Abstract
In the central domain of fission yeast centromeres the kinetochore is assembled upon CENP-ACnp1 nucleosomes. Normally, siRNAs generated from flanking outer repeat transcripts direct histone H3 lysine 9 methyltransferase Clr4 to homologous loci to form heterochromatin. Outer repeats, RNAi and centromeric heterochromatin are required to establish CENP-ACnp1 chromatin. We demonstrate that tethering Clr4 via DNA binding sites at euchromatic loci induces heterochromatin assembly, with or without active RNAi. This synthetic heterochromatin completely substitutes for outer repeats on plasmid-based minichromosomes, promoting de novo CENP-ACnp1 and kinetochore assembly, to allow their mitotic segregation, even with RNAi inactive. Thus, the role of outer repeats in centromere establishment is simply the provision of RNAi substrates to direct heterochromatin formation; H3K9 methylation-dependent heterochromatin is alone sufficient to form functional centromeres.
It is unclear what features define the chromosomal location where histone H3 is replaced by the centromere specific histone H3 variant CENP-A, to allow kinetochore assembly (1). Kinetochores in many organisms are surrounded by heterochromatin (2). In fission yeast (Schizosaccharomyces pombe), heterochromatin formed on the outer repeats (otr, composed of dg and dh elements), flanks the central domain chromatin, in which canonical H3 is replaced by CENP-ACnp1 that promotes kinetochore assembly (3-7). Outer repeat heterochromatin on minichromosomes is necessary to allow de novo establishment of CENP-ACnp1 chromatin and kinetochore protein recruitment (8); it also contributes to centromere function by ensuring robust cohesion between sister-centromeres (9, 10). Heterochromatin is formed by the action of RNAi-directed chromatin modification on non-coding outer repeat transcripts to create methyl-H3K9 binding sites for the chromo-domain proteins Swi6, Chp1, Chp2 and Clr4. Thus, RNAi components, histone deacetylases (HDACs), the Clr4 H3K9 methyltransferase and chromo-domain proteins all contribute to heterochromatin integrity (reviewed (11, 12).
Active RNAi, Clr4 methyltransferase and the Swi6 (HP1) chromo-domain protein are necessary to form heterochromatin on outer repeats and establish CENP-ACnp1 chromatin over the adjacent central domain on newly introduced centromeric DNA plasmids (8). To test if artificially tethering Clr4 to a euchromatic locus can promote heterochromatin assembly, and if this alone is sufficient to establish a functional centromere, the DNA binding domain of the Saccharomyces cerevisiae Gal4 protein (GBD) was fused to (Fig. 1A): the N-terminus of the gene encoding wild-type Clr4 (GBD-clr4), Clr4-Δcd lacking the chromo-domain (GBD-clr4-Δcd) and, Clr4-H410K a catalytically inactive SET-domain mutant (GBD-clr4-Δcd-H410K). These GBD-to-Clr4 fusions are produced, providing the only source of Clr4 protein (fig. S1A) and were tested in combination with the following reporters: three Gal4 binding sites (3xgbs) upstream of ade6+ inserted at the ura4 (Fig. 1A: ura4::3xgbs-ade6+) or arg3 loci, or ten gbs sites upstream of ura4+ (fig. S1B, C).
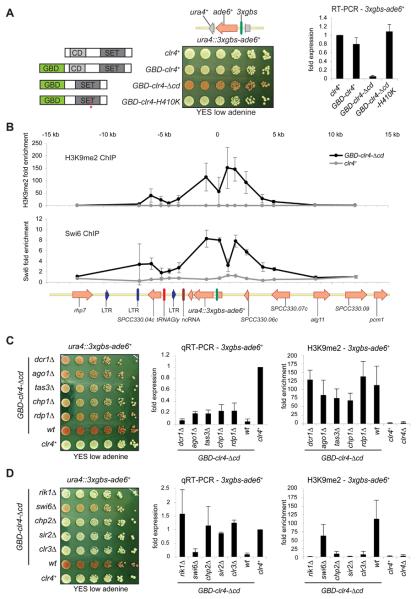
Fig. 1.
