expression during neuronal differentiation (Sauka-Spengler and Bronner-Fraser, 2008). These developmental gene networks must be tightly controlled in order to ensure that each neuron is capable of performing the biological functions required for neuronal communication (Eisen, 1991, Francis and Landis, 1999, Groves and Bronner-Fraser, 1999, Flores et al., 2000, Jessell, 2000). Acetylcholine (ACh) is an excitatory neurotransmitter that participates in signaling through both ionotropic and G-protein coupled receptors. Cholinergic signaling mediated by ionotropic nicotinic acetylcholine receptors (nAChR)2 is involved in an array of physiological functions including learning, memory and attention (Albuquerque et al., 2009). Interestingly, substantial alterations in cholinergic signal transduction are observed in numerous neurological disorders including Alzheimer’s disease, schizophrenia, epilepsy and nicotine addiction (Whitehouse et al., 1988, Steinlein et al., 1995, De Fusco et al., 2000, Perry et al., 2001, Isacson et al., 2002, Perl et al., 2003, Teaktong et al., 2003). Hence, further elucidating the molecular mechanisms that act to control expression of the nAChR subunit genes will contribute not only to our understanding of neuronal development but to a greater understanding of the pathophysiology of several neurological disorders as well.
Neuronal nAChRs are ligand-gated cation channels encoded by a family of genes designated CHRNA2 – 10 and CHRNB2 – B4. Mature nAChRs exist as heteropentamers containing a combination of α (α2 – α6) and β (β2 – β4) subunits, while a subset of receptors are homopentamers containing α subunits alone (α7 – α10) (Albuquerque et al., 2009). Neuronal populations in both the central (CNS) and peripheral nervous systems (PNS) express multiple nAChR subtypes (Goldman et al., 1987, Corriveau and Berg, 1993, Vernallis et al., 1993, Conroy and Berg, 1995, Gotti et al., 1997, Zoli et al., 1998, Genzen et al., 2001). Each distinct nAChR subtype displays unique biophysical and pharmacological properties, which are determined by subunit composition (McGehee and Role, 1995). The specific subunits assembled into a particular receptor subtype is dictated, at least in part, by the cell-type specific transcription of the individual subunit genes (Albuquerque et al., 2009).
Three of the 12 neuronal nicotinic subunit genes, those encoding the α5, α3 and β4 subunits, are located in a highly conserved genomic cluster (Fig. 1A; also see (Boulter et al., 1990)). These three receptor subunit genes are co-expressed in a variety of cell types and tissues in both the PNS and CNS (Gotti et al., 2006). The co-expression of these three genes is thought to result from the coordinate regulation of the CHRNA5/A3/B4 locus (Albuquerque et al., 2009). As a likely consequence of their co-expression, the α5α3β4 receptor subtype is the predominant nAChR expressed in the PNS where they play a crucial role in mediating fast synaptic transmission in autonomic ganglia (Rust et al., 1994, Conroy and Berg, 1995, Flores et al., 1996). Despite their largely overlapping patterns of expression, there are a small subset of regions where these genes appear to be uniquely regulated (Gotti et al., 2007, Grady et al., 2009).
Fig. 1.
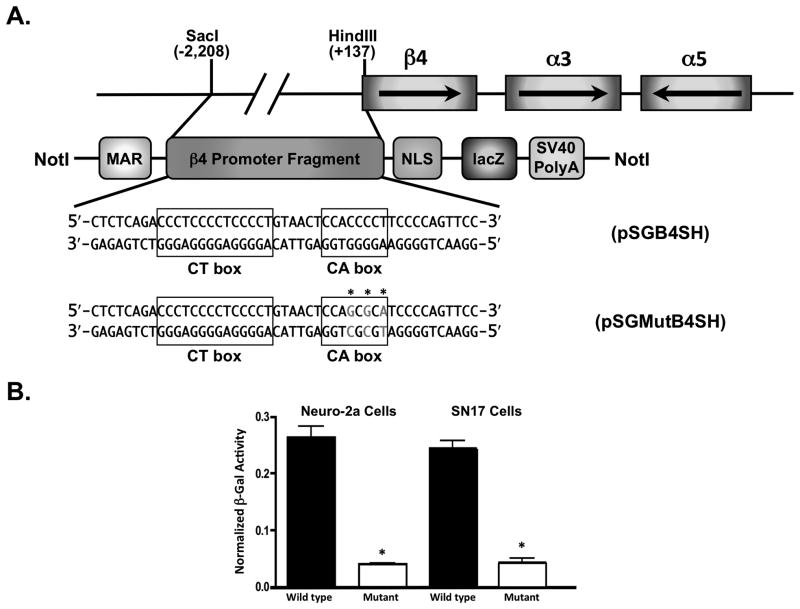
WT and mutant CA box β4 promoter transgenic constructs. (A) WT and mutant β4 promoter/lacZ transgene architecture. The β4/α3/α5 gene cluster is depicted as boxes with arrows depicting the direction of transcription. Below the clustered nAChR subunit genes is a schematic of the linearized construct used to generate the transgenic animals. MAR, matrix attachment region; NLS, nuclear localization sequence. Shown below this schematic are the nucleotide sequences of the WT and mutant CA boxes. Mutations made to the CA box in the mutant transgenic construct are shown in grey with asterisks over the mutated nucleotides. (B) β4 promoter/lacZ transcriptional activity in vitro. DNA from either the WT (black bars) or mutant (white bars) transgenic constructs was transfected into Neuro-2a (left) or SN17 cells (right) along with a luciferase construct in which the SV40 promoter drives expression of the firefly luciferase gene. β-Gal activity was normalized to luciferase activity in order to correct for differences in transfection efficiencies. The data shown here are an average of 3 independent experiments, error bars represent standard deviations of the means. Student t test indicated that CA box mutation significantly decreased the β-gal activity of the mutant transgenic construct in both cell lines, p<0.05.
Recently, a number of genome-wide association studies have linked single nucleotide polymorphisms in the clustered nAChR subunit genes to an increased likelihood of nicotine dependence and lung cancer (Greenbaum and Lerer, 2009). These results highlight the significance of the clustered subunit genes with respect to tobacco related illness. These data have brought to light the importance of the α5α3β4 subtype and shifted the focus from the more commonly studied α7 and α4β2 nAChR subtypes (Sciamanna et al., 1997, Picciotto et al., 1998, Wang et al., 2001, Walters et al., 2006, Breitling et al., 2009, Paleari et al., 2009, Sun et al., 2009).
nAChR subunit gene knock-out mice have proven to be a valuable tool for deciphering the function of specific nAChR subtypes in the nervous system. The β4 knock-out animals display significantly milder somatic symptoms of nicotine withdrawal (Salas et al., 2004). These results demonstrate that the β4 subunit plays a major role in the molecular mechanism underlying nicotine dependence. There is also some indication that blockade of α3β4 nAChRs results in a reduction of opioid and stimulant self-administration, suggesting that nAChRs that contain the β4 subunit are involved in mediating withdrawal syndromes elicited by other drugs of abuse (Glick et al., 2002). Thus, elucidating the molecular mechanisms underlying expression of the β4 gene will improve our understanding of nicotine addiction and withdrawal as well as lung cancer, and other tobacco-related diseases.
