Abstract
Background
Searching optima is one of the most challenging tasks in clustering genes from available experimental data or given functions. SA, GA, PSO and other similar efficient global optimization methods are used by biotechnologists. All these algorithms are based on the imitation of natural phenomena.
Results
This paper proposes a novel searching optimization algorithm called Gravitation Field Algorithm (GFA) which is derived from the famous astronomy theory Solar Nebular Disk Model (SNDM) of planetary formation. GFA simulates the Gravitation field and outperforms GA and SA in some multimodal functions optimization problem. And GFA also can be used in the forms of unimodal functions. GFA clusters the dataset well from the Gene Expression Omnibus.
Conclusions
The mathematical proof demonstrates that GFA could be convergent in the global optimum by probability 1 in three conditions for one independent variable mass functions. In addition to these results, the fundamental optimization concept in this paper is used to analyze how SA and GA affect the global search and the inherent defects in SA and GA. Some results and source code (in Matlab) are publicly available at http://ccst.jlu.edu.cn/CSBG/GFA.
Background
Two of the most challenging tasks of optimization algorithms are to search the global optimum and to find all local optima of the space of solutions in clustering genes from available experimental data [1], e.g. the gene expression profiles, or given functions. In view of recent technological developments for large-scale measurements of DNA expression level, these two problems can often be formulated and many methods have been proposed. In particular, the heuristic searches are more promising than other kinds of searching approaches. These approaches include GA (genetic algorithm) [2], SA (simulated annealing) [3], PSO (Particle Swarm Optimization) [4] etc. But some inherent drawbacks, especially the inability to the multi-modal functions optimization, can be found from the traditional heuristic search algorithms above. Each of these concepts allows for several modifications, which multiplies the number of possible models for data analysis we can change the algorithm themselves, to find all the valleys of given functions. But we still have a lot of parameters to consider, as known as the number of valleys and the valley distance etc, and the performances of the modifications are not good enough.
GA is a traditional searching-optimization algorithm, this algorithm can search global optimal solution with probability criterion [5], but it can't converge to the global optimal solution in the theory [6]. So GA always traps in local optima or genetic drift. Anyway, the run-time of this algorithm is acceptable for most cases of searching-optimization problems.
SA is a generic probabilistic meta-algorithm for global optimization problems, namely locating a good approximation to the global optimum of a given function in a large search space. If we search enough time with SA, it will converge in the global optimal solution with probability 1 [7]. But the biggest drawback of SA is that the running-time of SA is so long that the efficiency is not tolerant to us.
Recently, some other searching algorithms have been proposed and discussed, such as PSO etc. These algorithms can search solution like GA. But actually, most of them will not converge to the global optimal solution either.
In this paper, we propose a new heuristic search approach Gravitation Field Algorithm (GFA). It not only can handle unimodal functions optimization, which traditional heuristic searches can do the same, but also can deal with multimodal functions optimization, which traditional heuristic search algorithms can't do. The experiments of the benchmark functions demonstrate that. The idea of GFA is derived from the modern widely accepted theory of planetary formation-- the Solar Nebular Disk Model (SNDM) [8] in the astronomy.
The complex astronomy theory can be introduced simply as follows:
Several billions years ago, there wasn't any planet in the Solar System; just dusts rounded the whole world. Then the dusts assembled by their own gravitations. For a long time, the rocks had come out. It was the threshold time of the whole Solar System. From then, the rocks moved fast to assemble together, and the bigger rocks arrested the smaller rocks. And they became larger rocks. Finally, the planets came out, and the rocks around them were absorbed out.
GFA is derived from the point of view of the hypothesis theory described above. To start with, all the solutions, which are the dusts in the algorithm model, are initialized randomly, or based on the prior knowledge; what's more, we assign every dust (solution) a weight, we call it mass, whose values are based on the mass function generated from the space of the problem solution; finally, the GFA begins. The power of attraction, which belongs to a certain dust and exists between every two dusts, pulls other dusts, which have the same influence to other dusts. Hence, the dusts assemble together, and the planets come out in the end--they are the optima. If you want to find global optimal solution, the planets assemble again, and the biggest planets will come out. To give a penalty of that the highest mass dust rules the whole space of solution, we propose a distance which can reduce the effect of gravitation field.
Methods
Description of GFA
Before all the works start, we design a mass function on the basis of special problem. The mass function is similar to the fitness function in GA. Both of them are score functions which are the criterions for a special solution. We can make the mass function be proportional to, or inversely proportional to, the extreme values of the problem. It all depends.
