Abstract
Virtually all organisms exposed to light are capable of sensing this environmental signal. In recent years the photoreceptors that mediate the ability of fungi to “see” have been identified in diverse species, and increasingly characterized. The small sizes of fungal genomes and ease in genetic and molecular biology manipulations make this kingdom ideal amongst the eukaryotes for understanding photosensing. The most widespread and conserved photosensory protein in the fungi is White collar 1 (WC-1), a flavin-binding photoreceptor that functions with WC-2 as a transcription factor complex. Other photosensory proteins in fungi include opsins, phytochromes and cryptochromes whose roles in fungal photobiology are not fully resolved and their distribution in the fungi requires further taxon sampling. Additional unknown photoreceptors await discovery. This review discusses the effects of light on fungi and the evolutionary processes that may have shaped the ability of species to sense and respond to this signal.
Keywords: Chromophore, circadian clock, evolution, LOV domain, photobiology, signal transduction
1. Introduction
The ability to see is considered a key sense in humans and, as such, extensive research has addressed the mechanisms and evolution of the photosensory properties in humans and animals. In plants and other photosynthetic organisms, sensing light is required to adapt to the available energy source provided by the sun, and photosensory proteins are also well studied in these groups (Briggs and Spudich, 2005). The fungi represent the third group of macroscopic eukaryotes, and they are the sibling kingdom to the animals. Fungi have also been studied for their ability to sense light, their responses, and the molecular basis of light-sensing. With small genome sizes relative to plants and animals, advances in genome sequencing provide gene content information on numerous and phylogenetically-diverse fungal species. In addition, genetic transformation protocols are available for many fungal species, enabling gene functions to be tested specifically by the creation of mutant strains and observing the phenotypes of these mutants. The application of these resources and recent studies enable a glimpse into the evolution of the molecular basis of vision in this eukaryotic kingdom.
A common question is why fungi would benefit from sensing light, as they do not use light for photosynthesis or to see adjacent objects. The main selective pressures are likely protection against DNA damage caused by ultraviolet wavelengths, timing spore release, directional discharge of spores, and as a clue for nutrient availability in the environment. These original roles have been elaborated upon such that the responses of fungi to light are now diverse and often species dependent. No general assumptions can be made about the effects of light on a species being newly investigated. However, one common feature is changes in spore production in response to light. Given the role of spores in fungal dissemination in the wild, understanding light-sensing is of practical interest because of the potential to control spread of fungi in nature. Additional practical applications of studying fungal photobiology include (i) the link between light-sensing and virulence in pathogenic species, (ii) photoregulation of secondary metabolite biosynthesis and enzyme production, (iii) the effects of light on commercial mushroom cultivation, and (iv) the use of fungal photoreceptors for experimentation in other organisms.
The molecular basis behind light-sensing in fungi is an active area of research, and the subject has been reviewed recently (Corrochano, 2007; Herrera-Estrella and Horwitz, 2007; Purschwitz et al., 2006). This issue of Fungal Genetics and Biology provides an additional set of reviews on specific topics of fungal photobiology. Collectively, these articles demonstrate the high level of research productivity. Here, we update developments on known photoreceptors with a focus on evolution of light-sensing, and place an emphasis on White collar 1 because of its widely established conserved role in blue-light sensing. We highlight areas that are unresolved, but potentially important, for further understanding light-sensing in fungi.
2. The effects of light on fungi
The fungi last shared a common ancestor perhaps more than a billion years ago, as based on fossil and molecular clock evidence, and had certainly diverged into the current extant major lineages 460 million years ago (Taylor and Berbee, 2006). How many or what percentage of the estimated 1.5 million fungal species sense light is unknown. Interpolating reports in the literature indicates that the majority of species, with some notable and major exceptions like the Saccharomycotina yeasts, respond to light. Indeed, a review on the topic of light-responses in fungi from fifty years ago already includes more than 250 species (Marsh et al., 1959), and research on fungal photobiology has continued in the intervening half-century. Changes in emphases since the 1050s were towards discovering effective wavelengths, which revealed that responses often occur with blue light but can occur over the full visible spectrum, the analysis of physiological and biochemical responses, and identification of the genes and proteins required for light-sensing (Corrochano and Avalos, 2010; Tisch and Schmoll, 2010).
2.1. Sensing and signal transduction vs. indirect effects of light
Photosensory proteins are defined as those that regulate a signal transduction pathway (Briggs and Spudich, 2005). As other non-sensory proteins use the same cofactors found in photoreceptors, these proteins can be affected by light and elicit a response to light, and yet not function in a sensory fashion (Hug, 1978; Hug, 1981). However, this distinction can be a grey area, as for example in distinguishing between overlapping sensory and DNA repair roles of some of the cryptochrome-photolyase proteins. A second ambiguous area of photosensor function is whether they have both light and dark regulatory roles. For example, mutation of the wc-1 homolog in Fusarium fujikuroi impairs secondary metabolism (Estrada and Avalos, 2008) and in Fusarium oxysporum alters hydrophobin production (Ruiz-Roldán et al., 2008), with both effects seen in the absence of light. Other considerations are that light can cause changes in the composition of growth medium that may affect fungal behavior, and the possible role of damaging wavelengths or photon intensities on the response of species.
An example of light affecting behavior not associated with an “established” photosensor comes from reports on Saccharomyces cerevisiae from 30 years ago. The genome of S. cerevisiae does not encode any protein similar to known photoreceptors beyond the opsin-related proteins (Fig. 1). However, light affected S. cerevisiae in these studies in a number of ways when grown under low temperature conditions. Light in the blue wavelengths inhibited respiration, protein synthesis and membrane integrity to inhibit growth, and this was dependent on cytochrome function because respiration-deficient rho mutants were no longer sensitive to light exposure (Ułaszewski et al., 1979). Even more striking was the ability to entrain S. cerevisiae wild type strains, but not rho mutant strains, with light to generate a free-running circadian rhythm as measured by histidine uptake (Edmunds et al., 1979). Effects of light on Saccharomycotina relatives of S. cerevisiae have also been reported (eg. Andrieu et al., 1977; Lapeña et al., 2006), and these are also unlikely to represent sensory responses given the absence of photosensory proteins in the sequences of their genomes.
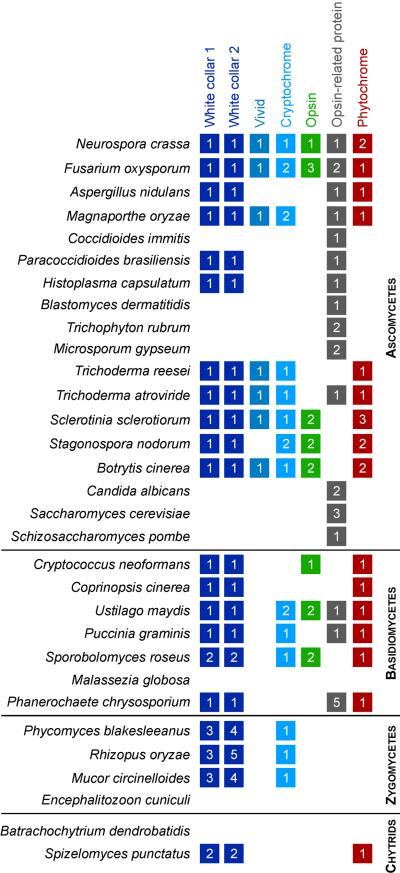
Fig. 1.
Distribution of photosensory proteins and their copy numbers in species across the fungal kingdom, based on whole-genome sequence information. Wherever possible, a representative example is provided rather than all sequenced members of a clade (eg. Fusarium, the Saccharomycotina yeasts, Schizosaccharomyces, Microsporum and Trichophyton). The terms zygomycete and chytrid do not refer to monophyletic groups. Presence or absence is inferred primarily by BLASTp analysis against GenBank and fungal genome databases at the Broad Institute and Department of Energy. For the cryptochrome category, the CPD-photolyase class is not included (but see Bayram et al., 2008a), and opsins consider type I only.
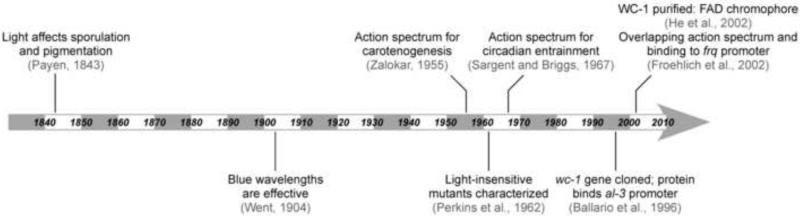
The gold standard for demonstrating a photosensory function is a multistep process. First, a light-response is observed. Second, the efficiency of that response is measured over a range of wavelengths to generate an action spectrum of the response either crudely into broad color groups (eg. blue, green, or red), or more accurately over narrow wavelengths. Third, the photosensor gene and protein are identified (eg. from cloning, by complementation, bioinformatic analysis of genomes, mutant screens, protein analysis). Fourth, the light response must be impaired by mutating the gene. Fifth, the photosensory protein is purified and characterized to demonstrate that it has the same absorption spectrum as the phenotypic action spectrum in step 2, as well as enabling the identification of the associated chromophore. Note that all known photosensory proteins function by interacting with a photoreactive molecule (eg. flavins, tetrapyrroles, retinals). Sixth, the photosensor function must explain the phenotypes observed, in that it should regulate genes or proteins specifically required for the responses. Clearly, this is a multidisciplinary process and often only partially achieved. For the best-studied fungus, Neurospora crassa, 160 years passed from the first observation of a response to light to characterization of purified WC-1 protein (Fig. 2).
Fig. 2.
From response to receptor: 160 years to demonstrate WC-1 is a photosensor in N. crassa. These events demonstrate the multidimensional process of demonstrating a protein is a photoreceptor.
[Additional references from figure (Froehlich et al., 2002; He et al., 2002; Payen, 1843; Perkins et al., 1962; Sargent and Briggs, 1967; Went, 1904; Zalokar, 1955)].
2.2. Fungal behavior affected by light
Different properties of fungi are influenced by light exposure to enable their success in the wild. The original selective pressure for light-sensing was protection against damaging light, as illustrated by induction of photolyase genes or pigmentation by light in highly-diverged species. However, the benefit of avoiding harmful irradiation must be balanced by the chemoorganoheterotrophic physiology of fungi. Members of this kingdom acquire nutrients from organic material, and this material relies directly or indirectly on light as an energy source and to fix carbon through photosynthesis.
The most commonly reported effect of light on fungi is change in sporulation, either as stimulation or as repression of asexual/sexual spore production (Corrochano and Galland, 2006; Marsh et al., 1959). A second process often regulated by light is an increase in pigments such as carotenoids or melanins. In some fungi the spores themselves are pigmented, and sporulation may also be co-regulated with the biosynthesis of secondary metabolites. The role of pigment is likely protection against ultraviolet damage, as mutant strains affected in genes required for pigment biosynthesis are more sensitive than wild type to UV light (eg. Moliné et al., 2009).
