Abstract
Objective
To conduct postmortem human brain research into the neuropathological basis of schizophrenia, it is critical to establish cohorts that are well-characterised and well-matched. Our objective was to determine if specimen characteristics, including: diagnosis, age, postmortem interval (PMI), brain acidity (pH), and/or the agonal state of the subject at death related to RNA quality, and to determine the most appropriate reference gene mRNAs.
Methods
We selected a matched cohort of 74 cases (37 schizophrenia / schizoaffective disorder cases and 37 controls cases). Middle frontal gyrus tissue was pulverised, tissue pH was measured, RNA isolated for cDNA from each case, and RNA integrity number (RIN) measurements were assessed. Using RT-PCR, we measured nine housekeeper genes and calculated a geomean in each diagnostic group.
Results
We found that the RINs were very good (mean 7.3) and all nine housekeeper control genes were significantly correlated with RIN. Seven of nine housekeeper genes were also correlated with pH, and two clinical variables, agonal state and duration of illness did have an effect on some control mRNAs. No major impact of PMI or freezer time on housekeeper mRNAs was detected. Our results show that people with schizophrenia had significantly less PPIA, and SDHA and tended to have less GUSB and B2M mRNA suggesting that these control genes may not be good candidates for normalisation.
Conclusions
In our cohort, less than 10% variability in RIN values was detected and the diagnostic groups were well matched overall. Our cohort was adequately powered (0.80–0.90) to detect mRNA differences (25%) due to disease. Our study suggests that multiple factors should be considered in mRNA expression studies of human brain tissues. When schizophrenia cases are adequately matched to control cases subtle differences in gene expression can be reliably detected.
Keywords: postmortem brain, pH, RIN, PMI, housekeeping genes
INTRODUCTION
Schizophrenia research on postmortem human tissue has matured in the past decade, largely due to two factors 1) the widespread availability of larger cohorts that are more extensively screened and 2) the readily available quantitative molecular tools especially for gene expression studies. In the past, lack of replication of basic biological findings in brains of people with schizophrenia across laboratories hampered forward progress. The renewed research effort on postmortem brains is finally yielding numerous replicable findings across independent laboratories and independent cohorts, which is a critical advance for postmortem brain research on schizophrenia. Studies of postmortem human brain provide a valuable avenue for elucidating the underlying cellular and molecular mechanisms of schizophrenia that are not accessible through live imaging of people or through animal models. Thus, there is a need for directly studying postmortem human brains from people with schizophrenia, although collecting and characterizing human brains is methodologically challenging [1–3] and standards regarding inclusion criteria necessary for rigorous postmortem research are not universally agreed upon and continue to change [4–11]. Numerous premortem, antemortem and postmortem factors can influence the quality of human brain tissue and need to be considered and the relative importance of confounding factors for specific molecules are often not known.
Despite these limitations, postmortem human studies of psychiatric illnesses such as schizophrenia remain an extremely valuable tool, as they have already begun to shed light on a number of molecular mechanisms that contribute to the aetiology and development of schizophrenia [12–17]. The establishment of multiple large cohorts of well characterised brains of people with schizophrenia and controls from around the world contribute to our understanding of the biological basis for schizophrenia and will be critical to advancing the field. Therefore, given our experience, and with access to a fairly large collection of postmortem tissue samples, we set out to establish a new cohort of schizophrenia and control brains from Sydney, Australia.
The assembly and characterization of this cohort has been driven by the Schizophrenia Research Institute (SRI), a `virtual institute' supporting the collaboration of Australian scientists and clinicians who bring a variety of knowledge, skills and techniques together to find ways to prevent and cure schizophrenia [18]. In 2007, through the Institute's Developmental Neurobiology Panel, SRI commenced a process of actively facilitating the development of a large-scale collaborative research program based on the use of a common collection of postmortem brains for schizophrenia research from the Tissue Resource Centre at the University of Sydney. The results from this multifaceted investigation will be collected and collated, thereby allowing for testing of relationships among a variety of measures. This report represents the first step in this collaborative process whereby the collection and quality control processes for this new SRI tissue cohort are described and the selection of reference genes is considered.
METHODS AND MATERIALS
Subjects
All research was approved and conducted under the guidelines of the Human Research Ethics Committee at the University of New South Wales (HREC 07261). The study cohort included 37 schizophrenia/schizoaffective disorder cases and 37 matched controls (Table 1). Controls were selected prior to RNA extraction based on the following parameters (in descending order of importance for matching), brain pH (±1), age at death (within 10 years), and PMI (within 10 hours). Other factors such as hemisphere, gender, time in freezer and agonal state were matched on a stratum (group-wise) basis. Details of brain collection and determination of clinical and tissue factors can be found in Supplementary Materials and Methods, and in Tables 5S and 6S.
Table 1.
Overview of cohort demographic measures.
| Demographic Measures | Control Cases | Schizophrenia Cases |
|---|---|---|
| Age | 51.1 +/− 2.40 | 51.3 +/− 2.32 |
| pH (DLPFC) | 6.66 +/− 0.29 | 6.61 +/− 0.30 |
| PMI | 24.8 +/− 10.97 | 28.8 +/− 14.07 |
| RIN | 7.3 +/− 0.57 | 7.3 +/− 0.58 |
| Gender | 7F, 30M | 13F, 24M |
| Hemisphere | 23R, 14L | 17R, 20L |
PMI=postmortem interval; RIN=RNA integrity number
RNA extraction and quality assessment
Tissue from dorsal lateral prefrontal cortex (DLPFC) was pulverized over dry ice. Approximately 300 mg was weighed while frozen and stored at −80°C until extraction. Total RNA was extracted with TRIzol Reagent (Invitrogen Life Science, Cat. No. 15596-018, Mount Waverley, Victoria), using a polytron as previously described [19]. Total RNA pellets were re-suspended in DEPC treated water (Sigma-Aldrich, Cat. No. 95284-100ML, Castle Hill NSW). The yield of total RNA was then analysed using a spectrophotometer (Nanodrop ND-1000, Thermo Scientific). Total RNA was not further purified through a column. The quality of extracted total RNA was determined by high resolution capillary electrophoresis using the Agilent Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA, USA). Approximately 100–200ng RNA was applied to an RNA 6000 Nano LabChip, without heating prior to loading, and subsequent steps performed according to the manufacturer's protocol. Two internal controls (samples with established high quality RNA RIN of 10) were loaded on each chip, and all samples analysed on the same day. The RNA Integrity Number (RIN) was calculated by an algorithm incorporating information from the entire electrophoretic trace and used as an indicator of RNA quality, ranging from 1 (lowest quality) to 10 (highest quality) [20]. Three separate aliquots of total RNA were used to synthesise cDNA from 3μg of total RNA in a 26.25μl reaction using the SuperScript® First-Strand Synthesis kit (Invitrogen, Carlsbad, CA, USA) and were pooled to reduce any potential variability in cDNA reactions prior to diluting for RT-PCR.
Quantitative Real-Time PCR
Transcript levels for nine housekeeper genes – glucuronidase β (GUSB), cyclophilin (PPIA), ubiquitin C (UBC), porphobilinogen deaminase (PBGD), succinate dehydrogenase complex subunit A (SDHA), β-actin (ACTB), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), TATA box binding protein (TBP) and β-2-microglobulin (B2M) – were measured by quantitative real-time PCR (qPCR) in each cohort sample using an ABI Prism 7900HT Fast Real Time PCR system with a 384-well format and TaqMan Gene Expression Assays (Applied Biosystems, Inc) (Table 2). Samples were run with a seven point standard curve using serial dilutions of pooled cDNA derived from a representative sample of subjects (three controls and three patients). The `no template control' did not produce a signal in any assay. All amplifications from each subject were performed in triplicate and relative quantities were determined from the standard curve. Outliers due to measurement errors were omitted if the percent variance of the triplicates was greater than 30% of the quantity mean value as previously described ([21], [22],[23]) and the mean re-calculated based on two values (this occurred in less than 5% of the samples). Stability of mRNA expression was measured using the program geNorm VBA applet for Microsoft Excel v3.5, developed by Vandesompele and colleagues (2002), where more stable genes will have a lower M–value [24]
Table 2.
