Abstract
Meiotic recombination events typically cluster within narrow regions of the genome termed hotspots. A series of recent papers reveals that PRDM9, a C2H2-type zinc-finger protein with histone H3 lysine 4 methyltransferase activity, plays a major role in the specification of hotspots. The zinc fingers that contact DNA in a sequence-dependent manner evolve rapidly and are under positive selection, leading to differences in the location of recombination hotspots as well as hybrid sterility.
Introduction and context
Meiotic recombination is an essential biological process among eukaryotes, forming physical connections between homologous chromosomes required for their orderly segregation during the first meiotic division [1]. In addition to this mechanical role, meiotic recombination creates genetic variation by reshuffling alleles between homologous chromosomes. Meiotic recombination events are not randomly distributed along the chromosomes, but occur at specialized sites, 1-2 kb long, called hotspots [2].
Accumulated evidence suggests that three categories of factors are involved in controlling the hotspots’ location and activity: local DNA sequence, epigenetic mechanisms, and trans-acting loci. For example, human hotspots are enriched for a degenerate 13 base pair (bp) motif, suggesting that underlying DNA sequence is a major determinant in hotspot specification [3]. Genetic factors also control meiotic recombination in the major histocompatibility complex of the mouse genome [4]. In this context, then, it is surprising that despite nearly 99% identity at aligned bases, humans and chimpanzees share almost no recombination hotspots [5,6], suggesting that epigenetic mechanisms are also important in hotspot specification. Consistent with this interpretation, individual chromatids can differ in their ability to recombine according to their parental origin [7] and human imprinted chromosomal regions are enriched in recombination hotspots [8,9]. Furthermore, markers of germline DNA methylation are positively correlated with regional levels of recombination [10], despite the fact that recombination takes place when maternal/paternal differences in DNA methylation are assumed to be erased (reviewed in [11]). The importance of trans-acting factors is best exemplified in mice, in which a region on chromosome 17 controls the genome-wide distribution of meiotic hotspots [12]. Inter-individual variation in the presence/absence of specific hotspots in the human can also be independent of DNA sequence variation, suggesting differences in epigenetic marking and/or trans-acting factors [13].
A flurry of recent studies [14-18] has now placed PRDM9 at center stage in this process, providing a unifying view of meiotic hotspot specification. PRDM9 is a rapidly evolving zinc finger protein with sequence-specific DNA binding and histone methyltransferase activity. These characteristics of PRDM9 neatly wrap genetic, epigenetic, and trans-acting factors known to influence recombination into one intriguing package.
Major recent advances
Several independent groups have obtained compelling evidence that PRDM9 is a major determinant of meiotic recombination hotspots in mammalian species.
Myers et al. [14] demonstrated that the degenerate 13-bp motif associated previously with 40% of human hotspots does not function in chimpanzees. This group was able to predict, bioinformatically, the proteins that are able to bind to the human motif. Using a complex computational algorithm, they narrowed down the list of potential candidates to a single C2H2-type zinc-finger protein: PRDM9. The predicted binding sequence for chimpanzee PRDM9 is completely different from the predicted human binding sequence. The reason for this divergence is that the DNA sequence that encodes the zinc-finger array of PRDM9 constitutes a minisatellite and is predicted to have a high intrinsic mutation rate [15,16]. Consistent with this prediction, they demonstrate that PRDM9 is the most divergent of all human-chimpanzee pairs of orthologous zinc-finger proteins [15,16].
Parvanov et al. [17] identified Prdm9 as the most likely candidate to control the genome-wide distribution of meiotic hotspots in the mouse by narrowing the candidate region of chromosome 17 to 181 kb through meiotic mapping in an inter-subspecific mouse cross. The candidate region contained only four protein-coding genes, of which Prdm9 is the only one expressed exclusively during early meiosis in both males and females. By sequencing exon 12, which encodes the entire zinc-finger repeat domain, in 20 mouse strains and 64 human samples, Parvanov et al. found multiple alleles, again suggesting accelerated evolution of this gene.
Baudat et al. [18] found that two mouse strains that had different recombination activities at two individual hotspots exhibited DNA polymorphisms in the sequence encoding the zinc-finger domain of Prdm9. As in humans, these allelic variants were predicted to recognize different binding motifs. Using southwestern and band-shift assays, this group provided the first direct evidence that PRDM9 variants bind to specific hotspot motifs.
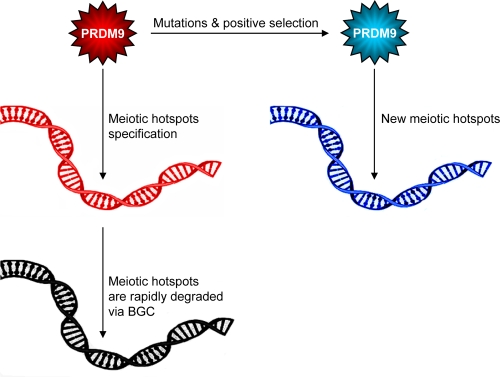
Taken together, these studies suggest a new model that neatly solves the ‘hotspot conversion paradox’ [19] (Figure 1).
