Abstract
Molecular diversity among clinical isolates of Madurella mycetomatis, the prime fungal agent of human mycetoma in Sudan, could possibly explain the diverse clinical presentations of this severely debilitating infectious disease. In addition, culture-independent DNA-mediated typing tests need to be developed for this organism, since M. mycetomatis DNA, but not the organism itself, can be identified in soil, the material from which infections are thought to originate. A collection of 38 different clinical M. mycetomatis isolates was characterized by large-scale random amplification of polymorphic DNA using 20 different primer species. These analyses, involving at least 2,600 annealing sites, showed a complete lack of DNA fingerprint variation among the various isolates. From the resulting homogeneous DNA fingerprints, seven fragments were cloned and sequenced, and novel, species-specific PCR restriction fragment length polymorphism (RFLP) tests were designed. The seven PCR RFLP tests were successfully performed on the 38 different M. mycetomatis strains. However, again all M. mycetomatis DNA patterns obtained appeared to be identical, whereas patterns produced using DNAs from other fungal species were clearly discriminatory. These results suggest that there is little genetic variation among clinically relevant M. mycetomatis strains from Sudan. The data tentatively imply that different manifestations of mycetoma are due to differences in host susceptibility rather than differential virulence of the causative agent.
Mycetoma presents as a chronic, relatively painless, subcutaneous granulomatous lesion which is characterized by the formation of multiple sinuses. In ∼40% of all infections, a fungus is the causative agent, the sclerotia of which are shed through these sinuses in the form of fungal grains (10, 13). The color of these grains has diagnostic value. Green grains identify Aspergillus flavus as the most likely causative agent, whereas white grains are usually produced by Pseudallescheria boydii, Aspergillus nidulans, or Acremonium kiliense. Brownish grains are produced by Neotestudina rosatii, while deep-black grains are produced by species such as Curvularia lunata, Exophiala jeanselmei, Pyrenochaeta romeroi, Leptosphaeria senegalensis, Madurella grisea, and Madurella mycetomatis. The last fungus is the most prevalent mycetoma agent in Sudan (10). Although this agent has been shown to occur in various geographic regions, most cases of M. mycetomatis mycetoma occur in a relatively comprehensive “mycetoma belt” (1). The precise mechanism of infection remains enigmatic, but it is frequently suggested that traumatic inoculation of fungus-containing soil, assisted by the presence of plant materials such as thorns, provides a likely route of inoculation (10, 11, 13, 19). However, successful cultivation of the organism from soil has been documented only sparsely, although recent molecular detection has revealed that fungal DNA can be detected quite easily in soils from different regions in the mycetoma belt (1). Because of the apparent impossibility of culturing the fungus directly from soil, it is very hard to study the precise transmission routes. Since M. mycetomatis DNA can be easily amplified from soil, development of methods for further analysis of the genetic variation in this soil-embedded material has a high priority. Tools for the assessment of strain-specific characteristics may also be helpful for distinguishing relapsing disease from reinfection events.
For the reasons outlined, above we have tried to develop direct DNA identification assays suited to discrimination between Sudanese M. mycetomatis strains. Using a large collection of clinical isolates of M. mycetomatis, we tried to identify polymorphic genome fragments suited to direct DNA characterization. Strains were initially typed by high-throughput random amplification of polymorphic DNA (RAPD). Possible sequence variability within individual RAPD fragments was subsequently investigated by the application of PCR restriction fragment length polymorphism (RFLP) tests.
MATERIALS AND METHODS
Strains and culture conditions.
The Sudanese M. mycetomatis strains (n = 38) and two clinical isolates from Mali (p1 and p2) were identified by PCR as previously described by Ahmed et al. (2). The strains were isolated by direct culture of black grains obtained from deep biopsies of patient lesions at the Mycetoma Research Center, Khartoum, Sudan. The patients originated from a large geographic region, essentially covering the entire Sudanese mycetoma belt. The strains were maintained on Sabouraud agar (Difco Laboratories, Detroit, Mich.) with gentamicin at an incubation temperature of 37°C. Cultures of (distantly) M. mycetomatis-related organisms were obtained from the Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands (CBS). These fungi were Alternaria infectoria (CBS 160.79), Alternaria tenuissima (CBS 160.52), Alternaria alternata (CBS 137.90), Curvularia geniculata (CBS 731.96), Curvularia verruculosa (CBS 444.70), P. boydii (two different strains, CBS 883.71 and CBS 1003.92), Leptosphaeria tompkinsii (CBS 201.79), Bipolaris hawaiiensis (CBS 727.96), E. jeanselmei (CBS 635.69), Phialophora verrucosa (CBS 839.69), Aspergillus fumigatus (dH 12472), Penicillium crustaceum (CBS 581.67), Fusarium oxysporum (CBS 1098.98), Fusarium solani (CBS 1022.56), Fusarium lichenicola (CBS 623.92), and M. grisea (CBS 172.22).