Housekeeper genes with ABI Taqman Gene Expression assay part numbers.
| Gene Symbol | Gene Name | Taqman Assay |
|---|---|---|
| GUSB | Glucuronidase, beta | Hs99999908_m1 |
| PBDG | Hydroxymethylbilane synthase | Hs00609297_m1 |
| PPIA | Peptidylprolyl isomerase A (Cyclophilin A) | Hs99999904_m1 |
| UBC | Ubiquitin C | Hs00824723_m1 |
| SDHA | Succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | Hs00188166_m1 |
| ACTB | Actin, beta | Hs99999903_m1 |
| TBP | TATA box binding protein | Hs00427620_m1 |
| B2M | Beta-2-microglobulin | Hs99999907_m1 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | Hs99999905_m1 |
Statistical analysis
Statistical analyses were conducted using Statistica (StatSoft Inc., 2005, STATISTICA version 7.1. www.statsoft.com). After measurement errors were removed, population outliers were determined and samples greater than two standard deviations from the mean for each group were removed (on average 1–2 subjects/group). We also defined population outliers using the Grubb's test which yielded similar results. Both paired and non-paired Student's t-tests were used to determine if the diagnostic groups differed on individual housekeepers or geomean. To maximize power, the analyses in this study were carried out with the schizophrenia patients combined with the schizoaffective patients as a single diagnostic category; however, it is recognized that other analyses focusing only on the schizophrenia group are also planned. Correlations were run on continuous variables to determine if any of the demographic factors (age, pH, PMI, or RIN) related to housekeeper mRNA levels or their geometric mean. If this was the case, analysis of covariance (ANCOVA) analyses were used. Independent t-tests were conducted to examine whether gender or hemisphere were related to housekeeper gene expression. Factorial ANOVA was used to determine the effect of gender or hemisphere on gene expression in the diagnostic groups. Statistical significance level was set at p= or < 0.05. When we report significance level, we rounded all p values three significant figures.
RESULTS
Matching of diagnostic groups
The patients with schizophrenia and schizoaffective disorder (n=37) were on average 51 years old, with a brain pH of 6.61 and a PMI of 28.5 hours (Table 1). The normal control subjects selected to match these cases (n=37) had an average age of 51 years, a brain pH of 6.66 and a PMI of 25.0 hours. The mean RIN values were 7.3 for both groups and had a low coefficient of variation (between 7.8 and 8.0%). As anticipated age, pH, PMI, freezer storage time, and RIN did not differ significantly between the groups (all t<1.3, all p>0.210). Freezer storage time of brain tissue for patients averaged 81 months and did not differ statistically as compared to normal control cases at 70 months. The number of males was higher than females in both groups although the number of males was slightly lower in the schizophrenia group (n=24) as compared to the normal control group (n=30). Females with schizophrenia (n=13) outnumbered their control group counterparts (n=7). The number of right and left hemispheres studied was comparable for both the control (L=14, R=23) and patient (L=20, R=17) groups. One patient with a comorbid diagnosis for alcohol dependency and another schizophrenia patient with comorbid drug abuse at the time of death were included in the cohort.
Relationship of demographic variables to each other and RIN
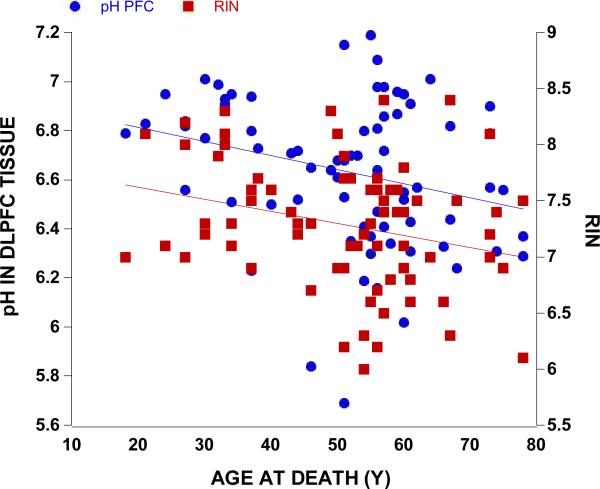
Our subjects spanned a considerable age range (18–78 years) and we found that age at death negatively correlated with the overall brain weight (r=−0.32, p=0.005, n=74), overall brain volume (r=−0.26, p=0.026, n=74), and age correlated with DLPFC pH (r=−0.27, p=0.018, n=74) and RIN (r=−0.27, p=0.020, n=74) (Figure 1). As expected, females (n=20, 1300 mLs) had smaller brains as compared to males (n=54, 1468 mLs, t=5.13 df= 72, p<0.0001) and females (1288 g) had a lighter brain as compared to males (1469 g), (t=5.55 df=72, p<0.0001). Patients with schizophrenia (1394 g) had a lighter brain weight compared to non-affected controls (1446 g) though this difference was not statistically significant (t=−1.53 df=72, p=0.131). People with schizophrenia tended to smoke (n=23) rather than not smoke (n=7) and were more likely to be confirmed smokers than controls (n=9). Smokers tended to have lower brain pH values in both frontal cortex (6.63 versus 6.66) and cerebellum (6.45 versus 6.57) and frontal cortex RIN was marginally lower in the smokers (7.2 versus 7.3). However, the pH and RIN values in the smokers and non-smokers did not differ statistically.
Figure 1.
Overall pH and RIN values similarly decreased with increasing age (both r=−0.27, p<0.020, n=74).
As expected, the RIN measurement for DLPFC total RNA positively correlated with brain pH taken from the same tissue (r=0.39, p<0.001) and also correlated with cerebellar pH (r=0.31, p=0.007) (Figure 6A-Supp). We found that the pH measurements from the cerebellum and middle frontal gyrus correlated significantly with each other (r=0.48, p<0.001, n=74). PMI did not correlate with RIN (r=0.02, p=0.873) (Figure 6B-Supp) or cerebellar pH (r=0.17, p=0.216). Additionally, we extracted the same amount of RNA per gram of frozen tissue in people with schizophrenia and unaffected controls (both, 0.66 μg/mg, t=−0.06, df=72, p=0.958).
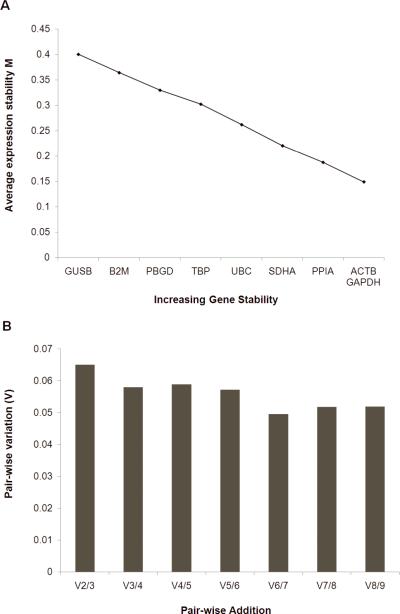
Transcript stability and calculation of geometric mean
We determined that ACTB and GADPH were the two most stable genes generating the lowest M value, and that B2M and GUSB were the least stable genes with the highest M values (Figure 2A). Pair-wise variation analysis suggests that using the two most stable housekeeping genes (ACTB and GADPH) is sufficient to determine the normalisation factor as the V2/3 value is below the suggested threshold of 0.15 (Figure 2B) [24]. However, for the purposes of normalising our data to housekeeper control genes, we chose a further two genes as it has also been recommended that genes used for normalisation are not significantly altered in schizophrenia compared to controls (see below), and represent transcripts of different levels of expression. We chose 4 genes, two (ACTB and GAPDH) with higher expression levels, one (UBC) being in the mid-range of expression, and one (TBP) with lower expression levels [25]. When the geomean was recalculated using these four housekeeping genes [herein geomean(4)] and outliers removed (1 case removed) we did not detect a significant difference between our normalizing factor geomean in patients with schizophrenia compared to controls (unpaired t-test, t=0.82 df=71, p=0.415; paired t-test, t=−0.79, df=35, p=0.420) (Figure 3A).
