Abstract
Partial-volume effects (PVE) in stroke imaging could hinder proper delineation of normal, ischemic, and at-risk tissues. Cerebral-blood-flow (CBF) and apparent diffusion coefficient (ADC) were measured at high and low resolution (HR = 128 × 128, LR = 64 × 64) in focal ischemia in rats during the acute phase. The data were evaluated for PVE on ischemic tissue classification on a pixel-by-pixel basis and the misclassified pixels were quantified as ischemia progressed. The main drawbacks of high-resolution imaging are reduced temporal resolution and/or signal-to-noise ratio. The high- versus low-resolution scatterplots and histograms of pixels along the normal–abnormal boundaries in the ADC and CBF maps showed marked ischemia-related PVE. By comparison with the homologous regions in the contralateral normal hemisphere, the effect of increased noise and intrinsic tissue heterogeneity due to high resolution could be distinguished from ischemia-related PVE. Degrading the high-resolution (128 × 128) data to a 64 × 64 or 32 × 32 matrix increased the severity of PVE. Zero-filling of low-resolution (64 × 64) data to 128 × 128 also increased PVE. It was concluded that PVE: (1) misclassified substantial pixels along the normal–abnormal boundaries, (2) overestimated abnormal volumes at the expense of mostly “at-risk” and some “normal” tissues, (3) were more severe at the early time points postischemia, and (4) confounded the interpretation of the operationally defined ischemic penumbra.
Keywords: high-resolution imaging, columnar resolution, layer specificity, ADC, CBF, perfusion-diffusion mismatch, arterial spin labeling, penumbra
Magnetic resonance imaging (MRI) has inherently poor signal-to-noise ratio (SNR) and spatial and/or temporal resolution relative to many invasive, histologic, and electrophysiological techniques. The advantages of MRI are its superior ability to image the entire brain noninvasively at a reasonably high combined spatiotemporal resolution and to longitudinally assess anatomy, physiology, and function in a single setting. Improving spatial resolution to minimize partial-volume effect (PVE) is desirable because it yields improved parcellation of anatomic, physiologic, and functional information.
In stroke imaging, PVE could hamper proper delineation of normal, ischemic, and at-risk tissues by blurring the boundaries among different tissue types and tissue viability. Visual delineation of ischemic lesions by manually drawing regions of interest (ROI) on the diffusion- and perfusion-weighted images is a common clinical practice (1) and the presence of PVE could lead to significant errors in identifying ischemic tissue fates (2). In addition, it is conceivable that a substantial number of pixels with mild apparent diffusion coefficient (ADC) or cerebral blood flow (CBF) reduction could arise simply from the physical effect of partial voluming, thereby confounding the interpretation of the operationally defined ischemic penumbra (2). High-resolution imaging could minimize tissue classification errors. Other advantages of high-resolution imaging include finer delineation of anatomic structures and marked increase in pixel density, which increases the statistical power of pixel-by-pixel cluster analysis, and reducing signal loss due to intravoxel dephasing. The drawbacks are longer acquisition time and/or reduced SNR.
Perfusion and diffusion imaging of animal stroke models in the acute phase (3–7) remains limited to low spatial resolution (64 × 64 matrix, 400 × 400 × 2000 μm) due to its high temporal resolution requirements. With the dynamic contrast imaging technique, stroke perfusion imaging requires high temporal resolution to detect the passage of an exogenous contrast agent. Furthermore, only a single measure of the CBF index can be acquired due to contrast-reagent recirculation and potential toxicity. Comparison with the homologous regions in the contralateral normal left hemisphere is commonly made. With the advent of the continuous arterial spin-labeling technique (8–10), there is no recirculation of the magnetically labeled water since its “half life” is on the order of tissue water T1, CBF can be measured quantitatively, and interpretation can be made without reference to the contralateral hemisphere. This approach is well suited for high-resolution stroke imaging where signal averaging is needed.
In this study, the PVE on stroke imaging were systematically analyzed. Quantitative perfusion and diffusion imaging was carried out at both high and low resolution in an experimental rat stroke model during the acute phase. Perfusion imaging was acquired using the two-coil, continuous arterial spin-labeling (ASL) technique. PVE on the classification of ischemic tissue fates were quantitatively evaluated on a pixel-by-pixel basis. Analysis included the high-resolution (128 × 128) data, low-resolution (64 × 64) data, downgraded resolution data by taking the central 64 × 64 or 32 × 32 matrix from the high-resolution (128 × 128) data, and zero-filled low-resolution data to achieve a higher nominal resolution.