Several species of fungi have phototropic responses such that they bend towards or away from a source of light. In others, development is controlled such that fruiting structures are positioned towards the light for spore release, like the perithecial beaks in N. crassa that orient towards blue wavelengths (Backus, 1937; Harding and Melles, 1983). Some of the best and most spectacular examples of phototropism are in the Mucormycotina in which the asexual sporangiophores bend towards blue wavelengths. For Pilobolus species, the sporangiophore formation is also under circadian clock control (Schmidle, 1951; Uebelmesser, 1954) and the spore mass is forcibly discharged towards the rising sun. Similar phototropism occurs in Phycomyces blakesleeanus and Mucor circinelloides, and wc-1 homologs mediate the photosensory process in these two species and probably in Pilobolus (Idnurm et al., 2006; Kubo, 2009; Page, 1956; Silva et al., 2006).
An unexplained link exists between light-sensing and fungal pathogenicity (Idnurm and Crosson, 2009). In human pathogens, virulence of Histoplasma capsulatum is affected by prior exposure to light (Campbell and Berliner, 1973), while wc-1 mutant strains of Cryptocococcus neoformans and F. oxysporum have reduced virulence in mouse models (Idnurm and Heitman, 2005; Ruiz-Roldán et al., 2008). Other studies suggest links between light exposure and virulence in insect pathogens. Spores of Metarhizium anisopliae formed under blue light are more pathogenic towards Galleria mellonella wax moth larvae (Alves et al., 1980). Surprisingly, M. anisopliae spores produced under white light were non-pathogenic, which may imply wavelength-specificity or multiple and contra-acting photosensors. The role of light-sensing in regulating disease development post-inoculation is harder to interpret, since this could be due to either the host and/or the pathogen. For example, in the pathogenic interaction of Rhizophydium planktonicum on Asterionella formosa, the diatom is responsible for the attachment of chytrid zoospores in response to light. This was illustrated by killing the diatoms and showing that the chytrid zoospores no longer attached to the dead diatoms under light conditions (Canter and Jaworski, 1981). The underlying basis for light-sensing (or wc-1) affecting fungal virulence remains to be elucidated.
2.3. My fungus shows no response to light!
Although light-sensing is common in fungi, the effects may not always be obvious and may be difficult to identify. Passaging and specific selection for conveniently-behaved laboratory strains can be a factor. For example, the effects of light on the circadian clock are enhanced in N. crassa by the band mutation (Belden et al., 2007). The function of phytochrome in light-sensing in Aspergillus nidulans was masked by the veA mutation that causes constitutive conidiation under all light regimes (Blumenstein et al., 2005; Mooney and Yager, 1990). Some effects of light may be subtle, counteracted by exposure to the full visible spectrum, or best detected at specific developmental stages. Finally, there appears to be good evidence for the absence of light-sensing in some fungal species, most notably in ascomycete yeast species (eg. S. cerevisiae and S. pombe, Fig. 1). As such, the absence of a light response may be a real biological property.
In searching for an effect of light, it is worth testing sporulation or pigmentation in a set of wild type strains, rather than use a single laboratory-preferred wild type. An alternative approach is to compare the transcript profile of the fungus grown in the dark or exposed to light: this may be performed if microarray resources are available or using a small gene set such as common light-regulated genes (eg. ferrochelatase or photolyase). A third approach is to explore changes in protein abundance or post-translational modifications such as phosphorylation.
3. Three photosensors with emerging roles in fungal photobiology
The Mycota encode in their genomes representatives of major classes of photosensor known from other organisms, that is the opsins, phytochromes and cryptochromes. Fungi also contain proteins with light-sensing LOV domains that are discussed in section 4. While some species encode all types in their genome, most photoreceptors show a sporadic distribution across the kingdom (Fig. 1). Access to genome sequencing information enables a broad search for candidate photosensors in fungi. Similar analyses identified photosensors in ¼ of prokaryote genomes even excluding opsins (Krauss et al., 2009; Losi and Gärtner, 2008). The broad distribution of photosensing across the major domains of life underscores the importance light-sensing has in biology. In the following section what is known about three photoreceptors type in fungi will be described.
3.1. Opsins
Opsins are membrane-bound proteins with seven transmembrane α-helices that bind retinal via a conserved lysine residue. Opsins use light either for sensory purposes or to transport ions like H+ or Cl- across membranes (reviewed by Spudich et al., 2000). First reported from a fungus a decade ago (Bieszke et al., 1999a), the role of opsins in fungal photosensing is still unclear. Biochemical analysis of heterologously-expressed fungal opsins shows that these proteins could function as either sensors in the case of N. crassa or ion transporters in the case of Leptosphaeria maculans (Bieszke et al., 1999b; Waschuk et al., 2005). Phenotypes associated with mutating the N. crassa nop-1 gene are subtle, such as a growth defect in the presence of light when mitochondrial ATP synthesis is impaired or evident by changes in transcript levels for genes required for carotenoid biosynthesis (Bieszke et al., 1999a; Bieszke et al., 2007). A whole genome transcript profile in N. crassa does not suggest, at least at a transcriptional level, that NOP-1 mediates a light response (Chen et al., 2009). However, mutation of the nop-1 gene derepresses the N. crassa transcriptional response of several genes to light (Olmedo et al., 2010). An analysis of opsins in F. fujikuroi showed that mutation of the opsA gene had no growth or morphological phenotype, but did show changes in expression of genes for carotenoid biosynthesis (Estrada and Avalos, 2009). This research also raises the question of whether opsins could play a chemosensory role to regulate carotene production (the retinal chromophore derives from the β-carotene) particularly as another opsin gene carO is found within the cluster of genes for biosynthesis of β-carotene (Prado et al., 2004).
An opsin is clearly implicated in phototaxis of the chytrid Allomyces reticulatus because the action spectrum of phototaxis shifts away from the normal green wavelengths when the organism is grown in retinal analogs (Saranak and Foster, 1997). Surprisingly, the draft genome sequence of Allomyces macrogynus does not contain an obvious type I (for classification see Spudich et al., 2000) microbial opsin homolog. Rather, this chytrid and others encode candidate animal-like type II opsins, which are not found in other fungal lineages. To complicate the situation, blue-light dependent phototaxis, rather than green as in A. reticulatus, is seen in another chytrid Rhizophydium littoreum (Muehlstein et al., 1987). This observation and the presence of a copy of wc-1 in the A. macrogynus genome could also implicate a WC-1 homolog in chytrid phototaxis.
The distribution of opsins in fungi is sporadic. Based on a phylogenetic analysis, the fungal opsins were proposed to be of a common origin from a horizontal transfer event, being most similar to opsins involved in chloride ion transport (Sharma et al., 2006). A confounding factor in the analysis of opsin evolution is the presence of opsin-related proteins in many species (Brown, 2004; Spudich et al., 2000). These are distinguished primarily on the absence of the conserved lysine residue for retinal linkage in transmembrane domain seven. Such proteins are found in the Saccharomycotina, as well as in some species that contain conventional opsins (Fig. 1). The origins of fungal opsins are worth exploring further when the sequences of more basal fungi and bacterial genomes are released. Despite the lack of a demonstrated major function in photosensing, fungal opsins have biological activity when expressed in other organisms, and may potentially yield new insights into other biological problems (Chow et al., 2010).
3.2. Phytochromes
Phytochromes combine a tetrapyrrole chromophore to sense red and far-red wavelengths usually with a kinase domain for signal transduction (Rockwell et al., 2006). Phytochrome distribution in fungi is also sporadic (Fig. 1). The fungal phytochromes are similar to bacterial counterparts except that they include a response regulator domain at the C-terminal end of the protein, in contrast to the bacteria in which these modules are encoded by two separate genes. A sensory function has been elucidated for a phytochrome of A. nidulans, in which the FphA protein regulates the asexual-sexual transition and secondary metabolite biosynthesis in response to light. Purified FphA protein has phytochrome properties (Blumenstein et al., 2005; Brandt et al., 2008), but it is not known how the signal is transmitted in the cell to affect physiology. Components influencing the pathway include the COP9 signalosome and protein kinase ImeB (Bayram et al., 2009; Busch et al., 2003).
In A. nidulans the phytochrome protein forms a complex with the blue light sensory system controlled by the White collar proteins (Purschwitz et al., 2009; Purschwitz et al., 2008). FphA physically interacts with LreB (a WC-2 homolog), which in turn interacts with LreA (a WC-1 homolog). FphA also physically binds the Velvet A (VeA) protein, whose homologs are emerging as global regulators for secondary metabolism and sporulation in fungi (Calvo, 2008). Velvet proteins (VeA and VelB) also interact with LaeA, another global transcriptional regulator for secondary metabolism (Bayram et al., 2008b). Thus, if all these interactions occur simultaneously a large complex could be present in the cell with a wide range of photosensor capabilities and extensive regulatory inputs and outputs. To add to the complexity, the CryA photolyase protein is also responsible for regulation of veA expression (Bayram et al., 2008a). The interconnections between these pathways and relationships are under investigation. For instance, the possible regulation of LaeA via VelB-VeA, controlled by light, would explain the regulation of secondary metabolism, but not morphogenesis since sporulation is unaffected in laeA mutants. A simple model would place light-sensing upstream of VeA function, with outputs via LaeA and other unknown components downstream: however, the phosphorylation state of VeA is unaffected by mutation of FphA or LreA-LreB (Purschwitz et al., 2009).
Although red light responses have been reported for other fungi, phytochrome functions remain uncharacterized in other species and phenotypes are unknown for the mutant strains in N. crassa and C. neoformans (Froehlich et al., 2005; Idnurm and Heitman, 2005). In N. crassa, mutation of the phy-2 phytochrome derepress the con-10 gene’s transcriptional response to light (Olmedo et al., 2010). Nevertheless, A. nidulans remains the current model for understanding phytochrome signaling in fungi, and its interaction between light and global regulatory proteins.
3.3. Cryptochromes and photolyases
Cryptochromes and photolyases make up a family of blue/UV-A light photoreceptors traditionally differentiated by the ability of photolyases to repair damaged DNA and cryptochromes to be unable to repair damage (Sancar, 2004). Cryptochromes consist of an N-terminal photolyase-related region (PHR) which binds two chromophores, flavin adenine dinucleotide (FAD) and 5,10-methenyltetrahydrofolate (MTHF)/Pterin, and a variable C-terminal domain. Cryptochromes are reported and characterized from prokaryotes to eukaryotes where they are implicated in photosensory regulation of growth, development, cell signaling and circadian rhythm, and even as magnetoreceptors (Liedvogel and Mouritsen, 2010). Based on phylogenetic and functional analysis, the cryptochrome-photolyase family is divided into as many as six subfamilies: class I CPD photolyases, class II CPD photolyases, plant cryptochromes, animal cryptochromes, 6-4 CPD photolyases and CRY-DASH proteins (Bayram et al., 2008a; Daiyasu et al., 2004). The class II CPD photolyases are not found in fungi, with the exception of a Microsporidia species Antonospora locustae probably representing an example of horizontal gene transfer (Slamovits and Keeling, 2004). Evolutionary comparisons indicate that cryptochromes diverged from CPD photolyases before the divergence of eukaryotes and prokaryotes, as evident by the existence of CRY-DASH, the only known cryptochrome within bacterial members (Brudler et al., 2003). CRY-DASH proteins are the most recently characterized members of cryptochrome/photolyase family (Brudler et al., 2003). Structurally, they most closely resemble photolyases as they lack a characteristic C-terminal domain of cryptochromes (Daiyasu et al. 2004). CRY-DASH has very weak to no photolyase activity for dsDNA, but a high degree of specificity and photolyase activity for cyclobutane pyrimidine dimers in ssDNA (Selby and Sancar, 2006). A set of reports on the function of the cryptochrome-photolyase family in fungi reveals potential signaling and DNA repair roles, thus providing additional evidence that this family originated from an ancient light-dependent DNA repair enzyme.