DNA isolation.
After 4 weeks of incubation at 37°C, the fungal material was excised from the agar and transferred into 6 ml of a 0.9% NaCl solution. The material was sonified for 2 min at 30 μm (Soniprep 150; Beun de Ronde, Abcoude, The Netherlands) to fully disrupt the mycelium. Five hundred microliters of this suspension was taken, and DNA was isolated according to the DNA extraction method described by Boom et al. (7). Because of the small amount of DNA isolated by this method, the following alternative method was employed. After sonification, the suspension was frozen in liquid nitrogen, thawed, and ground in a mortar using a porcelain pestle. DNA was extracted from this emulsified sample using the Wizard Genomic DNA purification kit (Promega Corp., Leiden, The Netherlands) employing the yeast protocol, which starts with the addition of the nuclear lysis solution.
RAPD analysis.
The RAPD reactions were performed in 50-μl reaction volumes containing 5 μl of 10× Supertaq PCR buffer 1 (HT Biotechnology Ltd., Cambridge, United Kingdom), 10 μl of 1 mM PCR nucleotide mix (Amersham Life Sciences, Roosendaal, The Netherlands), 0.5 μl of primer at 50 pmol per μl, 1.2 U of Supertaq (HT Biotechnology Ltd.), and distilled water to complete the volume. The 20 primers used are listed in Table 1. The PCR was performed in a model 60 Thermocycler (Biomed, Theres, Germany). PCR consisted of a predenaturation step of 4 min at a temperature of 94°C and 40 cycles each of a denaturation step of 1 min at 94°C, an annealing step of 1 min at 25°C, and an extension step of 2 min at 74°C. The RAPD patterns were analyzed on 1% agarose gels (Hispanagar; Sphaero Q, Leiden, The Netherlands) after 3 h of electrophoresis in 0.5× Tris-borate-EDTA at a constant current of 110 mA.
TABLE 1.
Primers used for high-throughput RAPD analysis of 38 clinical M. mycetomatis isolates
| Primer no. | Name | Sequence | Sizes (nt) of amplimers synthesized |
|---|---|---|---|
| 13 | ERIC-2 | AAGTAAGTGACTGGGGTGAGCG | 550, 900, 1,100 |
| 26 | Rep-21 | ICGICTTATCCIGGCCTAC | 950, 1,300, 1,500 |
| 46 | RAPD1 | GGTTGGGTGAGAATTGCACG | 200, 300, 500, 550, 700 |
| 47 | Random | GGCCATAGAGTGTTGCAGACAAACTGC | Nothing amplified |
| 51 | Random | GCGATCCCA | 500, 550, 720 |
| 52 | RAPD7 | GTGGATGCGA | 500, 550, 600, 800, 820, 1,500, 1,550 |
| 174 | VanC2 | CTTCCGCCATCATAGCT | Nothing amplified |
| 214 | CEP PA480 | GTTACCAACAGAATAAGC | Nothing amplified |
| 312 | PC2 | GCCTTCTCCAATGCAGCGAC | 450, 900 |
| 449 | 28.3b | TCCGCGGGGCGTCCGCCGGA | 650, 710, 950, 1,100 |
| 548 | wlaclus1 | ATCAGATCGTTCCTATACAG | Nothing amplified |
| 683 | uspA2repeat | TGGATATGATAGAGATTTTTCCAT | 600, 620, 630, 750, 800, 900, 1,100, 1,400, 1,500, 1,550 |
| 689 | campcoli1 | AGGCAAGGGAGCCTTTAATC | 600, 620, 800 |
| 695 | CalE1 | TATGACAAACACAGGGACAAC | 600, 1,100, 1,150, 1,400, 1,500, 1,550 |
| 699 | McCAACrep | GTCGGTATTATGGGCAG | 700, 900, 950, 1,000, 1,100, 1,300, 1,500 |
| 701 | repeatF1 | AGGATTTGGCGAAGTTTGG | 700, 800, 950, 960, 1,000, 1,100, 1,250, 1,260, 1,350, 1,550, 1,560, 1,600, 1,650 |
| 714 | PstBseq1 | TAGTCTGGTGGCTGGTGTGGGC | Nothing amplified |
| 718 | PstSprer | AACTTTCATAATGTCTCCTG | 500 |
| 729 | 13,1VNTR | TGAACCATGGGTAAATTTGA | Nothing amplified |
| 786 | AW-14.2 | GCTTATCGTAAAGTAAACGA | Nothing amplified |
Cloning, sequencing, and primer design.