Initialization
The algorithm begins with the initial step. We generate and select n dusts di (i = 1, ..., n) in the mass function domain [xbegin, xend] to build the initial solution set. The positions of the dusts can be random or based on the prior knowledge (i.e. there will be greater probability to exist peak values in some positions). Then we assign a mass value to every dust. The mass values, which are described above, are associated with their positions, and are calculated by the mass function. So when the dusts move as we described below, they have a certain probability to find a new position, in which there is a new mass value that is bigger than the mass value of the centre dust. This initialization approach was used because the extension of the space solution could be considered. And the density of the dusts is proportional to the probability of existence of extreme value.
Strategy of division
To decompose the solution space, we divide the space into pieces called groups. In one group, the special dust called centre dust corresponds to the max mass value in the group. The other dusts called surrounding dusts whose mass values are smaller than the mass value of the centre dust are in the power distance of the centre dust. The power distance is the space of the group in which the centre dust pulls other surrounding dusts toward it. De-signing an effective strategy of division is a challenging work in the GFA. When the number of in-dependent variables is greater than one, an appropriate strategy of division always needs a smart method. There are many criterions of division. For example, we can make a power weight strategy: the size of each group is decided by the mass value of the centre dust in the group, proportional or inversely proportional to the peak values. Fig. 1 was shown to explain this strategy.
Figure 1.
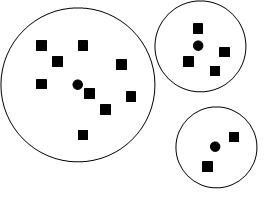
Graph of power weight strategy in solution space. The round black nodes are the centre dusts. The rectangle black nodes are surrounding dusts. The big white round charts with other nodes in them are the power distance domains of their internal centre dusts.
Here, another simple average strategy of division in the form of two independent variables is given. The task of this strategy is to find the method of division in which the areas of all groups are all the same. First, we decide the number of groups m based on the mass function and the prior knowledge. Then we factorize m with two maximum factors, like Eq. (1):
| (1) |
The sizes of all groups are the same in this strategy. The power distance of each centre dust in the solution space is defined by Eq. (2):
| (2) |
Where x is the domain of x axis of the mass function; y is the domain of y axis of the mass function.
The rules of the motion of the dusts and strategy of absorption
The rules of the motion of dusts are the kernel part of GFA. This part of GFA decides which one is the peak value among all the dusts in one group. And this step of the algorithm also can generate new mass values which can have bigger mass values than the mass values of centre dust with certain probability.
Each group contains only one dust, called centre dust, that doesn't move within an epoch (but may move in other epochs). The rest dusts in this group move toward the centre dust within the epoch. Formally, centre (di), where i = 1,..., n, is used to represent whether the dust di is the centre dust or not: centre (di) is set to true if di is the centre dust; centre (di) is set to false otherwise.
There are many kinds of strategies of motion. In this paper, the pace of the motion is set as Eq. (3):
| (3) |
Eq. (3) is easy but efficient. disi is the distance between the moving surrounding dust and the centre dust. M is a weight value for distance. 0.618/10 is chosen as M for this paper. Actually, many other weight values were tested in our experiment. But the speed and efficiency of GFA with the Golden Ration (0.618) are best. Maybe the value of it is not coincidence. But the reason, which is beyond our comprehension, will not mention in the paper. When the space of a group is small enough, the surrounding dusts will move with small pace calculated using Eq. (3); when the space of the group is big, surrounding dusts will move with big pace. Big pace will appear in two ways: One appears in the late algorithm. In this period, every dust in each group is quasi-optimal, big pace will not affect the efficiency of the GFA. So we make a big pace for fast convergence. The other one appears that we set a small number of dusts in the initial step. In this condition, maybe we want to make a fast convergence. And big pace could accelerate the convergence.
When the surrounding dusts move toward the centre dust, the positions of the moving dusts will be changed. So the mass values of the surrounding dusts will also be changed. The diagrams of the motion of surrounding dusts in the form of one in-dependent variable are shown in Fig. 2. When the mass value of a moving surrounding dust becomes bigger than the centre dust and any other surrounding dusts, this surrounding dust will become a new centre dust as shown in Fig. 2 (b) and 2(c).
Figure 2.