Figure 1. The role of PRDM9 in mammalian hotspot specification.
The minisatellite-like region that encodes the zinc-finger array of PRDM9 is highly mutable and the amino acid substitutions in the array are positively and recurrently selected during evolution to change DNA-binding specificity. The DNA motifs bound by PRDM9 are sites at which meiotic recombination occurs. However, as they recombine, these motifs are eroded by biased gene conversion (BGC) and, as a result, they are no longer recognized by PRDM9. Meanwhile, newly arisen PRDM9 alleles are positively selected and create a new family of hotspots, genome-wide.
The rapid evolution of PRDM9 and the associated between-species differences found in sites designated as recombination hotspots may be due to more than simple association. The first indication that mutations in PRDM9 could cause reproductive isolation came more than a year ago with the finding that the mouse Hst1 (Hybrid sterility 1) locus and the Prdm9 gene were one and the same [20] (it is difficult to imagine the joy, sense of achievement, and satisfaction that its discoverer [Jiri Forejt] must have felt in publishing this finding, 35 years after the initial discovery! [21]). So-called ‘hybrid sterility’ genes should be an important component of the pathway by which different species may be created. Recently published surveys of the sequences of PRDM9 zinc fingers from multiple species of mammals [15,16] and diverse metazoans [16] suggest that PRDM9 may have been involved in hybrid sterility in multiple lineages on many occasions.
Future directions
The precise molecular mechanisms of action of PRDM9 remain unknown. It is likely that the histone methyltransferase activity has an important role in this process. For example, studies on two mouse hotspots (Psmb9 on chromosome 17 and Hlx1 on chromosome 1), which we now know are controlled by Prdm9, have shown that the H3K4me3 mark (trimethylation of histone H3 at lysine 4) is specifically enriched in the chromatids that carry the active initiation sites, even before the double-strand breaks (DSBs) occur [22]. Local changes in the chromatin structure may be needed to recruit proteins involved in the initiation of DSBs. However, the current evidence is only correlative and the requirement for the histone methyltransferase activity of Prdm9 will need to be experimentally demonstrated by generating a knock-in mouse in which only the SET domain of the protein is defective, without alteration of the DNA-binding function.
It seems unlikely that PRDM9 is the only protein involved in mammalian hotspot specification. Firstly, the 13-bp motif recognized by PRDM9 was previously associated with the activity of 40% of human hotspots [3]. Secondly, spermatocytes from Prdm9-/- mice still have DSBs detectable [23]. This observation suggests that many meiotic recombination events can occur at sites not bound by Prdm9.
The discovery of the extraordinary evolutionary divergence of the DNA-binding capacity of PRDM9 seems to ignite controversy on its previously reported role as a transcription factor. Spermatocytes isolated from Prdm9-/- mice had dysregulated expression of 125 genes [23], suggesting that Prdm9 acts as a master transcriptional regulator of entry into meiosis. However, the evidence for a direct causal role in transcription regulation is limited [20,23]. Prdm9 encodes several protein isoforms, out of which only the long version contains the zinc-finger array [23]. It is plausible, therefore, that the regulatory role on transcription is played by the shorter Prdm9 isoforms, which contain only the KRAB and the SET domains, possibly via interactions with additional transcriptional factors. On the other hand, these studies shed new light on the previously discovered role of the Prdm9 gene in speciation [20]. Notably, the hybrid sterility phenotype attributed to Prdm9 depends on polymorphisms in the zinc-finger array of this gene and on several independently segregating genes [20]. However, the detailed mechanism by which hybrid sterility occurs (possibly arising from incompatibility between altered binding site-specificity and inability to suppress ‘centromere drive’ [24]) may be unrelated to the mechanism by which PRDM9 is involved in hotspot specification.
The uncovered properties of PRDM9 provide a new framework for interpreting the divergence of the fine-scale genetic maps between closely related species and even among individuals of the same species. Future studies on the roles of PRDM9 are likely to lead to new discoveries in the biology of meiotic recombination and to clinical applications in human fertility.
Acknowledgments
Work in the Sapienza laboratory is supported by grants from the National Institutes of Health (R01HD048730 and R01HD34508).
Abbreviations
- bp
base pair
- DSB
double-strand break
Competing Interests
The authors declare that they have no competing interests.
