Abstract
Background
Bacterial inclusion bodies are submicron protein clusters usually found in recombinant bacteria that have been traditionally considered as undesirable products from protein production processes. However, being fully biocompatible, they have been recently characterized as nanoparticulate inert materials useful as scaffolds for tissue engineering, with potentially wider applicability in biomedicine and material sciences. Current protocols for inclusion body isolation from Escherichia coli usually offer between 95 to 99% of protein recovery, what in practical terms, might imply extensive bacterial cell contamination, not compatible with the use of inclusion bodies in biological interfaces.
Results
Using an appropriate combination of chemical and mechanical cell disruption methods we have established a convenient procedure for the recovery of bacterial inclusion bodies with undetectable levels of viable cell contamination, below 10-1 cfu/ml, keeping the particulate organization of these aggregates regarding size and protein folding features.
Conclusions
The application of the developed protocol allows obtaining bacterial free inclusion bodies suitable for use in mammalian cell cultures and other biological interfaces.
Background
Bacterial inclusion bodies (IBs) are water-insoluble protein aggregates formed in the bacterial cytoplasm (and eventually periplasm) during the overproduction of recombinant proteins, especially those from viral or mammalian origin [1]. The diameter of these insoluble proteinaceous particles range from about 50 nm to 500 nm, depending on the background of the producer strain, harvesting time, culture conditions and recombinant protein [2]. Although IBs have traditionally been described as biologically inert protein clusters, recent insights show that these nanoparticles have not only an important level of molecular organization, but also that they are formed by a considerable extent of functional polypeptides [3-7]. In fact, IBs are pure [8], structurally organized [9], mechanically stable and biocompatible protein deposits [2], formed through stereospecific protein-protein cross-molecular interactions under amyloid-like schemes [9,10]. This turn in the understanding of IB biological nature has prompted to explore potential applications of such aggregates as straightforward bacterial products. One of the major IB applications is their use as particulate catalysts for different bioprocesses when formed by enzymes. It has been successfully proved with different enzyme-based IBs that these aggregates efficiently catalyse bioprocesses, becoming a promising alternative to classical enzyme immobilization [7,11]. In the completely different context of tissue engineering and regenerative medicine, it is widely accepted that the nano- and micro-modification of flat surfaces by different procedures, such as etching, lithography and particle decoration, can not only favour mammalian cell binding but also improve cell proliferation and substrate colonization [12]. In this regard, we have recently explored the performance of IBs as biocompatible particulate materials suitable for engineering surfaces roughness at a micro- and nano-scale level to stimulate, by mechano-transduction events, the growth of cultured mammalian cells [2,13,14].
On the other hand, a wide range of protocols for the purification of cytoplasmic IBs are available, all of them including bacterial lysis and IB washing steps. Bacterial lysis is achieved using either mechanical or non mechanical methods, or a combination of both, while washing steps include, among others, detergent and/or DNase treatments. These protocols have been mainly aimed to obtain IBs suitable for in vitro protein refolding attempts [15]. Since most of them permit the recovery of high amounts of pure IBs (usually representing between 90 and 95% of the total aggregated proteins), the presence of viable bacteria in the final sample is not routinely verified. Residual bacterial contamination may not be a critical issue when using IBs as the starting material for protein refolding procedures [16-18] (Table 1[19-58]). However, for applications in which IBs act as materials in biological interfaces, for instance in tissue engineering [2] or as biocatalysts [11], the presence of living bacteria would be not acceptable.
Table 1.
Bacterial lysis methods for IB purification.
| Lysis method | IB protein | DNase | Detergents | Tested viability | Reference | |
|---|---|---|---|---|---|---|
| NON MECHANICAL LYSIS METHODS | LYSOZYME | LACVP1 | Yes | Yes | No | [19-22] |
| VP1LAC | Yes | Yes | No | [7-9,19-27] | ||
| V2LAC | Yes | Yes | No | [22] | ||
| TSP | Yes | Yes | No | [23] | ||
| VP1GFP | Yes | Yes | No | [7,28-30] | ||
| hDHFR | Yes | Yes | No | [7] | ||
| Aβ42-BFP | Yes | Yes | No | [7] | ||
| β-lactamase | Yes | Yes | No | [31] | ||
| Prochymosin | No | Yes | No | [32] | ||
| HET-s fungal prion | Yes | Yes | No | [33] | ||
| Aβ42-GFP | Yes | Yes | No | [30,34] | ||
| Aβ42-BFP | Yes | Yes | No | [30] | ||
| MalE-Bla and MalE31-Bla | No | No | No | [35] | ||
| MalE-PhoA and MalE31-PhoA | No | No | No | [35] | ||
| NON-IONIC DETERGENTS | CBDclosN-SAA | No | Yes | No | [36] | |
| SAA-6HisC | No | Yes | No | [36] | ||
| Maltodextrin phosphorylase | No | Yes | No | [37] | ||
| CBDclosSabA | No | Yes | No | [38] | ||
| MECHANICAL LYSIS METHODS | HOMOGENIZER | rhBMP-2 | No | Yes | No | [17,39] |
| G-CSF | No | No | No | [40] | ||
| rhMCSF | No | No | No | [41] | ||
| rHEWL | No | Yes | No | [42] | ||
| GFP | No | No | No | [43] | ||
| FRENCH PRESS | EGD | No | No | No | [44] | |
| TvDAO | No | Yes | No | [45,46] | ||
| NS3 protein | No | Yes | No | [47] | ||
| SONICATION | IFN-α | No | No | No | [48] | |
| β-galactosidase | No | No | No | [49] | ||
| pGH | No | No | No | [50] | ||
| Pre-β-lactamase | No | No | No | [51] | ||
| Procathepsin B | No | Yes | No | [52] | ||
| COMBINED LYSIS METHODS |
SONICATION + HOMOGENIZER |
Npro fusion proteins | No | Yes | No | [53] |
|
SONICATION + LYSOZYME or LYSOZYME + SONICATION |
Prochymosin | No | Yes | Yes | [54] | |
| Cro-β-gal | Yes | Yes | Yes | [55] | ||
| His-GST-GFP | Yes | No | No | [56] | ||
| Prochymosin B | No | Yes | No | [54] | ||
| CLIPB14 Serine protease | Yes | Yes | No | [57] | ||
|
FRENCH PRESS + LYSOZYME or LYSOZYME + FRENCH PRESS |
PHA synthase | Yes | Yes | No | [58] | |
| Class II PHA synthase | Yes | Yes | No | [58] | ||
The aim of this study has been the development of an IB purification protocol rendering bacterial-free protein particles, which could not compromise the applicability of IBs as biomaterials for biomedical applications.