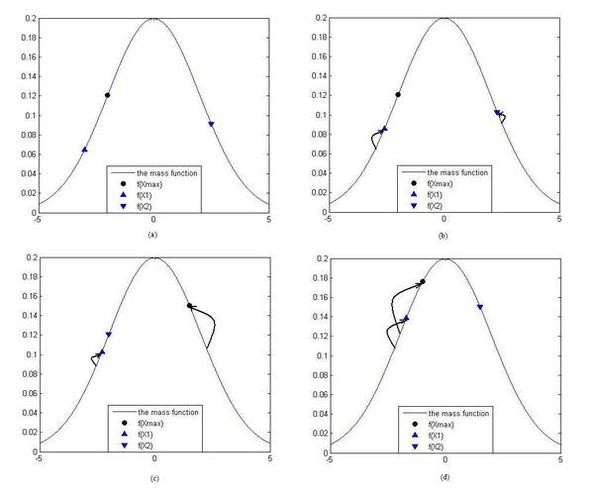
The figures of the rules of the motion of the dusts. The bell curve is the mass function. The round node is the mass value of the centre dust f (xmax). The up triangle node is the mass value of surrounding dust x1 f(x1). The down triangle node is the mass value of surrounding dust x2 f(x2). Sub-figure (a) is a small group which has 3 dusts. Sub-figure (b) is the figure of motion of the surrounding dusts toward the centre dust. In the sub-figure (c), the mass value of the surrounding dust x2 is bigger than the mass value of the centre dust. And the centre dust becomes a surrounding dust which is coded x2; the surrounding dust x2 becomes the centre dust. In the sub-figure (d), the mass value of the surrounding dust x2 is bigger than the mass value of the centre dust. The surrounding dust x2 becomes the centre dust again.
When all the distances between the surrounding dusts and the centre dust are small enough in one small group, such as smaller than a threshold, the surrounding dusts will be absorbed by the centre dust. Based on these rules of motion, we design the strategy of absorption as follows:
All the surrounding dusts are deleted, but the centre dust will not be. We set centre (di) = false, it represents the small group for the next step. When the number of surrounding dusts is bigger than a threshold, we will absorb all the other surrounding dusts for saving running time.
After the absorption of the small groups is complete, the next epoch begins if the algorithm has not converged to the optimal solution. We will divide the space of the solution again, and compute for searching peak values using the survive dusts.
The complete pseudo-code of a simple GFA is shown as Fig. 3.
Figure 3.

The pseudo-code of a simple GFA. In Fig 3, the number of dusts in one group is n, the number of groups is m, and the number of times of moving a dust is s.
Mathematical framework
Mathematical proof
The most important advantage of GFA is the ability to deal with the multimodal objective functions (i.e. the mass functions). GFA can converge for one independent variable mass functions with probability 1. We give a theorem and its strict mathematical proof as follows.
We define the mass function as f(x), and the dusts in one group of variable space are x1, x2, ..., xmax, ..., xn. The mass functions are subject to Eq. (4) as follows:
| (4) |
Where m = 1, 2, ..., n. After moving toward xmax in the group, the dusts become x1', x2', ..., xmax', ..., xn'.
First of all, we will give a description of groups and define two conceptions in the graph theory as follows:
Definition 1. In Two Side (ITS): In the line segment of the xm and xmax in the variable space, iff ∃ xm', such that: f (xm')≥ f (xmax'), then we call xm and xmax are ITS.
Definition 2. In One Side (IOS): In the line segment of the xm and xmax in the variable space, iff ∃ xm', so that: f (xm')≥ f (xmax'), then we call xm and xmax are IOS.
Now we give a theorem and prove it:
Theorem 1. GFA for one independent variable mass functions could converge in the global maximum with probability 1, when the three conditions as below are satisfied:
1) The scale of group is small enough; such that the number of peaks is at most one.
2) The motion of the surrounding dusts is smoothing.
3) The number of dusts in one group is big enough.
Proof. The formula form of condition 1) is Eq. (5):
| (5) |
The number of dusts of group g is D(g). The max value of mass function in group g is f(xmax), the real peak of the mass function in group g is f(xpeak) (maybe the xpeak isn't the dust in group g). Because the xmax moves toward itself, it doesn't move. So Eq. (6) can be established.
| (6) |
The convergence of one group can be proved as follows:
1) When xmax = xpeak, the group is convergent obviously, f(xmax) = f(xpeak).
2) When the xmax ≠ xpeak, then:
a) If exists xm, max and xm are ITS, the group will be convergent obviously according to condition 2), as Fig. 4 (a). The peak value calculated in the group by the GFA is the real peak value. We call this kind of condition is Real-Peak Condition (RPC).
Figure 4.
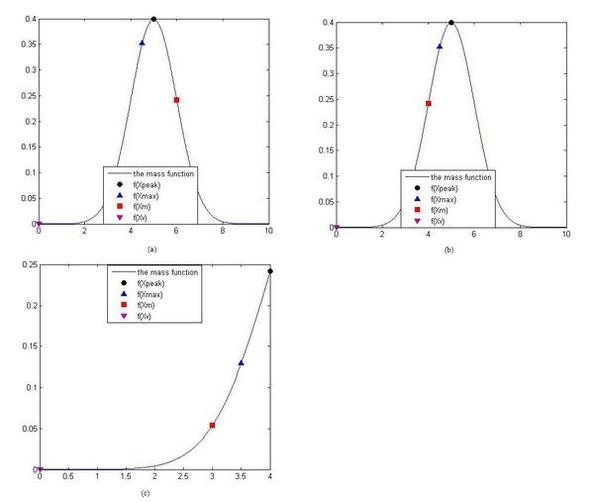
The diagrams of the moving step. f(Xpeak) is the peak value of the function in the group, the corresponding value in x axis is xpeak. f(Xmax) is the max value in the group, the corresponding value in x axis is xmax. f(Xm) is a mass value of a surrounding dust in the group, the corresponding value in x axis is xm. f(Xv) is the valley which is nearer than other valleys in the group, the corresponding value in x axis is xv. The sub-figure (a) is shown as the kind of the peak value must be found in the group. The sub-figure (b) and (c) are shown as the kind of the xmax will be the pseudo peak value with probability p.
