Abstract
Once thought to be transcriptional noise, large non-coding RNAs (lncRNAs) have recently been demonstrated to be functional molecules. Cell-type specific expression patterns of lncRNAs suggest that their transcription may be regulated epigenetically. Using a custom-designed microarray, here we examine the expression profile of lncRNAs in embryonic stem (ES) cells, lineage-restricted neuronal progenitor cells (NPC), and terminally differentiated fibroblasts. In addition, we also analyze the relationship between their expression and their promoter H3K4 and H3K27 methylation patterns. We find that numerous lncRNAs in these cell types undergo changes in the levels of expression and promoter H3K4me3 and H3K27me3. Interestingly, lncRNAs that are expressed at lower levels in ES cells exhibit higher levels of H3K27me3 at their promoters. Consistent with this result, knockdown of the H3K27me3 methyltransferase Ezh2 results in derepression of these lncRNAs in ES cells. Thus, our results establish a role for Ezh2-mediated H3K27 methylation in lncRNA silencing in ES cells and reveal that lncRNAs are subject to epigenetic regulation in a similar manner to that of protein coding genes.
Introduction
Recent studies have revealed that the mammalian “transcriptome” is much more substantial and complex than originally believed. The ENCODE project estimates that over 90% of all nucleotides within the human genome can be transcribed (1). Although transcription appears to be quite pervasive, only 15% of the genome experiences stable transcription in human cell lines (2). Nevertheless, protein coding genes account for just 1–2% of the genome (3) suggesting that cells dedicate a considerable amount of energy to produce transcriptional “dark matter” (4).
Although small RNAs (microRNAs, PIWI-associated RNAs, and small interfering RNAs) and housekeeping RNAs (transfer RNAs and ribosomal RNAs) account for some non-protein coding transcripts, the vast majority are long non-coding RNAs (lncRNAs) that are greater than 200 nucleotides in length (5). Although lncRNAs were once considered to be transcriptional noise, several features suggest that lncRNAs may play important roles in various processes. These features include: 1) cell-type specific expression patterns (6–8); 2) distinct subcellular localizations (7); 3) link to various diseases (9); and 4) evolutionary selection of the lncRNA sequence (10,11). Indeed, recent studies have uncovered functional roles for lncRNAs in diverse biological processes including X-inactivation, imprinting, Hox gene regulation, and the development of certain diseases (9,12–15).
Epigenetic modifications, including DNA methylation, histone modifications, and chromatin remodeling are believed to contribute to cell-type specific expression patterns of protein-coding genes (16,17). Interestingly, a previous study has identified many genomic regions that contain the active histone modification marks H3K4me3-H3K36me3, which could not be attributed to protein-coding genes (10). While this suggests that lncRNAs may be regulated epigenetically in a similar way to that of protein coding genes, this has not been tested directly.
To begin to understand the mechanism by which lncRNAs exhibit cell-type specific expression, we compared the expression profiles of lncRNAs in mouse ES cells, neuronal progenitors (NPCs), and terminally differentiated fibroblasts and analyzed the relationship between their expression and their promoter H3K4me3 and H3K27me3 levels. We found that these histone modifications undergo dynamic changes in a cell-type specific manner. In addition, we found that Ezh2-mediated H3K27 methylation plays an important role in silencing lncRNAs in ES cells. Our results suggest that lncRNAs are epigenetically regulated in a similar manner to that of protein coding genes.
Results
Expression profiling of lncRNAs in ES cells, NPCs, and fibroblasts
Of the 34,030 lncRNAs annotated in the Fantom3 database, 32,213 (95%) were mapped to the UCSC mm8 assembly of the mouse genome. To determine whether lncRNAs are differentially expressed in cells at different differentiation stages, we analyzed their expression in mouse ES cells, lineage-restricted neuroprogenitors (NPCs), and terminally differentiated fibroblasts using a custom-designed microarray containing oligonucleotide probes targeting these lncRNAs (Supplementary Table 1). Given that multiple transcriptional units can coexist within the same genomic space (18–22), lncRNAs may be regulated indirectly by their location within the same transcriptional unit of a protein coding gene. To simplify the analysis, we focused on the subset of lncRNAs that did not overlap with the promoter (defined as −1kb to +1kb surrounding the transcription start site) or the coding region of annotated genes. We found that the majority (65%, n=20949) of mapped lncRNAs annotated in the Fantom3 database overlapped with coding regions of genes annotated in the RefSeq database. A small fraction (1%, n=327) overlapped with gene promoters. The remaining lncRNAs (34%, n=10937) showed no overlap with annotated promoters or coding regions (Fig. 1A). These lncRNAs were used for all subsequent analyses.