Results
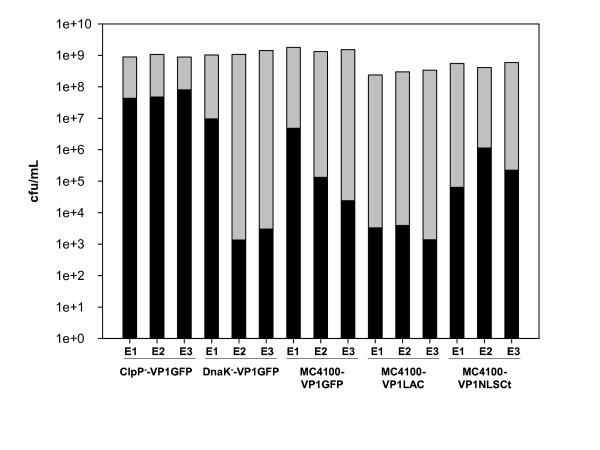
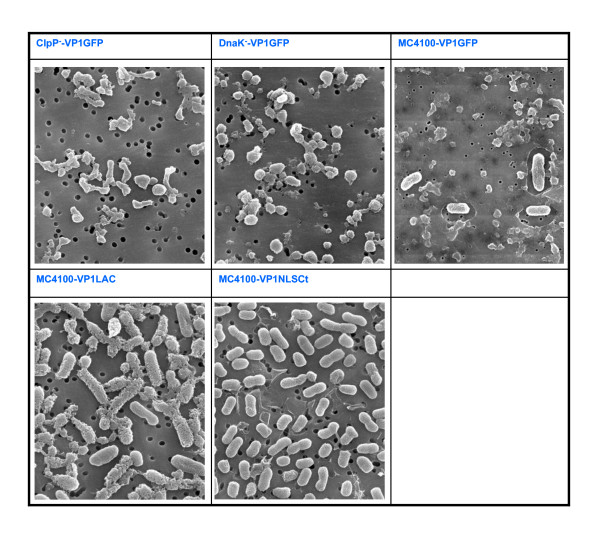
To explore the efficiency of conventional IB purification protocols based on lysozyme treatment combined with repeated detergent washing steps, we have determined the number of viable bacterial cells before and after cell lysis, using different bacterial strains (MC4100, DnaK- and ClpP-) carrying plasmids encoding different recombinant proteins (VP1GFP, VP1LAC and VP1NLSCt) (Figure 1). Our results indicate that the used standard protocol is inefficient concerning the complete removal of viable bacteria (Figure 1). The integrity of bacterial cells upon IB purification was confirmed by scanning electron microscopy (SEM) (Figure 2). Comparing the results obtained with Escherichia coli MC4100 carrying different plasmids, we noticed that, being the initial amount of bacteria around 1·109 cfu/ml in all cases, remaining viable cells after the protocol application ranged from 5·107 to 1·103 cfu/ml (Figure 1). In particular, we noted that the ClpP- strain carrying pTVP1GFP plasmid was the most resistant to cell lysis, since more than 107 cfu/ml remained after IB purification (Figure 1). Intriguingly, replicas of ClpP-/pTVP1GFP and MC4100/pTVP1LAC cultures gave quite similar viable cell counts, but the lysis of DnaK-/pTVP1GFP, MC4100/pTVP1GFP and MC4100/pTVP1NLSCt showed an important variability (Figure 1).
Figure 1.
Viable cells counts before (grey bars) and after (black bars) cell lysis, using a standard protocol based on lysozyme and detergent treatment. E1, E2 and E3 correspond to three different replicas.
Figure 2.
Scanning Electron Microscopy (SEM) images of MC4100, DnaK- and ClpP- overexpressing VP1GFP (top) and of MC4100 overexpressing VP1LAC and VP1NLSCt (bottom).