b) If doesn't exist xm, such that the xmax and xm are ITS, the group will not be convergent, the xmax will be the pseudo peak value of the group, as Fig. 4 (b) and Fig. 4 (c). Pseudo peak value exists with probability p. we call this kind of condition is Pseudo-Peak Condition. (PPC)
The probability p can be calculated as follow:
We define the power distance of the center dust (see Description of GFA section) in the space of the solution in the group for one independent variable is ltotal. ltotal is shown as the domain of x axis in Fig. 4 (a), (b) or 4(c). And the distance between the xmax and the nearest valley in the group for one independent variable is lmax. lmax is shown as the distance between xmax and the xv in Fig. 4 (a), (b) or 4(c). f(xv) is the valley which is nearest xmax in all valleys in the group.
When the PPC is on, there are n surrounding dusts in this group. So the n-1 dusts are in the space between xmax and the nearest valley (i.e. the space of lmax), and the locations of n-1 dusts are mutually independent. So when the strategy of initialization is used, i.e. all the dusts have equal possibility to exist anywhere, the probability of one dust is IOS with xmax is lmax/ltotal So the formula of p is described as Eq. (7):
| (7) |
Where P is the probability of all n-1 surrounding dusts are IOS with xmax. From the Fig. 4, we know that lmax < ltotal, i.e. Eq. (8) can be established as follows:
| (8) |
So when condition 3) is satisfied, i.e. n → ∞, Eq. (7) can be:
| (9) |
So when PPC is on, one group will converge in the real peak, like RPC is on. So every group will converge with probability 1.
When every group converges, the surrounding dusts are absorbed out. The real peak and pseudo peak will be the new dusts in the following step.
When Xmax becomes a dust in a new group of a new epoch, the global peak value could be Xmax in one group, obviously.
Hence in a certain domain of variable x, GFA will converge in the global maximum.
Peaks versus valleys
GFA maximizes the objective or mass function f(x). That is, they solve problems of the form:
| (10) |
If you want to minimize f(x), like the experiments below, you can do so by maximizing -f(x), because the point at which the maximum of -f(x) occurs is the same as the point at which the minimum of f(x) occurs [9].
Computation complexity of GFA
Computing the mass function value once needs processing time Tf. Computing motion of every surrounding dust is Tm. Computing the size relationship of every dust once needs processing time Tg. We set the number of dusts in one group is n, the number of groups is m, and the number of times of moving a dust is s.
The computation complexity of processing mass function of every dust can be expressed by Eq. (11):
| (11) |
The computation complexity of Eq. (11) is o(mns).
The computation complexity of processing motion of every surrounding dust can be expressed by Eq. (12):
| (12) |
The computation complexity of Eq. (13) is o(mns).
| (13) |
The computation complexity of Eq. (14) is o(mns).
The computation complexity of GFA can be described as follows:
| (14) |
Then we will find the smallest dust with all groups. The computation complexity of this process can be o(m). It's smaller than others. It can be ignored. So the computation complexity of GFA is:
| (15) |
If the complexity of mass function and the dimensionality of f(x) are big enough, we can say T = Tf. The number of dusts is m × n, so the computation complexity of GFA is the product of the number of dusts and the number of times of moving of surrounding dusts. That is, the computation complexity is o(mns).
Results and Discussion
Test Method
To test the efficiency of GFA, we assessed the performance of the GFA by employing a suite of different benchmark mathematical functions (Eqs. (16-20)) and by comparing the performance with GA and SA. For each of the five test functions and the each method, 500 minimization runs were performed and mean squared error, standard deviation and mean gauss error values were calculated.
Benchmark functions
In the following benchmark functions (Eqs. (16-20)), D donates the number of independent variables, and we defined D = 50. The benchmark functions were selected as following:
Sphere:
| (16) |
Rosenbrock:
| (17) |
Rastrigin:
| (18) |
Griewangk:
| (19) |
Ackley:
| (20) |
Error functions
Mean squared error (MSE) [10] for benchmark functions was calculated as Eq. (21):
| (21) |
Where n is the number of runs, f(xi) is the performance for run i and the fopt(xi) is the function value at the global minimum.