Figure 2.
Average expression stability and optimal number of housekeeper genes for normalisation determined using the program geNorm. (A) Genes were ranked according to average expression stability. ACTB and GAPDH were the most stable genes (lowest M value), while GUSB was the least stable gene (highest M value). (B) Pairwise variation with the stepwise addition of more housekeeping genes shows that the use of the two most stable housekeeping genes will be sufficient for accurate normalisation as this satisfies the cutoff (0.15M) recommended by Vandesompele et al (2002).
Figure 3.
(A) The calculated geomean(4) was comparatively similar in both diagnostic groups whether paired(t=−0.79, df=35, p=0.420) or unpaired (t=0.82 df=71, p=0.415); (B) The geomean(4) decreased with an increase in age (r=−0.28, p=0.018); (C) Overall pH in DLPFC tissue was positively correlated geomean(4) for controls (r=0.41, p=0.012) and patients (r=0.52, p=0.001); (D) Agonal state 1 versus 3 showed a significant decrease in the geomean(4) (p=0.031 by planned post-hoc LSD); (E) RIN values strongly correlated with the geomean(4) (r=0.57, p<0.0001); (E) PMI had no effect on the geomean(4) (r=0.006, p=0.96).
Correlation of demographic variables and housekeeper mRNAs
Age
When considering the whole cohort, the expression of 6 of 9 housekeeping genes correlated negatively with age at death (SDHA r=−0.26, p=0.026; ACTB r=−0.28, p=0.018; GAPDH r=−0.28, p=0.016; PBGD r=−0.26, p=0.030, UBC r=−0.26, p=0.03, PPIA r=−0.26, p=0.025) as did the geomean(4) (r=−0.28, p=0.018) (Figure 3B). When we explored which diagnostic group contributed more to these overall correlations, we found that the normal group did not show any significant correlations with any housekeeper control genes or geomean(4) and age at death (all p>0.08). In contrast, the expression of 5 of 9 housekeeping genes and geomean(4) correlated negatively with age in schizophrenia patients only (SDHA r=−0.45, p=0.006; ACTB r=−0.47, p=0.004; GAPDH r=−0.37, p=0.024; UBC r=−0.37, p=0.028, PPIA r=−0.40, p=0.016, geomean(4) r=−0.45, p=0.006).
pH
Most (7 of 9) reference genes showed a significant positive correlation with DLPFC tissue pH (range r=0.26 to 0.58, p=0.02 to p<0.001). The only two potential reference genes that did not show a significant relationship with tissue pH were those deemed least stable, B2M (r=0.14, p=0.237) and GUSB (r=0.11, p=0.364). In general, most reference mRNAs were positively correlated to tissue pH in both control cases and in cases with schizophrenia. Additionally, geomean(4) showed a significant correlation to pH in both diagnostic groups (r=0.41, p=0.012 in controls, r=0.52, p=0.001 in patients) (Figure 3C).
Agonal State
Agonal state had a significant influence on DLPFC pH (F=3.07. df=2,71, p=0.051) and but this did not reach statistical significance with cerebellar pH (F=3.01, df=2,71, p=0.057). In both the prefrontal cortex and the cerebellum, the individuals with the highest agonal state rating had the lowest pH (6.44 and 6.32 respectively). Each agonal state rating had significantly different DLPFC pH values by post-hoc LSD (all p< or = 0.05). Although individuals with the highest agonal state scores had the lowest RNA quality as assessed by RIN, this did not reach statistical significance. Those individuals with the longest agonal state in our study did show a 27% decrease (F=2.68, df=2,33, p=0.086) in expression of the geomean(4) of our housekeeper normaliser genes (Agonal state,1 vs 3, p=0.031 by planned post-hoc LSD) (Figure 3D).
RIN
All nine potential reference genes showed a significant positive correlation with DLPFC RNA quality as determined by RIN. Most of the genes (8 of 9) and the geomean(4) (Figure 3E) showed a strong and statistically significant correlation [range r=0.43 to 0.63, all p<0.001, except GUSB mRNA (r=0.24, p=0.042)]. All nine housekeeper mRNAs in the control group displayed a correlation to RIN (all r>0.46, all p<0.006). Similarly, 8 of 9 housekeeping genes showed a correlation to RIN in the schizophrenia group (excluding GUSB) (all r>0.36, all p<0.029).
PMI
Of the housekeeper control mRNAs, most of the housekeeper genes and the geomean(4) were not correlated with PMI (Figure 3F), (all r<0.067, all p>0.572 except B2M r=−0.20, p=0.088). The expression of GUSB in schizophrenia patients only (r=−0.35, p=0.041) was significantly negatively correlated with PMI.
Diagnostic differences in housekeeper control mRNA
T-tests
In our study, we found ~30% coefficient of variation in housekeeper mRNA in both groups (Table 3), which is less variable than previously found [6]. Surprisingly, we found that some housekeeping control genes were reduced in the DLPFC of patients with schizophrenia by as much as 10–15% (Figure 4 A). We found that SDHA (t=−2.21, df=69, p=0.030 unpaired; t=−2.55, df=33, p=0.016 paired), PPIA mRNA (t=−2.32, df=71, p=0.023 unpaired; t=−2.55, df=35, p=0.015 paired) and GUSB (t=1.93, df=68, p=0.057 unpaired; t=2.02, df=32, p=0.052 paired) were significantly lower in the schizophrenia and schizoaffective disorder patient group by 14.4%, 14.9%, and 12.4% respectively. The five other housekeeper genes examined – UBC, ACTB, GAPDH, TBP, PBGD – and the geomean(4) did not significantly differ in patients compared to controls (all p>0.343, Figure 4B). After ANCOVA analysis, the significant reduction in SDHA mRNA (F=4.54, df=69, p=0.036) and PPIA mRNA (F=5.78, df=68, p=0.018) was maintained. No significant differences were found in any of the other mRNAs or the geomean(4) by ANCOVA (p>0.05).
Table 3.
Housekeeper coefficiencies of variance for the diagnostic groups.
| GENE | % CON | CON % CV | SCZ % CV |
|---|---|---|---|
| SDHA | 85.6 | 30.7 | 27.3 |
| ACTB | 93.6 | 34.2 | 32.1 |
| GAPDH | 93.4 | 36.6 | 36.3 |
| B2M | 88.7 | 29.1 | 25.5 |
| TBP | 103.1 | 35.7 | 38.5 |
| GUSB | 87.6 | 29.6 | 26.7 |
| PPIA | 85.1 | 29.5 | 29.5 |
| PBGD | 93.2 | 28.1 | 34.3 |
| UBC | 96.1 | 34.9 | 34.4 |
| GEOMEAN(4) | 93.5 | 36.2 | 33.2 |
Figure 4.
mRNA expression of housekeeping genes in control and schizophrenic DLPFC. mRNA expression was measured by quantitative RT-PCR and all cases expressed as a percentage of the control mean for that gene. (A) A significant reduction in expression of SDHA, and cyclophilin (PPIA) and strong trend in GUSB mRNAs was found in schizophrenia cases compared to controls (14.4%, p=0.016; 14.9%, p=0.015; and 12.4%, p=0.052 respectively by paired t-test). (B) mRNA expression levels for all remaining housekeeping genes that did not significantly differ between diagnostic groups, including the geomean(4) (all p>0.34).