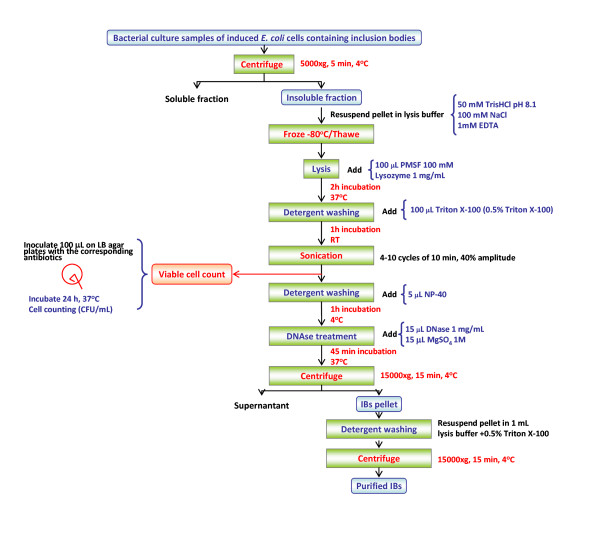
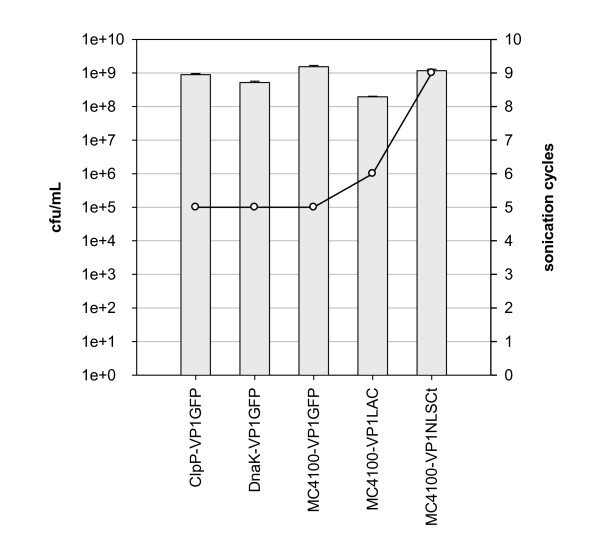
Because of the poor performance of the standard protocol cell lysis based on lysozyme, we decided to explore the effectiveness of other cell disruption protocols (Table 1). Viable bacteria were still observed after the application of lysis methods such as the French Press and freeze-thawing (data not shown). Specifically, we determined the viable cell concentration after using the French Press up to 7 rounds at 2,000 psi, observing a non significant decrease in the viable cell counts. Additionally, the effectiveness of freeze-thawing rounds was also not relevant. Thus, neither the physical nor the chemical methods tested were effective enough to obtain IBs free from bacterial contamination. Therefore, as the bacterial lysis was a bottleneck of the whole IB purification process, we decided to develop an improved protocol by combining both sonication and lysozyme treatment. After testing several combinations of these procedures and determining cell counts at the end of each process, the best protocol (Figure 3) combined both physical and chemical lysis methods with some washing steps and a DNase treatment (Figure 3). After the sonication step (Figure 3), no viable bacteria were observed in the sample containing purified IBs, regardless of the strain and plasmid used. However, the number of sonication cycles needed to eliminate all viable bacteria clearly depended on the particular protein encoded in the plasmid (Figure 4). Full lysis of Escherichia coli strains overproducing VP1GFP needed 5 sonication rounds of 10 min at 40% amplitude under 0.5 s cycles, independently of the genetic background (Figure 4). However, strains overproducing VP1LAC or VP1NLSCt proteins required 6 and 9 disruption cycles, respectively (Figure 4).
Figure 3.
IB purification protocol: lysozyme-detergent, sonication and repeated detergent washing treatment.
Figure 4.
Viable cells counts in initial bacterial cultures (grey bars) and sonication cycles needed to eliminate all viable bacteria (white circles).
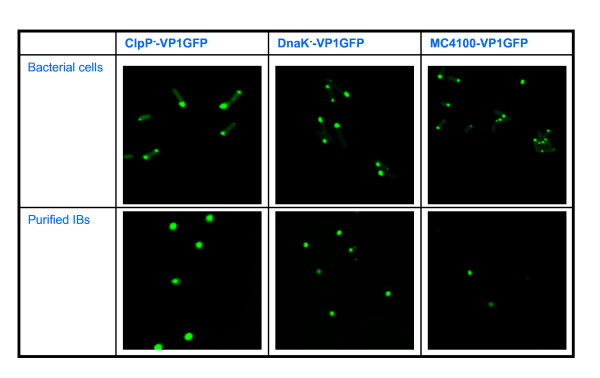
Since the IB-forming VP1GFP protein, encoded in all the plasmids used here, is a suitable model protein to easily determine functionality, IB architecture and mechanical stability [2,28], we evaluated the degree of IB purity as well as the functionality of the embedded protein, after IB isolation from MC4100/pTVP1GFP, DnaK-/pTVP1GFP and ClpP-/pTVP1GFP cells, by confocal microscopy. The obtained images confirmed that isolated IBs were still fluorescent and their morphology fully preserved. This demonstrates that the developed protocol does not significantly alter the final protein quality of highly pure IBs (Figure 5).
Figure 5.
Confocal microscopy images of ClpP-, DnaK- and MC4100 cells overproducing VP1GFP (top). Confocal microscopy images of IBs purified from these strains (bottom).