The standard deviation (STD) [11] was calculated as Eq. (22):
| (22) |
The gauss error (GE) function [12,13], which can be in the form of Eq. (23), is different from MSE:
| (23) |
GE is a partial differential equation which is twice the integral of the Gaussian distribution with 0 mean and variance of 1/2. And this function only can be used for a scalar. So we used it with average form called mean gauss error (MGE):
| (24) |
Simulation Result
Searching global minimum
The implementation of GA and SA we used to compare with GFA is the Genetic Algorithm and Direct Search Toolbox™, which can be found from [14]. This toolbox was written with Matlab. Both the maximum number of epochs of GA and the initial temperature of SA were fixed to 1,000. For some functions, like Eq. (16), if the start point is fixed, the SA results of all the runs are all the same. So the initial start point of SA was randomly around the global minimum. A table that summarizes the parameter setting for each algorithm in each dataset was given as Table 1.
Table 1.
Configuration of parameter setting for GFA, GA and SA.
| Algorithm parameters | GFA | GA | SA |
|---|---|---|---|
| Max. numbers of iterations | 1000 | 1000 | 1000 |
| Population size | 50 | 50 | - |
| Number of polulations | 200 | 200 | - |
| Initial temperature | - | - | 0~5.0 |
| Mutation rate | 0.02 | 0.02 | - |
To compare GFA with GA and SA, 500 minimization runs were performed on our suite of benchmark functions. And the domain of each dimension was defined as [-2, 2]. The comparison results were shown in Table 2.
Table 2.
MSE, STD, and MGE of GA, GFA, and SA, Best performance (i.e., lowest error) for each function is highlighted in bold underline letters.
| Sphere | Rosenbrock | Rastrigin | Griewank | Ackley | |
|---|---|---|---|---|---|
| GA | |||||
| MSE | 7.2747 | 0.0054 | 7054.2 | 6.7709e-007 | 14.6001 |
| STD | 0.7409 | 0.0556 | 16.2621 | 6.8194e-004 | 0.1891 |
| MGE | 0.9927 | 0.0546 | 1 | 5.2149e-004 | 1 |
| GFA | |||||
| MSE | 0.3347 | 0.0156 | 152.0279 | 5.3590e-007 | 16.9460 |
| STD | 0.2940 | 0.0156 | 6.5840 | 5.0375e-004 | 16.9460 |
| MGE | 0.4839 | 0.1181 | 0.9756 | 6.0026e-004 | 1 |
| SA | |||||
| MSE | 1619.2 | 0.0069 | 745.7810 | 0.0030 | 26.9123 |
| STD | 7.1102 | 0.0827 | 12.5530 | 0.0385 | 26.9123 |
| MGE | 0.9967 | 0.0061 | 0.9952 | 0.0439 | 1 |
From Table 2, we found that sphere's, Rastrigin's and Griewank's functions could be optimized better by GFA than GA and SA, especially for the sphere's and Rastrigin's function.
Mean numbers of epochs were not same. It was an important criterion of the algorithm efficiency. The mean error served as success criterions along with a maximal number of epochs. If the threshold was not reached by an optimization method within 1,000 epochs, the run was judged as failure. No matter whether it was reached or not, the number of epochs that were needed to reach the threshold was rounded off and recorded in Table 3.
Table 3.
Mean numbers of epochs until the minimization threshold was reached and mean number of failures.
| Sphere | Rosenbrock | Rastrigin | Griewank | Ackley | |
|---|---|---|---|---|---|
| GA | |||||
| mean number of epochs | 51 | 57 | 51 | 51 | 92 |
| number of failures | 0 | 0 | 0 | 0 | 0 |
| GFA | |||||
| mean number of epochs | 31 | 46 | 30 | 49 | 97 |
| number of failures | 0 | 0 | 0 | 0 | 0 |
| SA | |||||
| mean number of epochs | 816 | 46024 | 6349 | 21634 | 1001 |
| number of failures | 108 | 500 | 314 | 126 | 234 |
Best performance (i.e., lowest number) for each function is highlighted in bold underline letters.
GFA was able to outperform GA and SA in four of five benchmark functions in terms of epochs needed and least failures. Only in one of the five functions, GFA did not outperform in both speed and robustness. The minimization of the Ackley's function took slightly more epochs with the GFA (97) than with the GA (92). From the Table 3, we know that GA could do better for optimizing the complex function, such as Ackley's function. But for some simple function, such as sphere's function, GFA could do better.
For more accurate computation, we defined the initial number of dusts 10,000, the max power distance of center dust 5, and the epochs 1,000, which was the same as used in GA and SA (for more information of detail parameters, please see description of GFA).