Effects of hemisphere and gender on geomean(4)
We found that there was a main effect of hemisphere on the geomean(4) by 2–way ANOVA (F=6.26, df=1, 69, p=0.021), but no overall effect of diagnosis (F=6.26, df=1, 69, p=0.228) and no interaction effect (F=6.26, df=1,69, p=0.989). Both people with schizophrenia and controls had significantly more expression of the housekeeper geomean(4) on the left hemisphere as compared to the right. When we co-varied for factors which correlated with the geomean(4) (age, pH and RIN), we found that the main effect of hemisphere was similar (F=3.86, df 1,66, p=0.053) and the effect of diagnosis and the diagnosis x hemisphere interaction remained non-significant. We did not detect a main effect of gender or diagnosis or an interaction between diagnosis and gender on the levels of geomean(4) (all F<1.3, all p>0.260).
Consideration of clinical variables
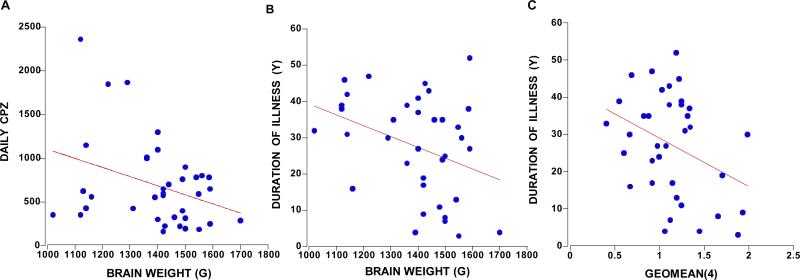
The patients with schizophrenia in our cohort had 4–45 years duration of illness which was positively correlated with daily chlorpromazine equivalent neuroleptic dose (CPZ) (r=0.38, p=0.020, n=37) and the last recorded dose of CPZ (r=0.38, p=0.027, n=37). Total brain weight of people with schizophrenia and/or schizoaffective disorders was negatively correlated with daily CPZ (r=−0.34, p=0.038, n=37) (Figure 5A), duration of illness (r=−0.35, p=0.031, n=37) (Figure 5B), and lifetime CPZ estimates (r= −0.39, p=0.018, n=37). The relationship of brain volume with duration of illness and lifetime CPZ estimates reached trend levels of significance (p=0.060–0.084, n=37). The only control mRNA that showed a statistically significant correlation with any medication measure was UBC with the last recorded CPZ (r=−0.36, p=0.048, n=31). Duration of illness did, however, negatively correlate with SDHA and ACTB (r>−0.37 p=0.028, n=35; r=−0.38, p=0.022, n=36 respectively) mRNA expression, and with the geomean(4) (r=−0.35, p=0.034, n=36) (Figure 5C). Since these same dependent variables correlated with age in the people with schizophrenia only, we ran partial correlations with age and found that only SDHA mRNA retained a relationship with duration of illness at a trend level (r=−0.32, p=0.066).
Figure 5.
The clinical variables that correlated with brain weight were daily CPZ (r=−0.34, p=0.038, n=37) (A) and duration of illness (B) (r=−0.35, p=0.031, n=37). Duration of illness negatively correlated with the geomean(4) shown in panel C (r=−0.35, p=0.034, n=36).
Power analysis
Power calculations, performed using means and standard deviations of housekeeping genes in controls and patients with schizophrenia, were used to determine the sample sizes required to reject a null hypothesis of no difference in housekeeping gene expression between diagnostic groups. Power analysis indicated that our cohort contains a sufficient sample size to detect a 1.5–fold difference in the mRNA expression of all housekeeping genes between patients and controls, with 90% statistical power (α=0.05), requiring between 6–12 samples per group to detect this fold change. Additionally, we have sufficient sample size to detect a 1.25–fold change in expression for all of our genes at 80% power and for 6 of 9 genes at 90% power (Table 4). To detect a 1.1–fold difference with 90% statistical power would require 141–299 samples per group, or relaxing stringency to 70% statistical power, 83–176 samples per group (Table 4).
Table 4.
Calculations of sample sizes sufficient to detect 1.1, 1.12, and 1.5 fold differences in gene expression at a=0.05 and 70%, 80%, and 90% statistical power.
| Power 70% |
Power 80% |
Power 90% |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 1.1 fold | 1.25 fold | 1.5 fold | 1.1 fold | 1.25 fold | 1.5 fold | 1.1 fold | 1.25 fold | 1.5 fold | |
| SDHA | 91 | 15 | 4 | 116 | 19 | 5 | 154 | 25 | 7 |
| PPIA | 92 | 15 | 4 | 118 | 19 | 5 | 157 | 26 | 7 |
| B2M | 83 | 14 | 4 | 106 | 17 | 5 | 141 | 23 | 6 |
| GUSB | 87 | 14 | 4 | 111 | 18 | 5 | 148 | 24 | 6 |
| ACTB | 127 | 21 | 6 | 163 | 26 | 7 | 217 | 35 | 9 |
| GAPDH | 154 | 25 | 7 | 196 | 32 | 8 | 262 | 42 | 11 |
| TBP | 176 | 29 | 8 | 225 | 36 | 9 | 299 | 48 | 12 |
| PBGD | 112 | 18 | 5 | 143 | 23 | 6 | 190 | 31 | 8 |
| UBC | 143 | 23 | 6 | 183 | 30 | 8 | 244 | 39 | 10 |
DISCUSSION
Some consensus concerning the effects and relative importance of pre and postmortem factors on human brain RNA quality is emerging. We find, as others do, that RIN is a powerful indicator of mRNA levels and appears to be the most critical factor for matching schizophrenia cases and controls [6, 26, 27]. However, since RIN measures are only available after RNA isolation, we found that tissue pH can be used as a general guide to select potential controls to match to cases prior to RNA isolation [26, 27]. Since cerebellar and cortical pH correlate, determining cerebellar pH may be a valuable first step in evaluating the suitability of cases for RNA analysis [26, 28]; however, high tissue pH does not always indicate good RNA [26, 27] and low pH does not necessarily yield low quality RNA. The current cut off for what is an acceptable RIN value varies considerably, with some studies suggesting that a moderate amount of degradation and a RIN as low as 3.8 to 4.0 may be usable [6, 8, 29]. Also, it should be mentioned that RIN values are typically higher after total RNA is purified by columns, however, this “purification” step would exclude some small RNAs of interest like microRNAs, and thus columns were not used in our study. Ultimately, the cut off for a usable RIN value will depend on criteria set by the end user depending on the assay employed and the molecule of interest, and thus, will vary. Earlier studies emphasize the importance of matching for tissue pH and RIN, and we have prioritized tissue pH, RIN, and age as variables to control for in assembling a cohort of schizophrenia and normal brains that are pair-selected within ranges, creating a closely matched and well-powered cohort.