Figure 1. lncRNAs are differentially expressed in ES cells, NPCs, and fibroblasts.
(A) Pie chart representation of the distribution of lncRNAs in the mouse genome. Independent Fantom3 lncRNA entries were scanned for overlap with promoters, defined as −1kb to +1kb surrounding the TSS, and coding regions of annotated genes from the REFSEQ database.
(B) Heat map presentation of the expression profile of lncRNAs in ES cells, NPCs, and fibroblasts. The data presented for each cell type was compared to universal reference RNA and was subjected to uncentered hierarchical clustering. ES: embryonic stem cells, NPC: neuroprogenitors, FIB: fibroblasts.
(C) RT-qPCR validation of microarray data for randomly chosen lncRNAs that exhibited at least 2-fold elevated transcription in ES versus both NPCs and fibroblasts.
We found that 3270 (10.2%) unique lncRNAs exhibit at least a two-fold change when compared from one cell type to another (Fig. 1B). lncRNAs that exhibited at least a two-fold change in ES cells compared to both NPCs and fibroblasts were randomly selected and confirmed by quantitative PCR analysis (Fig. 1C). These studies indicate that at least a subset of lncRNAs exhibit cell type-specific expression.
H3K4me3 and H3K27me3 distribution in lncRNA promoters in ES, NPC, and fibroblasts
Given that epigenetic mechanisms contribute to cell-type specific expression of protein coding genes, we asked whether lncRNAs are epigenetically regulated. The Trithorax and Polycomb group of proteins are key developmental players responsible for lineage-specific gene expression (23). Some of these proteins participate in multi-protein complexes that are responsible for the tri-methylation of lysine 4 and 27 of histone H3 (H3K4me3 and H3K27me3) with H3K4me3 and H3K27me3 correlating with transcriptional activation and repression, respectively (17). Specific genomic regions in ES cells are marked with both H3K4me3 and H3K27me3 and are termed “bivalent” domains, which are transcriptionally inactive regions that are poised for transcriptional activation (24).
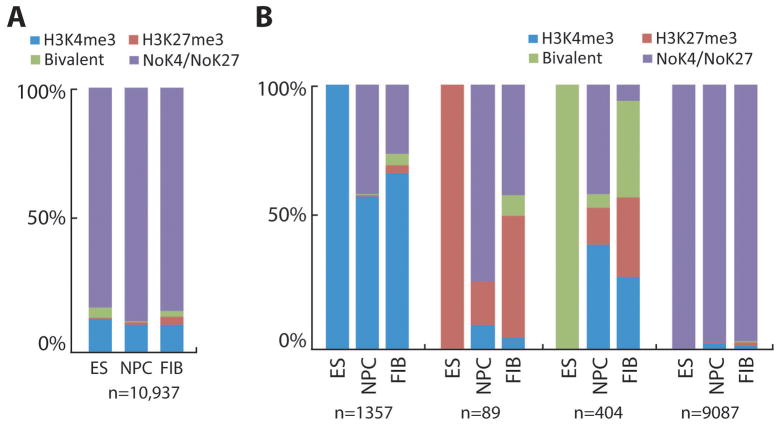
Taking advantage of previously published genome-wide location data for H3K4me3 and H3K27me3 (25), we examined these two histone methylation marks at the promoters of lncRNAs in mouse ES cells, NPCs, and fibroblasts (Fig. 2A). We found a small group of unique lncRNAs (17%) that contain these modifications. In ES cells, roughly 80% of all H3K27me3 positive lncRNA promoters also contain H3K4me3, which is similar to previous reports on protein coding genes (24). In contrast to H3K27me3, less than a quarter of all H3K4me3 positive promoters are bivalent suggesting that most H3K4me3 positive lncRNAs may have housekeeping functions or are important for ES cell identity. Overall, ES cells contained more bivalent lncRNA promoters when compared to NPCs and fibroblasts. In addition, H3K27me3 alone is notably enriched in fibroblasts compared to ES cells and NPCs indicating that this subset of lncRNAs may have to be silenced in terminally differentiated cells.