Overall, The GFA achieved a large decrease on the MSE compared to the GA for some functions (De Jong's Sphere: 21.735-fold; Rastrigin: 46.4007-fold). Relatively simple functions could be optimized better than GA and SA through the phenomenon of the experiment. The efficiency of SA was poorly in the experiment. And the run-time of GFA and GA were in the same magnitude, but the run-time of SA is much longer than the two others. We can see the total running-time from Table 4.
Table 4.
total running-time of GFA, GA and SA with 500 runs.
| Sphere | Rosenbrock | Rastrigin | Griewank | Ackley | |
|---|---|---|---|---|---|
| GA | 187.13 | 63.29 | 157.33 | 63.98 | 101.30 |
| GFA | 201.37 | 67.90 | 113.46 | 69.42 | 84.59 |
| SA | 14211.08 | 11463.29 | 52914.75 | 15536.02 | 6406..68 |
Smallest running-time for each function is highlighted in bold underline letters. 2.66 GHz 2 cores Intel CPU and 2 G memories are in the computer used to calculate Eqs. (16-20) with GA, GFA and SA. The unit of the running-time is second.
Searching multi-minima
Although minimum of certain given function could be optimized to resolve the problem for most situations, minimum is not always what we want only. Sometimes a part of minima is also needed. Bayesian network inferring, for example, is a stochastic approach. So the minimum of Bayesian criterion function is not always fit the realistic network best. It is important to find other minima for the problems as cases described above. A number of lowest minima can be retained in the search.
To run multimodal optimization algorithm, we must track more information than the global optimization. Searching exactly a certain number of minima or even all of the minima in the domain is a challenging work, which is also meaningful for some problems, e.g. gene cluster. In the experiment, top 5 minima were searched with GFA. It's beyond GA's and SA's ability.
Because sphere's function is a unimodal function, we could not use it for search multi-minima. We use the rest of the four benchmark function. To complete the experiment, 500 optimization runs were performed on each benchmark functions. The settings were shown as Table 5.
Table 5.
GFA settings configurations of multi-minima optimization
| Rosenbrock | Rastrigin | Griewank | Ackley | |
|---|---|---|---|---|
| Domain of each variable | [-2,2] | [-1.5,1.5] | [-1,1] | [-2,2] |
| Max numbers of iterations | 1000 | 1000 | 1500 | 2000 |
| number of dusts | 10000 | 10000 | 15000 | 20000 |
| Number of groups | 200 | 200 | 300 | 400 |
The direct result values of the multi-valleys searching with GFA are shown in Table 6. From Table 6, it can be seen that for all multimodal functions the searching results can reach high precision. It means that the stability of GFA for multi-minima optimization is very good.
Table 6.
Top 5 mean minima value of 500 runs for each benchmark function
| Minima Index | Rosenbrock | Rastrigin | Griewank | Ackley |
|---|---|---|---|---|
| 1 | 1.0029 | 1.0470 | 0.0112 | 3.9736 |
| 2 | 0.1018 | 1.2767 | 0.0083 | 3.524 |
| 3 | 0.0030 | 0.0706 | 0.0006 | 0.0032 |
| 4 | 0.1038 | 1.0313 | 0.0079 | 3.623 |
| 5 | 1.0033 | 1.0200 | 0.0102 | 3.6535 |
Application to gene cluster
GFA could be applied in many science research areas, especially bioinformatics. We used it for gene cluster. The clustering algorithm we used was K-means. The dataset used in this paper was from the experiment of Spellman [15]. And the data which was MIAME compliant [16] could be downloaded from GEO with accession number GDS38. The number of cases was 7,680, of which the 17 missing values were excluded. We computed SMBS correlation coefficient [17] and excluded missing values with Matlab. We excluded cases when the respondent was dropped only on analyses involving variables that had missing values. The data with 7663 cases and 16 samples was divided into 20 parts, as known as clusters. So we distributed the cases into 20 groups. And our mission was that make sure the mean distance was smallest or the number of runs exceeded 1,000. To compare with GFA, 3 methods, GA, SA and Cluster 3.0 [18], were used to test the efficiency of the novel algorithm. K-means was used in Cluster 3.0 with correlation (centered) coefficient. For there four clustering methods, 500 runs were performed. And we got the mean value.
To visualize the result of the gene cluster, we used the free software TreeView [19] from Eisen's Lab. A part results computed by the novel algorithm and other cluster methods of gene cluster were showed in a picture which is Additional file 1. This picture shows the C8.txt and result.txt with Group = 7, which should be opened with excel software.