In fungi, the first hints of photosensory functions for this family come from the photolyase gene phr1 of Trichoderma atroviride (Berrocal-Tito et al., 2007). The phr1 gene autoregulates its own photoinduction, as assessed through the transcript regulation of the phr1 promoter fused to gfp expressed in wild type or phr1 mutant backgrounds. phr1 also contributes to the level of induction of four blu blue-light regulated genes (Berrocal-Tito et al., 2007).
A. nidulans encodes no separate photolyase or cryptochrome genes, but instead encodes a dual function photolyase-like protein CryA that functions both as a UV-A dependent photorepair protein and as a regulator of sexual development (Bayram et al., 2008a). Phylogenetic analysis and amino acid alignment studies cluster CryA with class I CPD photolyases, leading to the proposal that CryA represents a missing link in the evolutionary transition of cryptochromes from photolyases. Deletion of cryA in A. nidulans leads to altered development and toxin biosynthesis, not dependent on light. However, dose response curves over four wavelengths also revealed a near UV-blue light dependent role for CryA in repressing sexual development. Genes such as veA, nsdA and rosA that regulate sexual development are overexpressed in cryA mutant strains, providing an explanation for altered reproduction (Bayram et al., 2008a).
In Cercospora zeae-maydis an ortholog of 6-4 photolyase (PHL1) has been characterized and may also represent an evolutionary intermediate between DNA repair enzymes and regulatory cryptochromes (Bluhm and Dunkle, 2008). This role is implicated in mutant strains by the loss of photoreactivation after UV irradiation and accompanying lack of induction of the gene CPD1 that encodes the cyclobutane pyrimidine dimer photolyase, as well as two genes RAD2 and RVB2 required for DNA repair. However, these three genes have low expression levels in the dark in phl1 mutant strains and light responses are still observed, suggesting the presence of other photosensors. Additional non-light dependent changes were also observed in the mutant, included enhanced conidiation and reduced production of the phytotoxin cercosporin (Bluhm and Dunkle, 2008).
For CRY-DASH proteins in fungi, roles in light-sensing at present remain modest. In Sclerotinia sclerotiorum, minor UV-A specific effects of CRY-DASH on development have been described (Veluchamy and Rollins, 2008). In N. crassa, cry encodes a CRY-DASH protein that itself is regulated by blue light in a WC-1 dependent manner. Disruption has a slight effect on the entrainment of the circadian clock (Froehlich et al., 2010). Deletion of N. crassa cry also derepresses light-dependent gene expression of at least three genes, con-10, al-1 and vvd (Olmedo et al., 2010).
In summary, the cryptochrome-photolyase family consists of related but distinct proteins with either DNA repair or sensory properties, or both. At present these proteins may play a modest role in the biology of these fungi in response to light, although it is difficult to rule out the effects seen by mutation in the absence of light, and the potential of nucleic acid damage being sensed, rather than light, and this leading to a response.
3.4. Dst2: a new flavin-binding photosensor?
A recent and surprising discovery was the identification of the dst2 gene of Coprinopsis cinerea (Kuratani et al., 2010). Strains of this basidiomycete fungus had been identified, by screening UV and insertional mutants, that did not develop a normal mushroom cap in response to light, hence being designated “dark stipe” dst strains. The dst1 gene had a base pair substitution leading to a premature stop codon in the homolog of wc-1 (Terashima et al., 2005), and it seemed likely that dst2 would be the homolog of wc-2. However, dst2 encodes a protein with a flavin-binding domain unlike the LOV domain of WC-1 or the BLUF domain proteins seen in some bacteria, as well as a berberine domain (pfam08031: found usually in proteins involved in isoquinoline alkaloid biosynthesis). DST2 homologs are limited to other basidiomycetes, although structurally-equivalent proteins are found more widely. Double dst1 dst2 mutants have the same phenotypes as single mutant strains, suggesting that they function in the same pathway although the mechanism is unclear (Kuratani et al., 2010). The presence of an FAD-binding domain in the DST2 protein is a particularly interesting finding: this protein or protein domain may function in photosensing more widely, or could represent an example of recent de novo evolution of a photosensor.
4. White collar 1 is a conserved blue-light sensor in fungi
In addition to opsins, phytochromes and cryptochromes, many fungi encode copies of White collar 1 in their genomes for sensing blue and near UV wavelengths via a flavin chromophore (Fig. 1). The role and mechanism of the White Collar 1-White Collar 2 complex in light-sensing has been well-studied and reviewed (Corrochano, 2007; Liu et al., 2003; Purschwitz et al., 2006). WC-1 has been intensively investigated because of its integral role in the circadian clock machinery of N. crassa, from where wc-1 was first cloned as a gene required for light responses (Ballario et al., 1996), and in which it also regulates sporulation, pigmentation, and phototropism. White collar 1 is a protein containing three PAS domains (named after the proteins Per, Arnt and Sim). The most N-terminal PAS domain is specialized into a LOV form (for light, oxygen, voltage) to which a flavin chromophore binds. LOV domain proteins also function in light-sensing in plants and bacteria (Krauss et al., 2009). Most WC-1 homologs also have a C-terminal zinc finger DNA-binding domain of the GATA class and thus function as transcription factors. Evidence that WC-1 of N. crassa is a light-sensor has been established through the multistep process described in section 2.1, with these events illustrated in the timeline of Fig. 2.
4.1. wc-1 is an ancient gene: discovery of wc-1 homologs and their function beyond Neurospora crassa
As illustrated in figure 1, wc-1 homologs are found in the genomes of many fungal species. Subsequent to the discovery of wc-1 in N. crassa, homologs have been implicated in light-sensing in diverse fungal species through functional studies in which mutation of the gene impairs responses to light (Table 1). Other analyses have focused on light-responses by fungi and the photoregulation and cloning of wc-1 homologs (Ambra et al., 2004; Sano et al., 2007). Indeed, with analysis of wc-1 function in Mucormycotina species, light-sensing is the best characterized signal transduction system in the kingdom from the perspective of evolutionary depth. In other basal members of the kingdom (Fig. 3), wc-1 homologs are evident in the genomes of two chytrids A. macrogynus (phylum Blastocladiomycota) and Spizellomyces punctatus (phylum Chytridiomycota). A potential homolog is also present in Conidiobolus conatus (subphylum Entomophthoromycotina) represented by an expressed sequence tag (Freimoser et al., 2003), thus implicating the gene in a lineage in addition to the Mucormycotina of what was previously the zygomycetes.
Table 1.
wc-1 homologs and their functions in fungi inferred from studying mutant strains.
| Species | Gene name | Light-regulated functions | Reference for gene cloning/identification |
|---|---|---|---|
| Ascomycota | |||
| Neurospora crassa | wc-1 | Circadian clock entraintment and phase shifting, carotenogenesis, sporulation, induction of protoperithecia, phototropism of the perithecial beaks | (Ballario et al., 1996) |
| Trichoderma atroviride | blr-1 | Conidiation | (Casas-Flores et al., 2004) |
| Magnaporthe oryzae | mgwc-1 | Conidiation, conidia release | (Lee et al., 2006) |
| Bipolaris oryzae | BLR1 | Conidial development | (Kihara et al., 2007) |
| Aspergillus nidulans | lreA | Asexual/sexual reproduction, mycotoxin biosynthesis | (Purschwitz et al., 2008) |
| Fusarium fujikuroi | wcoA | Conidiation, mycotoxin biosynthesis | (Estrada and Avalos, 2008) |
| Fusarium oxysporum | wc1 | Conidiation, hydrophobicity, photoreactivation, virulence in mouse model | (Ruiz-Roldán et al., 2008) |
| Trichoderma reesei | blr1 | Conidiation, growth rate | (Castellanos et al., 2010) |
| Basidiomycota | |||
| Cryptococcus neoformans | BWC1 / CWC1 | Inhibition of mating, UV sensitivity, virulence | (Idnurm and Heitman, 2005; Lu et al., 2005) |
| Coprinopsis cinerea | dst1 | Mushroom cap development | (Terashima et al., 2005) |
| Mucormycotina | |||
| Phycomyces blakesleeanus | madA | Phototropism, induction of carotene biosynthesis, sporulation | (Idnurm et al., 2006) |
| Mucor circinelloides | mcwc-1a | Phototropism | (Silva et al., 2006) |
| Mucor circinelloides | mcwc-1c | Induction of carotene biosynthesis | (Silva et al., 2006) |
| Mucor circinelloides | mcwc-1b | Interaction with crgA to regulate carotenogenesis | (Silva et al., 2008) |
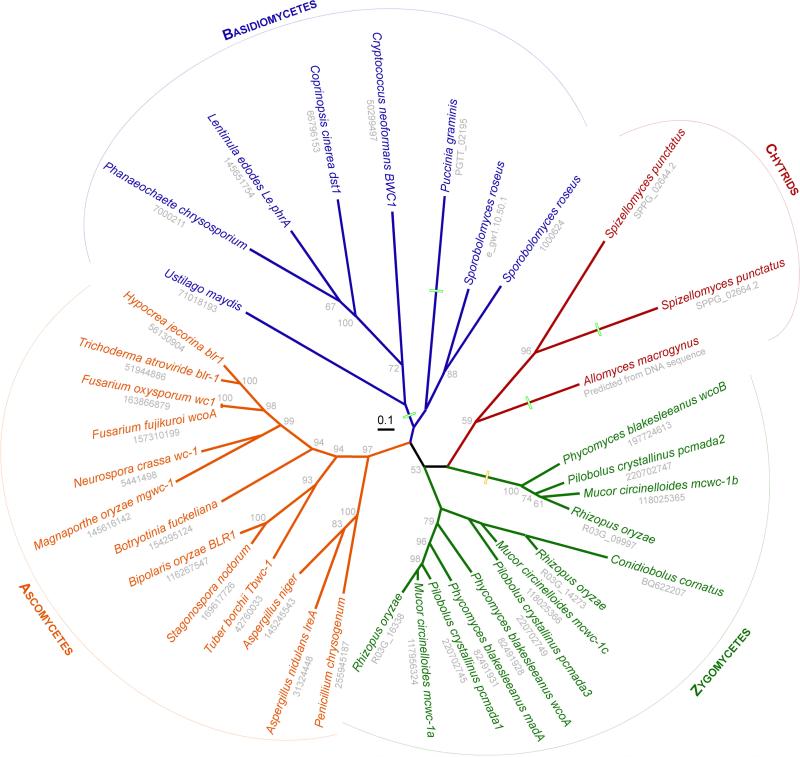
Fig. 3.