The electronic version of this article is the complete one and can be found at: http://f1000.com/reports/b/2/37
References
- 1.Roeder GS. Chromosome synapsis and genetic recombination: their roles in meiotic chromosome segregation. Trends Genet. 1990;6:385–9. doi: 10.1016/0168-9525(90)90297-J. [DOI] [PubMed] [Google Scholar]
- 2.Arnheim N, Calabrese P, Tiemann-Boege I. Mammalian meiotic recombination hot spots. Annu Rev Genet. 2007;41:369–99. doi: 10.1146/annurev.genet.41.110306.130301. [DOI] [PubMed] [Google Scholar]
- 3.Myers S, Freeman C, Auton A, Donnelly P, McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet. 2008;40:1124–9. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- 4.Shiroishi T, Sagai T, Hanzawa N, Gotoh H, Moriwaki K. Genetic control of sex-dependent meiotic recombination in the major histocompatibility complex of the mouse. EMBO J. 1991;10:681–6. doi: 10.1002/j.1460-2075.1991.tb07997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ptak SE, Hinds DA, Koehler K, Nickel B, Patil N, Ballinger DG, Przeworski M, Frazer KA, Pääbo S. Fine-scale recombination patterns differ between chimpanzees and humans. Nat Genet. 2005;37:429–34. doi: 10.1038/ng1529. [DOI] [PubMed] [Google Scholar]
- 6.Winckler W, Myers SR, Richter DJ, Onofrio RC, McDonald GJ, Bontrop RE, McVean GA, Gabriel SB, Reich D, Donnelly P, Altshuler D. Comparison of fine-scale recombination rates in humans and chimpanzees. Science. 2005;308:107–11. doi: 10.1126/science.1105322. [DOI] [PubMed] [Google Scholar]; F1000 Factor 3.3 RecommendedEvaluated by Jonathan Pritchard 13 Apr 2005, Karin Schmitt 18 Apr 2005, Tony Long 27 Apr 2005
- 7.Paigen K, Szatkiewicz JP, Sawyer K, Leahy N, Parvanov ED, Ng SH, Graber JH, Broman KW, Petkov PM. The recombinational anatomy of a mouse chromosome. PLoS Genet. 2008;4:e1000119. doi: 10.1371/journal.pgen.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lercher MJ, Hurst LD. Imprinted chromosomal regions of the human genome have unusually high recombination rates. Genetics. 2003;165:1629–32. doi: 10.1093/genetics/165.3.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandovici I, Kassovska-Bratinova S, Vaughan JE, Stewart R, Leppert M, Sapienza C. Human imprinted chromosomal regions are historical hot-spots of recombination. PLoS Genet. 2006;2:e101. doi: 10.1371/journal.pgen.0020101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sigurdsson MI, Smith AV, Bjornsson HT, Jonsson JJ. HapMap methylation-associated SNPs, markers of germline DNA methylation, positively correlate with regional levels of human meiotic recombination. Genome Res. 2009;19:581–9. doi: 10.1101/gr.086181.108. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 3.2 RecommendedEvaluated by Carmen Sapienza 18 Feb 2009, Michael Zhang 15 Jul 2009
- 11.Paigen K, Petkov P. Mammalian recombination hot spots: properties, control and evolution. Nat Rev Genet. 2010;11:221–33. doi: 10.1038/nrg2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grey C, Baudat F, de Massy B. Genome-wide control of the distribution of meiotic recombination. PLoS Biol. 2009;7:e35. doi: 10.1371/journal.pbio.1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neumann R, Jeffreys AJ. Polymorphism in the activity of human crossover hotspots independent of local DNA sequence variation. Hum Mol Genet. 2006;15:1401–11. doi: 10.1093/hmg/ddl063. [DOI] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Molly Przeworski 28 Apr 2006
- 14.Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, Macfie TS, McVean G, Donnelly P. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876–9. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oliver PL, Goodstadt L, Bayes JJ, Birtle Z, Roach KC, Phadnis N, Beatson SA, Lunter G, Malik HS, Ponting CP. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 2009;5:e1000753. doi: 10.1371/journal.pgen.1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Hopi Hoekstra 22 Dec 2009
- 16.Thomas JH, Emerson RO, Shendure J. Extraordinary molecular evolution in the PRDM9 fertility gene. PLoS One. 2009;4:e8505. doi: 10.1371/journal.pone.0008505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parvanov ED, Petkov PM, Paigen K. Prdm9 controls activation of mammalian recombination hotspots. Science. 2010;327:835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–40. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 9.0 ExceptionalEvaluated by Mohamed Noor 09 Feb 2010
- 19.Boulton A, Myers RS, Redfield RJ. The hotspot conversion paradox and the evolution of meiotic recombination. Proc Natl Acad Sci U S A. 1997;94:8058–63. doi: 10.1073/pnas.94.15.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mihola O, Trachtulec Z, Vlcek C, Schimenti JC, Forejt J. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science. 2009;323:373–5. doi: 10.1126/science.1163601. [DOI] [PubMed] [Google Scholar]
- 21.Forejt J, Iványi P. Genetic studies on male sterility of hybrids between laboratory and wild mice (Mus musculus L.) Genet Res. 1974;24:189–206. doi: 10.1017/S0016672300015214. [DOI] [PubMed] [Google Scholar]
- 22.Buard J, Barthès P, Grey C, de Massy B. Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. EMBO J. 2009;28:2616–24. doi: 10.1038/emboj.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayashi K, Yoshida K, Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438:374–8. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- 24.Henikoff S, Malik HS. Centromeres: selfish drivers. Nature. 2002;417:227. doi: 10.1038/417227a. [DOI] [PubMed] [Google Scholar]