The expression patterns of the β4 subunit gene have been characterized by several groups, (Dineley-Miller and Patrick, 1992, Zoli et al., 1995, Winzer-Serhan and Leslie, 1997) yet the mechanisms that control expression are not completely understood. We previously identified in vitro an 8-base pair transcriptional regulatory element in the β4 promoter that is critical for promoter activity (Bigger et al., 1996). This regulatory element, deemed the CA box due to its nucleotide composition, is also the binding site for several transcription factors in vivo (Scofield et al., 2008). We recently demonstrated that a 2.3-kb fragment of the β4 promoter region containing the CA box is capable of directing cell-type specific and developmentally regulated expression of a reporter gene in vivo (Bruschweiler-Li et al., 2010).
In this study we sought to determine if the CA box regulatory element within the β4 promoter region is indeed critical for the temporally and spatially restricted transcriptional activity of the β4 promoter in vivo.
Experimental Procedures
Construction of the mutant CA box β4 promoter/β-galactosidase transgene
The 2,346-base pair SacI/HindIII fragment of the rat β4 subunit gene was excised from the pSGB4SH construct (Bruschweiler-Li et al., 2010) and used as the template for mutagenic polymerase chain reaction (PCR). For mutagenesis, the primers were designed to make 3 base substitutions in the CA box within the context of the 2.3-kb promoter fragment (Fig. 1A). After mutagenesis the resulting fragment was ligated back into the pSG-MAR backbone in order to generate the pSGMutB4SH construct. The SacI/HindIII fragment in the resulting construct was sequenced in order to ensure that only the expected mutations were made (Genewiz, California, USA).
Cell culture and transfection
The mouse neuroblastoma cell line Neuro-2a (Olmsted et al., 1970b) and the mouse septal cholinergic cell line SN17 (Hammond et al., 1990) were cultured and transfected as previously described (Liu et al., 1999). Briefly, transfections were done using a liposome-mediated approach (Lipofectamine 2000, Invitrogen, California, USA). The cells were transfected with either WT or mutant CA box construct and a luciferase expression construct, pGL-Promoter, in which the SV40 promoter drives expression of the firefly luciferease gene. (Promega Corporation, Wisconsin, USA). The cells were allowed to incubate in transfection medium for 2 days and then harvested and assayed for beta-galactosidase (β-gal) (Galacto-Star, Applied Biosystems, California, USA) and luciferase (Luciferase Assay System, Promega) activities in a Lumimark microplate luminometer (Bio-Rad, California, USA). To correct for differences in transfection efficiencies between dishes, the β-gal activity in each sample was normalized to the luciferase activity in that same sample. The student t test was used for statistical analysis.
Generation of transgenic mice
pSGMutB4SH was digested with NotI to release the mutated β4/β-gal transgene. Following agarose gel electrophoresis, the transgene fragment was excised and the DNA was extracted from the gel using a QIAquick Gel Extraction Kit (QIAGEN, California, USA). The purified DNA was injected into pronuclei followed by implantation into pseudopregnant females. The C57BL/6 × SJL F2 hybrid mouse strain was used for all transgenic experiments. Injection of DNA and all subsequent steps up to and including the generation of founder animals were performed by the University of Massachusetts Medical School Transgenic Animal Modeling Core. Transgenic founders were identified by PCR. Founders were mated with C57BL/6 × SJL F2 hybrid mice to establish transgenic lines. Adequate measures were taken to minimize pain and discomfort to the animals. All procedures were conducted in accordance with the rules of the Institutional Animal Care and Use Committee of the University of Massachusetts Medical School.
Determination of transgene copy number
Transgene copy number of the mutant CA box transgenic lines was determined using absolute quantification-based real-time PCR as described previously (Bruschweiler-Li et al., 2010).
Histochemical analysis of transgenic mice
Two ages of transgenic mice were studied: embryonic day (ED) 18.5 and postnatal day (PD) 30. Mice were anesthetized with pentobarbital and perfused transcardially with cold 0.1 M sodium phosphate buffer/2 mM MgCl2 followed by fixative (cold 4% paraformaldehyde). Tissues were then dissected and post-fixed for 5–6 hours (ED18.5) or 4 hours (PD30). Fixed tissues were transferred to 30% sucrose/2 mM MgCl2, in 1X phosphate-buffered saline (PBS) and incubated at 4°C overnight. Tissues were embedded in Tissue-Tek (Miles, Indiana, USA) and quick frozen on dry ice. If not used immediately, the samples were stored at −80°C. Sectioning was done on a Leica CM3050S cryostat at −28°C generating either 14 μm (ED18.5) or 25 μm (PD30) thick sections that were transferred directly onto Superfrost glass slides (Fisher, Pennsylvania, USA). Slides were air-dried at room temperature, washed with sodium phosphate buffer and then incubated overnight at 37°C with β-gal staining solution (0.1 M NaHPO4, 0.1 M NaH2PO4, 2 mM MgCl2, 0.1% sodium deoxycholate, 0.02% NP-40, 10 mM K3(Fe)CN6, 10 mM K4(Fe)CN6, 1 mg/ml X-gal). In order to minimize any variability in the β-Gal staining results, sections from mutant CA box transgenic lines and the corresponding WT transgenic and non-transgenic lines were stained at the same time and in the same batch of staining solution. Two animals from each transgenic founder line were studied in depth. Following β-gal staining, slides were washed with 1X PBS and incubated in distilled water either for 1 h (ED18.5) or overnight (PD30). Slides were then counter-stained with Neutral Red (1% w/v in 37 mM sodium acetate), dehydrated through a graded series of ethanol solutions (50%, 70%, 90% and 100%) and cleared with xylene. The slides were air-dried overnight at room temperature in a fume hood followed by the application of cover slips. Microscopy was done using a Zeiss Axiovert 200M microscope with a high resolution Retiga 1300R CCD camera and Slidebook image analysis software. Anatomical analysis was done with the aid of the Paxinos and Franklin mouse brain atlas (Franklin and Paxinos, 2001) and the Kaufman atlas of mouse development (Kaufman, 1998).
Chromatin immunoprecipitation (ChIP)
Brain tissue ChIP experiments were performed as described previously (Scofield et al., 2008). In short, frozen transgenic brain tissue was ground to a powder and subsequently titurated in PBS with 1% formaldehyde. Cross-linking was carried out at 37°C for 10 minutes. Cells were collected by centrifugation, washed twice with PBS containing protease inhibitors and then collected again by centrifugation. The resulting pellet was re-suspended in lysis buffer with protease inhibitors and then sonicated in order to shear chromatin into an average size of 500 base pairs. Fragmented chromatin was spun down at 4°C in order to eliminate cellular debris. The resulting chromatin samples were diluted in ChIP dilution buffer and used for immunoprecipitation.