Phylogentic tree of WC-1 proteins. Homologs were downloaded from GenBank (gi numbers in grey; EST accession for C. coriobolus) or genome sequencing centers (P. chrysosporium, S. roseus, S. punctatus, A. macrogynus, P. graminis), aligned in ClustalW, the alignment edited in MacClade, and this alignment analyzed by maximum likelihood in PHYLIP. Bootstrap values above 50% adjacent to the nodes represent 100 replicates. The names of the genes encoding these proteins are provided after the species when known. Boxes represent lineages without the zinc finger DNA-binding domain or, in the case of the zygomycete group, a divergence from canonical form. Note that the terms zygomycete and chytrid no longer refer to monophyletic groups.
WC-1 is the only photosensory protein found in chytrids, zygomycetes, basidiomycetes and ascomycetes, which suggests a single origin and the subsequent losses in certain lineages (Fig. 1). A comparison of the proteins encoded by wc-1 homologs is illustrated in a phylogenetic study (Fig. 3). For this study, protein sequences were downloaded from GenBank or genome sequencing centers, aligned, and analyzed by maximum likelihood with proml under default settings in PHYLIP. One caveat of this analysis is that not all WC-1 homologs have cDNA sequence confirmation, and as such the tree topology and node support could differ if the predicted protein sequences changed from representatives. The WC-1 tree branches into the four major divisions of fungi (Fig. 3), and within each phylum follows the relationships provided by a six gene phylogeny of fungi (James et al., 2006). Congruence between WC-1 and house-keeping gene phylogenies thus further supports a single origin for this photosensor in the fungi.
Examination of other species would help reconstruct the evolution of wc-1 more fully in the fungi. For instance, is wc-1 present in the Glomeromycota? There is a report of blue-light controlling hyphal branching in Gigaspora gigantea, with an action spectrum showing peaks consistent with a flavin-based sensory system (Nagahashi and Douds, 2003), suggesting that a wc-1 (or cryptochrome) homolog could control photosensing in this lineage. Completion of the Glomus intraradices genome may reveal wc-1 homologs. Second, what happened to wc-1 in the basal ascomycetes? Two species, Yarrowia lipolytica and Schizosaccharomyces japonica, have LOV domain proteins in their genomes but they are unlike WC-1 in domain organization. These may reflect the remnants of the loss of wc-1 from these genomes, or horizontal transfer events. Here, genome availability from additional Taphrinomycotina species could clarify these evolutionary events. Of note, the genome sequence will be completed by JGI for Taphrina deformans. This species has a reported diurnal cycle of ascospore release (Yarwood, 1941), but this was observed on infected peach leaves and may reflect host, rather than fungal, physiology or temperature effects. Also, the new set of related species known only as “Soil Clone Group I” is worth examining if a species can be cultured, because the group represents an early branch in the Ascomycota (Porter et al., 2008).
In some fungi wc-1 has duplicated in the genome (Fig. 1, 3). This is most evident in the Mucormycotina, where three copies of wc-1 show similarity at the DNA sequence level, conserved intron-exon boundaries, and a conserved upstream gene in the case of P. blakesleeanus (Idnurm et al., 2006). The duplication events must have occurred early in the evolution of the subphylum, because all four species in figure 3 encode three copies of wc-1, and these form clades based on protein similarity rather than species similarity. Duplications have also occurred in the genomes of the Pucciniomycotina species Sporobolomyces roseus and the chytrid species S. punctatus. For the Mucormycotina, comparison of the functions of the three wc-1 copies demonstrates different evolutionary events following duplication. In P. blakesleeanus, a wc-1 homolog named madA regulates photoinduction of carotenogenesis, photomorphogenesis and phototropism (Idnurm et al., 2006). In M. circinelloides, one copy mcwc-1a regulates phototropism and a second copy mcwc-1c regulates photoinduction of carotenogenesis (Silva et al., 2006). This represents an example of diversification in gene function after gene duplication, in this case the downstream regulatory function after light-preception. In M. circinelloides, a function has also been identified for the third wc-1 gene, mcwc-1b, as a carotenoid regulator in certain mutant strain backgrounds (Silva et al., 2008). The Mucormycotina species also contain duplicated copies of wc-2, one of which functions with MADA in P. blakesleeanus (Sanz et al., 2009). It will be interesting to elucidate the functions of these copies in M. circinelloides to see if their functions correspond to the duplicated wc-1 genes.
Given the evidence for a single origin of wc-1 in fungal evolution, the absence of wc-1 genes from species and lineages implies gene loss. The entire Saccharomycotina clade is missing wc-1, and other photosensors (Fig. 1). This is also the situation for a number of pathogenic species in the genera Candida, Malassezia, the Microsporidia (now clearly part of the Mucormycotina; (Lee et al., 2008)), dermatophytes (Microsporum gypsum, M. canis, Trichophyton rubrum, T. tonsurans, T. equinum), and the chytrid amphibian-pathogen Batrochochytrium dendrobatidis. These species are obligate pathogens with reduced genomes or are tightly-associated with an animal host. Nevertheless, other fungal pathogens showed a mixed distribution of wc-1 (or other photosensors), such as present in Paracoccidioides brasiliensis and Histoplasma capsulatum, but absent from close relatives Coccidioides immitis and Blastomyces dermatatidis (Fig. 1). The lack of regular exposure to light on these species could have selected against maintaining photoreceptor genes. Loss of vision in the absence of light is known from the animal kingdom, eg. blind moles or sightless fish. A similar trend has been suggested in the prokaryotes for loss of LOV-mediated photosensing in obligate pathogens, anaerobes, or extremophiles that thrive under conditions in which there is no or little light (Krauss et al., 2009).
4.2. WC-2 is the theoretically unnecessary, yet essential, interacting partner for WC-1
Compared to the complexity of signal transduction for other environmental signals, the machinery behind WC-1 photosensing in fungi is remarkably efficient. Most homologs feature a light-sensing domain and a DNA-binding domain, and the transduction could be directly from the photon signal to a change in gene transcription. While research on WC-1 of N. crassa shows that the protein has a more complicated system of regulation, including its phosphorylation state and cycling in and out of the nucleus, in theory WC-1 should be capable of functioning in isolation without any accessory proteins. It is therefore surprising that a second protein, WC-2, is necessary for photosensing in all species in which it has been examined. WC-2 was first characterized from N. crassa (Linden and Macino, 1997), and subsequently in other ascomycetes such as A. nidulans, Bipolaris oryzae and T. atroviride (Casas-Flores et al., 2004; Moriwaki et al., 2008; Purschwitz et al., 2008). Furthermore, wc-2 homologs are required for blue-light responses in basidiomycetes C. neoformans and Lentinula edodes (Idnurm and Heitman, 2005; Sano et al., 2009; Yeh et al., 2009) and the Mucormycotina species P. blakesleeanus (Sanz et al., 2009) in which the proteins have been shown to interact physically with WC-1 homologs. A wc-2 homolog was also cloned, prior to the discovery of N. crassa wc-1 and wc-2, from a phage expression library of Fusarium solani f. sp. pisi used on a concatermerized 37 mer oligonucleotide from the promoter of a cutinase gene (Li and Kolattukudy, 1995). At present this gene is unlinked to photosensing. Of additional note, in all species indicated in figure 1, if a wc-1 homolog is in the genome then a candidate wc-2 homolog containing a PAS domain and zinc finger domain is also present, further implicating the essential role of WC-2 in signal transduction.
Studies in N. crassa that specifically address the requirement for WC-2 in photosensing have shown that in this species WC-2 controls stabilization of WC-1 (Cheng et al., 2002) and that the DNA-binding domain of WC-2 is required for light-induced transcription since this occurs in strains in which WC-1 has a deleted zinc finger domain (Cheng et al., 2003b). Similarly, for those species like many basidiomycetes that encode a WC-1 protein without a zinc finger, the zinc finger domain of the WC-2 protein would be essential for the WC-1/WC-2 complex to act as a transcriptional regulator.
4.3. VIVID and ENVOY
In addition to WC-1 homologs, some ascomycete species also encode a small LOV domain protein in their genomes (Fig. 1). The gene was first identified in N. crassa as VIVID (186 amino acids) based on bright orange pigmentation of mutants grown in the light (Heintzen et al., 2001). The VIVID protein physically binds flavin to act as a second photosensory protein in N. crassa (Schwerdtfeger and Linden, 2003). ENVOY is another small protein (207 amino acids) with a LOV domain that was identified in Hypocrea jecorina (Trichoderma reesei) based on its role in regulating cellulase (Schmoll et al., 2005). The two genes do not cross complement one another. The role of ENVOY in photobiology is complicated by functions in the presence and absence of light, and the fact that genes are either up or down regulated by its presence (Schuster et al., 2007). It is unclear how these small LOV-domain proteins modulate the output of the light signal as the immediate targets for either VIVID or ENVOY are unknown.
4.4. Genes regulated by light
Common changes in fungal biology by light are summarized in section 2: this diversity of responses indicates that between species there must be many different light-regulated genes (Tisch and Schmoll, 2010). Further advances in genomic resources will likely yield global insights into which genes and proteins are regulated by light. Microarrays have been performed on N. crassa, T. atroviride and C. neoformans to examine transcript differences in response to light. From these studies, the numbers of genes that are regulated by light vary between organisms and even research groups. For T. atroviride, an estimated 2.8 % of the genome is light-regulated (Rosales-Saavedra et al., 2006). Microarray studies on N. crassa suggest 3-7 % genes are light-regulated (Chen et al., 2009; Dong et al., 2008; Lewis et al., 2002). In contrast, for C. neoformans less than a quarter of a percent (0.25%) of the genome was light-regulated (Idnurm and Heitman, 2010). Each of these studies used different media and light regimes in different laboratories, thus preventing direct cross-species comparisons. Changing media conditions, developmental stages, light intensities, or wavelengths, may uncover alternative sets of light-regulated genes.
A recent analysis of the N. crassa transcriptome reveals that almost all transcript changes in response to light require wc-1 (Chen et al., 2009). The study also confirms previous observations for two classes of genes expressed soon after light-exposure and others at a later time point. The “early” genes are controlled directly by WC-1. A key new gene, sub-1, encodes a GATA-type transcription factor and is a major regulator of the “late” genes because their photoinduction is lost in Δsub-1 strains (Chen et al., 2009). Thus, signal transduction of light in N. crassa can include an additional transcriptional layer, in which light sensed by the WCC induces SUB1 production, which in turn regulates transcription of the late light-induced genes.
There are few genes known to be commonly photoregulated in fungi. Genes that are often light-regulated encode photolyase or enzymes for carotene biosynthesis, although it should be noted that photolyase is absent from some fungal genomes and in some species carotene biosynthesis is not induced by light. Both photolyase and carotene function in protection against UV damage. One gene that may represent a globally photoregulated gene is ferrochelatase, which catalyzes the final step in the biosynthesis of heme (Idnurm and Heitman, 2010). A microarray study on C. neoformans identified this transcript as the most upregulated in response to light. The gene is also light-regulated in N. crassa and P. blakesleeanus, and in all three species mutation of the White collar complex abolishes induction (Idnurm and Heitman, 2010). The hypothesis to explain this remarkable conservation is that porphyrin molecules are highly photoreactive, and their reduction by ferrochelatase helps alleviate the dangerous effects of light. However, this needs to be confirmed through biochemical measurements of porphyrin and heme levels in fungi under light-dark conditions. With a diversity of transcription responses and yet some conserved genes, more studies are clearly needed to understand the effects of light on transcription (or other regulated processes) in fungi.