Discussion
The presence of bacteria in purified IB samples can be a major drawback when using these nanoparticles for biomedical and industrial applications. Although many IB purification protocols have been developed, their effectiveness regarding residual cell viability had not been tested. In this study we have explored different IB purification methods, focusing our attention on the lysis step, which seems to be decisive to obtain bacteria-free samples. The obtained results clearly show that IB purification methods based on lysozyme treatment, French press or freezing-thawing cycles are not effective concerning complete bacterial cell lysis, while the combination of both sonication and lysozyme treatments was the most effective option (Figure 3). Even though these methods have already been combined in different protocols, cell lysis efficiency remained unproved. Menzella and co-workers and Schrodel and collaborators used 5 min of sonication in order to obtain pure IBs [54,56]. However, our data clearly prove that at least 5 sonication cycles of 10 min at 40% amplitude under 0.5 s cycles are needed to reach a sample completely free from contaminating bacteria. Furthermore, the obtained results indicated that sonication cycles, sonication time but also lysozyme concentration must be determined for each specific protein (Figure 4).
As mentioned before, our results showed a surprising variability among bacterial strains overproducing different recombinant proteins. Villa and collaborators have recently described that membrane lipids are dramatically influenced by the stress resulting from recombinant protein production [59]. Therefore, as the membrane protein composition and permeability in recombinant bacteria can be influenced by the specific produced protein [59,60], the observed variability regarding lysis efficiency could be accounted by different features of the recombinant polypeptide, that dissimilarly causes stress effects on the host cell.
Conclusion
Results presented here prove that the existing IB purification protocols may be not appropriate when those aggregates have to be used for both catalysis and biomedical purposes, due to residual but significant levels of metabolically active bacterial cells. In this context, a novel protocol developed in this study, which combines both sonication-lysozyme treatment with DNase and detergent washing steps, has proved to be highly efficient regarding cell lysis and useful to obtain preparations of cell-free IBs.
Materials and methods
Strains and plasmids
The Escherichia coli strains used in this work were MC4100 (araD139 Δ(argF-lac) U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR, StrepR) [61] and their derivatives JGT19 (clpP::cat StrepR) and JGT20 (dnak756 thr::Tn10, StrepR, TcR) [62]. The strain MC4100 was transformed with three different plasmids: pTVP1GFP, pTVP1LAC or pTVP1NLSCt encoding engineered versions of GFP and β-galactosidase [7] respectively. The three proteins were fused to the VP1 capsid protein of foot-and-mouth disease virus that dramatically reduces the solubility of the whole fusion, resulting in its aggregation as IBs [22]. JGT10 and JGT20 were only transformed with pTVP1GFP.
Culture conditions
Bacterial strains were cultured in shake flask cultures at 37°C and 250 rpm in LB rich medium [61] plus 100 μg/ml ampicillin for plasmid maintenance. Recombinant gene expression was induced when the optical density at 550 nm reached 0.5, by adding IPTG to 1 mM. Cell samples were taken at 3 h after induction of gene expression and were processed for bacterial counts, IB sampling and purification and microscopy analyses. Data for further analysis were obtained from three independent experiments.
Bacterial counts
The concentration of colony forming units (cfu/ml) was determined on LB plates with the corresponding antibiotics. After an appropriate dilution in Ringer 1/4, samples were inoculated on LB plates and incubated at 37°C o/n. Cell counting was always performed in triplicate.
IBs sampling and purification
Culture samples of 20 ml were taken 3 h after induction and IBs were purified by using two different purification protocols as follows.
Lysozyme and repeated detergent washing treatment: cells were harvested by centrifugation at 15,000 g at 4°C for 15 min and resuspended in 400 μl of lysis buffer (50 mM TrisHCl (pH 8.1), 100 mM NaCl and 1 mM EDTA,) and kept at -80°C o/n. After thawing, phenylmethanesulphonylfluoride (PMSF) (2.8 μl, 100 mM) and lysozyme (11.2 μl, 10 mg/ml) were added. After 45 min of incubation at 37°C, 4 μl of Nonidet P40 (NP-40) were added and the mixture incubated at 4°C for 1 h. Then, 12 μl of DNase I (from a 1 mg/ml stock) and 12 μl of 1 M MgSO4 were added and the resulting mixture was further incubated at 37°C for 45 min. Protein aggregates were separated by centrifugation at 15,000 g for 15 min at 4°C. Finally, IBs were washed once with 1 ml of the same lysis buffer containing 0.5% Triton X-100. After a final centrifugation at 15,000 g for 15 min at 4°C, pellets were stored at -80°C until analysis. All incubations were done under gentle agitation.