PCR
Each ChIP-derived DNA sample was used in PCR with the following primers designed to amplify a segment of the rat transgenic mutant β4 promoter, 5′ - TAAGCTGCCTCGGGTGAACTAAGA-3′, 5′ - TGTCTGGGGGAACCTGTGGCTAT-3′. ChIP-derived DNA was also amplified using the following primers designed to amplify a segment of the mouse endogenous β4 promoter, 5′ - TTGGGTAAGCCAGGCTAAGA-3′, 5′ - GGTCCCGAGACTTTCTCACA-3′. Following amplification, PCR products were electrophoresed through a 2% agarose gel. Densitometry values for ChIP-DNA derived PCR product were obtained using the Ultra-Violet Products EpiChemi3 Darkroom imaging system and UVP analysis software (UVP, California, USA). One-way ANOVA was used for statistical analysis followed by Tukey’s multiple comparison post-test.
RESULTS
Previously, we showed that mutagenesis of the CA box within the β4 promoter virtually eliminates promoter activity in vitro (Bigger et al., 1996). In order to investigate the importance of the CA box for β4 promoter activity in vivo, we generated transgenic mice that express a lacZ reporter gene under direction of a mutant β4 promoter in which the CA box sequence was changed from 5′ – CCACCCCT –3′, to 5′ – CCAGCGCA –3′ (Fig. 1A). When these constructs were tested in vitro, significantly reduced levels of β-Gal activity were observed in the two neuronal-like cell lines Neuro-2a (Olmsted et al., 1970a) and SN17 (Hammond et al., 1990) (Fig. 1B). These results indicated that the β4 promoter constructs were indeed suitable for the generation of transgenic animals.
A total of 6 β4 promoter CA box mutant transgenic lines (lines 19, 25, 28, 30, 33, and 83) were generated and analyzed for β-Gal activity at both ED18.5 and PD30. Using a quantitative PCR approach described previously (Bruschweiler-Li et al., 2010), transgene copy number was determined for each line and revealed that line 19 has approximately 10 copies, line 25 has 62 copies, line 28 has 69 copies, line 30 has 25 copies, line 33 has 23 copies and line 83 has 37 copies.
In the WT β4 promoter/lacZ transgenic lines, we observed a striking recapitulation of endogenous β4 gene expression patterns at ED18.5 in both the CNS and PNS, including the spinal cord, intestine, and cortex (Fig. 2A-C; also see (Bruschweiler-Li et al., 2010)). In contrast, no reporter gene expression was seen in any of these tissues at ED18.5 in any of the mutant CA box transgenic lines (Fig. 2E-G).
Fig. 2.
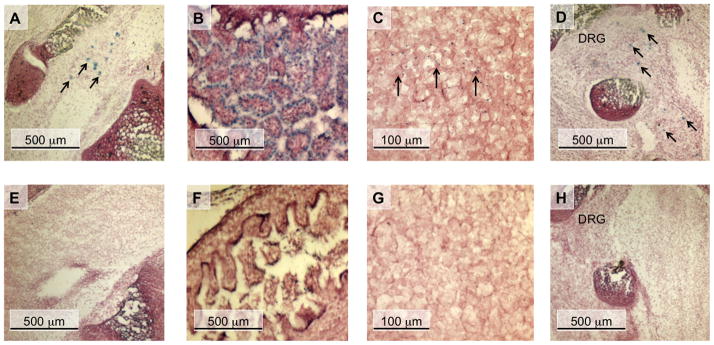
nAChR β4 subunit promoter activity in ED18.5 transgenic mice. Sagittal sections of WT transgenic (A - D) and mutant CA box transgenic (E - H) ED18.5 mouse embryos are shown. These sections were simultaneously stained for β-Gal activity and then counter-stained with neutral red. (A and E) lower lumbar region of the spinal cord; (B and F), intestine; (C and G) cortex; (D and H) lower lumbar dorsal root ganglion (DRG). Arrows in panels A, C and D indicate β-gal-expressing cells.
β4 gene expression is observed early during prenatal development in the dorsal root ganglia (DRG) where it co-assembles with the α3 and α5 subunits to form the α5α3β4 nAChR subtype. This receptor subtype has been shown previously to be the predominant subtype expressed in the PNS (Conroy and Berg, 1995) where it plays a role in nicotinic transmission at the synapses of autonomic ganglia (Vernallis et al., 1993). At ED18.5, we observed lacZ expression in the DRG of the WT transgenic lines (Fig. 2D), however no lacZ expression was observed in the DRG of the mutant CA box transgenic lines (Fig. 2H). Moreover, we did not observe lacZ expression in any cell type in any of the mutant CA box transgenic founder lines at ED18.5, suggesting that the CA box plays a critical role in mediating β4 promoter activity at this stage of development.
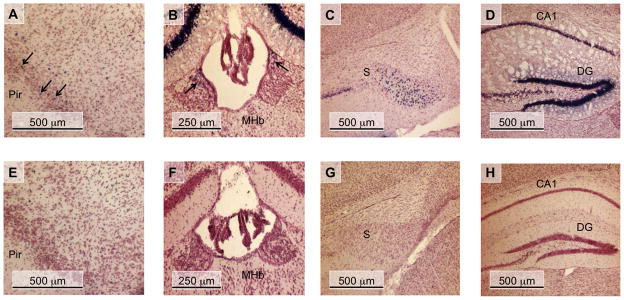
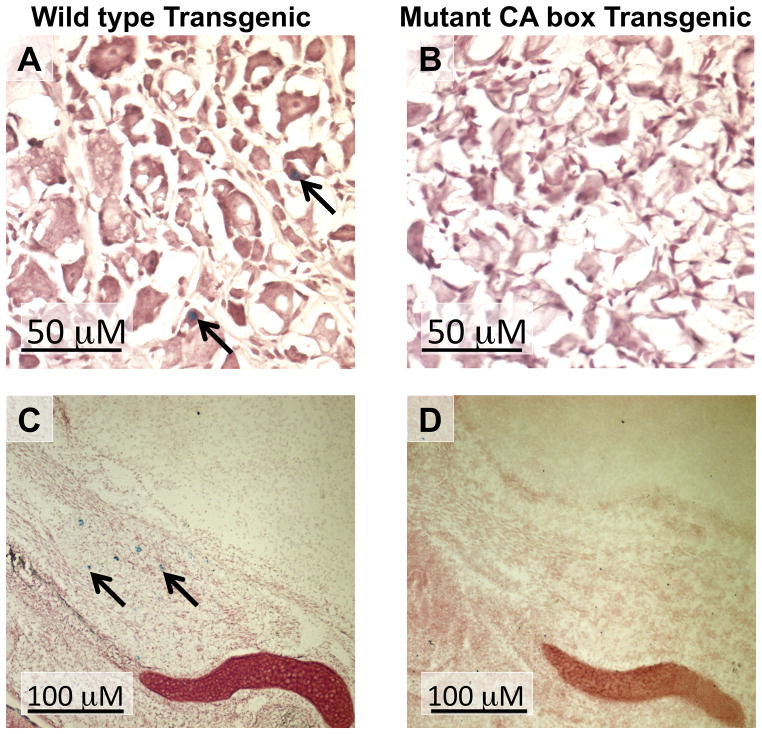
β-Gal activity was observed in PD30 WT transgenic lines in several areas that endogenously express the β4 gene (Dineley-Miller and Patrick, 1992, Winzer-Serhan and Leslie, 1997, Gahring et al., 2004) including the piriform cortex, medial habenula, and the subiculum (Table 1) (Fig. 3A-C) (Bruschweiler-Li et al., 2010). Out of the 6 mutant CA box transgenic founder lines investigated, no lacZ expression was observed in any region of the brain at PD30 in 4 of these lines, lines 25, 30, 33, and 83 (Table 1). The remaining 2 lines, mutant CA box transgenic lines 19 and 28, expressed extremely low levels of the lacZ reporter gene in a small subset of brain regions (Table 1). Interestingly, no expression of lacZ was observed in the mutant CA box transgenic animals in several areas of endogenous β4 expression including the piriform cortex, medial habenula, or the subiculum (Fig. 3E-G) (Dineley-Miller and Patrick, 1992, Winzer-Serhan and Leslie, 1997, Gahring et al., 2004). In addition, the mutant CA box transgenic lines had fewer lacZ positive cells when compared to the WT transgenic lines in several areas of endogenous β4 expression. For example, we observed a drastically reduced number of lacZ positive cells in the dentate gyrus of mutant CA box transgenic line 28 when compared to WT transgenic line 39 (Fig. 3D, H) (Table 1). Consistent with our previous observations at ED18.5, lacZ expression was detected in the DRG of WT transgenic lines at PD30 while no expression of lacZ was detected in the DRG in any of the 6 mutant CA box transgenic lines (Fig. 4A, B). In addition, we observed lacZ expression in the trigeminal ganglion in the WT transgenic line 54 at ED18.5 (Bruschweiler-Li et al., 2010), while no lacZ expression is present in any of the mutant transgenic lines at ED18.5 (Fig. 4C, D). Taken together, these data suggest that the CA box regulatory element plays a critical role in directing β4 promoter activity in vivo.