5. Unanswered questions and directions for future research
Despite decades of research on light-sensing in fungi (e.g. Fig. 2) there are abundant areas for future discoveries about the sensory properties of fungi and even for well-studied components like White collar 1 and 2. Four potential directions are raised for fungal photobiology research.
5.1. What are the functions of photosensors currently unassociated with a light response, and do the known photosensors interact with each other?
The genomes of many fungi contain multiple candidate photosensors (Fig. 1): what roles do these proteins have in light-sensing, if any? Even in situations in which genes have been studied, it is not clear what functions opsins, phytochromes or cryptochromes have in the majority of fungi. Likewise, the purpose of multiple wc-1 homologs in most Mucormycotina or Pucciniomycotina remains a mystery. Is it an opsin or a WC-1 homolog that controls chytrid phototaxis?
Sunlight consists of a range of wavelengths that are not uniform in intensity throughout the day. Related to photosensory function is to establish how exposure to different wavelengths influences fungal biology. This could reflect cross-talk between different photosensors, perhaps even as a protein complex as possible in A. nidulans, or by photoregulation of one photosystem by another, such as cryptochromephotolyases or opsins by WC-1. Another explanation is in the photochemical properties of the chromophores themselves at different energy states. For WC-1 homologs, the shift in absorbance spectra when flavins are exposed to light could provide red-light absorbing transient states, explaining blue-red interactions (Galland and Tölle, 2003). This hypothesis is most compelling in P. blakesleeanus in which it was elucidated since the reported effects of red light in this organism cannot be explained by a phytochrome protein given the absence of such a gene from the genome (Fig. 1). However, such changes have not been observed in analysis of flavins within the context of the LOV domain. Likewise, near UV wavelengths of high intensity can cause a rapid reversal of LOV domain-flavin adducts, potentially explaining some specific effects by a narrow range of blue light wavelengths (Kennis et al., 2004). One question is whether photosensor chromophores can adopt different absorbance profiles in different homologs as happens with opsins in animals via specific amino acid residues in spectral tuning sites (Hunt et al., 2007). In M. circinelloides, mcwc-1c regulates blue-light induction of carotenogensis, while mcwc-1a controls both blue- and green-light phototropism (Silva et al., 2006), indicating a response for WC-1 proteins beyond blue wavelengths.
Fungi respond to light over different intensity thresholds and in some species over as large a range as nine orders of magnitude of intensity (Corrochano, 2007). These two properties should be explained by the photosensor(s) itself controling the sensitivity of the proteins to photon intensity, changing photoreceptor concentration, or the influence of interacting proteins. These ideas are explored below while discussing photoadaptation.
5.2. Are there unknown photosensors in fungi?
Responses to light are observed in species in which the photoreceptors described above have been mutated, leading to the hypothesis for the existence of unknown photosensors. This evidence comes from multiple species, including F. fujikuroi, T. atroviride and P. blakesleeanus. In T. atroviride, mutation of either blr-1 or blr-2 causes a dramatic reduction in radial growth rate, but only under blue or red illumination (but not white light), suggesting the presence of additional photosensors (Casas-Flores et al., 2004). Furthermore, light-dependent transcript changes still occur in blr-1 or blr-2 mutants (Rosales-Saavedra et al., 2006). In F. fujikuroi, carotenogenesis remains light-induced in wcoA mutant strains (Estrada and Avalos, 2008). In P. blakesleeanus part of the light-response is mediated by the madA-madB genes, but another gene designated madC, known from phototropism mutants, also contributes (Lipson et al., 1980). Viewed from another perspective, the genome sequences of fungi contain potential yet uncharacterized new photosensors. A number of ascomycete species encode proteins comprising a Regulator of G protein Signaling domain and LOV domain (eg. Botrytis cinerea BC1G_08584.1, Magnaporthe oryzae MGG_08735.5, S. sclerotiorum SS1G_01379). Such proteins would be of high interest to analyze further to see if these mediate residual blue-light effects in wc-1 mutant strains, and could participate in cross talk between G protein and cAMP signaling and light-signaling. A third LOV-like domain has been reported in a small number of species (Krauss et al., 2009), but it is unknown if this protein is involved as a light-sensor or to detect an alternative signal. Other LOV domain proteins are found sporadically in fungal species, eg. Puccinia graminis PGTG_08913.2 and S. roseus (protein unannotated).
5.3. How does photoadaptation work?
Some species of fungi are capable of adapting to the intensity of light to which they are exposed. The trait is observed phenotypically as a transient increase and then decrease in a response or adaptation to ambient light conditions in which a step-up in light intensity induces a response. This may be evident at the transcript level in which species such as N. crassa or T. atroviride show a rise and fall in transcript abundance. Photoadaption may be a subtle phenotype to explore without an excellent set of experimental resources. Multiple mechanisms, which are not mutually exclusive, are or could be involved in photoadaptation.
First, accessory photosensors control photoadaptation, as known for N. crassa VVD (Heintzen et al., 2001). The mechanism behind their influence on WC-1/WC-2 is unresolved, although VVD causes hyperphosphorylation of WC-1. In species in which more than one copy of wc-1 is present, it is possible that one of these regulates the dampening response. A second mechanism for photoadaptation can be post-translational modifications of the photosensor proteins themselves blocking signaling, as for example VVD influencing the phosphoylation state of WC-1 (He and Liu, 2005). Upregulation of components of the machinery sensitizes the species to light, such as the wc-2 homologs BWC2/CWC2 in C. neoformans or blr-2 in T. atroviride (Esquivel-Naranjo and Herrera-Estrella, 2007; Lu et al., 2005; Yeh et al., 2009): these observations imply that protein abundance limits the signal response, and conversely that decreasing abundance could mediate down-regulation of a response. A reduction in the amount of the photoreceptor MADA has been proposed as the basis for gene photoadaptation in P. blakesleeanus (Rodríguez-Romero and Corrochano 2006). A third mechanism for the adaptation to light could be controlled by the covalent linkage of the flavin to the protein and the properties of this protein-chromophore complex. For the case of LOV domain proteins, LOV-flavin adduct is formed in response to photons and the signal is turned on. The rate of decay of this intermediate would influence the time of the response. For instance, long half-lives could prevent reactivation, while the LOV-flavin adduct form may reduce protein stability leading to rapid turn over. The resolution from LOV-flavin adduct to LOV domain+flavin varies enormously between LOV domains, as illustrated by analysis of the VVD protein photocycle (Zoltowski et al., 2009). The VVD LOV domain is considered a slow cycling form, with a 5 h half-life. Mutations of as few as one or two amino acid residues can dramatically alter this cycle, from a 28 s to 50 h half-life (Zoltowski et al., 2009). A slow decay and requirement for protein turn over would provide a different response compared to a protein that rapidly disassociates the flavin-cysteine adduct and could sense another photon.
5.4. What structural changes occur in photosensory proteins in response to light to transmit the signal?
With the exception of VIVID from N. crassa (Zoltowski et al., 2007; Zoltowski et al., 2009), no crystal or other structure is known for the fungal photosensory proteins. Such information is essential for understanding how photosensors physically change in response to light to transmit the light signal. It is possible to model such information for opsins, phytochromes or cryptochromes based on structures gained from homologs in other organisms. For these types of photosensors, research priorities in fungi may be more productive first focused on linking opsin function to a light response, or establishing the downstream targets of phytochromes and cryptochromes. In the case of WC-1, a set of crystal structures would be an informative research direction, first because WC-1 is the photosensor conserved in most fungi and second because of its unique composition. Structural information exists for the domains in WC-1 from other proteins, including LOV, other PAS domains and the GATA-type Zn finger: how these fit together in signaling is unknown. Gaining a comprehensive understanding is a challenge given not just the dark-light states, but also subcellular localization, interaction with other proteins and DNA, and the inherent limitations of purifying sufficient quantities of WC-1 proteins of N. crassa or other species (He et al., 2002; He and Liu, 2005).
6. Concluding comments on the evolution of vision in fungi
Writing at the close of the bicentennial year of Charles Darwin’s birth leads to consideration of the developments in understanding evolution since The Origin of Species was published 150 years ago. In addition to his interests in evolution, Darwin was a photobiologist (Darwin and Darwin, 1881), so much so that he is sometimes erroneously credited with discovering the “blue light effect”. At the time this book was published, blue light was already well-established as a plant signal (see Davenport, 1899; Whippo and Hangarter, 2006 for history) and even in fungi such as Phycomyces nitens (Vines, 1878).
The eye is often used in discussion, including by Darwin, about evolution from the perspective of how could something so “complex” evolve. This evolutionary process is explained in a multicellular organism as a transition from one to a few clustered photosensory cells, an invagination to provide information about the direction of the light source, and lastly the development of a lens. A more pertinent question, given the microbial dominance of life on earth, is not so much organ development which remarkably is regulated by genes like Pax-6 conserved from fruit flies to mammals, but on the origins of the first photosensory proteins. It is striking that only a small number of chemical chromophores participate in photosensing (Delbrück, 1976), with their binding domains derived independently in some cases like the flavins. One can hypothesize how gene fusion events brought about a photosensor if the two genes encoded a regulator and a protein that interacted with a chromophore. This is supported by the modular nature of bacterial photosensors in which similar sensory domains are fused to different transduction components (Losi and Gärtner, 2008). An eukaryotic example is phototropism in plants and fungi: both use a LOV domain hypothesized to have the same origin from an α-Proteobacterium (Krauss et al., 2009) and the plant phot1 LOV domain can replace that in WC-1 in fungi (Cheng et al., 2003a). In plants, however, LOV is fused to a kinase domain and modulates bending toward the light in a multicellular structure. In fungi like the Mucormycotina, LOV is fused to a transcription factor domain and modulates bending towards the light in a unicellular structure.
In summary, the discovery and characterization of photosensory proteins in fungi is important for understanding fungal biology for practical reasons, and also provides greater insight into the evolution of vision.