METHODS
Animal Preparations
Fourteen male Sprague–Dawley rats (300–350 g, Taconic Farms, NY) were initially anesthetized with chloral hydrate (400 mg/kg, i.p., Sigma, St. Louis, MO). The left femoral artery was catheterized. Permanent focal brain ischemia of the right hemisphere was induced using the intraluminal middle cerebral artery occlusion method (4,5). Rectal temperature was maintained at 36.5–37.5°C throughout. Heart rate and mean arterial blood pressure were measured via a pressure transducer connected to the arterial line and were recorded continuously onto a PC via the Biopac system (Santa Barbara, CA). Respiration rate was derived from the slow modulations on top of the cardiac waveforms. Anesthesia was switched to ~1.25% isoflurane once the animal was in the magnet and during imaging. Two groups of animals were studied. In Group I (n = 6), high-resolution (HR) data sets were acquired at 30, 60, 90, 120, and 180 min postocclusion followed by a low-resolution data set (~200 min). In Group II (n = 8), low-resolution (LR) data sets were acquired at 30, 60, 90, 120, and 180 min postocclusion followed by a HR data set (~200 min). Each block of HR and LR data set took 30 and 7.5 min to acquire, respectively; the time points quoted were at the middle of each data-acquisition period.
Histology
2,3,5-Triphenyltetrazolium chloride (TTC) staining was performed at 24 hr postischemia. Eight 1.5-mm slices corresponding to the MR slices were carefully sectioned coronally and incubated in 2% TTC solution at 37°C for 30 min and fixed in 10% buffered formalin solution. TTC infarct volumes with edema correction were derived as described elsewhere (11).
MR Experiments
MRI was performed on a Bruker 4.7 T/40 cm (Billerica, MA) and a 20 G/cm gradient insert (ID = 12 cm, 120-μs rise time). The animal was placed in an MR-compatible stereotaxic headset and onto an animal holder, which had a built-in neck coil for spin labeling. A surface coil (2.3-cm ID) was placed on top of the rat head for brain imaging. Coil-to-coil interaction was actively decoupled.
ADCav was obtained by averaging three ADC maps with diffusion-sensitive gradients separately applied along the x, y, or z direction. High-resolution data were acquired using four-segment, spin-echo, echo-planar images (EPI), matrix = 128 × 128, FOV = 2.56 cm × 2.56 cm, spectral width = 200 kHz, TR = 2 sec per segment, TE = 37.5 msec, b = 10 and 1270 sec/mm2, Δ = 17.53 msec, δ = 5.6 msec, eight 1.5-mm slices, and 16 averages (8.5 min). Low-resolution data were acquired using identical parameters, except a single-shot EPI and a matrix of 64 × 64 were used (2.5 min).
CBF was measured using the continuous ASL technique (4,9). High-resolution data were acquired using four-segment, gradient-echo EPI, spectral width = 200 kHz, matrix = 128 × 128, FOV = 2.56 × 2.56 cm, eight 1.5-mm slices, TE = 15 msec, and TR = 2 sec per segment. Seventy-six pairs of images (one with and the other without ASL preparation) were obtained for signal averaging with half acquired before and the other half after the ADC measurements (20 min). Low-resolution data were acquired using identical parameters, except a single-shot EPI and a matrix of 64 × 64 were used (5 min).
Data Analysis
Data analysis used code written in Matlab (MathWorks, Natick, MA) and STIMULATE (12) software. In addition to LR data acquired separately, LR data were also generated from the central 64 × 64 or 32 × 32 k-space lines of the HR data. The advantage of using the extracted LR for comparison was that both HR and LR data sets came from the same time points and from the same animals. Data are reported as means ± SD. Statistical comparisons used Student’s t test. PVE of LR was compared to the HR data throughout but the HR data were not implied to be the “gold standard.”
ADC and CBF Maps
ADC maps with intensity in units of square millimeters per second were calculated pixel-by-pixel by using (13) ADC = −ln(S2/S1)/(b2 − b1) where bi = γ2Gi2δ2(Δ − δ/3), where ln is the natural logarithm, S1 and S2 are the signal intensities obtained with b1 (10 sec/mm2) and b2 (1270 sec/mm2), respectively. The b-value is proportional to the gradient strength (G), magnetogyric ratio (γ), duration of each gradient pulse (δ), and the time (Δ) between applications of the two gradient pulses. CBF images (SCBF) with intensity in units of milliters per gram per minute were calculated (4,9) pixel by pixel using SCBF = λ/T1 (Sc − SL)/(SL + (2α − 1)Sc), where SC and SL are signal intensities of the control and labeled images, respectively. λ, the water brain–blood partition coefficient, was 0.9 (14). T1 was 1.5 sec at 4.7 T.α, the arterial spin-labeling efficiency, was 0.75 (4,5).
Iterative Self-Organizing Data Analysis (ISODATA) Lesion Volumes
Lesion volumes were also calculated using the modified ISODATA based on ADC and CBF characteristics as described elsewhere (15). The ISODATA-derived lesion volumes of HR and LR data were compared with TTC.
Defining the ROI for Analysis
A standard-deviation index (κ) of each pixel with respect to its eight neighbors on each two-dimensional image was calculated for the ADC and CBF maps,
| [1] |
Pixels with high standard deviations were localized along the boundaries of different tissue types. ROIs were drawn based on these standard-deviation maps along the normal–abnormal boundaries (2–4 pixels thick) on the HR ADC and CBF maps. ROIs on the ADC and CBF maps were generally not the same. For comparisons, homologous ROIs were obtained on the contralateral normal left hemisphere by symmetrically reflecting the right-hemisphere ROIs along the midline to the left hemisphere.