Table 1.
β4/lacZ transgene expression in the central nervous system of PD30 transgenic animals.
| FOUNDER LINE: | 39 | 54 | 19 | 25 | 28 | 30 | 33 | 83 |
|---|---|---|---|---|---|---|---|---|
| Brain Region | WT Transgenic | Mutant CA box Transgenic | ||||||
| Piriform Cortex | +++ | ++++ | − | − | − | − | − | − |
| Caudate Putamen | + | ++ | − | − | − | − | − | − |
| Medial Habenula | +++ | ++ | − | − | − | − | − | − |
| Dentate Gyrus | ++++ | ++ | + | − | + | − | − | − |
| Subiculum | +++ | +++ | − | − | − | − | − | − |
| Locus Coeruleus | ++ | − | − | − | − | − | − | − |
| Cerebellum | − | − | − | − | − | − | − | − |
Expression levels were scored as follows: −, no expression; +, low level; ++, intermediate level; +++, high level; ++++, very high level.
Fig. 3.
nAChR β4 subunit promoter activity of in the CNS of PD30 transgenic mice. Coronal sections of WT transgenic (A - D) and mutant CA box transgenic (E - H) PD30 mouse brains are shown. These sections were simultaneously stained for β-Gal activity, and then counter-stained with neutral red. (A and E) piriform cortex (Pir); (B and F) medial habenula (Mhb); (C and G) subiculum (S); (D and H) hippocampus, corpus ammon layer 1 (CA1), dentate gyrus (DG); Arrows in panels A and B indicate β-Gal-expressing cells.
Fig. 4.
nAChR β4 subunit promoter activity of in the DRG of PD30 transgenic mice and in the Trigeminal ganglion of ED18.5 transgenic mice. Sections of WT transgenic line 54 (A) and mutant CA box transgenic line 28 (B) PD30 DRG are shown as well as WT transgenic line 54 (C) and mutant CA box transgenic line 28 (d) ED18.5 trigeminal ganglion. These sections were simultaneously stained for β-Gal activity and then counter-stained with neutral red. Arrows in panels A and C indicate β-Gal-expressing cells.
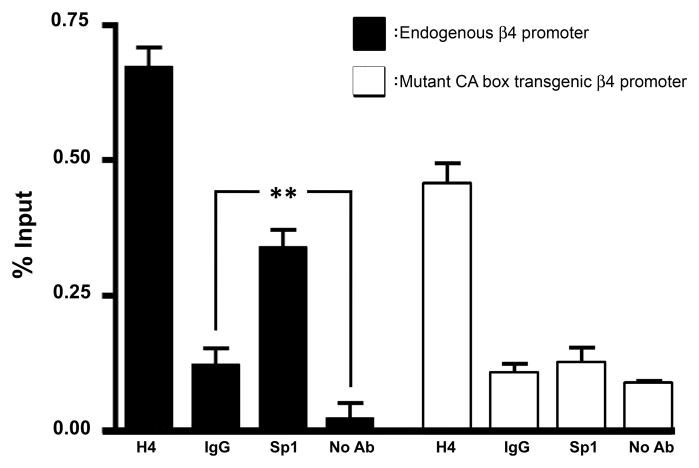
We previously demonstrated that, in addition to virtually eliminating promoter activity, mutation of the β4 promoter CA box substantially reduced binding and transactivation by the transcription factor Sp1, in vitro (Bigger et al., 1996, Bigger et al., 1997). In order to test the hypothesis that CA box mutation results in reduced association of the Sp1 transcription factor with the β4 promoter in vivo, brain tissue from mutant CA box transgenic line 28 was used as a source of chromatin for ChIP assays. Consistent with our previous work (Scofield et al., 2008), PCR using Sp1 ChIP-derived DNA as template resulted in amplification of the endogenous mouse β4 promoter to levels that were approximately 3-fold higher than controls (Fig. 5, black bars), indicating an interaction of Sp1 with the endogenous β4 promoter. Conversely, PCR reactions using the same Sp1 ChIP-derived DNA as template did not result in amplification of the mutant CA box transgenic β4 promoter above background levels (Fig. 5, white bars), indicating that there was no or significantly reduced interaction between Sp1 and the mutant β4 promoter.
Fig. 5.
CA box mutation results in decreased association of Sp1 with the β4 promoter. ChIP experiments were performed using brain tissue from mutant β4 promoter/lacZ transgenic line 28. Transgenic brain ChIP-derived DNA was used as template for PCR designed to amplify either the WT endogenous mouse β4 promoter (black bars), or the CA box mutant transgenic rat β4 promoter (white bars). (H4) anti-acetylated histone protein H4; (IgG) normal rabbit IgG; (Sp1) Transcription factor Sp1; (No Ab) Mock IP. The data shown here are an average of 4 independent experiments, expressed as a percentage of a 2% un-precipitated input sample. One-way ANOVA statistical analysis performed on the WT β4 promoter values (black bars) indicated that ChIP values differed significantly from the total mean (ANOVA F3, 7 = 148.4, p<0.001). Tukey’s multiple comparison post-test resulted in a significant difference between the Sp1 ChIPs and the WT IgG and mock ChIP controls, p <0.01. One-way ANOVA statistical analysis performed on the transgenic mutant β4 promoter values (white bars) indicated that ChIP values differed significantly from the total mean (ANOVA F3, 7 = 94.62, p<0.001). Tukey’s multiple comparison post-test resulted in no significant difference between Sp1 ChIPs and the IgG and mock ChIP controls at the mutant β4 promoter. Error bars represent standard deviation of the means.