After electrophoresis, DNA-containing agarose plugs were excised from the gel. The DNA was extracted from this gel with the QIAquick gel extraction kit (Qiagen Gmbh, Hilden, Germany). The extracted DNA fragments were cloned into pCR2.1-TOPO (TOPO TA cloning kit; Invitrogen, Leek, The Netherlands) according to the protocol supplied. The cloned fragments were sequenced (BaseClear, Leiden, The Netherlands), and the sequences were aligned and compared to other sequences in the National Center for Biotechnology Information database using BLASTN version 2.2.2 and BLASTX version 2.2.3 as the analytical software. Primers suited for the specific reamplification of internal elements of the RAPD clones were designed using the program PrimerSelect version 4.00 (DNASTAR, Madison, Wis.). These primers are listed in Table 2.
TABLE 2.
RAPD fragments cloned and used for PCR-RFLP procedure
| Fragment no. | RAPD primer | Fragment size (nt) | Sequence length (nt) | Sequence homol- ogy | Forward primer
|
Reverse primer
|
Annealing temp (°C) | ||
|---|---|---|---|---|---|---|---|---|---|
| No. | Sequence | No. | Sequence | ||||||
| 1 | 695 | 600 | 585 | None | 989 | AGAAAGCGGACGGAGAGCAAAATG | 990 | TATGACAAACACAGGGACAACGAG | 54 |
| 5 | 701 | 1,550 | 1,467 | None | 1144 | AGGGTGAATTTGAGCCAGTTGAGT | 1145 | AGTGGGGCGAGTACAGGGATAATA | 55 |
| 7 | 701 | 1,000 | 811 | None | 995 | TGCTGCTGCTGCTCCTCCTGTTAC | 996 | CGACTACCACCACCGCCACTACT | 59 |
| 8 | 701 | 800 | 682 | None | 998 | AGTTTGGCACGAGGAAGGTCATTG | 999 | TTTGGCGAAGTTTGGGTAGGATGG | 54 |
| 15 | 52 | 1,050 | 1,061 | cumA | 813 | TTTCAGGCTCAGGGTGCGTTCGTG | 814 | GTTTAGGTGATGCCGGTGTATGGT | 56 |
| 16 | 52 | 800 | 793 | None | 810 | CGGTCGGCGGTTAGAGAAAT | 811 | TCGGGGGCCACTGCTGCTA | 50 |
| 17 | 46 | 450 | 439 | None | 1192 | TGCACGTGGAATCGCATAATAATA | 1193 | GAGAATTGCACGGAAGTTTGGTAA | 55 |
Detection of polymorphism in the reamplified RAPD fragments.
The fragments mentioned above were amplified by PCR in a Biomed thermocycler model 60 according to the following program: predenaturation (4 min at 94°C), 40 cycles of amplification (94°C for 1 min, annealing for 2 min, and 74°C for 2 min), and 5 min of postamplification at 74°C. For individual PCRs, a specific annealing temperature was calculated (Table 2) and the PCR fragments were digested using six different enzymes or enzyme combinations and selected using the computer program MapDraw (DNASTAR). The enzymes selected generate complex mixtures of restriction fragments for each of the individual amplicons during RFLP analysis. The restriction enzymes used were AluI, BsmAI, BstUI, DdeI, HaeIII, HincII, HinfI, HinP1I, MspI, RsaI (all New England Biolabs, Beverley, Mass.), CfoI, NciI, XhoI (Boehringer Mannheim, Mannheim, Germany), NlaIII (Amersham Life Sciences), and TaqI (Amersham Biosciences, Roosendaal, The Netherlands). For each restriction enzyme, the buffer recommended by the manufacturer was used. Reactions were performed overnight at the appropriate incubation temperature. The resulting fragments were analyzed on 3% agarose gels (Metaphor Agarose; BioWhittaker Molecular Applications, Rockland, Maine) by 1.5 h of electrophoresis in 0.5× Tris-borate-EDTA at a constant current of 110 mA.