Acknowledgments
We thank the Broad Institute and Department of Energy for access to genome information and continued generation of fungal genome sequences. For research support on fungal photobiology, we gratefully acknowledge the United States National Institutes of Health (K22 AI073917), National Science Foundation (MCB-0920581), UMKC (Faculty Research Grant), the European Regional Development Fund, Spanish Ministerio de Ciencia e Innovación (BIO2009-12486), and Junta de Andalucía (P06-CVI-01650, P09-CVI-5027).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alves SB, Forti LC, Cividanes FJ. Influence of light color on some biological activities Metarhizium anisopliae (Metsch) Sorok. an entomopathogenic fungus. Rev. Bras. Ent. 1980;24:123–125. [Google Scholar]
- Ambra R, Grimaldi B, Zamboni S, Filetici P, Macino G, Ballario P. Photomorphogenesis in the hypogeous fungus Tuber borchii: isolation and characterization of Tbwc-1, the homologue of the blue-light photoreceptor of Neurospora crassa. Fungal Genet. Biol. 2004;41:688–697. doi: 10.1016/j.fgb.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Andrieu S, Biguet J, Jaques R, Dehorter B, Lacoste L. Photoinhibition de la Chlamydosporulation de Candida albicans. Sabouraudia. 1977;15:207–214. [PubMed] [Google Scholar]
- Backus MP. Phototropic response of perithecial necks in Neurospora. Mycologia. 1937;29:383–386. [Google Scholar]
- Ballario P, Vittorioso P, Magrelli A, Talora C, Cabibbo A, Macino G. White collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein. EMBO J. 1996;15:1650–1657. [PMC free article] [PubMed] [Google Scholar]
- Bayram Ö, Biesemann C, Krappmann S, Galland P, Braus GH. More than a repair enzyme: Aspergillus nidulans photolyase-like CryA is a regulator of sexual development. Mol. Biol. Cell. 2008a;19:3254–3262. doi: 10.1091/mbc.E08-01-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayram Ö, Krappmann S, Ni M, Bok JW, Helmstaedt K, Valerius O, Braus-Stromeyer S, Kwon N-J, Keller NP, Yu J-H, Braus GH. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science. 2008b;320:1504–1506. doi: 10.1126/science.1155888. [DOI] [PubMed] [Google Scholar]
- Bayram Ö, Sari F, Braus GH, Irniger S. The protein kinase ImeB is required for light-mediated inhibition of sexual development and for mycotoxin production in Aspergillus nidulans. Mol. Microbiol. 2009;71:1278–1295. doi: 10.1111/j.1365-2958.2009.06606.x. [DOI] [PubMed] [Google Scholar]
- Belden WJ, Larrondo LF, Froehlich AC, Shi M, Chen CH, Loros JJ, Dunlap JC. The band mutation in Neurospora crassa is a dominant allele of ras-1 implicating RAS signaling in circadian output. Genes Dev. 2007;21:1494–1505. doi: 10.1101/gad.1551707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal-Tito GM, Esquivel-Naranjo EU, Horwitz BA, Herrera-Estrella A. Trichoderma atroviride PHR1, a fungal photolyase responsible for DNA repair, autoregulates its own photoinduction. Eukaryot. Cell. 2007;6:1682–1692. doi: 10.1128/EC.00208-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieszke JA, Braun EL, Bean LE, Kang S, Natvig DO, Borkovich KA. The nop-1 gene of Neurospora crassa encodes a seven transmembrane helix retinal-binding protein homologous to archaeal rhodopsins. Proc. Natl. Acad. Sci. USA. 1999a;96:8034–8039. doi: 10.1073/pnas.96.14.8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieszke JA, Li L, Borkovich KA. The fungal opsin gene nop-1 is negatively-regulated by a component of the blue light sensing pathway and influences conidiation-specific gene expression in Neurospora crassa. Curr. Genet. 2007;52:149–157. doi: 10.1007/s00294-007-0148-8. [DOI] [PubMed] [Google Scholar]
- Bieszke JA, Spudich EN, Scott KL, Borkovich KA, Spudich JL. A eukaryotic protein, NOP-1, binds retinal to form an archaeal rhodopsin-like photochemically reactive pigment. Biochemistry. 1999b;38:14138–14145. doi: 10.1021/bi9916170. [DOI] [PubMed] [Google Scholar]
- Bluhm BH, Dunkle LD. PHL1 of Cercospora zeae-maydis encodes a member of the photolyase/cryptochrome family involved in UV protection and fungal development. Fungal Genet. Biol. 2008;45:1364–1372. doi: 10.1016/j.fgb.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Blumenstein A, Vienken K, Tasler R, Purschwitz J, Veith D, Frankenberg-Dinkel N, Fischer R. The Aspergillus nidulans phytochrome FphA represses sexual development in red light. Curr. Biol. 2005;15:1833–1838. doi: 10.1016/j.cub.2005.08.061. [DOI] [PubMed] [Google Scholar]
- Brandt S, von Stetten D, Günther M, Hildebrandt P, Frankenberg-Dinkel N. The fungal phytochrome FphA from Aspergillus nidulans. J. Biol. Chem. 2008;283:34605–34614. doi: 10.1074/jbc.M805506200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs WR, Spudich JL, editors. Handbook of Photosensory Receptors. Wiley-VCH; Weinheim: 2005. [Google Scholar]
- Brown LS. Fungal rhodopsins and opsin-related proteins: eukaryotic homologues of bacteriorhodopsin with unknown functions. Photochem. Photobiol. Sci. 2004;3:555–565. doi: 10.1039/b315527g. [DOI] [PubMed] [Google Scholar]
- Brudler R, Hitomi K, Daiyasu H, Toh H, Kucho K.-i., Ishiura M, Kanehisa M, Roberts VA, Todo T, Tainer JA, Getzoff ED. Identification of a new cryptochrome class: structure, function, and evolution. Mol. Cell. 2003;11:59–67. doi: 10.1016/s1097-2765(03)00008-x. [DOI] [PubMed] [Google Scholar]
- Busch S, Eckert SE, Krappmann S, Braus GH. The COP9 signalosome is an essential regulator of development in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 2003;49:717–730. doi: 10.1046/j.1365-2958.2003.03612.x. [DOI] [PubMed] [Google Scholar]
- Calvo AM. The VeA regulatory system and its role in morphological and chemical development in fungi. Fungal Genet. Biol. 2008;45:1053–1061. doi: 10.1016/j.fgb.2008.03.014. [DOI] [PubMed] [Google Scholar]
- Campbell CC, Berliner MD. Virulence differences in mice of type A and B Histoplasma capsulatum yeasts grown in continuous light and total darkness. Infect. Immun. 1973;8:677–678. doi: 10.1128/iai.8.4.677-678.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canter HM, Jaworski GHM. The effect of light and darkness upon infection of Asterionella formosa Hassall by the chytrid Rhizophydium planktonicum Canter emend. Ann. Bot. 1981;47:13–30. [Google Scholar]
- Casas-Flores S, Rios-Momberg M, Bibbins M, Ponce-Noyola P, Herrera-Estrella A. BLR-1 and BLR-2, key regulatory elements of photoconidiation and mycelial growth in Trichoderma atroviride. Microbiology. 2004;150:3561–3569. doi: 10.1099/mic.0.27346-0. [DOI] [PubMed] [Google Scholar]
- Castellanos F, Schmoll M, Martínez P, Tisch D, Kubicek CP, Herrera-Estrella A, Esquivel-Naranjo EU. Crucial factors of the light perception machinery and their impact on growth and cellulase gene transcription in Trichoderma reesei. Fungal Genet. Biol. 2010;47:468–476. doi: 10.1016/j.fgb.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Chen C-H, Ringelberg CS, Gross RH, Dunlap JC, Loros JJ. Genome-wide analysis of light-inducible responses reveals hierarchical light signalling in Neurospora. EMBO J. 2009;28:1029–1042. doi: 10.1038/emboj.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, He Q, Yang Y, Wang L, Liu Y. Functional conservation of light, oxygen, or voltage domains in light sensing. Proc. Natl. Acad. Sci. USA. 2003a;100:5938–5943. doi: 10.1073/pnas.1031791100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, Yang Y, Gardner KH, Liu Y. PAS domain-mediated WC-1/WC-2 interaction is essential for maintaining the steady-state level of WC-1 and the function of both proteins in circadian clock and light responses of Neurospora. Mol. Cell. Biol. 2002;22:517–524. doi: 10.1128/MCB.22.2.517-524.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, Yang Y, Wang L, He Q, Liu Y. WHITE COLLAR-1, a multifunctional Neurospora protein involved in the circadian feedback loops, light sensing, and transcription repression of wc-2. J. Biol. Chem. 2003b;278:3801–3808. doi: 10.1074/jbc.M209592200. [DOI] [PubMed] [Google Scholar]
- Chow BY, Han X, Dobry AS, Qian X, Chuong AS, Li M, Henninger MA, Belfort GM, Lin Y, Monahan PE, Boyden ES. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrochano LM. Fungal photoreceptors: sensory molecules for fungal development and behaviour. Photochem. Photobiol. Sci. 2007;6:725–736. doi: 10.1039/b702155k. [DOI] [PubMed] [Google Scholar]
- Corrochano LM, Avalos J. Light sensing. In: Borkovich KA, Ebbole DJ, editors. Cellular and molecular biology of filamentous fungi. ASM Press; Washington, DC: 2010. [Google Scholar]
- Corrochano LM, Galland P. Photomorphogenesis and gravitropism in fungi. In: Kües U, Fischer R, editors. The Mycota I. Springer-Verlag; Berlin Heidelberg: 2006. [Google Scholar]
- Daiyasu H, Ishikawa T, Kuma K, Iwai S, Todo T, Toh H. Identification of cryptochrome DASH from vertebrates. Genes Cells. 2004;9:479–495. doi: 10.1111/j.1356-9597.2004.00738.x. [DOI] [PubMed] [Google Scholar]
- Darwin C, Darwin F. The power of movement in plants. D. Appleton and Company; New York: 1881. [Google Scholar]
- Davenport CB. Experimental Morphology. MacMillan Company; New York: 1899. [Google Scholar]
- Delbrück M. Light and Life III. Carlsberg Res. Commun. 1976;41:299–309. [Google Scholar]
- Dong W, Tang X, Yu Y, Nilsen R, Kim R, Griffith J, Arnold J, Schüttler H-B. Systems biology of the clock in Neurospora crassa. PLoS ONE. 2008;3:e3105. doi: 10.1371/journal.pone.0003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds LN, Jr., Apter RI, Rosenthal PJ, Shen W-K, Woodward JR. Light effects in yeast: persisting oscillations in cell division activity and amino acid transport in cultures of Saccharomyces cerevisiae entrained by light-dark cycles. Photochem. Photobiol. 1979;30:595–601. doi: 10.1111/j.1751-1097.1979.tb07186.x. [DOI] [PubMed] [Google Scholar]
- Esquivel-Naranjo EU, Herrera-Estrella A. Enhanced responsiveness and sensitivity to blue light by blr-2 overexpression in Trichoderma atroviride. Microbiology. 2007;153:3909–3922. doi: 10.1099/mic.0.2007/007302-0. [DOI] [PubMed] [Google Scholar]