Lysozyme-detergent, sonication and repeated detergent washing treatment: samples of bacterial cultures (20 ml) were centrifuged at 4°C at 5,000 g for 5 min and resuspended in lysis buffer (20 ml, 50 mM TrisHCl (pH 8.1), 100 mM NaCl, and 1 mM EDTA) and frozen at -80°C o/n. After thawing, phenylmethanesulphonylfluoride (PMSF) (100 μl, 100 mM) and 1 mg/ml lysozyme (400 μl, 50 mg/ml) were added. After 2 h of incubation at 37°C, 100 μl of Triton X-100 were added (0.5% Triton X-100) and incubated at room temperature for 1 h. Then, the mixture was ice-jacketed, and sonicated between 4 and 10 cycles of 10 min at 40% amplitude under 0.5 s cycles. After sonication, an aliquot of 100 μl of the suspensions were inoculated on LB plates with the corresponding antibiotics and incubated at 37°C o/n. After that, 5 μl of Nonidet P40 (NP-40) were added to the rest of the suspension, and samples incubated at 4°C for 1 h. Then, DNA was removed with DNase (15 μl, 1 mg/ml) and MgSO4 (15 μl, 1 M) for 45 min at 37°C. Finally, samples were centrifuged at 4°C at 15,000 g for 15 min, and the pellet containing pure IBs was washed once with 1 ml of lysis buffer containing Triton X-100 (0.5%). After a final centrifugation at 15,000 g for 15 min at 4°C, pellets were stored at -80°C until analysis. All incubations were done under agitation.
Microscopy analyses of bacteria and IBs
Fluorescence microscopy
At 3 h post-induction, VP1GFP-producing cells were fixed with 0.1% formaldehyde in phosphate buffered saline (PBS) and purified IBs were also resuspended in PBS and stored at 4°C until observed. Samples of bacterial cells or IBs were placed on a glass slide, fixed with a slide cover and observed with a Leica TCS SP2 AOBS confocal fluorescence microscope (Leica Microsystems Heidelberg GmbH, Mannheim, Germany) using a Plan-Apochromat objective (zoom 4 or 8; 1024 × 1024 pixels) and optical lens magnification (63×, NA 1.4 oil). Photomicrographs were obtained after excitation at 488 nm and at emission wavelengths between 500 and 600 nm.
Scanning Electron Microscopy
Bacterial samples and purified IBs were retained on a nuclepore membrane (Nuclepore Polycarbonate Track-etched Membrane, 0.2 μm pore size, Whatman Ltd.) and fixed with 2.5% phosphate buffered glutaraldehyde (Na2HPO4 0.9 M, Na2H2PO4 0.06 M, pH 8.0) for 1 h at 4°C. After that, the samples were dehydrated with increasing concentrations of ethanol in water (30, 50, 70, 90 and 100%) by consecutive 5 min washing steps. Ethanol was finally evaporated using the critical point method in a K850 CPD desiccator (Emitech, Ashford, UK). The dried membranes were sputtered with gold using a K550 Sputter Coater (Emitech, Ashford, UK) for observation. Microscopy was performed with a scanning microscope Hitachi S-570 (Hitachi LTD. Tokyo, Japan) using an acceleration between 0.5-30 kV.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
ERC performed most of the experiments and prepared the final data and figures. OCG and JSF purified inclusion bodies and analysed samples by fluorescence microscopy. AV and EGF conceived of the study. EGF directed the work and prepared the manuscript. All authors read and approved the final manuscript.
Contributor Information
Escarlata Rodríguez-Carmona, Email: escarlata.rodriguez@uab.cat.
Olivia Cano-Garrido, Email: olivia.cano@campus.uab.es.
Joaquin Seras-Franzoso, Email: joaquin.seras@gmail.com.
Antonio Villaverde, Email: Antoni.Villaverde@uab.cat.
Elena García-Fruitós, Email: elena.garcia.fruitos@uab.cat.
Acknowledgements
The authors appreciate the financial support through MEC (BIO2007-61194, BFU2010-17450) and AGAUR (2009SGR-108). OCG is a recipient of a scholarship for initiation in research from CIBER-BBN, Spain. JSF is a recipient of a doctoral fellowship from UAB, Spain. We also appreciate the support from The Biomedical Research Networking Centre in Bioengineering, Biomaterials and Nanomedicine (CIBER-BBN, Spain), an initiative funded by the VI National R&D&i Plan 2008-2011, Iniciativa Ingenio 2010, Consolider Program, CIBER Actions and financed by the Instituto de Salud Carlos III with assistance from the European Regional Development Fund. Antonio P. Villaverde has been granted with an ICREA ACADEMIA award (from ICREA, Catalonia, Spain).