Figure 2. lncRNA promoter H3K4me3 and H3K27me3 patterns in ES cells, NPCs, and fibroblasts.
Distribution of H3K4me3 and H3K27me3 at lncRNA promoters in ES cells, NPCs, and fibroblast cells for (A) all lncRNAs examined and (B) lncRNAs grouped by their epigenetic modification state in ES cells and then assessed for modification in both NPC and fibroblast cells. The presence or absence of H3K4me3 and H3K27me3 peaks was assessed within the promoters of lncRNAs that did not overlap with annotated REFSEQ genes (n=10937) in mouse ES cells, NPCs, and fibroblasts. H3K4me3 only, blue; H3K27me3 only, red; H3K4me3 and H3K27me3 (bivalent), green; no H3K4me3 and no H3K27me3, purple.
We then asked whether lncRNAs containing these modifications retain the same modification or if they undergo epigenetic changes when ES cells become lineage-restricted (NPC) or terminally differentiated (fibroblasts). Unlike those lacking modifications, many lncRNAs that possess H3K4me3, H3K27me3, or both do undergo dynamic changes (Fig. 2B). Interestingly, while more than half of lncRNAs possessing H3K4me3 in ES cells retain this mark, the majority of the bivalent lncRNAs in ES cells commit to either the active or repressive mark in NPCs and fibroblasts. This suggests that the lncRNAs retaining H3K4me3 may have housekeeping functions, while those with the bivalent mark in ES cells, but commit to the active or repressive mark in NPCs or fibroblasts may have important roles in cell lineage commitment. On the other hand, the group of lncRNAs that loses the H3K4me3 mark in NPCs and fibroblasts may be interesting for future studies as these may be important for ES cell identity. It is interesting to note that most lncRNAs containing only H3K27me3 in ES cells appear to lose this modification in NPCs and fibroblasts, particularly in NPCs (Fig. 2B).
Promoter H3K4me3 and H3K27me3 correlate with lncRNA transcription levels
We next set out to examine whether changes in H3K4 and H3K27 methylation of lncRNA promoters correlate with expression of lncRNAs. To this end, lncRNAs were categorized into two groups based on their expression levels in ES cells compared to both NPCs and fibroblasts (at least a 2-fold change in ES versus NPCs/fibroblasts). The average tag density for H3K4me3 and H3K27me3 was examined over a region +/− 5kb from the transcription start site using previously published data (25). Similar to previous reports on protein coding genes, levels of H3K4me3 peak at transcription start sites (TSS) and are independent of expression status (Fig. 3A). However, lncRNAs that were expressed higher in ES cells compared to NPCs/fibroblasts tended to have slightly higher levels of H3K4me3. On the other hand, lncRNAs that were expressed at lower levels in ES cells exhibited a broad peak of H3K27me3 that sharply dropped after the transcription start site (Fig. 3B). Levels of H3K27me3 remained consistently low in the same region when the opposite group was examined.
Figure 3. Changes in promoter H3K4me3 and H3K27me3 levels are correlated with lncRNA expression.