- Estrada AF, Avalos J. The White Collar protein WcoA of Fusarium fujikuroi is not essential for photocarotenogenesis, but is involved in the regulation of secondary metabolism and conidiation. Fungal Genet. Biol. 2008;45:705–718. doi: 10.1016/j.fgb.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Estrada AF, Avalos J. Regulation and targeted mutation of opsA, coding for the NOP-1 opsin orthologue in Fusarium fujikuroi. J. Mol. Biol. 2009;387:59–73. doi: 10.1016/j.jmb.2009.01.057. [DOI] [PubMed] [Google Scholar]
- Freimoser FM, Screen S, Hu G, St Leger R. EST analysis of genes expressed by the zygomycete pathogen Conidiobolus coronatus during growth on insect cuticle. Microbiology. 2003;149:1893–1900. doi: 10.1099/mic.0.26252-0. [DOI] [PubMed] [Google Scholar]
- Froehlich AC, Chen C-H, Belden WJ, Madeti C, Roenneberg T, Merrow M, Loros J, Dunlap J. Genetic and molecular characterization of a cryptochrome from the filamentous fungus Neurospora crassa. Eukaryot. Cell. 2010 doi: 10.1128/EC.00380-09. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froehlich AC, Liu Y, Loros JJ, Dunlap JC. White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science. 2002;297:815–819. doi: 10.1126/science.1073681. [DOI] [PubMed] [Google Scholar]
- Froehlich AC, Noh B, Vierstra RD, Loros J, Dunlap JC. Genetic and molecular analysis of phytochromes from the filamentous fungus Neurospora crassa. Eukaryot. Cell. 2005;4:2140–2152. doi: 10.1128/EC.4.12.2140-2152.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galland P, Tölle N. Light-induced fluorescence changes in Phycomyces: evidence for blue light-receptor associated flavo-semiquinones. Planta. 2003;217:971–982. doi: 10.1007/s00425-003-1068-6. [DOI] [PubMed] [Google Scholar]
- Harding RW, Melles S. Genetic analysis of phototropism of Neurospora crassa perithecial beaks using white collar and albino mutants. Plant Physiol. 1983;72:996–1000. doi: 10.1104/pp.72.4.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Cheng P, Yang Y, Wang L, Gardner KH, Liu Y. White collar-1, a DNA binding transcription factor and a light sensor. Science. 2002;297:840–843. doi: 10.1126/science.1072795. [DOI] [PubMed] [Google Scholar]
- He Q, Liu Y. Molecular mechanism of light responses in Neurospora: from light-induced transcription to photoadaptation. Genes Dev. 2005;19:2888–2899. doi: 10.1101/gad.1369605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzen C, Loros JJ, Dunlap JC. The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell. 2001;104:453–464. doi: 10.1016/s0092-8674(01)00232-x. [DOI] [PubMed] [Google Scholar]
- Herrera-Estrella A, Horwitz BA. Looking through the eyes of fungi: molecular genetics of photoreception. Mol. Microbiol. 2007;64:5–15. doi: 10.1111/j.1365-2958.2007.05632.x. [DOI] [PubMed] [Google Scholar]
- Hug DH. The activation of enzymes with light. In: Smith KC, editor. Photochemical and Photobiological Reviews. Plenum Press; New York and London: 1978. pp. 1–33. [Google Scholar]
- Hug DH. Photoactivation of enzymes. In: Smith KC, editor. Photochemical and Photobiological Reviews. Plenum Press; New York and London: 1981. pp. 87–138. [Google Scholar]
- Hunt DM, Carvalho LS, Cowing JA, Parry JWL, Wilkie SE, Davies WL, Bowmaker JK. Spectral tuning of shortwave-sensitive visual pigments in vertebrates. Photochem. Photobiol. 2007;83:303–310. doi: 10.1562/2006-06-27-IR-952. [DOI] [PubMed] [Google Scholar]
- Idnurm A, Crosson S. The photobiology of microbial pathogenesis. PLoS Pathog. 2009;5:e1000470. doi: 10.1371/journal.ppat.1000470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Heitman J. Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol. 2005;3:615–626. doi: 10.1371/journal.pbio.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Heitman J. Ferrochelatase is a conserved downstream target of the blue light-sensing white collar complex in fungi. manuscript submitted. 2010. [DOI] [PMC free article] [PubMed]
- Idnurm A, Rodríguez-Romero J, Corrochano LM, Sanz C, Iturriaga EA, Eslava AP, Heitman J. The Phycomyces madA gene encodes a blue-light photoreceptor for phototropism and other light responses. Proc. Natl. Acad. Sci. USA. 2006;103:4546–4551. doi: 10.1073/pnas.0600633103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TY, Kauff F, Schoch C, Matheny PB, Hofstetter V, Cox C, Celio G, Gueidan C, Fraker E, Miadlikowska J, Lumbsch HT, Rauhut A, Reeb V, Arnold AE, Amtoft A, Stajich JE, Hosaka K, Sung G-H, Johnson D, O'Rourke B, Crockett M, Binder M, Curtis JM, Slot JC, Wang Z, Wilson AW, Schüßler A, Longcore JE, O'Donnell K, Mozley-Standridge S, Porter D, Letcher PM, Powell MJ, Taylor JW, White MM, Griffith GW, Davies DR, Humber RA, Morton JB, Sugiyama J, Rossman AY, Rogers JD, Pfister DH, Hewitt D, Hansen K, Hambleton S, Shoemaker RA, Kohlmeyer J, Volkmann-Kohlmeyer B, Spotts RA, Serdani M, Crous PW, Hughes KW, Matsuura K, Langer E, Langer G, Untereiner WA, Lücking R, Büdel B, Geiser DM, Aptroot A, Diederich P, Schmitt I, Schultz M, Yahr R, Hibbett D, Lutzoni F, McLaughlin D, Spatafora J, Vilgalys R. Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- Kennis JTM, van Stokkum IHM, Crosson S, Gauden M, Moffat K, van Grondelle R. The LOV2 domain of phototropin: a reversible photochromic switch. J. Am. Chem. Soc. 2004;126:4512–4513. doi: 10.1021/ja031840r. [DOI] [PubMed] [Google Scholar]
- Kihara J, Moriwaki A, Tanaka N, Ueno M, Arase S. Characterization of the BLR1 gene encoding a putative blue-light regulator in the phytopathogenic fungus Bipolaris oryzae. FEMS Microbiol. Lett. 2007;266:110–8. doi: 10.1111/j.1574-6968.2006.00514.x. [DOI] [PubMed] [Google Scholar]
- Krauss U, Minh BQ, Losi A, Gärtner W, Eggert T, von Haeseler A, Jaeger K-E. Distribution and phylogeny of light-oxygen-voltage-blue-light-signaling proteins in the three kingdoms of life. J. Bacteriol. 2009;191:7234–7242. doi: 10.1128/JB.00923-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo H. Isolation of madA homologs in Pilobolus crystallinus. Mycoscience. 2009;5:400–406. [Google Scholar]
- Kuratani M, Tanaka K, Terashima K, Muraguchi H, Nakazawa T, Nakahori K, Kamada T. The dst2 gene essential for photomorphogenesis of Coprinopsis cinerea encodes a protein with a putative FAD-binding-4 domain. Fungal Genet. Biol. 2010;47:152–158. doi: 10.1016/j.fgb.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Lapeña MA, Vicente-Soler J, Soto T, Madrid M, Núñez A, García E, Cansado J, Gacto M. Light-induced rhythmic changes in thermotolerance in stationary-phase cells of Candida utilis. Int. Microbiol. 2006;9:61–64. [PubMed] [Google Scholar]
- Lee K, Singh P, Chung W-C, Ash J, Kim TS, Hang L, Park S. Light regulation of asexual development in the rice blast fungus, Magnaporthe grisea. Fungal Genet. Biol. 2006;43:694–706. doi: 10.1016/j.fgb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Lee SC, Corradi N, Byrnes EJ, 3rd, Torres-Martinez S, Dietrich FS, Keeling PJ, Heitman J. Microsporidia evolved from ancestral sexual fungi. Curr. Biol. 2008;18:1675–1679. doi: 10.1016/j.cub.2008.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis ZA, Correa A, Schwerdtfeger C, Link KL, Xie X, Gomer RH, Thomas T, Ebbole DJ, Bell-Pedersen D. Overexpression of White Collar-1 (WC-1) activates circadian clock-associated genes, but is not sufficient to induce most light-regulated gene expression in Neurospora crassa. Mol. Microbiol. 2002;45:917–931. doi: 10.1046/j.1365-2958.2002.03074.x. [DOI] [PubMed] [Google Scholar]
- Li D, Kolattukudy PE. Cloning and expression of cDNA encoding a protein that binds a palindromic promoter element essential for induction of fungal cutinase by plant cutin. J. Biol. Chem. 1995;270:11753–11756. doi: 10.1074/jbc.270.20.11753. [DOI] [PubMed] [Google Scholar]
- Liedvogel M, Mouritsen H. Cryptochromes-a potential magnetoreceptor: what do we know and what do we want to know? J. R. Soc. Interface. 2010;7:S147–S162. doi: 10.1098/rsif.2009.0411.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden H, Macino G. White collar 2, a partner in blue-light signal transduction, controlling expression of light-regulated genes in Neurospora crassa. EMBO J. 1997;16:98–109. doi: 10.1093/emboj/16.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson ED, Terasaka DT, Silverstein PS. Double mutants of Phycomyces with abnormal phototropism. Mol. Gen. Genet. 1980;179:155–162. [Google Scholar]
- Liu Y, He Q, Cheng P. Photoreception in Neurospora: a tale of two White Collar proteins. Cell Mol. Life Sci. 2003;60:2131–2138. doi: 10.1007/s00018-003-3109-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losi A, Gärtner W. Bacterial bilin- and flavin-binding photoreceptors. Photochem. Photobiol. Sci. 2008;7:1168–1178. doi: 10.1039/b802472c. [DOI] [PubMed] [Google Scholar]
- Lu Y-K, Sun K-H, Shen W-C. Blue light negatively regulates the sexual filamentation via the Cwc1 and Cwc2 proteins in Cryptococcus neoformans. Mol. Microbiol. 2005;56:280–291. doi: 10.1111/j.1365-2958.2005.04549.x. [DOI] [PubMed] [Google Scholar]
- Marsh PB, Taylor EE, Bassler LM. A guide to the literature on certain effects of light on fungi: reproduction, morphology, pigmentation, and phototropic phenomena. Plant Dis. Rep. Suppl. 1959;261:251–312. [Google Scholar]
- Moliné M, Libkind D, del Carmen Diéguez M, van Broock M. Photoprotective role of carotenoids in yeasts: response to UV-B of pigmented and naturally-occurring albino strains. J. Photochem. Photobiol. B. 2009;95:156–161. doi: 10.1016/j.jphotobiol.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Mooney JL, Yager LN. Light is required for condiation in Aspergillus nidulans. Genes Dev. 1990;4:1473–1482. doi: 10.1101/gad.4.9.1473. [DOI] [PubMed] [Google Scholar]
- Moriwaki A, Katsube H, Ueno M, Arase S, Kihara J. Cloning and characterization of the BLR2, the homologue of the blue-light regulator of Neurospora crassa WC-2, in the phytopathogenic fungus Bipolaris oryzae. Curr. Microbiol. 2008;56:115–121. doi: 10.1007/s00284-007-9080-x. [DOI] [PubMed] [Google Scholar]