Most brain banks attempt to collect cases with the shortest PMI possible, although researchers have challenged the importance of PMI in determining RNA quality [6, 30–33]. A number of studies suggest that PMI has no correlation or only a weak correlation with RNA quality [10, 26–28]. Studies which have examined human tissue collected during neurosurgery for epilepsy with minimal delay shows that the freshly acquired brain tissue pH and RIN are comparable or even lower to that collected postmortem [10, 26]. In agreement with these published findings, we find no correlation between PMI and most of our RNA quality measures in the DLPFC [26, 34] with only one slight negative correlation with SDHA as previously described [6]. A microarray study in mouse brain tissue identified over 1,000 transcripts that are degradation susceptible (labile) with increasing PMI, but they found that increased PMI does not impact GADPH mRNA levels [35]. Other studies in human brain suggest that another housekeeper control gene, B2M, is highly labile [36, 37]. On one hand, B2M may serve to “correct” for degradation levels if used to normaliser genes of interest; on the other hand, we found that B2M is a fairly unstable housekeeper control gene and thus, we would not recommend it as a good control for schizophrenia. Other studies in humans suggest that some housekeeper transcripts are slightly influenced by PMI, especially in the hippocampus [6], though this may not be the case for all hippocampal housekeeper RNAs [28]. Thus, while effects of PMI need to be evaluated for particular transcripts of interest through correlational analyses, PMI is unlikely to be a major determinant of RNA or transcript quality and may be helpful but not critical to consider when matching samples.
In contrast, it has been fairly well demonstrated that tissue pH influences mRNA preservation in postmortem tissue [6, 32, 38, 39]. In our study, we found that the majority of housekeeper RNAs assayed and the geomean(4) correlated quite strongly with tissue pH. Studies that have measured the same housekeeper controls have also found positive correlations with brain tissue pH [6, 40]. Some previous reports suggest that a pH cut off (< pH 5.9–6.0) can be used as a screen to choose which brains from which to extract RNA and analyse further [4], or that tissue pH could be used as a critical lower threshold for transcriptome analysis (pH range 6.54–6.8) [28, 41]. Since most postmortem samples used here and in other current cohorts span a fairly large pH range [6, 26–28] with as many as 39–56% of subjects from psychiatric groups below 6.5 [8] and as many as 28% of our current sample, it would not be reasonable to exclude this large number of cases. Thus it is of critical importance to either pair match for, or to group (stratum) match for tissue pH. Several research groups have pointed out that pH is not always a reliable predictor of RNA quality and we find that the individual with the lowest brain pH (5.7) had a RIN of 6.2. Hence, we agree with the recommendation that RNA isolation and RIN quality assessment should be completed on all samples before precious material is excluded from cohorts based on tissue pH value alone [6, 27].
The importance of agonal state in postmortem research has been increasingly recognized over the past several decades [36, 41–44]. We found that the longer the agonal state (prolonged death) the lower the brain pH; however we did not find an overall effect of agonal state on RIN. This agrees with studies that show that as agonal state scores increase, the tissue pH decreases [27, 28, 45], but RIN may not differ significantly with agonal state [27]. We did find a lower value for geomean(4) with higher agonal state scores. Prolonged agonal state can down-regulate some mRNAs (energy metabolism genes), while increasing others (stress response and inflammation control genes) [45], suggesting that agonal state is an important factor to consider when matching samples or groups of samples.
Studies of the effects of aging on housekeeper gene expression in the human brain have led to the conclusion that age at death influences mRNA expression, especially when individuals span a large age range [6, 40]. In our study, individuals spanned six decades of life with an average age of 51, which is comparable to several other cohorts used to study people with schizophrenia compared to controls [6, 46]. One of the surprising findings of our study is that 5 of 9 housekeeper control genes and the geomean(4) negatively correlated with age in the schizophrenia patients only. We also found that females but not males with schizophrenia had significantly smaller brains with increasing age. Lighter brains in both sexes with schizophrenia has been reported previously [47]; however the neurobiological underpinnings of this are not understood. We also found that overall brain weight and brain volume negatively correlated with the mean daily dose of neuroleptic medication and duration of illness, suggesting that the longer someone was ill or the higher the dose of daily neuroleptics, the lower the brain weight and the smaller the brain at death. Patients with a longer duration of illness tended to have higher average lifetime antipsychotic dose, higher antipsychotic dose at the time of death, and lower housekeeper mRNA expression. Since the clinical measure of duration of illness is linked to age at death, we attempted to control for the effect of age by using partial correlation and found that SDHA mRNA correlated negatively with duration of illness at a trend level. The fact that the correlations between duration of illness and housekeeper gene mRNAs is no longer statistically significant is difficult to interpret as age is not independent from duration of illness and thus some of the true variance coming from the duration of illness will be subtracted in the partial correlation for age. This is also true for earlier reports that partialled out the effect of age and failed to find an effect of duration of illness on brain weight by meta-analysis [47]. The fact that older people with schizophrenia with a longer duration of illness have smaller brains and have decreased control mRNAs suggests that the longer someone lives with schizophrenia the greater the chance they have accumulated deleterious impacts on their brain. This observation, while made on cross-sectional data across the lifespan, fits well with volumetric studies showing decreased grey matter volume over time in people suffering from chronic schizophrenia [48, 49]. Our data suggest that some of these deleterious effects may be linked to antipsychotic medication effects, or at the very least are not prevented by current treatment strategies. Recent reports suggest that chronic antipsychotic use (both typical and atypical) causes a 10% decrease in brain weight and does in fact decrease cortical brain volume in controlled stereological studies of non-human primates [50, 51]). This volume reduction was linked to a significant reduction in astrocytes and oligodendrocytes, a finding that parallels the reduction in astrocytic and oligodendrocyte markers found in some [52–55] but not all [56, 57] postmortem studies of people with schizophrenia.
In our study, we found that two housekeeper genes β-Actin and GADPH mRNAs which do not differ between people with schizophrenia compared to control as previously found ([58],[59]) and our data suggests that both are particularly stable mRNAs and useful housekeeping control genes for our postmortem human brain studies. However, if should be emphasized that the utility of GADPH as a control varies from brain collection to brain collection as some laboratories find GADPH to be stable and to not differ in people with schizophrenia as compared to controls as we do ([60], [59]), while others suggest that is one of the least stable and thus least reliable control genes ([22], [61]). We found that three putative housekeepers, GUSB, cyclophilin, and SDHA are lower in people with schizophrenia. Cyclophilin, and SDHA mRNA differences reached statistical significance and both of these housekeeper mRNAs may be regulated by cellular stress. Cyclophilin encodes a member of the peptidyl-prolyl cis-trans isomerase (PPIase) family which is a cyclosporin binding-protein and can accelerate the folding of proteins. Cyclophilin mRNA is not typically reduced in patients with schizophrenia in other cohorts [60, 62–64]. Thus, the reductions in cyclophilin mRNA that we find may represent increased power to detect a difference (~10% reduction) or may represent a finding specific to this cohort. The other transcript we found to be decreased, SDHA encodes a major catalytic subunit of succinate-ubiquinone oxidoreductase, part of the mitochondrial respiratory chain complex II on the mitochondrial inner membrane. Mutations in SDHA have been associated with a form of mitochondrial respiratory chain deficiency known as Leigh Syndrome and with late-onset optic atrophy [65, 66]. Dysfunction of mitochondrial respiration can lead to cognitive decline in attention and mental flexibility as respiratory chain diseases most often impact muscle and the CNS [67]. CNS symptoms can include epilepsy, demyelination, dementia and psychosis. Oxidative stress related to alterations in metabolism linked to mitochondrial distress has been observed in schizophrenia [43, 68–70]. GUSB, or β-glucuonidase, mRNA encodes a protein that is required to degrade glycosaminoglycans (i.e. heparin sulphate) and genetic changes in GUSB can result in deafness, behavioural deficits and mental retardation [71]. Thus, although certain genes are chosen to represent the integrity of the transcript pool and are thus used as housekeeper control or reference genes, they can have a critical function in brain as evidenced when mutated. The question regarding of whether slightly lowered reference gene transcript levels have any functional significance requires further investigation. Our study suggests that GUSB, cyclophilin and SDHA mRNAs are not optimal choices for reference genes in our schizophrenia studies.