Tethered Clr4 silences transcription and forms a 10 kb domain of heterochromatin and requires HDACs, Rik1 and Chp2, but not RNAi. (A) Left: Clr4 fusion proteins used. Gal4 DNA binding, chromo, and SET domains, and H410K (asterisk) mutation, are indicated. Centre: Plating assay on low adenine of cells expressing the indicated Clr4 proteins and containing the 3xgbs-ade6+ reporter inserted at ura4+. ade6+ expressing cells form white colonies; ade− and ade6+ repressed cells form red colonies. Right: qRT-PCR showing 3xgbs-ade6+ transcript levels (error bar is SD, n=3). (B) ChIP analysis with α-H3K9me2 or α-Swi6 antibodies of 25 kb region surrounding ura4::3xgbs-ade6+ in presence of Clr4 or GBD-clr4-Δcd. Genomic features are depicted below.
(C) Plating assay on low adenine of cells with indicated genes deleted expressing Clr4 or GBD-clr4-Δcd with the ura4::3xgbs-ade6+ reporter (left). qRT-PCR for 3xgbs-ade6+ transcript levels (middle) and qChIP for H3K9me2 levels on ura4::3xgbs-ade6+ in the indicated strains (Error bar: SD, n=3). (D) As above with indicated mutants.
Full ade6+ expression results in white colonies whereas silencing causes pink/red colonies to form. The GBD-Clr4-Δcd fusion protein clearly represses the ura4::3xgbs-ade6+ (Fig. 1A) and other reporters (fig. S1B, C) indicating that this silencing is reporter gene and locus independent. It is dependent on Clr4 catalytic activity as no repression occurred with the H410K mutant (Fig. 1A); therefore repression does not result from indirect recruitment of Clr4-associated repressive factors. Although the GBD-Clr4 full-length fusion protein is recruited to the reporter (fig. S2A), it is unable to mediate repression (Fig. 1A). Deletion of the Clr4 chromo-domain affects endogenous heterochromatin and this may release limiting factors for participation in silencing at the tethering site (fig. S2A). Consistent with this, full-length GBD-Clr4 fusion protein can silence the reporter in dcr1Δ cells (fig. S2B). Analyses of anti-H3K9me2 and Swi6 ChIP demonstrate that in cells expressing GBD-Clr4-Δcd protein, H3K9 is dimethylated on, and Swi6 recruited to, a region of approximately 10 kb including and surrounding the ura4::3xgbs-ade6+ reporter (Fig. 1B). Consequently, genes neighbouring the reporter are also repressed in cells expressing GBD-Clr4-Δcd (fig. S3). This synthetic heterochromatin is established de novo, and maintained, independently of RNAi as the reporter remains largely repressed and assembled in H3K9me2 chromatin in cells lacking Dcr1, Ago1, Tas3, Chp1, or Rdp1 (Fig. 1C; fig. S4). Moreover, no ade6+ or ura4+ reporter gene homologous siRNA were detected (fig. S5). Silencing and H3K9me2 levels are maintained in dcr1Δ cells (Fig. 1C); thus, although Ago1 associates with 3xgbs-ade6+ (fig. S5C), the contribution of it and other RNAi components may only be as bound physical entities. In contrast, silencing is highly dependent on chromatin factors Rik1 (Clr4-associated), Chp2 (a Swi6-related protein), and Sir2 and Clr3 HDACs which must act independently of RNAi (Fig. 1E; fig. S4). Synthetic heterochromatin is essentially insensitive to loss of Swi6; this is consistent the reported role for Swi6 in RNAi-dependent silencing, while Chp2 acts with Clr3 to mediate RNAi-independent transcriptional repression (13). In cells producing only mutant histone H3K9R or H3K9A proteins, 3xgbs-ade6+ is expressed, indicating that the Clr4 substrate, H3K9, is critical for silencing by tethered Clr4 activity (fig. S6).