But there is no single best criterion for obtaining a partition because no precise and workable definition of cluster exists. Clusters can be of any arbitrary shapes and sizes in a multidimensional pattern space [20]. It is impossible to objectively evaluate how good a specific clustering method is without referring to what the clustering will be used for [21]. So we evaluate the result with F test with only mathematical aspect. And P values of all the 16 samples are less than 0.01. The total results of the gene cluster with GFA could be downloaded from our website.
In the Bioinformatics, we could check the biologically meaning. Correlations between cis elements and expression profiles could be established within the novel algorithm cluster result. The cis elements we used were from [22]. It's generated by Li-Hsieh Lin etc [23]. We compare GFA with GA, SA and Cluster 3.0 in C8 and summarized it in Table 7.
Table 7.
cis-regulatory elements correspond each method
| Fkh1 | Fkh2 | Ndd1 | Mcm1 | Ace2 | Swi5 | Mbp1 | Swi4 | Swi6 | |
|---|---|---|---|---|---|---|---|---|---|
| GFA | 1 | 3 | 4 | 2 | 0 | 5 | 2 | 3 | 2 |
| Cluster 3.0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 3 | 2 |
| GA | 3 | 4 | 3 | 2 | 1 | 4 | 4 | 2 | 1 |
| SA | 0 | 3 | 2 | 1 | 1 | 2 | 3 | 4 | 1 |
From Table 7, the efficiency of GFA could be seen. The genes with the same cis-regulatory elements could be clustered better by GFA than Cluster 3.0 and SA. Only in the case of the Ace2 cis element, the GFA with optimized parameters could not outperform the Cluster 3.0. Some efficiency is very obvious, especially Ndd1 and Swi5. But it seams that the efficiency of GA and GFA are the same. The results of this cluster experiment indicate that GFA method does work in gene cluster with finding cis-regulatory element in the same cluster. Other clustering results have the same properties.
Software
The developed algorithm software of single-machine for GFA was implemented with matlab R2008a in the software package GFA, which is a short code file, freely available from our website: http://ccst.jlu.edu.cn/CSBG/GFA. You can implement the multi-machines parallel algorithm for GFA based on the detail in the conclusion section below.
Conclusions
In this paper we propose a generalization searching-optimization algorithm called GFA. This algorithm derives from the SNDM theory, and the efficiency of the algorithm is shown as above. We can summarize them into three parts:
In the form of one independent variable, the GFA will converge with probability 1. It gives us a balance level between time and efficiency. If you want to find exactly extreme value of the mass function, you can disperse more dusts to avoid the condition of the p (see mathematical proof). If you want to find the extreme value fast, you can define a small number of initial dusts.
GFA can find all needed the peaks of the solution. No matter the number of initial dusts is big or small.
The running-time of GFA is very short. The reasons are the strategy of the division and the rules of motion described as above. The space of solution is cut into small groups. It's the decomposition of any complex problem. Even more, it will support us a feasibility of parallel computing. In this view, this algorithm is similar to the parallel genetic algorithm [24]. But this mechanism of GFA can be faster than GA's. We could use a large number of idle computers to calculate a complex problem, and the running-time is inversely proportional to the number of the computers.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MZ designed the algorithms, carried out the experiments and drafted the manuscript. CGZ and GXL conceived and coordinated the research, participated in the design of the experiments and carried out parts of the experiments. YCL and helped to draft the manuscript. All authors read and approved the final manuscript.
Supplementary Material
A picture in which is a graph of a part of clusters with the real gene expression dataset GDS38. In the picture, there are four graphs which were computed by GFA(a), Cluster 3.0(b), GA(c) and SA(d) with the K-means clustering algorithm all. They were corresponded the same cluster in the 20 ones. Red represents positive, green represents negative, black represents zero and grey represents missing values.
Contributor Information
Ming Zheng, Email: zheng_mth@163.com.
Gui-xia Liu, Email: liugx@jlu.edu.cn.
Chun-guang Zhou, Email: cgzhou@jlu.edu.cn.
Yan-chun Liang, Email: ycliang@jlu.edu.cn.
Yan Wang, Email: wy6868@hotmail.com.
Acknowledgements
This work is supported by the NSFC (60873146, 60973092, 60903097); the National HighTech R&D Program of China (863) (2009AA02Z307); the Ph.D. Programs Foundation of Ministry of Education of China (20090061120094); the project of 200810026 support by Jilin University, "211" and "985" project of Jilin University.