References
- Marston FA. The purification of eukaryotic polypeptides synthesized in Escherichia coli. Biochem J. 1986;240:1–12. doi: 10.1042/bj2400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Fruitos E, Rodriguez-Carmona E, Diez-Gil C, Ferraz RM, Vazquez E, Corchero JL. et al. Surface Cell Growth Engineering Assisted by a Novel Bacterial Nanomaterial. Adv Mater. 2009;21:4249. doi: 10.1002/adma.200900283. [DOI] [Google Scholar]
- Ventura S, Villaverde A. Protein quality in bacterial inclusion bodies. Trends Biotechnol. 2006;24:179–185. doi: 10.1016/j.tibtech.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Montalban N, Garcia-Fruitos E, Villaverde A. Recombinant protein solubility-does more mean better? Nat Biotechnol. 2007;25:718–720. doi: 10.1038/nbt0707-718. [DOI] [PubMed] [Google Scholar]
- Peternel S, Grdadolnik J, Gaberc-Porekar V, Komel R. Engineering inclusion bodies for non denaturing extraction of functional proteins. Microb Cell Fact. 2008;7:34. doi: 10.1186/1475-2859-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Alonso M, Gonzalez-Montalban N, Garcia-Fruitos E, Villaverde A. Learning about protein solubility from bacterial inclusion bodies. Microb Cell Fact. 2009;8:4. doi: 10.1186/1475-2859-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Fruitos E, Gonzalez-Montalban N, Morell M, Vera A, Ferraz RM, Aris A. et al. Aggregation as bacterial inclusion bodies does not imply inactivation of enzymes and fluorescent proteins. Microb Cell Fact. 2005;4:27. doi: 10.1186/1475-2859-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrio MM, Corchero JL, Villaverde A. Dynamics of in vivo protein aggregation: building inclusion bodies in recombinant bacteria. FEMS Microbiol Lett. 1998;169:9–15. doi: 10.1016/S0378-1097(98)00444-3. [DOI] [PubMed] [Google Scholar]
- Carrio M, Gonzalez-Montalban N, Vera A, Villaverde A, Ventura S. Amyloid-like properties of bacterial inclusion bodies. J Mol Biol. 2005;347:1025–1037. doi: 10.1016/j.jmb.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Speed MA, Wang DI, King J. Specific aggregation of partially folded polypeptide chains: the molecular basis of inclusion body composition. Nat Biotechnol. 1996;14:1283–1287. doi: 10.1038/nbt1096-1283. [DOI] [PubMed] [Google Scholar]
- Garcia-Fruitos E, Villaverde A. Friendly production of bacterial inclusion bodies. Korean J Chem Eng. 2010;27:385–389. [Google Scholar]
- Dalby MJ. Nanostructured surfaces: cell engineering and cell biology. Nanomedicine-UK. 2009;4:247–248. doi: 10.2217/nnm.09.1. [DOI] [PubMed] [Google Scholar]
- Diez-Gil C, Krabbenborg S, Garcia-Fruitos E, Vazquez E, Rodriguez-Carmona E, Ratera I. et al. The nanoscale properties of bacterial inclusion bodies and their effect on mammalian cell proliferation. Biomaterials. 2010;31:5805–5812. doi: 10.1016/j.biomaterials.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Garcia-Fruitos E, Seras-Franzoso J, Vazquez E, Villaverde A. Tunable geometry of bacterial inclusion bodies as substrate materials for tissue engineering. Nanotechnology. 2010;21:205101. doi: 10.1088/0957-4484/21/20/205101. [DOI] [PubMed] [Google Scholar]
- Vallejo LF, Rinas U. Strategies for the recovery of active proteins through refolding of bacterial inclusion body proteins. Microb Cell Fact. 2004;3:11. doi: 10.1186/1475-2859-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph R, Lilie H. In vitro folding of inclusion body proteins. FASEB J. 1996;10:49–56. [PubMed] [Google Scholar]
- Vallejo LF, Brokelmann M, Marten S, Trappe S, Cabrera-Crespo J, Hoffmann A. et al. Renaturation and purification of bone morphogenetic protein-2 produced as inclusion bodies in high-cell-density cultures of recombinant Escherichia coli. J Biotechnol. 2002;94:185–194. doi: 10.1016/S0168-1656(01)00425-4. [DOI] [PubMed] [Google Scholar]
- Clark ED. Protein refolding for industrial processes. Curr Opin Biotechnol. 2001;12:202–207. doi: 10.1016/S0958-1669(00)00200-7. [DOI] [PubMed] [Google Scholar]
- Carrio MM, Corchero JL, Villaverde A. Proteolytic digestion of bacterial inclusion body proteins during dynamic transition between soluble and insoluble forms. Biochim Biophys Acta. 1999;1434:170–176. doi: 10.1016/s0167-4838(99)00177-6. [DOI] [PubMed] [Google Scholar]
- Carrio MM, Cubarsi R, Villaverde A. Fine architecture of bacterial inclusion bodies. FEBS Lett. 2000;471:7–11. doi: 10.1016/S0014-5793(00)01357-0. [DOI] [PubMed] [Google Scholar]
- Cubarsi R, Carrio MM, Villaverde A. In situ proteolytic digestion of inclusion body polypeptides occurs as a cascade process. Biochem Biophys Res Commun. 2001;282:436–441. doi: 10.1006/bbrc.2001.4583. [DOI] [PubMed] [Google Scholar]
- Corchero JL, Viaplana E, Benito A, Villaverde A. The position of the heterologous domain can influence the solubility and proteolysis of beta-galactosidase fusion proteins in E. coli. J Biotechnol. 1996;48:191–200. doi: 10.1016/0168-1656(96)01508-8. [DOI] [PubMed] [Google Scholar]