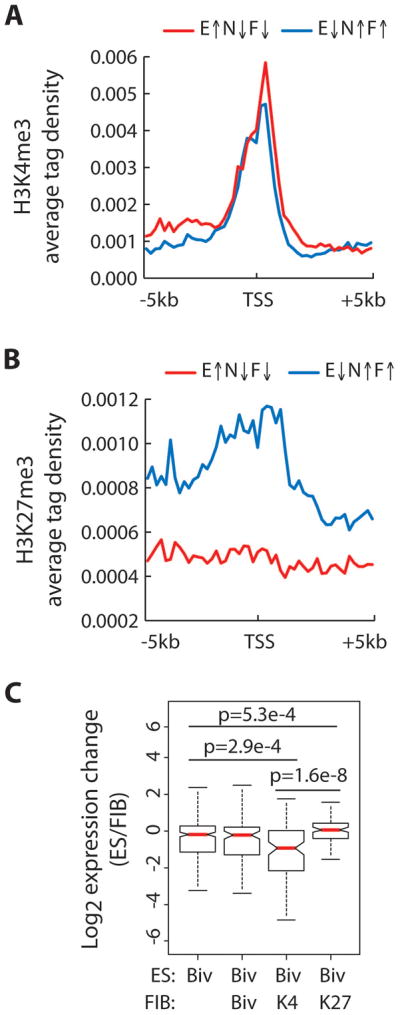
The average H3K4me3 (A) and H3K27me3 (B) tag density across the promoters of lncRNAs in ES cells grouped by relative expression in ES versus NPC and fibroblast. X-axis represents the region spanning −5kb to +5kb away from the lncRNA transcription start site (TSS). Red line indicates lncRNAs that exhibit at least 2-fold higher expression in ES cells relative to both NPCs and fibroblast cells, E↑N↓F↓ (n=1511). Blue line indicates lncRNAs that exhibit at least 2-fold lower expression in ES cells relative to both NPCs and fibroblast cells, E↓N↑F↑ (n=1696). (C) Box plot presentation of expression change (ES/FIB) in bivalent-marked lncRNAs in ES cells (n=321), which are further divided into three groups according to their promoter H3K4 and H3K27 methylation state in fibroblast cells: H3K4me3 and H3K27me3, Biv (n=115); H3K4me3 only, K4 (n=98); H3K27me3 only, K27 (n=89). Significant p values as determined by Wilcoxon signed-rank test are indicated.
As mentioned above, previous studies of protein coding genes in ES cells suggest that gene promoters marked by bivalent domains are thought to be poised for activation or repression upon differentiation (24). To determine whether the same trend holds true for lncRNAs, we categorized lncRNAs that were bivalent in ES cells based on their modification state in fibroblast cells and examined their expression change upon differentiation. On average, bivalent lncRNAs that become H3K4me3 in fibroblasts are expressed significantly higher in fibroblasts compared to ES cells, while those that become H3K27me3 are expressed significantly lower (Fig. 3C). Collectively, the above results demonstrate that H3K4me3 and H3K27me3 can be correlated with the activation and repression of lncRNAs, respectively. Therefore, the relationship between transcription levels and promoter H3K4me3 and H3K27me3 levels previously discovered for protein coding genes holds true for lncRNAs.
Ezh2-mediated H3K27 methylation represses lncRNAs in ES cells
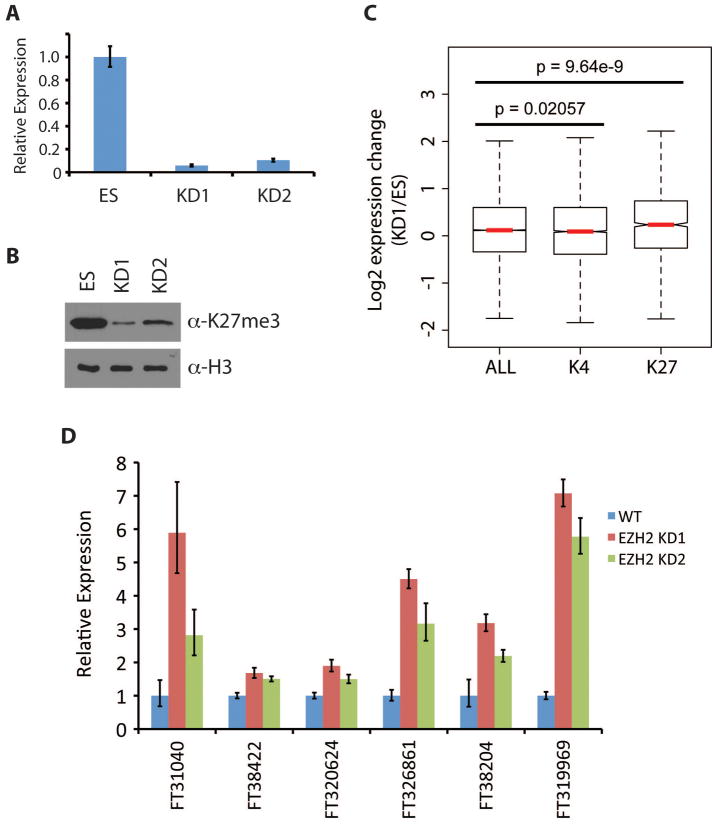
We next asked whether the presence H3K27me3 at the promoter affects expression of lncRNAs in ES cells. To this end, we attempted to manipulate levels of H3K27me3 by knocking down Ezh2, the catalytic subunit of the Polycomb Repressive Complex 2 (PRC2) (26). We generated lentiviruses expressing two independent shRNAs targeting Ezh2, which we used for transduction of ES cells. Following puromycin selection, knockdown efficiency was confirmed by RT-qPCR (Fig. 4A). Consistent with down-regulation of Ezh2, global levels of H3K27me3 were dramatically reduced (Fig. 4B).