DISCUSSION
The biophysical diversity of nAChRs is a result of the large variety of subunit combinations that can assemble into functional receptors (Gotti et al., 2006). While the exact molecular mechanisms controlling the incorporation of a particular subunit into a mature receptor are not completely understood, regulation at the level of transcription likely plays a critical role. Consistent with this hypothesis, the α5, α3 and β4 genes are co-expressed in a variety of cell types in the PNS where the α5α3β4 receptor subtype is the most predominant nicotinic receptor subtype expressed (Rust et al., 1994, Conroy and Berg, 1995). The co-expression of these subunits is likely the result of coordinate regulation of the α5α3β4 locus by shared transcriptional regulatory features. An example of which is the positive regulation of these three genes by the transcription factor Sp1 (Yang et al., 1995, Bigger et al., 1996, Bigger et al., 1997, Campos-Caro et al., 1999, Melnikova et al., 2000a, Terzano et al., 2000, Campos-Caro et al., 2001, Melnikova and Gardner, 2001, Valor et al., 2002). It is most likely that Sp1 acts in concert with cell-type specific regulatory factors in order to direct expression of the clustered subunit genes to the appropriate cell types.
Several groups, including our own, have intensively studied the transcriptional regulatory mechanisms that act to control expression of these three subunit genes. These studies have uncovered several regulatory elements (discussed below) that are involved in directing cell-type specific expression of the clustered nAChR subunit genes. Here, we have used mutant β4 promoter transgenic mice to demonstrate that the CA box is a critical mediator of β4 promoter activity in vivo. Mutation of the CA box resulted in absent or substantially decreased expression of the lacZ reporter gene in an array of brain regions where lacZ expression is observed in the WT β4 promoter/lacZ transgenic lines (Bruschweiler-Li et al., 2010) and β4 is endogenously expressed (Winzer-Serhan and Leslie, 1997, Gahring et al., 2004). Taken together, these results demonstrate that the CA box plays a pivotal role in the transcriptional activity of the β4 promoter in vivo.
A previous study done by the Deneris group uncovered an additional transcriptional regulatory element that mediates expression of the β4 gene (Xu et al., 2006). This element, deemed the conserved non-coding region #4 (CNR4), is located approximately 30-kb upstream of the β4 gene. CNR4 appears to contain regulatory information needed to direct expression of the β4 gene to the pineal gland and interpeduncular nucleus (Xu et al., 1999), two areas of endogenous β4 gene expression where we do not see lacZ expression in the WT or mutant CA box β4 transgenic lines. Additional experiments demonstrated that CNR4 is critical for the coordinate expression of the β4 and α3 genes in the pineal gland as well as in the superior cervical ganglion. Based on its high degree of conservation and its distal location from the genes it acts to regulate, CNR4 is thought to be a locus control region-like regulatory domain (Xu et al., 2006).
In addition to CNR4, mutation of several E26 transformation-specific sequence (ETS) factor binding sites in an enhancer located in the β4 3′-untranslated region (the β43′ element) can severely diminish expression of the α5, α3 and β4 genes in the adrenal gland (Xu et al., 2006). Furthermore, mutation of ETS binding sites within the β43′ enhancer also diminished expression of these genes in the superior cervical ganglion (Xu et al., 2006). It is clear from these data as well as data from our own lab that several transcriptional regulatory elements located around the clustered subunit genes act in concert to provide the necessary regulatory information to drive expression of the required subunits in the appropriate cell type (Yang et al., 1997, Francis and Deneris, 2002, Xu et al., 2006, Fuentes Medel and Gardner, 2007, Bruschweiler-Li et al., 2010).
ChIP experiments done on mutant CA box transgenic brain tissue demonstrated a reduced interaction of Sp1 with the CA box mutant β4 promoter. These data are in agreement with previous studies from our lab indicating that CA box mutation abrogates Sp factor binding and transactivation of the β4 promoter in vitro (Bigger et al., 1996, Bigger et al., 1997). Because the β4 promoter lacks a TATA box, Sp1 could play a crucial role in tethering the basal transcription machinery to the β4 promoter, as it does in other TATA-less promoters (Pugh and Tjian, 1991) The lack of Sp1 interaction with the CA box mutant promoter provides a plausible molecular mechanism for the severely reduced β4 promoter activity observed in the CA box mutant transgenic lines. CA box mutation may also impede the transactivation ability of additional transcriptional regulatory factors that associate with the β4 promoter at this region by virtue of protein-protein interactions with Sp1 (Melnikova et al., 2000b, Scofield et al., 2008). This is likely the case for transcription factor c-Jun, which physically interacts with Sp1 and is capable of activating transcription from the β4 promoter in vitro, despite the fact that the β4 promoter lacks a consensus AP-1 binding site. At the WT β4 promoter, Sp1 and c-Jun synergistically activate transcription, however, when the CA box is mutated, the β4 promoter is no longer responsive to either Sp1 or c-Jun (Melnikova and Gardner, 2001). In addition, the transcription factor Sox10 also synergistically transactivates the β4 promoter when supplied in concert with Sp1 (Melnikova et al., 2000a). Similar to what is observed with c-Jun, CA box mutation also drastically reduces Sox10’s ability to activate transcription from the β4 promoter (Liu et al., 1999). It is possible then that Sp1 may act to nucleate a complex of positive-acting transcription factors needed for cell-type specific expression for the β4 gene in vivo.
Apart from transcriptional regulation, it is important to note that other modes of regulation may act to mediate the temporally- and spatially-regulated transcriptional activity of the β4 subunit gene promoter. Possible modes of regulation include chromatin remodeling, post-translational modifications and regulation of β4 mRNA translation by microRNAs.
Although there was no 4 promoter activity in the mutant CA box transgenic animals at ED18.5, it remains unclear if the CA box simply plays a role in directing basal promoter activity or if it is involved in mediating the developmentally and temporally regulated expression of the β4 gene. Future studies will be focused on addressing these questions.
In conclusion, our results demonstrate that the CA box is a key determinant of β4 promoter activity at both ED18.5 and PD30 in both the PNS and CNS. Furthermore, these data also indicate that Sp1 plays a critical role in directing the positive transcriptional regulatory effect of the CA box in vivo. Positive regulation of the β4 promoter by Sp1 at the CA box is a crucial aspect of the intricate regulatory cascade that ensures accurate expression of the β4 gene, allowing nAChRs containing the β4 subunit to participate in both normal physiological processes (Dani and Bertrand, 2007, Albuquerque et al., 2009), as well as tobacco-related pathological conditions (Salas et al., 2004, Salas et al., 2009).