Scatterplots
HR versus LR scatterplots were evaluated for the ADC and CBF data in the left and right hemispheres from the above-prescribed ROIs. For each LR pixel, there are four HR pixels. A dispersion index (η) about the unity line on the HR versus LR scatterplots was computed pixel by pixel using
| [2] |
where m is the LR pixel number, and ln and hi,n are, respectively, the LR and HR ADC or CBF value. The dispersion indices were calculated for the ADC and CBF data in the left and right hemispheres.
ADC and CBF Histograms
ADC and CBF histograms of the left and right hemispheres at high and low resolution were obtained from the above-prescribed ROIs. Their distributions, means, standard deviations, and full-width at half maximum (FWHM) were evaluated. The tissue-volume ratios of pixels below to above the viability thresholds were quantified for the HR and LR data at each time point postischemia (right hemisphere only). Different ROIs were used at different time points as necessitated by the ischemic evolution. The ADC and CBF viability thresholds, derived previously using TTC correlation (4), were 0.53 × 10−3 mm2/sec and 0.3 mL/g/min, respectively.
PVE on Tissue Fates
Pixel-by-pixel tracking was performed to evaluate the PVE on the fates of different pixels and whether they were misassigned due to partial voluming. The 30- and 180-min time points were analyzed. For this analysis: (i) the “abnormal” pixels were defined as pixels with ADC values below the ADC viability threshold (0.53 × 10−3 mm2/sec (4)) or pixels with CBF values below the CBF viability threshold (0.3 mL/g/min (4)), (ii) the “in-between” (i.e., at-risk) pixels were defined as pixels below two standard deviations of the normal-left-hemisphere ADC mean (0.69 × 10−3 mm2/sec) but above the ADC viability threshold or pixels below 1 SD of the normal-left-hemisphere CBF mean (0.7 mL/g/min) but above the CBF viability threshold, and (iii) the “normal” pixels were defined as pixels with ADC or CBF values greater than the in-between pixels (4).
The abnormal, in-between, and normal pixels within the normal–abnormal boundary ROIs were quantified. The following misclassifications of pixels by PVE at low resolution were analyzed: (1) the total pixels that were misassigned, (2) the in-between pixels that were misassigned to abnormal and normal pixels, and (3) the normal pixels that were misassigned to abnormal and in-between pixels.
PVE of Further Reduced Resolution and of Zero-Filling
ADC histograms were analyzed for four data sets: (i) 128 × 128 true matrix size, (ii) 64 × 64 degraded from 128 × 128 data, (iii) 32 × 32 degraded from 128 × 128 data, and (iv) 64 × 64 degraded from 128 × 128 but then zero-filled to 128 × 128.
RESULTS
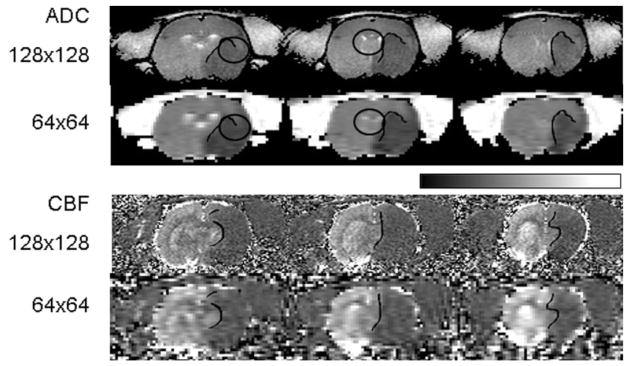
Visual inspection of the HR ADC and CBF maps (Fig. 1) showed clearer delineation of the border zones of different tissue types relative to the LR maps (Group II, 180 min postischemia). This effect was consistent and most evident at the tissue boundaries surrounding the cerebrospinal fluid, corpus callosum, and the ischemic lesion. Examples are emphasized by the ellipsoidal ROIs in Fig. 1. LR data derived from the central half of the HR k-space yielded similar results (Group I, not shown).
FIG. 1.
Representative low- and high-resolution ADC and CBF maps (three consecutive slices). ROIs along the normal–abnormal boundaries on the ADC and CBF maps in the ischemic right hemisphere are shown. Homologous ROIs were also obtained on the contralateral normal left hemisphere by symmetrically reflecting the right-hemisphere ROIs along the midline to the left hemisphere (not shown). ROIs on the ADC and CBF maps are generally not the same. The ellipsoidal ROIs call attention to specific examples of partial voluming between high and low resolution. The grayscale bar indicates ADC ranging from 0 to 1 × 10−3 mm2/sec and CBF ranging from 0 to 3 mL/g/min.
The ISODATA-derived total lesion volumes for HR and LR data at 3 hr were obtained. Although both were similar to TTC infarct volumes (182 ± 30 mm3), the lesion volumes at HR (184 ± 35 mm3) were closer to infarct volumes whereas those at LR (194 ± 33 mm3) consistently overestimated the infarct volumes slightly (pair-wise comparison). The ratio of HR to LR pixel numbers was 3.78 ± 0.2 to 1, below the expected 4-to-1 ratio. A similar ratio was reported when lesion volume was quantified using ROI analysis based on visual delineation or by taking pixels below the viability thresholds as lesions (4).