Figure 4. Ezh2-mediated H3K27 methylation represses lncRNA transcription in ES cells.
(A) Relative mRNA levels of Ezh2 in wildtype and Ezh2 knockdown ES cells as determined by RT-qPCR.
(B) Western blot analysis of H3K27me3 levels in Ezh2 KD ES cells.
(C) Box plot presentation of expression changes (KD1/ES) in all lncRNAs (n=24357) compared to lncRNAs marked by promoter H3K4me3 (n=6834) or H3K27me3 (n=1294) in ES cells. Significant p values were determined using the Student’s T test.
(D) Relative expression of lncRNAs upon Ezh2 knockdown in ES cells as determined by RT-qPCR. These lncRNAs were randomly selected from the list of lncRNAs that contain H3K27me3 in ES cells and are at least 2-fold upregulated upon Ezh2 knockdown as determined by microarray analysis.
We analyzed the lncRNA expression profile in Ezh2 KD ES cells using the custom-designed microarray described earlier and found that both knockdowns correlated well with each other (Pcc = 0.935). To determine whether H3K27me3 plays a role in silencing lncRNAs in ES cells, we grouped lncRNAs based on the presence of H3K4me3 or H3K27me3 at their promoter and examined their expression change upon Ezh2 knockdown (Fig. 4C). Not surprisingly, lncRNAs marked by H3K4me3 exhibited no significant changes in expression compared to all lncRNAs. On the other hand, we found that lncRNAs containing H3K27me3 tend to be expressed at a higher level upon Ezh2 knockdown when compared to those containing H3K4me3 (p = 1.415 × 10^-10) or all lncRNAs (p = 9.64 × 10^-9) indicating that Ezh2-mediated H3K27 methylation contributes to the repression of a subset of lncRNAs in ES cells.
To further analyze the effect of Ezh2 knockdown on lncRNA expression, we examined the microarray data and found 163 lncRNAs that were at least 2-fold upregulated in both knockdowns of Ezh2. Of these, 19 were also marked with H3K27me3 in ES cells. From this group, we randomly selected 6 lncRNAs and confirmed that their expression was de-repressed upon Ezh2 knockdown by RT-qPCR (Fig. 4D). The effect of Ezh2 on these lncRNAs appears to be direct as analysis of previously published genome-wide mapping data (27) indicated that the promoters of these lncRNAs are not only bound by Ezh2 but also by Suz12, another component of the PRC2 complex (26).
Discussion
In summary, our study examined the lncRNA expression profiles in ES, NPC, and fibroblasts and analyzed their relationship with their promoter H3K4me3 and H3K27me3 levels. Our results indicate that similar to protein coding genes, H3K4me3 and H3K27me3 are generally correlated to lncRNA activation and repression, respectively. In addition, through shRNA-mediated knockdown of Ezh2 in ES cells, we demonstrate that Ezh2-mediated H3K27 methylation also functions to repress the expression of lncRNAs, thus extending the observations from protein coding genes to lncRNAs.