Acknowledgments
We thank Steve Jones, Joe Gosselin and their colleagues in the UMMS Transgenic Animal Modeling Core for their outstanding services in generating the transgenic mice used in this study. We also thank Haley Melikian for her help with the microscopy, Stéphane Germain for the kind gift of pSG-MAR and Ricardo Medina for providing useful advice and reagents for determining transgene copy number. This work was supported by Grant Number R01NS030243 from the National Institute of Neurological Disorders and Stroke (PDG) as well as Grant Number R01AA017656 from the National Institute of Alcohol Abuse and Alcoholism (ART). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Neurological Disorders and Stroke, National Institute of Alcohol Abuse and Alcoholism or the National Institutes of Health.
Abbreviations
- β-Gal
beta-galactosidase
- CA box
the (5′ – CCACCCCT –3′) regulatory element in the β4 gene promoter
- ChIP
chromatin immunoprecipitation
- CNR4
conserved non-coding region 4
- CNS
central nervous system
- DRG
dorsal root ganglion
- ED18.5
embryonic day 18.5
- ETS
E26 transformation-specific sequence
- nAChR
nicotinic acetylcholine receptor
- PNS
peripheral nervous system
- PD30
postnatal day 30
- WT
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albuquerque EX, Pereira EF, Alkondon M, Rogers SW. Mammalian nicotinic acetylcholine receptors: from structure to function. Physiol Rev. 2009;89:73–120. doi: 10.1152/physrev.00015.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigger CB, Casanova EA, Gardner PD. Transcriptional regulation of neuronal nicotinic acetylcholine receptor genes. Functional interactions between Sp1 and the rat beta4 subunit gene promoter. J Biol Chem. 1996;271:32842–32848. doi: 10.1074/jbc.271.51.32842. [DOI] [PubMed] [Google Scholar]
- Bigger CB, Melnikova IN, Gardner PD. Sp1 and Sp3 regulate expression of the neuronal nicotinic acetylcholine receptor β4 subunit gene. J Biol Chem. 1997;272:25976–25982. doi: 10.1074/jbc.272.41.25976. [DOI] [PubMed] [Google Scholar]
- Boulter J, O’Shea-Greenfield A, Duvoisin RM, Connolly JG, Wada E, Jensen A, Gardner PD, Ballivet M, Deneris ES, McKinnon D, et al. Alpha 3, alpha 5, and beta 4: three members of the rat neuronal nicotinic acetylcholine receptor-related gene family form a gene cluster. J Biol Chem. 1990;265:4472–4482. [PubMed] [Google Scholar]
- Breitling LP, Dahmen N, Mittelstrass K, Rujescu D, Gallinat J, Fehr C, Giegling I, Lamina C, Illig T, Muller H, Raum E, Rothenbacher D, Wichmann HE, Brenner H, Winterer G. Association of nicotinic acetylcholine receptor subunit alpha 4 polymorphisms with nicotine dependence in 5500 Germans. Pharmacogenomics J. 2009;9:219–224. doi: 10.1038/tpj.2009.6. [DOI] [PubMed] [Google Scholar]
- Bruschweiler-Li L, Fuentes Medel YF, Scofield MD, Trang EBT, Binke SA, Gardner PD. Temporally- and spatially-regulated transcriptional activity of the nicotinic acetylcholine receptor β4 subunit gene promoter region. Neurosci. 2010;166:864–877. doi: 10.1016/j.neuroscience.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Caro A, Carrasco-Serrano C, Valor LM, Ballesta JJ, Criado M. Activity of the nicotinic acetylcholine receptor alpha5 and alpha7 subunit promoters in muscle cells. DNA Cell Biol. 2001;20:657–666. doi: 10.1089/104454901753340640. [DOI] [PubMed] [Google Scholar]
- Campos-Caro A, Carrasco-Serrano C, Valor LM, Viniegra S, Ballesta JJ, Criado M. Multiple functional Sp1 domains in the minimal promoter region of the neuronal nicotinic receptor alpha5 subunit gene. J Biol Chem. 1999;274:4693–4701. doi: 10.1074/jbc.274.8.4693. [DOI] [PubMed] [Google Scholar]
- Conroy WG, Berg DK. Neurons can maintain multiple classes of nicotinic acetylcholine receptors distinguished by different subunit compositions. J Biol Chem. 1995;270:4424–4431. doi: 10.1074/jbc.270.9.4424. [DOI] [PubMed] [Google Scholar]
- Corriveau RA, Berg DK. Coexpression of multiple acetylcholine receptor genes in neurons: quantification of transcripts during development. J Neurosci. 1993;13:2662–2671. doi: 10.1523/JNEUROSCI.13-06-02662.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani JA, Bertrand D. Nicotinic Acetylcholine Receptors and Nicotinic Cholinergic Mechanisms of the Central Nervous System. Annu Rev Pharmacol Toxicol. 2007;47:699–729. doi: 10.1146/annurev.pharmtox.47.120505.105214. [DOI] [PubMed] [Google Scholar]
- De Fusco M, Becchetti A, Patrignani A, Annesi G, Gambardella A, Quattrone A, Ballabio A, Wanke E, Casari G. The nicotinic receptor beta 2 subunit is mutant in nocturnal frontal lobe epilepsy. Nat Genet. 2000;26:275–276. doi: 10.1038/81566. [DOI] [PubMed] [Google Scholar]
- Dineley-Miller K, Patrick J. Gene transcripts for the nicotinic acetylcholine receptor subunit, beta4, are distributed in multiple areas of the rat central nervous system. Brain Res Mol Brain Res. 1992;16:339–344. doi: 10.1016/0169-328x(92)90244-6. [DOI] [PubMed] [Google Scholar]
- Eisen JS. Determination of primary motoneuron identity in developing zebrafish embryos. Science. 1991;252:569–572. doi: 10.1126/science.1708527. [DOI] [PubMed] [Google Scholar]
- Flores CM, DeCamp RM, Kilo S, Rogers SW, Hargreaves KM. Neuronal nicotinic rceptor expression in sensory neurons of the rat trigeminal ganglion: Demonstration of α3/β4, a novel subtype in the mammalian nervous system. J Neuroscience. 1996;16:7892–7901. doi: 10.1523/JNEUROSCI.16-24-07892.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores GV, Duan H, Yan H, Nagaraj R, Fu W, Zou Y, Noll M, Banerjee U. Combinatorial signaling in the specification of unique cell fates. Cell. 200;103:75–85. doi: 10.1016/s0092-8674(00)00106-9. [DOI] [PubMed] [Google Scholar]
- Francis N, Deneris ES. Retinal neuron activity of ETS domain-binding sites in a nicotinic acetylcholine receptor gene cluster enhancer. J Biol Chem. 2002;277:6511–6519. doi: 10.1074/jbc.M105616200. [DOI] [PubMed] [Google Scholar]
- Francis NJ, Landis SC. Cellular and molecular determinants of sympathetic neuron development. Annu Rev Neurosci. 1999;22:541–566. doi: 10.1146/annurev.neuro.22.1.541. [DOI] [PubMed] [Google Scholar]