Figure 2a shows the HR versus LR scatterplots (n = 14, Group I + II, 180 min) from the ROIs along the normal–abnormal boundaries on the ADC and CBF maps. For each LR pixel, there are four HR pixels. In the absence of increased “noise,” intrinsic tissue ADC or CBF heterogeneity and PVE due to ischemia at high resolution, all pixels on the scatterplots would fall onto the unity line. The dispersions about the unity line in the left hemisphere were indicative of reduced SNR and increased intrinsic tissue heterogeneity at high resolution. Dispersions about the unity line in the ischemic right-hemisphere ADC were larger relative to those in the left-hemisphere ADC. The right-hemisphere dispersions were asymmetric about the unity line, and the population of pixels located above the unity line was significantly larger than that under the unity line. These observations are indicative of the PVE due to ischemia. The average “ADC” distance (dispersion index η) to the unity line of the right hemisphere (0.066 ± 0.007 × 10−3 mm2/sec) was significantly larger (32%, P < 10−4) than that of the left hemisphere (0.050 ± 0.008 × 10−3 mm2/sec). Surprisingly, CBF dispersions about the unity lines between the left and right hemisphere were comparable. The average “CBF” distance to the unity line were not statistically difference between the right- and left-hemisphere data (0.25 ± 0.06 versus 0.23 ± 0.03 mL/g/min, P = 0.12). Nonetheless, the population of right-hemisphere CBF pixels was also asymmetrically distributed about the unity line with more pixels located above than under the unity line, indicative of the PVE due to ischemia.
FIG. 2.
(a) High- versus low-resolution scatterplots of the ADC and CBF data at 180 min (n = 14, Group I + II). Right-hemisphere data were obtained from an ROI of the normal–abnormal boundaries (2–4 pixels thick) on the ADC or CBF maps. Data from the homologous ROIs in the left hemisphere were also obtained for comparison. The diagonal dotted lines are lines of unity. (b) Low- and high-resolution ADC and CBF histograms (n = 14). The y-axis scales were independently adjusted such that the displayed heights of the high- and low-resolution left-hemisphere histograms were similar for ease of comparison. The vertical dash–dot lines are the ADC and CBF viability thresholds derived previously (4), and the dotted lines are the ADC value 2 SD below its normal left-hemisphere mean and the CBF value 1 SD below its normal left-hemisphere mean. Note that there was a small fraction (<5%) of pixels with negative CBF values; these arose from measurement noises from pixels with values close to 0 mL/g/min. LR, low resolution; HR, high resolution; LH, left hemisphere; RH, right hemisphere.
ADC and CBF histograms for the HR and LR data at 180 min postischemia are shown in Fig. 2b (n = 14, Group I + II, 180 min). In the left hemisphere, the ADC and CBF means, standard deviations, and FWHM were not statistically different between HR and LR data (P > 0.05), despite reduced SNR and increased tissue ADC and CBF heterogeneity at high resolution. The ADC histogram FWHM of the right hemisphere was significantly wider than that of the left hemisphere. In the ischemic right hemisphere, the FWHM of the HR ADC distribution was slightly wider than that of the LR ADC distribution, and the peak of the HR distribution was located at a higher ADC value. These observations suggested that HR imaging reduced partial voluming, and the effect of parcellating a single LR pixel into four HR pixels resulted in more normal or close to normal pixels, as visualized by the areas under curves (Fig. 2b). To quantify the PVE on potential tissue misclassification, the volume ratio of pixels below to above the ADC viability threshold (0.53 × 10−3 mm2/sec (4)) was derived. This volume ratio for the ADC HR and LR histograms was 1.2 and 2.1, respectively.
Contrary to expectation, the FWHM CBF distribution of the right hemisphere was narrower than that of the left hemisphere and the FWHM of the HR and LR CBF distributions were comparable (Fig. 2b). Nonetheless, the peak of the CBF distribution at high resolution was skewed toward a higher CBF value relative to that at low resolution, indicative of the presence of ischemia-related PVE. The volume ratios of pixels below to above the CBF viability threshold (0.3 mL/g/min (4)) were 0.22 and 0.53 for the HR and LR histogram, respectively.
The temporal characteristics of the PVE showed that both right-hemisphere ADC and CBF peaks between HR and LR data were more separated at early time points (Fig. 3, Group I only, n = 6). The right-hemisphere ADC and CBF distributions shifted to lower values as ischemia progressed.
FIG. 3.
Temporal evolution of low- and high-resolution (a) ADC and (b) CBF histograms of representative time points postischemia (Group I, n = 6). Different ROIs were used at different time points as necessitated by the ischemic evolution. The vertical dash–dot lines are the ADC and CBF viability thresholds (4), and the dotted lines are the ADC value 2 SD below its normal left-hemisphere mean and the CBF value 1 SD below its normal left-hemisphere mean. LR, low resolution; HR, high resolution; LH, left hemisphere; RH, right hemisphere.