Intact heterochromatin on the outer repeats is necessary for the establishment of CENP-ACnp1 chromatin on adjacent central domain DNA (8). Other features of centromeric outer repeats, such as protein binding sites (14) or non-coding RNA (15), could act in combination with heterochromatin to promote CENP-ACnp1 incorporation. To determine if heterochromatin alone is sufficient, we substituted the outer repeats on a plasmid with three Gal4-binding sites, or no sites, in close proximity to the central domain, generating the plasmids p3xgbs-cc2 and p0xgbs-cc2, respectively (Fig. 2, fig S7). GBD-Clr4-Δcd associates with the plasmid-borne Gal4-sites and H3K9me2 was detected over these and extends into nearby regions suggesting that a 6 kb domain of heterochromatin was formed (Fig. 2A; fig. S7). In support of this, the underlying plasmid-borne marker genes were silenced (fig. S8). Both CENP-ACnp1 and CENP-CCnp3 associated with the central core of the p3xgbs-cc2 plasmid, but only in cells expressing GBD-Clr4-Δcd (Fig. 2B), and CENP-ACnp1 was also detected at the extremities of the 8.5 kb central domain, but not on other regions of the plasmid (fig. S9). Thus, the synthetic heterochromatin domain formed by Clr4 methyltransferase bound to these Gal4-sites is sufficient to promote the assembly of CENP-ACnp1 across, and recruit other kinetochore proteins to, the central domain.
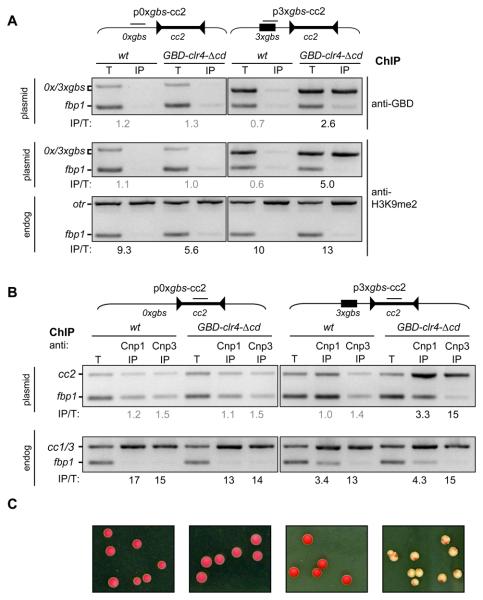
Fig. 2.
Tethered Clr4 promotes CENP-ACnp1 deposition, kinetochore assembly and centromere activity. (A) ChIP with anti-GBD (top) and anti-H3K9me2 (middle and bottom) of wt and GBD-clr4-Δcd cells transformed with p0xgbs-cc2 or p3xgbs-cc2 as indicated. Enrichment values (IP/T) were calculated from the intensity of the 0xgbs or 3xgbs band normalized to the control fbp1 locus. Grey values indicate no enrichment. H3K9me2 levels on endogenous centromeric otr repeats (bottom). (B) ChIP with anti-Cnp1 and anti-Cnp3 of wt and GBD-clr4-Δcd cells transformed with p0xgbs-cc2 or p3xgbs-cc2 as indicated. Enrichment values (IP/T) were calculated from the intensity of the plasmid cc2 band (top) or the endogenous cc1/3 central cores (middle), normalized to the control fbp1 locus (see SOM). (C) Colonies from strains indicated in C. transformed with p0xgbs-cc2 or p3xgbs-cc2 grown on low adenine. Plasmid mitotic stability is determined by colony colour: white/red sectored indicates stable; uniform red indicates complete loss (see SOM).