PCR specificity.
To test whether the M. mycetomatis sequence-based primers were susceptible to variation among different fungal species in general, DNAs from a phylogenetically diverse set of fungi were used as templates. The reaction conditions were identical to those mentioned above. The DNAs from the reference fungi were isolated in the same manner as for the M. mycetomatis isolates. The quality of the DNA was tested by a PCR with the universal primers ITS4 and ITS (results not shown) (2). To test whether the amplicons obtained from these fungi were different from the amplicons obtained with M. mycetomatis, an RFLP analysis was carried out with the same enzymes mentioned above for M. mycetomatis.
RESULTS
RAPD analysis of M. mycetomatis with different primers.
To produce DNA fingerprints for the different M. mycetomatis isolates, the efficacies of amplication by the primers were tested for a subgroup of the strains (n = 10). The results of these PCRs could be divided into two main groups. The first group was composed of the assays which resulted in no DNA amplification at all (primers 47, 174, 214, 548, 714, 729, and 786). For all of the other primers tested (Table 1), the PCRs resulted in DNA amplification but the fingerprints did not differentiate among the isolates (primers 13, 26, 46, 51, 52, 312, 449, 683, 689, 695, 699, 701, and 718). When primers 52, 683, 695, 699, and 701 were employed, minor differences among the fingerprints obtained for various isolates were sometimes observed. The problem was that these differences could not be adequately reproduced. We considered these products to be artifacts due to irregularities in cultivation and DNA isolation procedures. The identical core patterns generated by these PCRs, however, were highly reproducible. Overall, the RAPD analysis generated fingerprints based on 67 different DNA fragments. Using the 10 pilot strains, this involved scanning of 10 × 67 × 2 = 1,340 different primer annealing sites. Primers 46, 52, 683, and 701 (which generated the most complex fingerprints) were used to analyze the entire collection of M. mycetomatis isolates. The banding patterns obtained were compared to each other, and again the conclusion was that no reproducible banding pattern variability could be observed among the strains. Again, 38 × 35 × 2 = 2,660 different annealing sites were screened for polymorphism. In conclusion, despite all our experimental efforts, we were not able to detect significant differences among RAPD-generated fingerprints for any of the M. mycetomatis strains. The data obtained for strains p1 and p2 from Mali were identical to those obtained for the Sudanese isolates (results not shown).
Development of PCR RFLP tests.
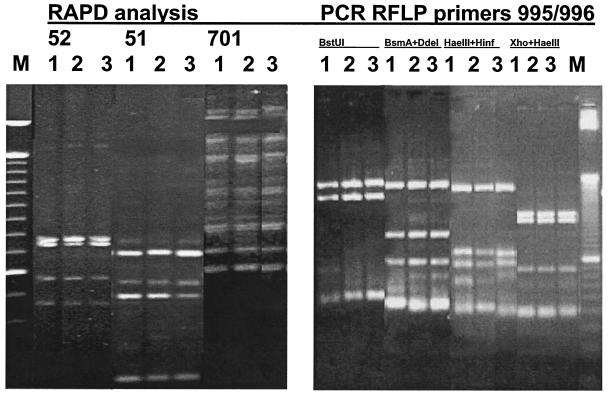
Since detection of genetic variability by RAPD failed, we decided to investigate DNA sequence variability in specific regions of the M. mycetomatis genome amplified by our RAPD primers. This should be considered a random approach for the identification of sequence heterogeneity. After the RAPD patterns were analyzed, seven consensus fragments present in the fingerprints of all strains were cut from the gel, cloned, and sequenced (see Table 2). The sequences were compared with all of the sequences in GenBank through BLAST analysis. Only one of the fragments showed homology with a known sequence. This fragment appeared to be partly homologous with the manganese-associated copper peroxidase-encoding cumA gene found in different Pseudomonas species (9, 12). Overall, 82 to 89% homology was found between the query sequence and its homologues. At the protein level, the homology appeared to be 97%. The seven sequences were reamplified from internal sequences (Table 2), which led to positive PCRs for all isolates. This is in perfect agreement with the ubiquitous presence of the cloned fragments in all of the RAPD fingerprints. To search for genetic variation within the amplicons, RFLP analysis was performed. Six enzymes or enzyme combinations were selected. After restriction with the chosen enzymes, all of the strains gave rise to identical patterns. Again, this approach failed to identify genetic variation among the M. mycetomatis strains, including those from Mali (Fig. 1).