Our finding that the Polycomb group of proteins regulates lncRNAs in addition to protein coding genes prompted us to investigate whether transcription factors are also involved in regulating lncRNAs. Taking advantage of previously published genome wide localization maps of transcription factors involved in ES cell biology (28), we examined the promoter regions of lncRNAs for the presence of transcription factors. Our analysis indicates that 30% (3232 out of 10937) of the lncRNAs in our dataset contain at least one transcription factor bound to its promoter (Fig. S1A). Furthermore, the promoter regions of almost 300 lncRNAs contain at least 3 of the 4 core transcription factors important for ES cell maintenance (Nanog, Oct4, Sox2, and Klf4) (Fig. S2B). These results suggest that transcription factors may play a role in regulating lncRNAs. It is also possible that other regulatory mechanisms employed by protein coding genes may apply to lncRNAs.
Several lncRNAs including Xist, HOTAIR, and AIR have recently been demonstrated to be functional molecules important for specific biological processes (12,13,15). Given that over 30,000 lncRNAs have been annotated in the FANTOM database, it is very likely that many more functional lncRNAs will be identified. In support of this prediction, we find that a subset of lncRNAs are differentially expressed and marked with different histone modifications suggesting that these molecules play specific roles in different cell types. However, characterizing their biochemical and biological function is no trivial matter. Based on the few number of characterized lncRNAs, it is clear that lncRNAs can participate in various biological processes and function through diverse mechanisms (9,14) thus making it difficult to predict their mechanism of action.
Despite these challenges, our demonstration that lncRNAs are regulated in a manner similar to that of protein coding genes allows for identification of functional lncRNAs through a candidate-based approach by analyzing expression data as well as the presence or absence of specific histone modifications. Because we find that a subset of lncRNAs are developmentally regulated, it will be of great interest to determine if lncRNAs play a crucial role in ES cell biology. Our analysis indicates that there are 13 lncRNAs that contain H3K4me3 (and no H3K27me3) in ES cells in addition to H3K27me3 (and no H3K4me3) in either NPCs or fibroblasts. Of these, 5 lncRNAs are expressed at least 2 fold higher in ES cells compared to NPCs or fibroblasts. These lncRNAs can be experimentally tested as candidates important for ES cell biology using a knockdown approach. A similar approach can be used to identify candidate lncRNAs that may be important for ES cell differentiation.
The production of diverse cell types in an organism has traditionally been attributed to differential gene expression that is controlled in part through epigenetic mechanisms such as histone modifications. However, emerging evidence demonstrating the differential expression of lncRNAs has indicated that lncRNAs may also contribute to this process. While many challenges remain with regards to determining the significance of these molecules, our work provides a resource for the scientific community to identify candidate lncRNAs that are important in ES cell biology. While only a fraction of annotated lncRNAs have currently been characterized, future work will undoubtedly uncover a larger biological role for lncRNAs than originally anticipated.
Methods
lncRNA definitions and analysis
The positions of lncRNAs from the Fantom3 database were extracted from http://jsm-research.imb.uq.edu.au/rnadb/ and used for all analyses in this work. Promoter regions were defined as 2kb surrounding the start site (TSS) of each transcription unit assessed. Overlap analysis was carried out using previously defined datasets: annotated REFSEQ gene lists were procured as described in (29), H3K4me3 and H3K27me3 peaks observed in ES cells, NPCs, and mouse embryonic fibroblasts as described in (25), genome wide locations of transcription factors in ES cells as described in (28). These analyses were carried out using the Python module FJOIN (30). Whole genome averaging of epigenetic mark enrichment was performed by normalizing H3K4me3 and H3K27me3 ChIP-seq reads against whole cell extract. The region from −5KB to +5KB relative to the TSS was divided into 200 bp windows and the average tag density of the subdivided promoters was computed. Unless otherwise stated, only Fantom3 entries that did not overlap with annotated REFSEQ genes or promoters were analyzed.
Microarray analysis
Total RNA was extracted from E14ps mES cells, mouse tail tip fibroblasts, and neurospheres, derived from E14ps mES cells using a previously published procedure (31). RNA quality was assessed via Agilent Bioanlayzer. mRNA was amplified using the Low RNA Linear Amplification Kit (Agilent Technologies) by standard manufacturers protocol. Hybridization of experimental RNA labeled with Cy5 and Cy3 labeled Universal Reference RNA (Ambion) was carried out by standard protocol on a custom-designed array manufactured by Agilent Technologies. Briefly, three unique 60nt probes were designed for each entry in the Fantom3 database and were synthesized along with lineage marker control probes on a 2x105K format array. Probe design files are available at the NCBI GEO database under accession number XXXX. Upon array scanning, data was quality filtered, normalized by the Lowess method, and all probes corresponding to a given lncRNA were averaged using the UNC Microarray Database (UNCMD). Intraclass correlation analysis was carried out in the UNCMD. All microarray data has been submitted to the NCBI GEO database under accession number XXXX.