- Carrio MM, Villaverde A. Protein aggregation as bacterial inclusion bodies is reversible. FEBS Lett. 2001;489:29–33. doi: 10.1016/S0014-5793(01)02073-7. [DOI] [PubMed] [Google Scholar]
- Carrio MM, Villaverde A. Role of molecular chaperones in inclusion body formation. FEBS Lett. 2003;537:215–221. doi: 10.1016/S0014-5793(03)00126-1. [DOI] [PubMed] [Google Scholar]
- Petersson L, Carrio MM, Vera A, Villaverde A. The impact of dnaKJ overexpression on recombinant protein solubility results from antagonistic effects on the control of protein quality. Biotechnol Lett. 2004;26:595–601. doi: 10.1023/B:BILE.0000021963.31863.a4. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Montalban N, Garcia-Fruitos E, Ventura S, Aris A, Villaverde A. The chaperone DnaK controls the fractioning of functional protein between soluble and insoluble cell fractions in inclusion body-forming cells. Microb Cell Fact. 2006;5:26. doi: 10.1186/1475-2859-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Montalban N, Natalello A, Garcia-Fruitos E, Villaverde A, Doglia SM. In situ protein folding and activation in bacterial inclusion bodies. Biotechnol Bioeng. 2008;100:797–802. doi: 10.1002/bit.21797. [DOI] [PubMed] [Google Scholar]
- Garcia-Fruitos E, Martinez-Alonso M, Gonzalez-Montalban N, Valli M, Mattanovich D, Villaverde A. Divergent genetic control of protein solubility and conformational quality in Escherichia coli. J Mol Biol. 2007;374:195–205. doi: 10.1016/j.jmb.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Martinez-Alonso M, Garcia-Fruitos E, Villaverde A. Yield, solubility and conformational quality of soluble proteins are not simultaneously favored in recombinant Escherichia coli. Biotechnol Bioeng. 2008;101:1353–1358. doi: 10.1002/bit.21996. [DOI] [PubMed] [Google Scholar]
- Morell M, Bravo R, Espargaro A, Sisquella X, Aviles FX, Fernandez-Busquets X. et al. Inclusion bodies: specificity in their aggregation process and amyloid-like structure. Biochim Biophys Acta. 2008;1783:1815–1825. doi: 10.1016/j.bbamcr.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Bowden GA, Paredes AM, Georgiou G. Structure and morphology of protein inclusion bodies in Escherichia coli. Nat Biotechnol. 1991;9:725–730. doi: 10.1038/nbt0891-725. [DOI] [PubMed] [Google Scholar]
- Taylor G, Hoare M, Gray DR, Marston FAO. Size and Density of Protein Inclusion-Bodies. Nat Biotechnol. 1986;4:553–557. doi: 10.1038/nbt0686-553. [DOI] [Google Scholar]
- Sabate R, Espargaro A, Saupe SJ, Ventura S. Characterization of the amyloid bacterial inclusion bodies of the HET-s fungal prion. Microb Cell Fact. 2009;8:56. doi: 10.1186/1475-2859-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espargaro A, Sabate R, Ventura S. Kinetic and thermodynamic stability of bacterial intracellular aggregates. FEBS Lett. 2008;582:3669–3673. doi: 10.1016/j.febslet.2008.09.049. [DOI] [PubMed] [Google Scholar]
- Arie JP, Miot M, Sassoon N, Betton JM. Formation of active inclusion bodies in the periplasm of Escherichia coli. Mol Microbiol. 2006;62:427–437. doi: 10.1111/j.1365-2958.2006.05394.x. [DOI] [PubMed] [Google Scholar]
- Nahalka J, Vikartovska A, Hrabarova E. A crosslinked inclusion body process for sialic acid synthesis. J Biotechnol. 2008;134:146–153. doi: 10.1016/j.jbiotec.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Nahalka J. Physiological aggregation of maltodextrin phosphorylase from Pyrococcus furiosus and its application in a process of batch starch degradation to alpha-D-glucose-1-phosphate. J Ind Microbiol Biotechnol. 2008;35:219–223. doi: 10.1007/s10295-007-0287-4. [DOI] [PubMed] [Google Scholar]
- Nahalka J, Mislovicova D, Kavcova H. Targeting lectin activity into inclusion bodies for the characterisation of glycoproteins. Mol Biosyst. 2009;5:819–821. doi: 10.1039/b900526a. [DOI] [PubMed] [Google Scholar]
- Vallejo LF, Rinas U. Optimized procedure for renaturation of recombinant human bone morphogenetic protein-2 at high protein concentration. Biotechnol Bioeng. 2004;85:601–609. doi: 10.1002/bit.10906. [DOI] [PubMed] [Google Scholar]
- Peternel S, Jevsevar S, Bele M, Gaberc-Porekar V, Menart V. New properties of inclusion bodies with implications for biotechnology. Biotechnol Appl Biochem. 2008;49:239–246. doi: 10.1042/BA20070140. [DOI] [PubMed] [Google Scholar]
- Tran-Moseman A, Schauer N, De Bernardez CE. Renaturation of Escherichia coli-derived recombinant human macrophage colony-stimulating factor. Protein Expr Purif. 1999;16:181–189. doi: 10.1006/prep.1999.1074. [DOI] [PubMed] [Google Scholar]
- Batas B, Schiraldi C, Chaudhuri JB. Inclusion body purification and protein refolding using microfiltration and size exclusion chromatography. J Biotechnol. 1999;68:149–158. doi: 10.1016/S0168-1656(98)00197-7. [DOI] [PubMed] [Google Scholar]
- Novak S, Maver U, Peternel S, Venturini P, Bele M, Gaberscek M. Electrophoretic deposition as a tool for separation of protein inclusion bodies from host bacteria in suspension. Colloid Surface A. 2009;340:155–160. doi: 10.1016/j.colsurfa.2009.03.023. [DOI] [Google Scholar]