Taken together, our data suggest that while that only a small amount of variance in RNA quality or housekeeper mRNA level is explained by any one demographic (age) or tissue characteristic (pH) their effects are consistent across many laboratories and represent known confounders/covariates which can be controlled for when constructing postmortem cohorts. We suggest that matching diagnostic groups minimally for age, pH and RIN, may help us to resolve differences in gene expression that occur in schizophrenia as either a cause, a compensation, or a consequence of the disease state [17].
Supplementary Material
Acknowledgements
Since its establishment, Schizophrenia Research Institute (SRI) has supported schizophrenia research across a broad range of disciplines and has also been a major supporter of the New South Wales Tissue Resource Centre, an infrastructure facility which collects, stores and distributes fixed and frozen post mortem human brain tissue for projects related to schizophrenia. This work was supported by Schizophrenia Research Institute, utilising infrastructure funding from NSW Health, the University of New South Wales School of Psychiatry, and the Prince of Wales Medical Research Institute. We thank Dusan Hadzi-Pavlovic for statistical advice.
References
- 1.Kleinman JE, Hyde TM, Herman MM. Methodological issues in the neuropathology of mental illness. In: Bloom FE, Kupfer DJ, editors. Psychopharmacology: The Fourth Generation of Progress. Raven Press, Ltd.; New York: 1995. pp. 859–864. [Google Scholar]
- 2.Lewis DA. The human brain revisited: opportunities and challenges in postmortem studies of psychiatric disorders. Neuropsychopharmacology. 2002;26:143–54. doi: 10.1016/S0893-133X(01)00393-1. [DOI] [PubMed] [Google Scholar]
- 3.Ravid R, Van Zwieten EJ, Swaab DF. Brain banking and the human hypothalamus--factors to match for, pitfalls and potentials. Prog Brain Res. 1992;93:83–95. doi: 10.1016/s0079-6123(08)64565-3. [DOI] [PubMed] [Google Scholar]
- 4.Bahn S, Augood SJ, Ryan M, Standaert DG, Starkey M, Emson PC. Gene expression profiling in the post-mortem human brain--no cause for dismay. J Chem Neuroanat. 2001;22:79–94. doi: 10.1016/s0891-0618(01)00099-0. [DOI] [PubMed] [Google Scholar]
- 5.Bunney WE, Bunney BG, Vawter MP, Tomita H, Li J, Evans SJ, Choudary PV, Myers RM, Jones EG, Watson SJ, Akil H. Microarray technology: a review of new strategies to discover candidate vulnerability genes in psychiatric disorders. Am J Psychiatry. 2003;160:657–66. doi: 10.1176/appi.ajp.160.4.657. [DOI] [PubMed] [Google Scholar]
- 6.Lipska BK, Deep-Soboslay A, Weickert CS, Hyde TM, Martin CE, Herman MM, Kleinman JE. Critical Factors in Gene Expression in Postmortem Human Brain: Focus on Studies in Schizophrenia. Biological Psychiatry. 2006;60:650–658. doi: 10.1016/j.biopsych.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 7.Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, Jaenisch R, Laird PW, Akbarian S. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS ONE. 2007;2:e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weis S, Llenos IC, Dulay JR, Elashoff M, Martinez-Murillo F, Miller CL. Quality control for microarray analysis of human brain samples: The impact of postmortem factors, RNA characteristics, and histopathology. J Neurosci Methods. 2007;165:198–209. doi: 10.1016/j.jneumeth.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Monoranu CM, Apfelbacher M, Grunblatt E, Puppe B, Alafuzoff I, Ferrer I, Al-Saraj S, Keyvani K, Schmitt A, Falkai P, Schittenhelm J, Halliday G, Kril J, Harper C, McLean C, Riederer P, Roggendorf W. pH measurement as quality control on human postmortem brain tissue: A Study of the BrainNet Europe Consortium. Neuropathol Appl Neurobiol. 2008 doi: 10.1111/j.1365-2990.2008.01003a.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chevyreva I, Faull RL, Green CR, Nicholson LF. Assessing RNA quality in postmortem human brain tissue. Exp Mol Pathol. 2008;84:71–7. doi: 10.1016/j.yexmp.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 11.Johnston NL, Cervenak J, Shore AD, Torrey EF, Yolken RH. Multivariate analysis of RNA levels from postmortem human brains as measured by three different methods of RT-PCR. Stanley Neuropathology Consortium. J Neurosci Methods. 1997;77:83–92. doi: 10.1016/s0165-0270(97)00115-5. [DOI] [PubMed] [Google Scholar]
- 12.Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. image 5. [DOI] [PubMed] [Google Scholar]
- 13.Mirnics K, Middleton FA, Marquez A, Lewis DA, Levitt P. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron. 2000;28:53–67. doi: 10.1016/s0896-6273(00)00085-4. [DOI] [PubMed] [Google Scholar]
- 14.Perlman WR, Weickert CS, Akil M, Kleinman JE. Postmortem investigations of the pathophysiology of schizophrenia: the role of susceptibility genes. J Psychiatry Neurosci. 2004;29:287–93. [PMC free article] [PubMed] [Google Scholar]
- 15.Pongrac J, Middleton FA, Lewis DA, Levitt P, Mirnics K. Gene expression profiling with DNA microarrays: advancing our understanding of psychiatric disorders. Neurochem Res. 2002;27:1049–63. doi: 10.1023/a:1020904821237. [DOI] [PubMed] [Google Scholar]
- 16.Tamminga CA, Holcomb HH. Phenotype of schizophrenia: a review and formulation. Mol Psychiatry. 2005;10:27–39. doi: 10.1038/sj.mp.4001563. [DOI] [PubMed] [Google Scholar]
- 17.Lewis DA. Gonzalez-Burgos G, Neuroplasticity of neocortical circuits in schizophrenia. Neuropsychopharmacology. 2008;33:141–65. doi: 10.1038/sj.npp.1301563. [DOI] [PubMed] [Google Scholar]
- 18.Draganic DM, Catts SV, Carr VJ. Neuroscience Institute of Schizophrenia and Allied Disorders (NISAD): 10 years of Australia's first virtual research institute. Aust N Z J Psychiatry. 2007;41:78–88. doi: 10.1080/00048670601057783. [DOI] [PubMed] [Google Scholar]
- 19.Kozlovsky N, Shanon-Weickert C, Tomaskovic-Crook E, Kleinman JE, Belmaker RH, Agam G. Reduced GSK-3beta mRNA levels in postmortem dorsolateral prefrontal cortex of schizophrenic patients. J Neural Transm. 2004;111:1583–92. doi: 10.1007/s00702-004-0166-3. [DOI] [PubMed] [Google Scholar]
- 20.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Law AJ, Kleinman JE, Weinberger DR, Weickert CS. Disease-associated intronic variants in the ErbB4 gene are related to altered ErbB4 splice-variant expression in the brain in schizophrenia. Hum Mol Genet. 2007;16:129–41. doi: 10.1093/hmg/ddl449. [DOI] [PubMed] [Google Scholar]