FIG. 1.
RAPD and PCR-RFLP analysis of DNA sequence variability in specific regions of the M. mycetomatis genome amplified by RAPD primers.
PCR specificity testing using DNAs from related fungal species.
To test whether the designed primers were sensitive to variation at the fungal species level, DNAs from other fungi were studied. DNA samples from four clinical M. mycetomatis isolates (p2, mm36, mm56, and mm83) were used as process controls. For instance, when primers 814 and 813 were used, A. infectoria, A. alternata, C. geniculata, P. boydii (CBS 1003.92), B. hawaiiensis, P. verruculosa, P. crustaceum, F. lichenicola, F. oxysporum, F. solani, and M. grisea gave rise to multiple PCR products, including (for A. infectoria, A. alternata, P. verruculosa, F. oxysporum, F. solani, and M. grisea) one of almost the same size as the amplicons obtained for M. mycetomatis. A single PCR product (each) was obtained for A. tenuissima, P. boydii (CBS 883.71), and L. tompkinsii. For the first two species, the PCR products had sizes similar to those of the fragments obtained for M. mycetomatis. DNAs from the fungi C. verrucolosa and E. jeanselmei did not result in amplification. The PCR products with sizes similar to those of M. mycetomatis were analyzed by RFLP involving two-enzyme combinations. Only in the case of the 814-813 PCR product of M. grisea did some restriction fragments have sizes identical to those of the restriction fragments obtained from M. mycetomatis. All species tested could be differentiated from the M. mycetomatis strains. The data for the other primers showed similar species specificity, although the DNAs from species other than those described above resulted in amplification of multiple DNA fragments or nonamplification (data not shown).
DISCUSSION
Classical high-throughput RAPD and newly developed PCR-RFLP tests were applied to detect possible genetic variation among different clinical M. mycetomatis strains from Sudan. The RAPD results were frustrating: out of 20 primer species employed, 25% did not result in any DNA amplification, whereas the remaining 75% generated completely identical patterns for all of the strains analyzed. Several thousands of annealing sites were scanned, and the complete lack of variability is at least remarkable. For most microorganisms, including eukaryotes, RAPD is a technique well suited for the detection of genetic diversity in and between different species. RAPD studies performed with other clinically relevant fungi, such as A. fumigatus, resulted in many different genotypes, even when only limited numbers of primers and fungal strains were employed. Bertout et al. defined eight different genotypes among 52 A. fumigatus isolates (4), while Bart-Delabesse et al. detected 31 different genotypes among 67 isolates (3). These are only two of the many examples which can be found. Examples for fungal species as diverse as Malassezia spp., Histoplasma capsulatum, Exophiala dermatitidis, and Blastomyces dermatitidis have been published, and all of these studies successfully demonstrated genetic diversity among strains (20, 21, 23, 24). On the other hand, past studies of other fungi, including the species Trichophyton rubrum and the varietal taxon Cryptococcus neoformans var. Grubii, and of amoebae such as Naegleria spp. revealed that for these species RAPD analysis was similarly homogeneous in its outcome (6, 8, 16, 18, 22), although generally far smaller numbers of primers were used. However, we feel confident in concluding that, based on the RAPD analysis, the M. mycetomatis isolates collected from infections in the Sudanese mycetoma belt are strongly clonal. The overall conclusion is that large clonal fungal clades causing human disease can be found.