Pixel-by-pixel analysis was used to track the misclassified pixels and these results are summarized in Table 1 for the 30 min time point. Approximately 30% of the total pixels were misassigned due to PVE, the majority of which were misassigned to abnormal pixels (Table 1, column d). Approximately half of the in-between pixels were misassigned to the abnormal zone (Tabel 1, column e), whereas only 3–15% were misassigned to the normal zone. Approximately 20% of the normal pixels were misassigned to the abnormal zone, whereas about half were misassigned to the in-between zone (Table 1, column f). Similar results were obtained at other time points.
Table 1.
Pixel Fractions within the Normal–Abnormal Boundary ROIs That Were Misassigned Due to Partial-Volume Effect (Group 1, n = 6) at 30 min postischemia
| (a) Abnormal | (b) In-between | (c) Normal | (d) Total to |
(e) In-between to |
(f) Normal, to |
||||
|---|---|---|---|---|---|---|---|---|---|
| Abnormal | Normal + in-between | Abnormal | Normal | Abnormal | In-between | ||||
| ADC | 0.32 | 0.50 | 0.18 | 0.30 | 0.03 | 0.52 | 0.03 | 0.21 | 0.57 |
| CBF | 0.16 | 0.34 | 0.50 | 0.24 | 0.05 | 0.44 | 0.15 | 0.18 | 0.41 |
Note. The two partitions that defined the normal, in-between, and abnormal pixels were, respectively, 0.69 × 10−3 and 0.53 × 10−3 mm2/sec for ADC and 0.3 and 0.7 mL/g/min for CBF (see Methods). There were ≈3000 HR pixels analyzed for ADC and ≈2000 HR pixels for CBF for all six animals. Pixel fractions of columns a–d are with respect to the total pixels, column e is with respect to the number of in-between pixels (b), and column f is with respect to the number of normal pixels (c).
PVE of Further Reduced Resolution and of Zero-Filling (Fig. 4)
FIG. 4.
ADC Histograms were plotted for four data sets: (i) 128 × 128 true matrix size, (ii) 64 × 64 degraded from 128 × 128 data, (iii) 32 × 32 degraded from 128 × 128 data, and (iv) 64 × 64 degraded from 128 × 128 but then zero-filled to 128 × 128. The four histograms displayed on the right with mean of ~0.72 × 10−3 mm2/sec were from the left normal hemisphere, and the four histograms on the left with mean of ~0.4 – 0.5 × 10−3 mm2/sec were from the right ischemic hemisphere. The vertical dash–dot lines are the ADC and CBF viability thresholds (4). All curves were smoothed slightly for clarity of display.
With further reduction in spatial resolution (32 × 32 reconstructed matrix), the right-hemisphere ADC distribution was similar to the 64 × 64 data set and its PVE was more severe than that of the 64 × 64 matrix. The volume ratio of pixels below to above the ADC viability threshold was 6.1, considerably larger than that at the 64 × 64 and 128 × 128 resolutions of 2.1 and 1.2, respectively. With zero-filling of the reconstructed 64 × 64 to 128 × 128, PVE was more severe relative to data without zero-filling, i.e., biased to more core pixels at the expense of at-risk pixels. As controls, left-hemisphere ADC histograms were analyzed and found to have similar shapes across four different resolutions.
DISCUSSION
The major findings of this study were as follows: (1) ADC and CBF standard deviations in the normal left hemisphere were comparable between high and low resolution, despite increased noise and tissue heterogeneity at high resolution, (2) substantial PVE was observed along the normal–abnormal boundaries on the ADC and CBF maps, (3) PVE resulted in overestimation of the abnormal tissue volumes at the expense of at risk and/or normal tissues, and misclassified pixels were quantitatively evaluated on a pixel-by-pixel basis, (4) PVE appeared to be more severe at the early time points postischemia, and (5) further reduction in spatial resolution and zero-filling resulted in more severe PVE.
Drawbacks of High-Resolution Imaging
The main drawbacks of HR imaging are: (1) increased likelihood of movement artifact associated with multiple segmented EPI acquisition, (2) longer acquisition time, and/or (3) reduced signal-to-noise ratio. Movement artifact from multishot EPI is unlikely to be a major issue in anesthetized animals although it could be a problem in awake animal and human studies if its effects were not corrected. The HR data took 4 times longer to acquire, which could lead to some temporal averaging of the ischemic evolution, particularly at the early time points. Herein, a 30-min temporal resolution was targeted for the HR data and the number of signal averages at HR was kept the same as LR. Nonetheless, the SNR at HR was sufficient for the analyses performed. A semi-quantitative comparison showed that SNR at HR was 2–3 times lower than that at LR, instead of the expected factor of 4. The reduced intravoxel dephasing and its effect on signal decays at HR could partially compensate the SNR loss. Remarkably, the standard deviations of the normal ADC and CBF values were not statistically different between HR and LR, likely compensated by the increased pixel density. With the expected improvement in MRI technologies, high spatiotemporal imaging of acute stroke will become routine.