- 22.Silberberg G, Baruch K, Navon R. Detection of stable reference genes for real-time PCR analysis in schizophrenia and bipolar disorder. Anal Biochem. 2009 doi: 10.1016/j.ab.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 23.Wong J, Weickert CS. Transcriptional interaction of an estrogen receptor splice variant and ErbB4 suggest convergence in gene susceptibility pathways in Schizophrenia. J Biol Chem. 2009 doi: 10.1074/jbc.M109.013243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bustin SA, Benes V, Nolan T, Pfaffl MW. Quantitative real-time RT-PCR--a perspective. J Mol Endocrinol. 2005;34:597–601. doi: 10.1677/jme.1.01755. [DOI] [PubMed] [Google Scholar]
- 26.Stan AD, Ghose S, Gao XM, Roberts RC, Lewis-Amezcua K, Hatanpaa KJ, Tamminga CA. Human postmortem tissue: what quality markers matter? Brain Res. 2006;1123:1–11. doi: 10.1016/j.brainres.2006.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Webster MJ. Tissue preparation and banking. Prog Brain Res. 2006;158:3–14. doi: 10.1016/S0079-6123(06)58001-X. [DOI] [PubMed] [Google Scholar]
- 28.Mexal S, Berger R, Adams CE, Ross RG, Freedman R, Leonard S. Brain pH has a significant impact on human postmortem hippocampal gene expression profiles. Brain Res. 2006;1106:1–11. doi: 10.1016/j.brainres.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 29.Schoor O, Weinschenk T, Hennenlotter J, Corvin S, Stenzl A, Rammensee HG, Stevanovic S. Moderate degradation does not preclude microarray analysis of small amounts of RNA. Biotechniques. 2003;35:1192–6. 1198–201. doi: 10.2144/03356rr01. [DOI] [PubMed] [Google Scholar]
- 30.Barton AJ, Pearson RC, Najlerahim A, Harrison PJ. Pre- and postmortem influences on brain RNA. J Neurochem. 1993;61:1–11. doi: 10.1111/j.1471-4159.1993.tb03532.x. [DOI] [PubMed] [Google Scholar]
- 31.Ervin JF, Heinzen EL, Cronin KD, Goldstein D, Szymanski MH, Burke JR, Welsh-Bohmer KA, Hulette CM. Postmortem delay has minimal effect on brain RNA integrity. J Neuropathol Exp Neurol. 2007;66:1093–9. doi: 10.1097/nen.0b013e31815c196a. [DOI] [PubMed] [Google Scholar]
- 32.Kingsbury AE, Foster OJ, Nisbet AP, Cairns N, Bray L, Eve DJ, Lees AJ, Marsden CD. Tissue pH as an indicator of mRNA preservation in human post-mortem brain. Brain Res Mol Brain Res. 1995;28:311–8. doi: 10.1016/0169-328x(94)00219-5. [DOI] [PubMed] [Google Scholar]
- 33.Schramm M, Falkai P, Tepest R, Schneider-Axmann T, Przkora R, Waha A, Pietsch T, Bonte W, Bayer TA. Stability of RNA transcripts in post-mortem psychiatric brains. J Neural Transm. 1999;106:329–35. doi: 10.1007/s007020050162. [DOI] [PubMed] [Google Scholar]
- 34.Trotter SA, Brill Ii LB, Bennett JP. Stability of gene expression in postmortem brain revealed by cDNA gene array analysis. Brain Research. 2002;942:120–123. doi: 10.1016/s0006-8993(02)02644-6. [DOI] [PubMed] [Google Scholar]
- 35.Catts VS, Catts SV, Fernandez HR, Taylor JM, Coulson EJ, Lutze-Mann LH. A microarray study of post-mortem mRNA degradation in mouse brain tissue. Brain Res Mol Brain Res. 2005;138:164–77. doi: 10.1016/j.molbrainres.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 36.Atz M, Walsh D, Cartagena P, Li J, Evans S, Choudary P, Overman K, Stein R, Tomita H, Potkin S, Myers R, Watson SJ, Jones EG, Akil H, Bunney WE, Jr., Vawter MP. Methodological considerations for gene expression profiling of human brain. J Neurosci Methods. 2007;163:295–309. doi: 10.1016/j.jneumeth.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auer H, Lyianarachchi S, Newsom D, Klisovic MI, Marcucci G, Kornacker K. Chipping away at the chip bias: RNA degradation in microarray analysis. Nat Genet. 2003;35:292–3. doi: 10.1038/ng1203-292. [DOI] [PubMed] [Google Scholar]
- 38.Harrison PJ, Heath PR, Eastwood SL, Burnet PW, McDonald B, Pearson RC. The relative importance of premortem acidosis and postmortem interval for human brain gene expression studies: selective mRNA vulnerability and comparison with their encoded proteins. Neurosci Lett. 1995;200:151–4. doi: 10.1016/0304-3940(95)12102-a. [DOI] [PubMed] [Google Scholar]
- 39.Miller CL, Diglisic S, Leister F, Webster M, Yolken RH. Evaluating RNA status for RTPCR in extracts of postmortem human brain tissue. Biotechniques. 2004;36:628–33. doi: 10.2144/04364ST03. [DOI] [PubMed] [Google Scholar]
- 40.Preece P, Cairns NJ. Quantifying mRNA in postmortem human brain: influence of gender, age at death, postmortem interval, brain pH, agonal state and inter-lobe mRNA variance. Brain Res Mol Brain Res. 2003;118:60–71. doi: 10.1016/s0169-328x(03)00337-1. [DOI] [PubMed] [Google Scholar]
- 41.Li JZ, Meng F, Tsavaler L, Evans SJ, Choudary PV, Tomita H, Vawter MP, Walsh D, Shokoohi V, Chung T, Bunney WE, Jones EG, Akil H, Watson SJ, Myers RM. Sample matching by inferred agonal stress in gene expression analyses of the brain. BMC Genomics. 2007;8:336. doi: 10.1186/1471-2164-8-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomita H, Vawter MP, Walsh DM, Evans SJ, Choudary PV, Li J, Overman KM, Atz ME, Myers RM, Jones EG, Watson SJ, Akil H, Bunney WE., Jr. Effect of agonal and postmortem factors on gene expression profile: quality control in microarray analyses of postmortem human brain. Biol Psychiatry. 2004;55:346–52. doi: 10.1016/j.biopsych.2003.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vawter MP, Tomita H, Meng F, Bolstad B, Li J, Evans S, Choudary P, Atz M, Shao L, Neal C, Walsh DM, Burmeister M, Speed T, Myers R, Jones EG, Watson SJ, Akil H, Bunney WE. Mitochondrial-related gene expression changes are sensitive to agonal-pH state: implications for brain disorders. Mol Psychiatry. 2006;11:615, 663–79. doi: 10.1038/sj.mp.4001830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hardy JA, Wester P, Winblad B, Gezelius C, Bring G, Eriksson A. The patients dying after long terminal phase have acidotic brains; implications for biochemical measurements on autopsy tissue. J Neural Transm. 1985;61:253–64. doi: 10.1007/BF01251916. [DOI] [PubMed] [Google Scholar]
- 45.Li JZ, Vawter MP, Walsh DM, Tomita H, Evans SJ, Choudary PV, Lopez JF, Avelar A, Shokoohi V, Chung T, Mesarwi O, Jones EG, Watson SJ, Akil H, Bunney WE, Jr., Myers RM. Systematic changes in gene expression in postmortem human brains associated with tissue pH and terminal medical conditions. Hum Mol Genet. 2004;13:609–16. doi: 10.1093/hmg/ddh065. [DOI] [PubMed] [Google Scholar]
- 46.Hashimoto T, Arion D, Unger T, Maldonado-Aviles JG, Morris HM, Volk DW, Mirnics K, Lewis DA. Alterations in GABA-related transcriptome in the dorsolateral prefrontal cortex of subjects with schizophrenia. Mol Psychiatry. 2008;13:147–61. doi: 10.1038/sj.mp.4002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harrison PJ, Freemantle N, Geddes JR. Meta-analysis of brain weight in schizophrenia. Schizophr Res. 2003;64:25–34. doi: 10.1016/s0920-9964(02)00502-9. [DOI] [PubMed] [Google Scholar]