- Franklin KBJ, Paxinos G. The Mouse Brain in Sterotaxic Coordinates. San Diego: Academic Press; 2001. [Google Scholar]
- Fuentes Medel YF, Gardner PD. Transcriptional repression by a conserved intronic sequence in the nicotinic receptor alpha3 subunit gene. J Biol Chem. 2007;282:19062–19070. doi: 10.1074/jbc.M702354200. [DOI] [PubMed] [Google Scholar]
- Gahring LC, Persiyanov K, Rogers SW. Neuronal and astrocyte expression of nicotinic receptor subunit beta4 in the adult mouse brain. J Comp Neurol. 2004;468:322–333. doi: 10.1002/cne.10942. [DOI] [PubMed] [Google Scholar]
- Genzen JR, Van Cleve W, McGehee DS. Dorsal root ganglion neurons express multiple nicotinic acetylcholine receptor subtypes. J Neurophysiol. 2001;86:1773–1782. doi: 10.1152/jn.2001.86.4.1773. [DOI] [PubMed] [Google Scholar]
- Glick SD, Maisonneuve IM, Kitchen BA, Fleck MW. Antagonism of alpha 3 beta 4 nicotinic receptors as a strategy to reduce opioid and stimulant self-administration. Eur J Pharmacol. 2002;438:99–105. doi: 10.1016/s0014-2999(02)01284-0. [DOI] [PubMed] [Google Scholar]
- Goldman D, Deneris E, Luyten W, Kochhar A, Patrick J, Heinemann S. Members of a nicotinic acetylcholine receptor gene family are expressed in different regions of the mammalian central nervous system. Cell. 1987;48:965–973. doi: 10.1016/0092-8674(87)90705-7. [DOI] [PubMed] [Google Scholar]
- Gotti C, Fornasari D, Clementi F. Human neuronal nicotinic receptors. Prog Neurobiol. 1997;53:199–237. doi: 10.1016/s0301-0082(97)00034-8. [DOI] [PubMed] [Google Scholar]
- Gotti C, Moretti M, Gaimarri A, Zanardi A, Clementi F, Zoli M. Heterogeneity and complexity of native brain nicotinic receptors. Biochem Pharmacol. 2007;74:1102–1111. doi: 10.1016/j.bcp.2007.05.023. [DOI] [PubMed] [Google Scholar]
- Gotti C, Zoli M, Clementi F. Brain nicotinic acetylcholine receptors: native subtypes and their relevance. Trends Pharmacol Sci. 2006;27:482–491. doi: 10.1016/j.tips.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Grady SR, Moretti M, Zoli M, Marks MJ, Zanardi A, Pucci L, Clementi F, Gotti C. Rodent habenulo-interpeduncular pathway expresses a large variety of uncommon nAChR subtypes, but only the alpha3beta4* and alpha3beta3beta4* subtypes mediate acetylcholine release. J Neurosci. 2009;29:2272–2282. doi: 10.1523/JNEUROSCI.5121-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum L, Lerer B. Differential contribution of genetic variation in multiple brain nicotinic cholinergic receptors to nicotine dependence: recent progress and emerging open questions. Mol Psychiatry. 2009 doi: 10.1038/mp.2009.59. [DOI] [PubMed] [Google Scholar]
- Groves AK, Bronner-Fraser M. Neural crest diversification. Curr Top Dev Biol. 1999;43:221–258. doi: 10.1016/s0070-2153(08)60383-x. [DOI] [PubMed] [Google Scholar]
- Hammond DN, Lee HJ, Tonsgard JH, Wainer BH. Development and characterization of clonal cell lines derived from septal cholinergic neurons. Brain Res. 1990;512:190–200. doi: 10.1016/0006-8993(90)90626-M. [DOI] [PubMed] [Google Scholar]
- Isacson O, Seo H, Lin L, Albeck D, Granholm AC. Alzheimer’s disease and Down’s syndrome: roles of APP, trophic factors and ACh. Trends Neurosci. 2002;25:79–84. doi: 10.1016/s0166-2236(02)02037-4. [DOI] [PubMed] [Google Scholar]
- Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Kaufman MH. The Atlas of Mouse Development. San Diego: Academic Press; 1998. [Google Scholar]
- Liu Q, Melnikova IN, Hu M, Gardner PD. Cell type-specific activation of neuronal nicotinic acetylcholine receptor subunit genes by Sox10. J Neurosci. 1999;19:9747–9755. doi: 10.1523/JNEUROSCI.19-22-09747.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGehee DS, Role LW. Physiological diversity of nicotinic acetylcholine receptors expressed by vertebrate neurons. Annu Rev Physiol. 1995;57:521–546. doi: 10.1146/annurev.ph.57.030195.002513. [DOI] [PubMed] [Google Scholar]
- Melnikova IN, Gardner PD. The signal transduction pathway underlying ion channel gene regulation by SP1-C-Jun interactions. J Biol Chem. 2001;276:19040–19045. doi: 10.1074/jbc.M010735200. [DOI] [PubMed] [Google Scholar]
- Melnikova IN, Lin HR, Blanchette AR, Gardner PD. Synergistic transcriptional activation by Sox10 and Sp1 family members. Neuropharm. 2000a;39:2615–2623. doi: 10.1016/s0028-3908(00)00125-8. [DOI] [PubMed] [Google Scholar]
- Melnikova IN, Yang Y, Gardner PD. Interactions between regulatory proteins that bind to the nicotinic receptor beta4 subunit gene promoter. Eur J Pharmacol. 2000b;393:75–83. doi: 10.1016/s0014-2999(99)00884-5. [DOI] [PubMed] [Google Scholar]
- Olmsted JB, Carlson K, Klebe R, Ruddle F, Rosenbaum J. Isolation of microtubule protein from cultured mouse neuroblastoma cells. Proc Natl Acad Sci U S A. 1970a;65:129–136. doi: 10.1073/pnas.65.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmsted JB, Carlson K, Klebe R, Ruddle F, Rosenbaum J. Isolation of microtubule protein from cultured mouse neuroblastoma cells. Proc Natl Acad Sci U S A. 1970b;65:129–136. doi: 10.1073/pnas.65.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paleari L, Negri E, Catassi A, Cilli M, Servent D, D’Angelillo R, Cesario A, Russo P, Fini M. Inhibition of nonneuronal alpha7-nicotinic receptor for lung cancer treatment. Am J Respir Crit Care Med. 2009;179:1141–1150. doi: 10.1164/rccm.200806-908OC. [DOI] [PubMed] [Google Scholar]
- Perl O, Ilani T, Strous RD, Lapidus R, Fuchs S. The alpha7 nicotinic acetylcholine receptor in schizophrenia: decreased mRNA levels in peripheral blood lymphocytes. Faseb J. 2003;17:1948–1950. doi: 10.1096/fj.03-0104fje. [DOI] [PubMed] [Google Scholar]
- Perry EK, Martin-Ruiz CM, Court JA. Nicotinic receptor subtypes in human brain related to aging and dementia. Alcohol. 2001;24:63–68. doi: 10.1016/s0741-8329(01)00130-6. [DOI] [PubMed] [Google Scholar]
- Picciotto MR, Zoli M, Rimondini R, Lena C, Marubio LM, Pich EM, Fuxe K, Changeux JP. Acetylcholine receptors containing the beta2 subunit are involved in the reinforcing properties of nicotine. Nature. 1998;391:173–177. doi: 10.1038/34413. [DOI] [PubMed] [Google Scholar]