The observations outlined above were further corroborated by PCR-RFLP, which again did not reveal any genetic variation among the isolates (1, 2). First, all the PCR primers selected from the sequences of the RAPD fragments that were cloned reacted positively with the DNAs from all 38 strains. Apparently, these randomly selected priming sites were well conserved. Secondly, all PCR products derived from the various M. mycetomatis isolates presented the same restriction pattern after digestion by six different enzymes, which is in line with at least species homogeneity. That PCR-RFLP can be useful to detect fungal genetic variability has been shown repeatedly as well (5, 14, 17, 24). To test whether the different primer combinations were species specific, they were tested on different fungal species. It appeared that when amplicons were obtained, the other fungi could be identified on the basis of a deviating RFLP pattern. Only for M. grisea and M. mycetomatis were similar restriction fragments found. These data demonstrate that significant interspecies variability exists and that this variation can be easily documented by PCR-RFLP.
The lack of molecular diversity among the M. mycetomatis strains that we document here seemingly contrasts with data of Lopes et al. (15). These authors were able to identify nine different genotypes among 17 M. mycetomatis isolates by RAPD and restriction endonuclease assays. These strains were obtained from geographically diverse locations. However, two isolates from Sudan could not be separated, which is in agreement with our findings. The Sudanese strains clustered with strains from Cameroon, Morocco, and Chad. Here, we included two strains from Mali (p1 and p2). Mali is in West Africa, while Sudan is in East Africa; climate and vegetation are completely different in the two countries. Still, the strains from Mali had the same genotype in the RAPD assay as the clinical isolates from Sudan. Although Lopes et al. (15) did not use molecular methods to identify M. mycetomatis to the precise species level, the conclusion, based on our and their observations, should be that at least in Sudan, and possibly even in other regions in Africa, certain genotypes of M. mycetomatis are preferentially found among patients with madura.
In conclusion, no sequence heterogeneity was encountered among a large number of clinical isolates of M. mycetomatis from Sudanese patients. This indicates that this geographically restricted population of M. mycetomatis is apparently clonal.
REFERENCES
- 1.Ahmed, A. O. A., D. Adelmann, A. H. Fahal, H. A. Verbrugh, A. van Belkum, and G. S. de Hoog. 2002. Environmental occurrence of Madurella mycetomatis, the major agent of human eumycetoma in Sudan. J. Clin. Microbiol. 40:1031-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed, A. O. A., M. M. Mukhtar, M. Kools-Sijmons, A. H. Fahal, G. S. de Hoog, A. H. G. Gerrits van den Ende, E. E. Zijlstra, H. A. Verbrugh, A. M. El Sir Abugroun, A. M. Elhassan, and A. van Belkum. 1999. Development of a species-specific PCR-restriction fragment length polymorphism analysis procedure for identification of Madurella mycetomatis. J. Clin. Microbiol. 37:3175-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bart-Delabesse, E., J. Sarfati, J. P. Debeaupuis, W. van Leeuwen, A. van Belkum, S. Bretagne, and J. P. Latge. 2001. Comparison of restriction fragment length polymorphism, microsatellite length polymorphism, and random amplification of polymorphic DNA analyses for fingerprinting Aspergillus fumigatus isolates. J. Clin. Microbiol. 39:2683-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertout, S., R. Renaud, and R. Barton. 2001. Genetic polymorphism of Aspergillus fumigatus in clinical samples from patients with invasive aspergillosis: investigation using multiple typing methods. J. Clin. Microbiol. 39:1731-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birch, M., M. J. Anderson, and D. W. Denning. 1995. Molecular typing of Aspergillus species. J. Hosp. Infect. 30:339-351. [DOI] [PubMed] [Google Scholar]
- 6.Boekhout, T., A. van Belkum, A. C. Leenders, H. A. Verbrugh, P. Mukmurangwa, D. Swinne, and L. Scheffers. 1997. Molecular typing of Cryptococcus neoformans: taxonomic and epidemiological aspects. Int. J. Syst. Bacteriol. 47:432-442. [DOI] [PubMed] [Google Scholar]