References
- Fang-xiang Wu. Genetic weighted k-means algorithm for clustering large-scale gene expression data. BMC Bioinformatics. 2008;9(Suppl 6):S12. doi: 10.1186/1471-2105-9-S6-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James T, Shuba G. Correction: genetic algorithm learning as a robust approach to RNA editing site site prediction. BMC Bioinformatics. 2006;7:406. doi: 10.1186/1471-2105-7-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui J, Hua Y, Fengzhu S. Searching for interpretable rules for disease mutations: a simulated annealing bump hunting strategy. BMC Bioinformatics. 2006;7:417. doi: 10.1186/1471-2105-7-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael M, Michael S, Gisbert S. Optimized Particle Swarm Optimization (OPSO) and its application to artificial neural network training. BMC Bioinformatics. 2006;7:125. doi: 10.1186/1471-2105-7-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pier-Luigi L, Santo M, Francesco P. Discovery of cancer vaccination protocols with a genetic algorithm driving an agent based simulator. BMC Bioinformatics. 2006;7:352. doi: 10.1186/1471-2105-7-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph G. Convergence properties of canonical genetic algorithms. IEEE Trans Neural Networks. 1994;5(1):96–101. doi: 10.1109/72.265964. [DOI] [PubMed] [Google Scholar]
- Geman S, Gemana D. Stochastic relaxation, Gibbs distributions and the Bayesian restoration of images. IEEE Trans. on Pattern Analysis and Machine Intelligence, PAMI. 1984. pp. 721–741. [DOI] [PubMed]
- Safronov VS. Evolution of the Protoplanetary Cloud and Formation of the Earth and the Planets. Jerusalem: Israel Program for Scientific Translations; 1972. [Google Scholar]
- Genetic Algorithm and Direct Search Toolbox User's Guide, copyright by the MathWorks, Inc. http://www.mathworks.co.uk/access/helpdesk/help/pdf_doc/gads/gads_tb.pdf
- Lohr SL, Rao JNK. Jackknife estimation of mean squared error of small area predictors in nonlinear mixed models. Biometrika. 2009;2:457. doi: 10.1093/biomet/asp003. [DOI] [Google Scholar]
- Petros EM, Philippe C. An EWMA chart for monitoring the process standard deviation when parameters are estimated. Computational statistics & data analysis. 2009;5:2653–2664. [Google Scholar]
- Cody WJ. Rational Chebyshev Approximations for the Error Function. Mathematics of Computation. 1969;23:631–637. [Google Scholar]
- Wan X, Karniadakis GE. A sharp error estimate for the fast Gauss transform. Journal of computational physics. 2006;11:7–12. doi: 10.1016/j.jcp.2006.04.016. [DOI] [Google Scholar]
- The website of Matlab tools for GA and SA. http://www.mathworks.com/products/gads/
- Paul TS, Gavin S, Michael QZ. etc. Comprehensive Identification of Cell Cycle-regulated Genes of the Yeast Saccharomyces cerevisiae by Microarray Hybridization. Molecular Biology of the Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tim FR, Philippe RS, Paul TS. etc. A simple spreadsheet-based, MIAME-supportive format for microarray data: MAGE-TAB. BMC Bioinformatics. 2006;7:489. doi: 10.1186/1471-2105-7-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Han, Liu Gui-xia, Zhou Chun-guang, etc. Measuring the Similarity of Co-regulated Genes by Integrating Quantity and Tendency of Gene Expression Changing. The 2nd International Conference on Bioinformatics and Biomedical Engineering. Shanghai, China: Bioinformatics and Biomedical Engineering. 2008. pp. 1896–1900.
- The website for Cluster 3.0. http://bonsai.ims.u-tokyo.ac.jp/~mdehoon/software/cluster
- The website for TreeView. http://rana.lbl.gov/downloads/TreeView/TreeView_vers_1_60.exe
- Jain AK, Dubes RC. Algorithms for Clustering Data. Prentice Hall, Englewood Cliffs, NJ; 1988. [Google Scholar]
- D'haeseleer P, Liang S, Somogyi R. Genetic network inference: from co-expression clustering to reverse engineering. Bioinformatics. 2000;8:707–726. doi: 10.1093/bioinformatics/16.8.707. [DOI] [PubMed] [Google Scholar]
- The website for Li-Hsieh Lin's cis elements. http://www.biomedcentral.com/content/supplementary/1471-2105-6-258-S1.xls
- Lin Li-Hsieh, Lee Hsiao-Ching, Li Wen-Hsiung. etc. Dynamic modeling of cis-regulatory circuits and gene expression prediction via cross-gene identification. BMC Bioinformatics. 2005;6:258. doi: 10.1186/1471-2105-6-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petty CC, League MR. A theoretical investigation of a parallel genetic algorithm. Proc. Of 3rd Int. Conf. On Genetic Algorithm, Morgan Kaufmanm. 1989. pp. 398–405.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A picture in which is a graph of a part of clusters with the real gene expression dataset GDS38. In the picture, there are four graphs which were computed by GFA(a), Cluster 3.0(b), GA(c) and SA(d) with the K-means clustering algorithm all. They were corresponded the same cluster in the 20 ones. Red represents positive, green represents negative, black represents zero and grey represents missing values.