Potential Issues with Analysis Approaches
The precise pixel fractions that were found to be misclassified were dependent on how the abnormal, normal, and in-between pixels were defined. The partitions that defined the abnormal pixels were the ADC and CBF viability thresholds, which were derived based on the TTC correlation reported previously (4). Additional data from different groups of animals following our initial reports (4,5) confirmed these viability thresholds. The partitions that separated the normal from the in-between pixels were 2 SD below the normal ADC mean and 1 SD below the CBF mean because the normal ADC distribution was sharper. Similar use of SD to describe these thresholds has been reported (16). Although not fully validated, these partitions make possible the quantitative evaluation of tissue fate misclassification due to the PVE. Results obtained with slight changes in the partitions did not alter the overall conclusions.
When an ROI of the entire right hemisphere was used in the histogram analysis, the ischemia-induced PVE was masked by the intrinsic tissue ADC and CBF heterogeneity. This is expected since the ischemia-induced PVE at the normal–abnormal boundaries constituted only a small fraction of the total brain tissue in this stroke model. Analysis enclosing only the normal–abnormal borders was, however, justified because these borders are clinically relevant and are the targets for therapeutic intervention. Using an automated segmentation algorithm would allow efficient identification of these border zones.
The use of the central k-space portion of the HR data to derive the LR data for comparison offered the advantage of having both HR and LR data sets derived from the same animals and from identical time points, resulting in more robust comparisons. Comparison with LR data acquired separately required the use of identical readout time and echo time (as employed herein). Both LR data acquired separately and LR data extracted from HR showed similar results at 3 hr where the ischemic lesion had stopped evolving (4), providing further justification to use the extracted LR data for comparison.
PVE analysis could be affected by differences in SNR between LR and HR. Although it is possible to match the SNR of the HR and LR data in principle, it is difficult in practice. The approach taken herein to compare the homologous regions in the normal left hemisphere could readily distinguish ischemia-related PVE from effects of SNR differences as well as intrinsic tissue heterogeneity. PVE analysis could also be affected by differences in number of pixels between LR and HR, as the ROIs drawn contained slightly different number of pixels between LR and HR data. This was addressed by analyzing the 64 × 64 data zero-filled to 128 × 128. The PVE with zero-filling or spatial interpolation (data not shown) from 64 × 64 to 128 × 128 was found to be even more severe. Nonetheless, analysis of the LR data without zero-filling remains appropriate as it is clinically relevant. Together, these results indicated that ischemia-related PVE was genuine, i.e., not an artifact of the choice of ROIs or the inclusion of different pixel number densities.
Origins and Consequences of the PVE
Data supporting the presence of PVE from ischemia were: (1) overestimated lesion volume by low resolution, (2) increased ADC dispersion about the unity line between the right and left hemisphere, (3) wider ADC distribution relative to the LR ADC in the right hemisphere, (4) the peaks of the HR ADC and CBF histograms in the right hemisphere were skewed toward higher values relative to those of the LR histograms. Surprisingly, the CBF distribution of the right hemisphere was narrower than that of the control left hemisphere. The most likely explanation, as supported by the experimental data, is that ischemia makes the CBF distribution more homogeneous, i.e., ischemia causes most pixels to have ~0 mL/g/min. This interpretation is consistent with current and previous observations (4,5) that “core” CBF pixels are more tightly distributed compared to the normal CBF pixels in the left hemisphere. Nonetheless, ischemia-related PVE in the CBF data were present because the peak of the right-hemisphere HR histogram was skewed toward higher CBF relative to that of the LR histogram.
Whether LR overestimated or underestimated lesion volumes depended on the ADC or CBF of the normal, in-between, and abnormal tissues and the partitions that defined these clusters. In principle, bias could occur in either direction. The ADC (or CBF) of a single LR pixel could be approximated by a weighted average of the four HR pixels. Along the normal–abnormal border, the ADC (or CBF) of a single LR pixel was heavily weighted by the low “ischemic” core ADC (or CBF). Experimental data showed that most affected pixels were those with mild ADC or CBF reduction and close to the viability thresholds, as expected. Thus, a single LR pixel classified as “ischemic core” might be parcellated into some normal or near-normal pixels at high resolution. This conclusion is further supported by similar results obtained using ROI, viability threshold, and ISODATA analysis to derive lesion volumes; these analyses demonstrated that PVE at LR overestimated lesion volumes when compared to TTC.
Potential Implications
When the entire right-hemisphere lesion volumes were quantified using ROI, viability threshold, and ISODATA analysis, the volume ratio of HR to LR pixel numbers was 3.7:1 to 3.8:1 instead of the expected 4:1. This volume ratio for the data obtained along the normal–abnormal ADC and CBF boundaries was even smaller, ca. 2:1 (i.e., HR and LR y-axes in Fig. 2b differed by a factor of ~2), consistent with the presence of PVE, which is expected to be more dramatic for tissues having larger borders with other tissue types. The implication is that the PVE is more severe for smaller infarct sizes with a larger surface-to-volume ratio. It is also more severe at earlier time points where the ADC and CBF gradients are likely to be sharper and where ADC and CBF are dynamically evolving.