Only plasmids carrying Gal4-sites formed white/sectored colonies when combined with GBD-Clr4-Δcd (Fig. 2C; see SOM), indicating that a functional centromere, capable of mitotic segregation, had formed. Similar results were obtained with Gal4-sites on the opposite side of cc2 (pcc2-3xgbs) (fig. S9). Both 3xgbs plasmids exhibit ~2 fold greater mitotic stability than pH'-cc2 which contains native outer repeat heterochromatin on a similarly sized functional minichromosome (fig. S9). This indicates that the function of natural outer repeat DNA at a plasmid-borne centromere can be substituted by artificially recruiting Clr4 to DNA. Using this sensitive plasmid-based assay, we conclude that centromeric outer repeats have no hidden unknown features, and apart from its role in directing heterochromatin assembly, the primary outer repeat DNA sequence is dispensable with respect to establishing centromere-kinetochore function.
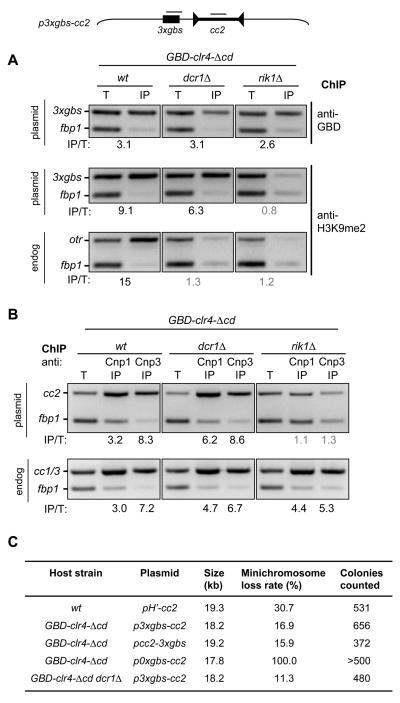
Centromeric outer repeats may just provide a dsRNA substrate for RNAi to direct H3K9 methylation and centromeric heterochromatin formation. To test if synthetic heterochromatin is completely RNAi independent, p3xgbs-cc2 was transformed into wild-type, dcr1Δ and rik1Δ cells expressing GBD-Clr4-Δcd. GBD-Clr4-Δcd was bound to the 3xgbs in all strains (Fig. 3A) whereas H3K9me2 was detected over this region in wild-type and dcr1Δ but not rik1Δ cells. This confirms that the establishment of H3K9me2 modified chromatin by tethered Clr4 occurs independently of RNAi, but depends on Rik1. In support of this, silencing with full-length GBD-Clr4 only occurred upon deletion of Dcr1 (fig. S2B). Anti-CENP-ACnp1 and CENP-CCnp3 ChIP indicated that recruitment of GBD-Clr4-Δcd to the plasmids' Gal4-sites allowed the establishment of CENP-ACnp1 chromatin on, and recruitment of CENP-CCnp3 to, the central core of p3xgbs-cc2 in wild-type and dcr1Δ cells, but not rik1Δ cells (Fig. 3B). The minichromosome with the Gal4-sites exhibited mitotic stability when combined with GBD-Clr4-Δcd in dcr1Δ cells (Fig. 3C; fig. S9). We conclude that the requirement for outer repeats and RNAi in the de novo establishment of functional heterochromatin to promote CENP-ACnp1 and kinetochore assembly, and form mitotically active centromeres, on plasmid-based minichromosomes can be fully substituted by Clr4-tethered synthetic heterochromatin formed adjacent to a central domain.
Fig. 3.