- 48.DeLisi LE, Sakuma M, Maurizio AM, Relja M, Hoff AL. Cerebral ventricular change over the first 10 years after the onset of schizophrenia. Psychiatry Res. 2004;130:57–70. doi: 10.1016/j.pscychresns.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 49.DeLisi LE, Sakuma M, Tew W, Kushner M, Hoff AL, Grimson R. Schizophrenia as a chronic active brain process: a study of progressive brain structural change subsequent to the onset of schizophrenia. Psychiatry Res. 1997;74:129–40. doi: 10.1016/s0925-4927(97)00012-7. [DOI] [PubMed] [Google Scholar]
- 50.Dorph-Petersen KA, Pierri JN, Perel JM, Sun Z, Sampson AR, Lewis DA. The influence of chronic exposure to antipsychotic medications on brain size before and after tissue fixation: a comparison of haloperidol and olanzapine in macaque monkeys. Neuropsychopharmacology. 2005;30:1649–61. doi: 10.1038/sj.npp.1300710. [DOI] [PubMed] [Google Scholar]
- 51.Konopaske GT, Dorph-Petersen KA, Pierri JN, Wu Q, Sampson AR, Lewis DA. Effect of chronic exposure to antipsychotic medication on cell numbers in the parietal cortex of macaque monkeys. Neuropsychopharmacology. 2007;32:1216–23. doi: 10.1038/sj.npp.1301233. [DOI] [PubMed] [Google Scholar]
- 52.Johnston-Wilson NL, Sims CD, Hofmann JP, Anderson L, Shore AD, Torrey EF, Yolken RH. Disease-specific alterations in frontal cortex brain proteins in schizophrenia, bipolar disorder, and major depressive disorder. The Stanley Neuropathology Consortium. Mol Psychiatry. 2000;5:142–9. doi: 10.1038/sj.mp.4000696. [DOI] [PubMed] [Google Scholar]
- 53.Steffek AE, McCullumsmith RE, Haroutunian V, Meador-Woodruff JH. Cortical expression of glial fibrillary acidic protein and glutamine synthetase is decreased in schizophrenia. Schizophr Res. 2008;103:71–82. doi: 10.1016/j.schres.2008.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Toro CT, Hallak JE, Dunham JS, Deakin JF. Glial fibrillary acidic protein and glutamine synthetase in subregions of prefrontal cortex in schizophrenia and mood disorder. Neurosci Lett. 2006;404:276–81. doi: 10.1016/j.neulet.2006.05.067. [DOI] [PubMed] [Google Scholar]
- 55.Webster MJ, O'Grady J, Kleinman JE, Weickert CS. Glial fibrillary acidic protein mRNA levels in the cingulate cortex of individuals with depression, bipolar disorder and schizophrenia. Neuroscience. 2005;133:453–61. doi: 10.1016/j.neuroscience.2005.02.037. [DOI] [PubMed] [Google Scholar]
- 56.Damadzic R, Bigelow LB, Krimer LS, Goldenson DA, Saunders RC, Kleinman JE, Herman MM. A quantitative immunohistochemical study of astrocytes in the entorhinal cortex in schizophrenia, bipolar disorder and major depression: absence of significant astrocytosis. Brain Res Bull. 2001;55:611–8. doi: 10.1016/s0361-9230(01)00529-9. [DOI] [PubMed] [Google Scholar]
- 57.Dean B, Gray L, Scarr E. Regionally specific changes in levels of cortical S100beta in bipolar 1 disorder but not schizophrenia. Aust N Z J Psychiatry. 2006;40:217–24. doi: 10.1080/j.1440-1614.2006.01777.x. [DOI] [PubMed] [Google Scholar]
- 58.Dean B, Keriakous D, Scarr E, Thomas EA. Gene expression profiling in Brodmann's area 46 from subjects with schizophrenia. Aust N Z J Psychiatry. 2007;41:308–20. doi: 10.1080/00048670701213245. [DOI] [PubMed] [Google Scholar]
- 59.Lauriat TL, Dracheva S, Chin B, Schmeidler J, McInnes LA, Haroutunian V. Quantitative analysis of glutamate transporter mRNA expression in prefrontal and primary visual cortex in normal and schizophrenic brain. Neuroscience. 2006;137:843–51. doi: 10.1016/j.neuroscience.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 60.O'Connor JA, Muly EC, Arnold SE, Hemby SE. AMPA receptor subunit and splice variant expression in the DLPFC of schizophrenic subjects and rhesus monkeys chronically administered antipsychotic drugs. Schizophr Res. 2007;90:28–40. doi: 10.1016/j.schres.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johansson S, Fuchs A, Okvist A, Karimi M, Harper C, Garrick T, Sheedy D, Hurd Y, Bakalkin G, Ekstrom TJ. Validation of endogenous controls for quantitative gene expression analysis: application on brain cortices of human chronic alcoholics. Brain Res. 2007;1132:20–8. doi: 10.1016/j.brainres.2006.11.026. [DOI] [PubMed] [Google Scholar]
- 62.Hashimoto R, Straub RE, Weickert CS, Hyde TM, Kleinman JE, Weinberger DR. Expression analysis of neuregulin-1 in the dorsolateral prefrontal cortex in schizophrenia. Mol Psychiatry. 2004;9:299–307. doi: 10.1038/sj.mp.4001434. [DOI] [PubMed] [Google Scholar]
- 63.Weickert CS, Straub RE, McClintock BW, Matsumoto M, Hashimoto R, Hyde TM, Herman MM, Weinberger DR, Kleinman JE. Human dysbindin (DTNBP1) gene expression in normal brain and in schizophrenic prefrontal cortex and midbrain. Arch Gen Psychiatry. 2004;61:544–55. doi: 10.1001/archpsyc.61.6.544. [DOI] [PubMed] [Google Scholar]
- 64.Weickert CS, Webster MJ, Hyde TM, Herman MM, Bachus SE, Bali G, Weinberger DR, Kleinman JE. Reduced GAP-43 mRNA in dorsolateral prefrontal cortex of patients with schizophrenia. Cereb Cortex. 2001;11:136–47. doi: 10.1093/cercor/11.2.136. [DOI] [PubMed] [Google Scholar]
- 65.Bayley JP, Devilee P, Taschner PE. The SDH mutation database: an online resource for succinate dehydrogenase sequence variants involved in pheochromocytoma, paraganglioma and mitochondrial complex II deficiency. BMC Med Genet. 2005;6:39. doi: 10.1186/1471-2350-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Horvath R, Abicht A, Holinski-Feder E, Laner A, Gempel K, Prokisch H, Lochmuller H, Klopstock T, Jaksch M. Leigh syndrome caused by mutations in the flavoprotein (Fp) subunit of succinate dehydrogenase (SDHA) J Neurol Neurosurg Psychiatry. 2006;77:74–6. doi: 10.1136/jnnp.2005.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Finsterer J. Cognitive decline as a manifestation of mitochondrial disorders (mitochondrial dementia) J Neurol Sci. 2008;272:20–33. doi: 10.1016/j.jns.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 68.Holmes E, Tsang TM, Huang JT, Leweke FM, Koethe D, Gerth CW, Nolden BM, Gross S, Schreiber D, Nicholson JK, Bahn S. Metabolic profiling of CSF: evidence that early intervention may impact on disease progression and outcome in schizophrenia. PLoS Med. 2006;3:e327. doi: 10.1371/journal.pmed.0030327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci. 2002;22:2718–29. doi: 10.1523/JNEUROSCI.22-07-02718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mimmack ML, Brooking J, Bahn S. Quantitative polymerase chain reaction: validation of microarray results from postmortem brain studies. Biol Psychiatry. 2004;55:337–45. doi: 10.1016/j.biopsych.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 71.Tomatsu S, Montano AM, Dung VC, Grubb JH, Sly WS. Mutations and polymorphisms in GUSB gene in mucopolysaccharidosis VII (Sly Syndrome) Hum Mutat. 2009;30:511–9. doi: 10.1002/humu.20828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.