The PVE also has other implications. In human stroke imaging, the PVE must be considered when deriving gray and white matter viability thresholds, which are different. In the clinical setting, mean-transit-time (MTT) maps have been reported to overestimate the operationally defined ischemic penumbra (17–19). Since MTT has a large dynamic range relative to ADC, it is relatively more susceptible to PVE. The PVE effect is potentially relevant to distinguishing the border between benign oligemia and penumbra as well as borders associated with the perfusion/diffusion mismatch; a sharper distinction between these borders will provide a more accurate delineation of ischemic tissue fates (2). The effect of partial voluming in fMRI could reduce the fMRI sensitivity and/or cause errors in mapping activated structures that are on the order of cortical columns (20–24). PVE is well known in spectroscopic imaging where the voxel size is considerably larger. In quantifying metabolite differences between gray and white matters, subvoxel correction of the gray and white matter volume fraction has been applied (25). Finally, penumbral tissue classification could be confounded by whether some pixels showing mild ADC and CBF reduction or MTT increase are simply the physical effect of partial voluming between normal and core tissues. This effect remains to be evaluated and is currently under investigation.
CONCLUSIONS
This study presents a quantitative and analytical evaluation of the PVE on the delineation of ischemic brain injury. The PVE has dramatic impact on the accuracy of tissue-volume classification of different ischemic tissue viability. Specifically, it overestimates the ischemic lesion volume at the expense of mostly at risk and some normal tissues. Marked reduction in PVE was achieved by reducing the voxel size by a factor of 4. It is suggested that a similar increase in spatial resolution in human stroke imaging is feasible with some time penalty. With the expected improvement in MRI technologies, high spatiotemporal resolution imaging of ischemic brain injury will be routine. These findings have important implications ranging from stroke to functional and spectroscopic imaging.
Acknowledgments
American Heart Association; Grant number: SDG-0430020N; Grant sponsor: NIH; Grant number: NINDS, R01-NS45879.
The authors thank Drs. Christopher H. Sotak of Worcester Polytechnic Institute for helpful discussions, Sheng-Kwei (Victor) Song of Washington University, and Essa Yacoub of University of Minnesota for critical comments on the manuscript.
References
- 1.Arenillas JF, Rovira A, Molina CA, Grive E, Montaner J, Alvarez-Sabin J. Prediction of early neurological deterioration using diffusion- and perfusion-weighted imaging in hyperacute middle cerebral artery ischemic stroke. Stroke. 2002;33:2197–2203. doi: 10.1161/01.str.0000027861.75884.df. [DOI] [PubMed] [Google Scholar]
- 2.Kidwell CS, Alger JR, Saver JL. Beyond mismatch: evolving paradigms in imaging the ischemic penumbra with multimodal magnetic resonance imaging. Stroke. 2003;34:2729–2735. doi: 10.1161/01.STR.0000097608.38779.CC. [DOI] [PubMed] [Google Scholar]
- 3.Li F, Silva MD, Sotak CH, Fisher M. Temporal evolution of ischemic injury evaluated with diffusion-, perfusion-, and T2-weighted MRI. Neurology. 2000;54:689–696. doi: 10.1212/wnl.54.3.689. [DOI] [PubMed] [Google Scholar]
- 4.Shen Q, Meng X, Fisher M, Sotak CH, Duong TQ. Pixel-by-pixel spatiotemporal progression of focal ischemia derived using quantitative perfusion and diffusion imaging. J Cereb Blood Flow Metab. 2003;23:1479–1488. doi: 10.1097/01.WCB.0000100064.36077.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen Q, Fisher M, Sotak CH, Duong TQ. Effect of reperfusion on ADC and CBF pixel-by-pixel dynamics in stroke: characterizing tissue fates using quantitative diffusion and perfusion imaging. J Cereb Blood Flow Metab. 2004;24:280–290. doi: 10.1097/01.WCB.0000110048.43905.E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoehn-Berlage M, Norris DG, Kohno K, Mies G, Leibfritz D, Hossmann K-A. Evolution of regional changes in apparent diffusion coefficient during focal ischemia of rat brain: the relationship of quantitative diffusion NMR imaging to reduction in cerebral blood flow and metabolic disturbances. J Cereb Blood Flow Metab. 1995;15:1002–1011. doi: 10.1038/jcbfm.1995.126. [DOI] [PubMed] [Google Scholar]
- 7.Kohno K, Hoehn-Berlage M, Mies G, Back T, Hossmann KA. Relationship between diffusion-weighted MR images, cerebral blood flow, and energy state in experimental brain infarction. Magn Reson Imag. 1995;13:73–80. doi: 10.1016/0730-725x(94)00080-m. [DOI] [PubMed] [Google Scholar]