- Pugh BF, Tjian R. Transcription from a TATA-less promoter requires a multisubunit TFIID complex. Genes Dev. 1991;5:1935–9145. doi: 10.1101/gad.5.11.1935. [DOI] [PubMed] [Google Scholar]
- Rust G, Burgunder JM, Lauterburg TE, Cachelin AB. Expression of neuronal nicotinic acetylcholine receptor subunit genes in the rat autonomic nervous system. Eur J Neurosci. 1994;6:478–485. doi: 10.1111/j.1460-9568.1994.tb00290.x. [DOI] [PubMed] [Google Scholar]
- Salas R, Pieri F, De Biasi M. Decreased signs of nicotine withdrawal in mice null for the beta4 nicotinic acetylcholine receptor subunit. J Neurosci. 2004;24:10035–10039. doi: 10.1523/JNEUROSCI.1939-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas R, Sturm R, Boulter J, De Biasi M. Nicotinic receptors in the habenulo-interpeduncular system are necessary for nicotine withdrawal in mice. J Neurosci. 2009;29:3014–3018. doi: 10.1523/JNEUROSCI.4934-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- Sciamanna MA, Griesmann GE, Williams CL, Lennon VA. Nicotinic acetylcholine receptors of muscle and neuronal (alpha7) types coexpressed in a small cell lung carcinoma. J Neurochem. 1997;69:2302–2311. doi: 10.1046/j.1471-4159.1997.69062302.x. [DOI] [PubMed] [Google Scholar]
- Scofield MD, Bruschweiler-Li L, Mou Z, Gardner PD. Transcription factor assembly on the nicotinic receptor beta4 subunit gene promoter. Neuroreport. 2008;19:687–690. doi: 10.1097/WNR.0b013e3282fbcef7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinlein OK, Mulley JC, Propping P, Wallace RH, Phillips HA, Sutherland GR, Scheffer IE, Berkovic SF. A missense mutation in the neuronal nicotinic acetylcholine receptor alpha 4 subunit is associated with autosomal dominant nocturnal frontal lobe epilepsy. Nat Genet. 1995;11:201–203. doi: 10.1038/ng1095-201. [DOI] [PubMed] [Google Scholar]
- Sun X, Ritzenthaler JD, Zheng Y, Roman J, Han S. Rosiglitazone inhibits alpha4 nicotinic acetylcholine receptor expression in human lung carcinoma cells through peroxisome proliferator-activated receptor gamma-independent signals. Mol Cancer Ther. 2009;8:110–118. doi: 10.1158/1535-7163.MCT-08-0719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teaktong T, Graham A, Court J, Perry R, Jaros E, Johnson M, Hall R, Perry E. Alzheimer’s disease is associated with a selective increase in alpha7 nicotinic acetylcholine receptor immunoreactivity in astrocytes. Glia. 2003;41:207–211. doi: 10.1002/glia.10132. [DOI] [PubMed] [Google Scholar]
- Terzano S, Flora A, Clementi F, Fornasari D. The minimal promoter of the human alpha 3 nicotinic receptor subunit gene. Molecular and functional characterization. J Biol Chem. 2000;275:41495–41503. doi: 10.1074/jbc.M006197200. [DOI] [PubMed] [Google Scholar]
- Valor LM, Campos-Caro A, Carrasco-Serrano C, Ortiz JA, Ballesta JJ, Criado M. Transcription factors NF-Y and Sp1 are important determinants of the promoter activity of the bovine and human neuronal nicotinic receptor beta 4 subunit genes. J Biol Chem. 2002;277:8866–8876. doi: 10.1074/jbc.M110454200. [DOI] [PubMed] [Google Scholar]
- Vernallis AB, Conroy WG, Berg DK. Neurons assemble acetylcholine receptors with as many as three kinds of subunits while maintaining subunit segregation among receptor subtypes. Neuron. 1993;10:451–464. doi: 10.1016/0896-6273(93)90333-m. [DOI] [PubMed] [Google Scholar]
- Walters CL, Brown S, Changeux JP, Martin B, Damaj MI. The beta2 but not alpha7 subunit of the nicotinic acetylcholine receptor is required for nicotine-conditioned place preference in mice. Psychopharmacology (Berl) 2006;184:339–344. doi: 10.1007/s00213-005-0295-x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Pereira EF, Maus AD, Ostlie NS, Navaneetham D, Lei S, Albuquerque EX, Conti-Fine BM. Human bronchial epithelial and endothelial cells express alpha7 nicotinic acetylcholine receptors. Mol Pharmacol. 2001;60:1201–1209. doi: 10.1124/mol.60.6.1201. [DOI] [PubMed] [Google Scholar]
- Whitehouse PJ, Martino AM, Marcus KA, Zweig RM, Singer HS, Price DL, Kellar KJ. Reductions in acetylcholine and nicotine binding in several degenerative diseases. Arch Neurol. 1988;45:722–724. doi: 10.1001/archneur.1988.00520310028012. [DOI] [PubMed] [Google Scholar]
- Winzer-Serhan UH, Leslie FM. Codistribution of nicotinic acetylcholine receptor subunit alpha3 and beta4 mRNAs during rat brain development. J Comp Neurol. 1997;386:540–554. doi: 10.1002/(sici)1096-9861(19971006)386:4<540::aid-cne2>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Xu W, Orr-Urtreger A, Nigro F, Gelber S, Sutcliffe CB, Armstrong D, Patrick JW, Role LW, Beaudet AL, De Biasi M. Multiorgan autonomic dysfunction in mice lacking the beta2 and the beta4 subunits of neuronal nicotinic acetylcholine receptors. J Neurosci. 1999;19:9298–9305. doi: 10.1523/JNEUROSCI.19-21-09298.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Scott MM, Deneris ES. Shared long-range regulatory elements coordinate expression of a gene cluster encoding nicotinic receptor heteromeric subtypes. Mol Cell Biol. 2006;26:5636–5649. doi: 10.1128/MCB.00456-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Fyodorov D, Deneris ES. Transcriptional analysis of acetylcholine receptor alpha 3 gene promoter motifs that bind Sp1 and AP2. J Biol Chem. 1995;270:8514–8520. doi: 10.1074/jbc.270.15.8514. [DOI] [PubMed] [Google Scholar]
- Yang X, Yang F, Fyodorov D, Wang F, McDonough J, Herrup K, Deneris E. Elements between the protein-coding regions of the adjacent beta 4 and alpha 3 acetylcholine receptor genes direct neuron-specific expression in the central nervous system. J Neurobiol. 1997;32:311–324. [PubMed] [Google Scholar]
- Zoli M, Le Novere N, Hill JA, Jr, Changeux JP. Developmental regulation of nicotinic ACh receptor subunit mRNAs in the rat central and peripheral nervous systems. J Neurosci. 1995;15:1912–1939. doi: 10.1523/JNEUROSCI.15-03-01912.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoli M, Lena C, Picciotto MR, Changeux JP. Identification of four classes of brain nicotinic receptors using beta2 mutant mice. J Neurosci. 1998;18:4461–4472. doi: 10.1523/JNEUROSCI.18-12-04461.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]