- Tokatlidis K, Dhurjati P, Millet J, Beguin P, Aubert JP. High activity of inclusion bodies formed in Escherichia coli overproducing Clostridium thermocellum endoglucanase D. FEBS Lett. 1991;282:205–208. doi: 10.1016/0014-5793(91)80478-L. [DOI] [PubMed] [Google Scholar]
- Nahalka J, Nidetzky B. Fusion to a pull-down domain: a novel approach of producing Trigonopsis variabilis D-amino acid oxidase as insoluble enzyme aggregates. Biotechnol Bioeng. 2007;97:454–461. doi: 10.1002/bit.21244. [DOI] [PubMed] [Google Scholar]
- Nahalka J, Dib I, Nidetzky B. Encapsulation of Trigonopsis variabilis D-amino acid oxidase and fast comparison of the operational stabilities of free and immobilized preparations of the enzyme. Biotechnol Bioeng. 2008;99:251–260. doi: 10.1002/bit.21579. [DOI] [PubMed] [Google Scholar]
- Li M, Poliakov A, Danielson UH, Su Z, Janson JC. Refolding of a recombinant full-length non-structural (NS3) protein from hepatitis C virus by chromatographic procedures. Biotechnol Lett. 2003;25:1729–1734. doi: 10.1023/A:1026050627117. [DOI] [PubMed] [Google Scholar]
- Babu KR, Swaminathan S, Marten S, Khanna N, Rinas U. Production of interferon-alpha in high cell density cultures of recombinant Escherichia coli and its single step purification from refolded inclusion body proteins. Appl Microbiol Biotechnol. 2000;53:655–660. doi: 10.1007/s002530000318. [DOI] [PubMed] [Google Scholar]
- Worrall DM, Goss NH. The formation of biologically active beta-galactosidase inclusion bodies in Escherichia coli. Aust J Biotechnol. 1989;3:28–32. [PubMed] [Google Scholar]
- Baranauskaite L, Sereikaite J, Gedminiene G, Bumeliene Z, Bumelis VA. Refolding of porcine growth hormone from inclusion bodies of Escherichia coli. Biocatal Biotransfor. 2005;23:185–189. doi: 10.1080/10242420500175820. [DOI] [Google Scholar]
- Rinas U, Bailey JE. Overexpression of bacterial hemoglobin causes incorporation of pre-beta-lactamase into cytoplasmic inclusion bodies. Appl Environ Microbiol. 1993;59:561–566. doi: 10.1128/aem.59.2.561-566.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhelj R, Dolinar M, Pungercar J, Turk V. The preparation of catalytically active human cathepsin B from its precursor expressed in Escherichia coli in the form of inclusion bodies. Eur J Biochem. 1995;229:533–539. doi: 10.1111/j.1432-1033.1995.0533k.x. [DOI] [PubMed] [Google Scholar]
- Kaar W, Ahrer K, Durauer A, Greinstetter S, Sprinzl W, Wechner P. et al. Refolding of Npro fusion proteins. Biotechnol Bioeng. 2009;104:774–784. doi: 10.1002/bit.22432. [DOI] [PubMed] [Google Scholar]
- Menzella HG, Gramajo HC, Ceccarelli EA. High recovery of prochymosin from inclusion bodies using controlled air oxidation. Protein Expr Purif. 2002;25:248–255. doi: 10.1016/S1046-5928(02)00006-2. [DOI] [PubMed] [Google Scholar]
- Kuczynska-Wisnik D, Zurawa-Janicka D, Narkiewicz J, Kwiatkowska J, Lipinska B, Laskowska E. Escherichia coli small heat shock proteins IbpA/B enhance activity of enzymes sequestered in inclusion bodies. Acta Biochim Pol. 2004;51:925–931. [PubMed] [Google Scholar]
- Schrodel A, de MA. Characterization of the aggregates formed during recombinant protein expression in bacteria. BMC Biochem. 2005;6:10. doi: 10.1186/1471-2091-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodel A, Volz J, de Marco A. Fusion tags and chaperone co-expression modulate both the solubility and the inclusion body features of the recombinant CLIPB14 serine protease. J Biotechnol. 2005;120:2–10. doi: 10.1016/j.jbiotec.2005.04.028. [DOI] [PubMed] [Google Scholar]
- Rehm BH, Qi Q, Beermann BB, Hinz HJ, Steinbuchel A. Matrix-assisted in vitro refolding of Pseudomonas aeruginosa class II polyhydroxyalkanoate synthase from inclusion bodies produced in recombinant Escherichia coli. Biochem J. 2001;358:263–268. doi: 10.1042/0264-6021:3580263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa R, Lotti M, Gatti-Lafranconi P. Components of the E. coli envelope are affected by and can react to protein over-production in the cytoplasm. Microb Cell Fact. 2009;8:32. doi: 10.1186/1475-2859-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ami D, Natalello A, Schultz T, Gatti-Lafranconi P, Lotti M, Doglia SM. et al. Effects of recombinant protein misfolding and aggregation on bacterial membranes. Biochim Biophys Acta. 2009;1794:263–269. doi: 10.1016/j.bbapap.2008.10.015. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular Cloning, A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY; 1989. [Google Scholar]
- Thomas JG, Baneyx F. Roles of the Escherichia coli small heat shock proteins IbpA and IbpB in thermal stress management: comparison with ClpA, ClpB, and HtpG In vivo. J Bacteriol. 1998;180:5165–5172. doi: 10.1128/jb.180.19.5165-5172.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]