Ezh2 knockdown and RT-qPCR
Hairpins targeting Ezh2 (KD1: 5′ GAACTGAAACCTTAAACCA 3′; KD2: 5′ GTATGTGGGCATCGAACGA 3′) were cloned into a lentiviral vector under the control of the U6 promoter using previously published methods (32). E14ps mES cells were infected with lentiviruses and subjected to puromycin selection. For RT-qPCR, 1μg of total RNA was subjected to reverse transcription using ImpromII kit (Promega). The resulting cDNA was diluted 1:5 and 5μl of the resulting solution was used for Real Time PCR analysis on an Applied Biosystems 7300 machine using SYBR GreenER (Invitrogen). Reactions were performed in triplicate. Gapdh was used as an endogenous control. Primer sequences can be found in Supplemental Table 2.
Boxplots and Statistics
Boxplots were generated in the R package for defined subsets of genes. RT-qPCR statistics were computed using student’s T test. Wilcoxon signed-rank test (Fig. 3) and Student’s T test (Fig. 4) were employed to generate the presented p values.
Supplementary Material
Acknowledgments
We thank Olena Taranova for providing the RNA and Jin He for generating the Ezh2 knockdown constructs. This work was supported by NIH (GM68804). Y.Z. is an Investigator of the Howard Hughes Medical Institute.
References
- 1.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, et al. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, et al. Science. 2005;308(5725):1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 3.Consortium IHGS. Nature. 2004;431(7011):931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 4.Johnson JM, Edwards S, Shoemaker D, Schadt EE. Trends Genet. 2005;21(2):93–102. doi: 10.1016/j.tig.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 5.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. Science. 2007;316(5830):1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 6.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, et al. Genome Res. 2008;18(9):1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Proc Natl Acad Sci U S A. 2008;105(2):716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, et al. Genome Res. 2006;16(1):11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ponting CP, Oliver PL, Reik W. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ponjavic J, Ponting CP, Lunter G. Genome Res. 2007;17(5):556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, et al. Science. 2008;322(5908):1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 13.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilusz JE, Sunwoo H, Spector DL. Genes Dev. 2009;23(13):1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Science. 2008;322(5902):750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cedar H, Bergman Y. Nat Rev Genet. 2009;10(5):295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 17.Martin C, Zhang Y. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 18.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. Science. 2005;309(5740):1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 19.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, et al. Nat Genet. 2006;38(6):626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 20.Ge X, Wu Q, Jung YC, Chen J, Wang SM. Bioinformatics. 2006;22(20):2475–2479. doi: 10.1093/bioinformatics/btl429. [DOI] [PubMed] [Google Scholar]
- 21.Kapranov P, Willingham AT, Gingeras TR. Nat Rev Genet. 2007;8(6):413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 22.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, et al. Science. 2005;309(5740):1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 23.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Cell. 2007;128(4):735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 24.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 25.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Nature. 2007;448(7153):553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Science. 2002;298(5595):1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 27.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, et al. PLoS Genet. 2008;4(10):e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, et al. Cell. 2008;133(6):1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 29.Kallin EM, Cao R, Jothi R, Xia K, Cui K, Zhao K, et al. PLoS Genet. 2009;5(6):e1000506. doi: 10.1371/journal.pgen.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richardson JE. J Comput Biol. 2006;13(8):1457–1464. doi: 10.1089/cmb.2006.13.1457. [DOI] [PubMed] [Google Scholar]
- 31.Ying QL, Smith AG. Methods Enzymol. 2003;365:327–341. doi: 10.1016/s0076-6879(03)65023-8. [DOI] [PubMed] [Google Scholar]
- 32.He J, Kallin EM, Tsukada Y, Zhang Y. Nat Struct Mol Biol. 2008;15(11):1169–1175. doi: 10.1038/nsmb.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.