- Muehlstein LK, Amon JP, Leffler DL. Phototaxis in the marine fungus Rhizophydium littoreum. Appl. Environ. Microbiol. 1987;53:1819–1821. doi: 10.1128/aem.53.8.1819-1821.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahashi G, Douds DD., Jr. Action spectrum for the induction of hyphal branches of an arbuscular mycorrhizal fungus: exposure sites versus branching sites. Mycol. Res. 2003;107:1075–1082. doi: 10.1017/s0953756203008232. [DOI] [PubMed] [Google Scholar]
- Olmedo M, Ruger-Herreros C, Luque EM, Corrochano LM. A complex photoreceptor system mediates the regulation by light of the conidiation genes con-10 and con-6 in Neurospora crassa. Fungal Genet. Biol. 2010;47:352–363. doi: 10.1016/j.fgb.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Page RM. Studies on the development of asexual reproductive structures in Pilobolus. Mycologia. 1956;48:206–224. [Google Scholar]
- Payen A. Extrain d'un rapport addressé à M. Le Maréchal Duc de Dalmatie, Ministre de la Guerre, Président du Conseil, sur une altération extraordinaire du pain du munition. Ann. Chim. Phys. 1843;3:5–21. [Google Scholar]
- Perkins DD, Glassey M, Bloom BA. New data on markers and rearrangements in Neurospora. Can. J. Genet. Cytol. 1962;4:187–205. [Google Scholar]
- Porter TM, Schadt CW, Rizvi L, Martin AP, Schmidt SK, Scott-Denton L, Vilgalys R, Moncalvo JM. Widespread occurrence and phylogenetic placement of a soil clone group adds a prominent new branch to the fungal tree of life. Mol. Phylogenet. Evol. 2008;46:635–644. doi: 10.1016/j.ympev.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Prado MM, Prado-Cabrero A, Fernández-Martin R, Avalos J. A gene of the opsin family in the carotenoid gene cluster of Fusarium fujikuroi. Curr. Genet. 2004;46:47–58. doi: 10.1007/s00294-004-0508-6. [DOI] [PubMed] [Google Scholar]
- Purschwitz J, Müller S, Fischer R. Mapping the interaction sites of Aspergillus nidulans phytochrome FphA with the global regulator VeA and the White Collar protein LreB. Mol Genet. Genomics. 2009;281:35–42. doi: 10.1007/s00438-008-0390-x. [DOI] [PubMed] [Google Scholar]
- Purschwitz J, Müller S, Kastner C, Fischer R. Seeing the rainbow: light sensing in fungi. Curr. Opin. Microbiol. 2006;9:566–571. doi: 10.1016/j.mib.2006.10.011. [DOI] [PubMed] [Google Scholar]
- Purschwitz J, Müller S, Kastner C, Schöser M, Haas H, Espeso EA, Atoui A, Calvo AM, Fischer R. Functional and physical interaction of blue- and red-light sensors in Aspergillus nidulans. Curr. Biol. 2008;18:255–259. doi: 10.1016/j.cub.2008.01.061. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Su YS, Lagarias JC. Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales-Saavedra T, Esquivel-Naranjo EU, Casas-Flores S, Martínez-Hernández P, Ibarra-Laclette E, Cortes-Penagos C, Herrera-Estrella A. Novel light-regulated genes in Trichoderma atroviride: a dissection by cDNA microarrays. Microbiology. 2006;152:3305–3317. doi: 10.1099/mic.0.29000-0. [DOI] [PubMed] [Google Scholar]
- Ruiz-Roldán MC, Garre V, Guarro J, Mariné M, Roncero MIG. Role of the white collar 1 photoreceptor in carotenogenesis, UV resistance, hydrophobicity, and virulence of Fusarium oxysporum. Eukaryot. Cell. 2008;7:1227–1230. doi: 10.1128/EC.00072-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A. Photolyase and cryptochrome blue-light photoreceptors. Adv. Protein Chem. 2004;69:73–100. doi: 10.1016/S0065-3233(04)69003-6. [DOI] [PubMed] [Google Scholar]
- Sano H, Kaneko S, Sakamoto Y, Sato T, Shishido K. The basidiomycetous mushroom Lentinula edodes white collar-2 homolog PHRB, a partner of putative blue-light photoreceptor PHRA, binds to a specific site in the promoter region of the L. edodes tyrosinase gene. Fungal Genet. Biol. 2009;46:333–341. doi: 10.1016/j.fgb.2009.01.001. [DOI] [PubMed] [Google Scholar]
- Sano H, Narikiyo T, Kaneko S, Yamazaki T, Shishido K. Sequence analysis and expression of a blue-light photoreceptor gene, Le.phrA from the basidiomycetous mushroom Lentinula edodes. Biosci. Biotechnol. Biochem. 2007;71:2206–2213. doi: 10.1271/bbb.70170. [DOI] [PubMed] [Google Scholar]
- Sanz C, Rodríguez-Romero J, Idnurm A, Heitman J, Christie JM, Corrochano LM, Eslava AP. A photoreceptor complex for fungal phototropism: the Phycomyces MADB protein is a WC-2 homolog that interacts with the MADA photoreceptor. Proc. Natl. Acad. Sci. USA. 2009;106:7095–7100. doi: 10.1073/pnas.0900879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saranak J, Foster KW. Rhodopsin guides fungal phototaxis. Nature. 1997;387:465–466. doi: 10.1038/387465a0. [DOI] [PubMed] [Google Scholar]
- Sargent ML, Briggs WR. The effects of light on a circadian rhythm of conidiation in Neurospora. Plant Physiol. 1967;42:1504–1510. doi: 10.1104/pp.42.11.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidle A. Die Tagesperiodizität der asexuallen Reproduktion von Pilobolus sphaerosporus. Arch. Mikrobiol. 1951;16:80–100. [Google Scholar]
- Schmoll M, Franchi L, Kubicek CP. Envoy, a PAS/LOV domain protein of Hypocrea jecorina (anamorph Trichoderma reesei), modulates cellulase gene transcription in response to light. Eukaryot. Cell. 2005;4:1998–2007. doi: 10.1128/EC.4.12.1998-2007.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster A, Kubicek CP, Friedl MA, Druzhinina IS, Schmoll M. Impact of light on Hypocrea jecorina and the multiple cellular roles of ENVOY in this process. BMC Genomics. 2007;8:449. doi: 10.1186/1471-2164-8-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwerdtfeger C, Linden H. VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO J. 2003;22:4846–4855. doi: 10.1093/emboj/cdg451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby CP, Sancar A. A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc. Natl. Acad. Sci. USA. 2006;103:17696–17700. doi: 10.1073/pnas.0607993103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma AK, Spudich JL, Doolittle WF. Microbial rhodopsins: functional versatility and genetic mobility. Trends Microbiol. 2006;14:463–469. doi: 10.1016/j.tim.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Silva F, Navarro E, Peñaranda A, Murcia-Flores L, Torres-Martínez S, Garre V. A RING-finger protein regulates carotenogenesis via proteolysis-independent ubiquitylation of a white collar-1-like activator. Mol. Microbiol. 2008;70:1026–1036. doi: 10.1111/j.1365-2958.2008.06470.x. [DOI] [PubMed] [Google Scholar]
- Silva F, Torres-Martínez S, Garre V. Distinct white collar-1 genes control specific light responses in Mucor circinelloides. Mol. Microbiol. 2006;61:1023–1037. doi: 10.1111/j.1365-2958.2006.05291.x. [DOI] [PubMed] [Google Scholar]
- Slamovits CH, Keeling PJ. Class II photolyase in a microsporidian intracellular parasite. J. Mol. Biol. 2004;341:713–721. doi: 10.1016/j.jmb.2004.06.032. [DOI] [PubMed] [Google Scholar]
- Spudich JL, Yang CS, Jung KH, Spudich EN. Retinylidene proteins: structures and functions from archaea to humans. Annu. Rev. Cell. Dev. Biol. 2000;16:365–392. doi: 10.1146/annurev.cellbio.16.1.365. [DOI] [PubMed] [Google Scholar]
- Taylor JW, Berbee ML. Dating divergences in the fungal tree of life: review and new analyses. Mycologia. 2006;98:838–849. doi: 10.3852/mycologia.98.6.838. [DOI] [PubMed] [Google Scholar]
- Terashima K, Yuki K, Muraguchi H, Akiyama M, Kamada T. The dst1 gene involved in mushroom photomorphogenesis of Coprinus cinereus encodes a putative photoreceptor for blue light. Genetics. 2005;171:101–108. doi: 10.1534/genetics.104.040048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisch D, Schmoll M. Light regulation of metabolic pathways in fungi. Appl. Microbiol. Biotechnol. 2010;85:1259–1277. doi: 10.1007/s00253-009-2320-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uebelmesser E-R. Über den endonomen Tagesrhythmus der Sporangienträgerbildung von Pilobolus. Arch. Mikrobiol. 1954;20:1–33. [PubMed] [Google Scholar]
- Ułaszewski S, Mamouneas T, Shen W-K, Rosenthal PJ, Woodward JR, Cirillo VP, Edmunds LN., Jr. Light effects in yeast: evidence for participation of cytochromes in photoinhibition of growth and transport in Saccharomyces cerevisiae cultured at low temperatures. J. Bacteriol. 1979;138:523–529. doi: 10.1128/jb.138.2.523-529.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veluchamy S, Rollins JA. A CRY-DASH-type photolyase/cryptochrome from Sclerotinia sclerotiorum mediates minor UV-A-specific effects on development. Fungal Genet. Biol. 2008;45:1265–1276. doi: 10.1016/j.fgb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Vines SH. The influence of light upon the growth of unicellular organs. Arb. Bot. Inst. Würzburg. 1878;II:133–147. [Google Scholar]
- Waschuk SA, Bezerra AG, Jr, Shi L, Brown LS. Leptosphaeria rhodopsin: bacteriorhodopsin-like proton pump from a eukaryote. Proc. Natl. Acad. Sci. USA. 2005;102:6879–6883. doi: 10.1073/pnas.0409659102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Went FAFC. Ueber den einfluss des lichtes auf die entstehung des carotins und auf die zersetzung der enzyme. Rec. Trav. Botan. Néerl. 1904;1:106–119. [Google Scholar]
- Whippo CW, Hangarter RP. Phototropism: bending towards enlightenment. Plant Cell. 2006;18:1110–1119. doi: 10.1105/tpc.105.039669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarwood CE. Diurnal cycle of ascus maturation of Taphrina deformans. Am. J. Bot. 1941;28:355–357. [Google Scholar]
- Yeh Y-L, Lin Y-S, Su B-J, Shen W-C. A screening for suppressor mutants reveals components involved in the blue light-inhibited sexual filamentation in Cryptococcus neoformans. Fungal Genet. Biol. 2009;46:42–54. doi: 10.1016/j.fgb.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Zalokar M. Biosynthesis of carotenoids in Neurospora; action spectrum of photoactivation. Arch. Biochem. 1955;56:318–325. doi: 10.1016/0003-9861(55)90252-6. [DOI] [PubMed] [Google Scholar]
- Zoltowski BD, Schwerdtfeger C, Widom J, Loros JJ, Bilwes AM, Dunlap JC, Crane BR. Conformational switching in the fungal light sensor Vivid. Science. 2007;316:1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoltowski BD, Vaccaro B, Crane BR. Mechanism-based tuning of a LOV domain photoreceptor. Nat. Chem. Biol. 2009;5:827–834. doi: 10.1038/nchembio.210. [DOI] [PMC free article] [PubMed] [Google Scholar]