- 8.Silva AC, Zhang W, Williams DS, Koretsky AP. Estimation of water extraction fractions in rat brain using magnetic resonance measurement of perfusion with arterial spin labeling. Magn Reson Med. 1997;37:58–68. doi: 10.1002/mrm.1910370110. [DOI] [PubMed] [Google Scholar]
- 9.Duong TQ, Silva AC, Lee S-P, Kim S-G. Functional MRI of calcium-dependent synaptic activity: cross correlation with CBF and BOLD measurements. Magn Reson Med. 2000;43:383–392. doi: 10.1002/(sici)1522-2594(200003)43:3<383::aid-mrm10>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 10.Calamante F, Thomas DL, Pell GS, Wiersma J, Turner R. Measuring cerebral blood flow using magnetic resonance imaging techniques. J Cereb Blood Flow Metab. 1999;19:701–735. doi: 10.1097/00004647-199907000-00001. [DOI] [PubMed] [Google Scholar]
- 11.Tatlisumak T, Carano RA, Takano K, Opgenorth T, Sotak CH, Fisher M. A novel endothelin antagonist, A-127722, attenuates ischemic lesion size in rats with temporal middle cerebral artery occlusion: a diffusion and perfusion MRI study. Stroke. 1998;29:850–858. doi: 10.1161/01.str.29.4.850. [DOI] [PubMed] [Google Scholar]
- 12.Strupp JP. Stimulate: A GUI based fMRI analysis software package. NeuroImage. 1996;3:S607. [Google Scholar]
- 13.Stejskal EO, Tanner JE. Spin diffusion measurements: spin echoes in the presence of a time-dependent field gradient. J Chem Phys. 1965;42:288–292. [Google Scholar]
- 14.Herscovitch P, Raichle ME. What is the correct value for the brain-blood partition coefficient for water? J Cereb Blood Flow Metab. 1985;5:65–69. doi: 10.1038/jcbfm.1985.9. [DOI] [PubMed] [Google Scholar]
- 15.Shen Q, Ren H, Bouley J, Fisher M, Duong TQ. Dynamic tracking of acute ischemic tissue fates using improved unsupervised ISODATA analysis of high-resolution quantitative perfusion and diffusion data. J Cereb Blood Flow Metab. 2004;24:887–897. doi: 10.1097/01.WCB.0000124321.60992.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin W, Lee J-M, Lee YZ, Vo KD, Pilgram T, Hsu CY. Temporal relationship between apparent diffusion coefficient and absolute measurements of cerebral blood flow in acute stroke patients. J Cereb Blood Flow Metab. 2002;34:64–70. doi: 10.1161/01.str.0000048151.28173.0d. [DOI] [PubMed] [Google Scholar]
- 17.Calamante F, Gadian DG, Connelly A. Delay and dispersion effects in dynamic susceptibility contrast MRI: simulations using singular value decomposition. Magn Reson Med. 2000;44:466–473. doi: 10.1002/1522-2594(200009)44:3<466::aid-mrm18>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 18.Calamante F, Gadian DG, Connelly A. Quantification of perfusion using bolus tracking magnetic resonance imaging in stroke: assumptions, limitations, and potential implications for clinical use. Stroke. 2002;33:1146–1151. doi: 10.1161/01.str.0000014208.05597.33. [DOI] [PubMed] [Google Scholar]
- 19.Wu O, Ostergaard L, Koroshetz WJ, Schwamm LH, O’Donnell J, Schaefer PW, Rosen BR, Weisskoff RM, Sorensen AG, Benner T. Tracer arrival timing-insensitive technique for estimating flow in MR perfusion-weighted imaging using singular value decomposition with a block-circulant deconvolution matrix. Magn Reson Med. 2003;50:856–864. doi: 10.1002/mrm.10522. [DOI] [PubMed] [Google Scholar]
- 20.Duong TQ, Kim D-S, Ugurbil K, Kim S-G. Localized blood flow response at sub-millimeter columnar resolution. Proc Natl Acad Sci USA. 2001;98:10904–10909. doi: 10.1073/pnas.191101098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duong TQ, Kim D-S, Ugurbil K, Kim S-G. Spatiotemporal dynamics of the BOLD fMRI signals in cat visual cortex: toward mapping columnar structures using the early negative response. Magn Reson Med. 2000;44:231–242. doi: 10.1002/1522-2594(200008)44:2<231::aid-mrm10>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 22.Menon RS, Ogawa S, Strupp JP, Ugurbil K. Ocular dominance in human V1 demonstrated by functional magnetic resonance imaging. J Neurophysiol. 1997;77:2780–2787. doi: 10.1152/jn.1997.77.5.2780. [DOI] [PubMed] [Google Scholar]
- 23.Cheng K, Waggoner RA, Tanaka K. Human ocular dominance columns as revealed by high-field functional magnetic resonance imaging. Neuron. 2001;32:359–397. doi: 10.1016/s0896-6273(01)00477-9. [DOI] [PubMed] [Google Scholar]
- 24.Kim D-S, Duong TQ, Kim S-G. High-resolution mapping of iso-orientation columns by fMRI. Nat Neurosci. 2000;3:164–169. doi: 10.1038/72109. [DOI] [PubMed] [Google Scholar]
- 25.Hetherington HP, Mason GF, Pan JW, Ponder SL, Vaughan JT, Twieg DB, Pohost GM. Evaluation of cerebral gray and white matter metabolite differences by spectroscopic imaging at 4.1T. Magn Reson Med. 1994;32:565–571. doi: 10.1002/mrm.1910320504. [DOI] [PubMed] [Google Scholar]