Synthetic heterochromatin allows establishment of functional centromeres in cells lacking RNAi. (A) ChIP with anti-GBD (top) and anti-H3K9me2 (middle and bottom) of wt, dcr1Δ and rik1Δ cells expressing GBD-clr4-Δcd cells transformed with p3xgbs-cc2. Enrichment values (IP/T) calculated as described in SOM. Grey values indicate no enrichment. H3K9me2 levels on endogenous centromeric otr repeats (bottom). (B) ChIP with anti-Cnp1 and anti-Cnp3 on identical cells as in A. Levels on endogenous centromeric cc1/3 sequences are shown (bottom). (C) Loss rate of indicated plasmid based minichromosomes in indicated host strains expressing Clr4 or GBD-clr4-Δcd. pH'-cc2 containing natural outer repeat heterochromatin is included for comparison (3). pcc2-3xgbs has the 3xgbs to the right of cc2 (see fig S9).
Three Gal4-binding sites, which directly recruit Clr4 methyltransferase activity, can replace the normal requirement for at least 2.1 kb of centromeric outer repeat DNA adjacent to a central domain on naïve plasmids to form active centromeres (3, 7, 8). This synthetic heterochromatin is therefore functional in that it generates sufficient sister–centromere cohesion and promotes assembly of CENP-ACnp1 in place of histone H3 to provide a foundation for kinetochore formation. This indicates that no other contribution of the outer repeats is required in terms of their primary DNA sequence in this plasmid-based establishment assay. RNAi components were previously shown in similar assays to be necessary for CENP-ACnp1 chromatin establishment (8). By artificially recruiting Clr4 activity we have bypassed RNAi and rule it out as being directly involved in promoting CENP-ACnp1 deposition. Thus the non-coding outer repeat transcripts themselves, and the resulting siRNA, are not required to form a mitotically functional minichromosomal centromere. It remains to be determined how heterochromatin promotes CENP-ACnp1 incorporation. Recent analyses suggest that the acetylated state of histones is important and HDAC inhibitors rescue CENP-ACnp1 chromatin assembly defects (4, 16). Synthetic heterochromatin placed close to a central domain may provide a favourable chromatin environment to attract key remodelling and modifying factors. Ultimately, similar manipulations as described here may improve the efficiency of human artificial chromosome formation.
Supplementary Material
Acknowledgements
We are grateful to the following colleagues for strains, and reagents: M. Baum, L. Clarke, K. Scott, J. Partridge, J. Ma, S. Grewal, D. Moazed, K. Takahashi, M. Yanagida, S.A. White. This research was supported by: EMBO-LTF (to FS), MRC Strategic Grant (G0301153 to RCA). Wellcome Trust Programme grant (065061/Z to RCA). RCA is a Wellcome Trust Principal Research Fellow.
References
- 1.Allshire RC, Karpen GH. Nat. Rev. Genet. 2008;9:923. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sullivan BA, Karpen GH. Nat. Struct. Mol. Biol. 2004;11:1076. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baum M, Ngan VK, Clarke L. Mol. Biol. Cell. 1994;5:747. doi: 10.1091/mbc.5.7.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayashi T, et al. Cell. 2004;118:715. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Pidoux AL, Richardson W, Allshire RC. J. Cell Biol. 2003;161:295. doi: 10.1083/jcb.200212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi K, Chen ES, Yanagida M. Science. 2000;288:2215. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi K, et al. Mol. Biol. Cell. 1992;3:819. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Folco HD, Pidoux AL, Urano T, Allshire RC. Science. 2008;319:94. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernard P, et al. Science. 2001;294:2539. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 10.Nonaka N, et al. Nat. Cell Biol. 2002;4:89. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- 11.Buhler M, Moazed D. Nat. Struct. Mol. Biol. 2007;14:1041. doi: 10.1038/nsmb1315. [DOI] [PubMed] [Google Scholar]
- 12.Grewal SI, Jia S. Nat. Rev. Genet. 2007;8:35. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 13.Motamedi MR, et al. Mol. Cell. 2008;32:778. doi: 10.1016/j.molcel.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakagawa H, et al. Genes Dev. 2002;16:1766. doi: 10.1101/gad.997702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Volpe TA, et al. Science. 2002;297:1833. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 16.Fujita Y, et al. Dev. Cell. 2007;12:17. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.