- 7.Boom, R., D. J. A. Sol, M. M. M. Salimans, C. L. Jansen, P. M. E. Wertheim van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 33:24-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandt, M. E., L. C. Hutwagner, L. A. Klug, W. S. Baughman, D. Rimland, E. A. Graviss, R. J. Hamill, C. Thomas, P. G. Pappas, A. L. Reingold, R. W. Pinner, et al. 1996. Molecular subtype distribution of Cryptococcus neoformans in four areas of the United States. J. Clin. Microbiol. 34:912-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brouwers, G. J., J. P. M. Vrind, P. L. Corstjens, P. Cornelis, C. Baysse, and E. W. de Vrind de Jong. 1999. CumA, a gene encoding a multicopper oxidase, is involved in Mn2+ oxidation in Pseudomonas putida GB1. Appl. Environ. Microbiol. 65:1762-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fahal, A. H., and M. A. Hassan. 1992. Mycetoma. Br. J. Surg. 79:1138-1141. [DOI] [PubMed] [Google Scholar]
- 11.Fahal, A. H., M. A. Hassan, and M. Sanhouri. 1994. Surgical treatment of mycetoma. Sudan Med. J. 32:98-104. [Google Scholar]
- 12.Francis, C. A., and B. M. Tebo. 2001. CumA multicopper oxidase genes from diverse Mn(II)-oxidizing and non-Mn(II)-oxidizing Pseudomonas strains. Appl. Environ. Microbiol. 67:4272-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gumaa, S. A. 1994. The aetiology and epidemiology of mycetoma. Sudan Med. J. 32:14-22. [Google Scholar]
- 14.Kemker, B. J., P. F. Lehmann, J. W. Lee, and T. J. Walsh. 1991. Distinction of deep versus superficial clinical and non-clinical isolates of Trichosporon beigelii by isoenzymes and restriction fragment length polymorphisms of rDNA generated by polymerase chain reaction. J. Clin. Microbiol. 29:1677-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopes, M. M., G. Freitas, and P. Boiron. 2000. Potential utility of random amplified polymorphic DNA (RAPD) and restriction endonuclease assay (REA) as typing systems for Madurella mycetomatis. Curr. Microbiol. 40:1-5. [DOI] [PubMed] [Google Scholar]
- 16.Mochizuki, T., N. Sugie, and M. Uehara. 1997. Random amplification of polymorphic DNA is useful for the differentiation of several anthropophilic dermatophytes. Mycoses 40:405-409. [DOI] [PubMed] [Google Scholar]
- 17.Semighini, C. P., G. Delmas, S. Park, D. Armstrong, D. Perlin, and G. H. Goldman. 2001. New restriction fragment length polymorphism (RFLP) markers for Aspergillus fumigatus. FEMS Immunol. Med. Microbiol. 31:15-19. [DOI] [PubMed] [Google Scholar]
- 18.Sugita, T., R. Ikeda, and T. Shinoda. 2001. Diversity among strains of Cryptococcus neoformans var. Gattii as revealed by a sequence analysis of multiple genes and a chemotype analysis of capsular polysaccharide. Microbiol. Immunol. 45:757-768. [DOI] [PubMed] [Google Scholar]
- 19.Suliman, E. N. 1994. Laboratory diagnosis of mycetoma. Sudan Med. J. 32:67-73. [Google Scholar]
- 20.Uijthof, J. M. J., G. S. de Hoog, A. W. A. M. de Cock, K. Takeo, and K. Nishimura. 1994. Pathogenicity of strains of the black yeast Exophiala (Wangiella) dermatitidis: an evaluation based on polymerase chain reaction. Mycoses 37:235-242. [DOI] [PubMed] [Google Scholar]
- 21.Van Belkum, A., T. Boekhout, and R. Bosboom. 1994. Monitoring spread of Malassezia infections in a neonatal intensive care unit by PCR-mediated genetic typing. J. Clin. Microbiol. 32:2528-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Belkum, A., J. de Jonckheere, and W. G. V. Quint. 1992. Genotyping Naegleria spp. and Naegleria fowleri isolates by interrepeat polymerase chain reaction. J. Clin. Microbiol. 30:2595-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woods, J. P., D. Kersulyte, W. E. Goldman, and D. E. Berg. 1993. Fast DNA isolation from Histoplasma capsulatum: methodology for arbitrary primer polymerase chain reaction-based epidemiological and clinical studies. J. Clin. Microbiol. 31:463-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yates-Siilata, K. E., D. M. Sander, and E. J. Keath. 1995. Genetic diversity in clinical isolates of the dimorphic fungus Blastomyces dermatitidis detected by a PCR-based random amplified polymorphic DNA assay. J. Clin. Microbiol. 33:2171-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]