Abstract
Opioid drugs such as heroin interact directly with opioid receptors whilst other addictive drugs, including marijuana, alcohol and nicotine indirectly activate endogenous opioid systems to contribute to their rewarding properties. The opioid system therefore plays a key role in addiction neurobiology and continues to be a primary focus for NIDA-supported research. Opioid receptors and their peptide ligands, the endorphins and enkephalins, form an extensive heterogeneous network throughout the central and peripheral nervous system. In addition to reward, opioid drugs regulate many functions such that opioid receptors are targets of choice in several physiological, neurological and psychiatric disorders. Because of the multiplicity and diversity of ligands and receptors, opioid receptors have served as an optimal model for G protein coupled receptor (GPCR) research. The isolation of opioid receptor genes opened the way to molecular manipulations of the receptors, both in artificial systems and in vivo, contributing to our current understanding of the diversity of opioid receptor biology at the behavioral, cellular and molecular levels. This review will briefly summarize some aspects of current knowledge that has accumulated since the very early characterization of opioid receptor genes. Importantly, we will identify a number of research directions that are likely to develop during the next decade.
1. Introduction: from receptor binding sites to receptor genes, a brief history
In 1973, three independent teams showed that opiates bind to membrane receptors in the brain (Pert and Snyder, 1973; Simon et al., 1973; Terenius, 1973), and opioid binding sites were named mu, delta and kappa receptors a few years later (Martin et al., 1976). At that time, receptors remained a concept in pharmacology, and opioid binding the only way to define the receptor site. Gene cloning and characterization was a necessary step to evolve our understanding of opioid receptors, as proteins that operate in the nervous system (Mansour et al., 1995) and control nociceptive, hedonic, emotional, as well as autonomic, neuroendocrine and immune responses in vivo (see for example Bodnar, 2007). The first opioid receptor gene was isolated by expression cloning in 1992 (Evans et al., 1992; Kieffer et al., 1992). Because of strong sequence homology across receptors, the entire opioid receptor gene family was readily cloned in the following two years. In the mid 90’s the entire endogenous opioid system, including peptides(see Akil et al., 1984)) and receptors (see Kieffer, 1995), was characterized at the molecular level. The receptor DNA sequences enabled cell lines and more recently animal models to be generated, in order to express mutant and tagged opioid receptors and characterize requirements for ligand trafficking, signaling, and selectivity for the different opioid receptors. The cloning also enabled the creation of mice lacking opioid receptors that could dissect out the role of each receptor in exogenous and endogenous opioid-mediated behaviors. The cloning finally launched genetics studies in humans, although this aspect will not be discussed here (see (LaForge et al., 2000; Ikeda et al., 2005; Mayer and Hollt, 2006).
2. From receptor genes to receptor proteins
Based upon the gene sequence, the primary amino-acid structure of the opioid receptor family placed these receptors into the large family of rhodopsin-like G-protein coupled receptors. Approximately 670 genes representing 2–3 percent of the human transcribed genome are dedicated to this family of receptors, and other members include the receptors for dopamine, serotonin, acetyl choline, epinephrine and many receptors for peptide neurotransmitters (Lagerstrom and Schioth, 2008).
2. 1. The opioid binding pocket
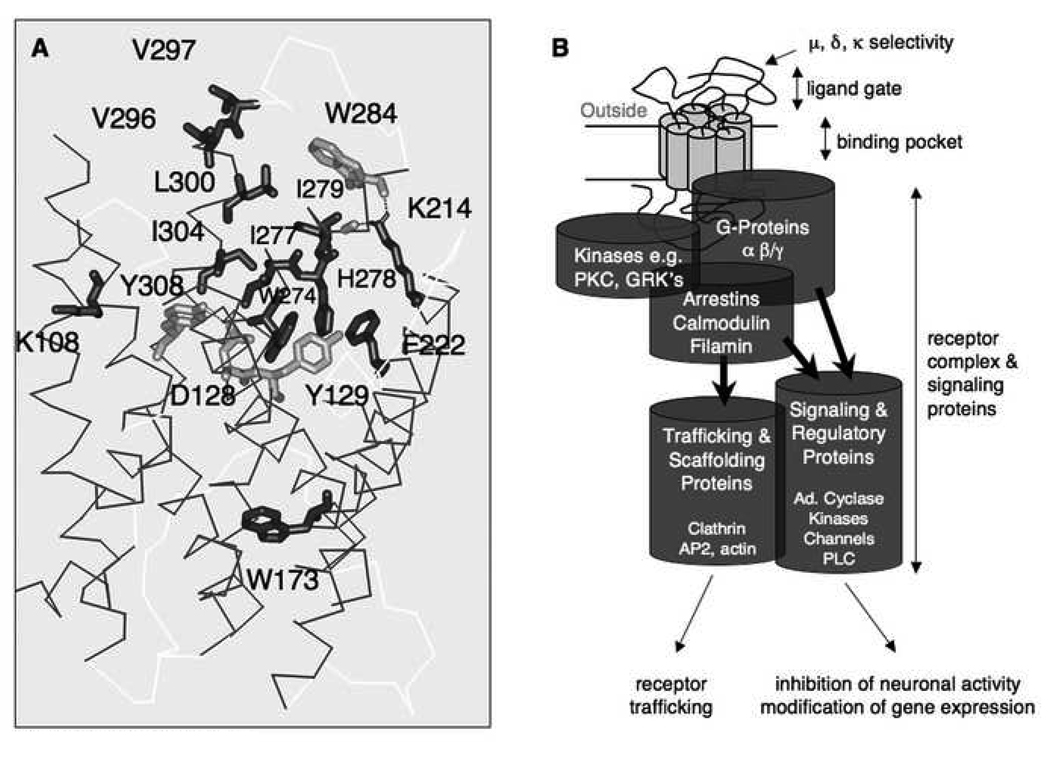
Overall mu, delta, kappa receptors show 60% amino acid sequence identity. Closest homology occurs within the seven-transmembrane helical core, which contains the opioid binding pocket (Fig. 1A). Extracellular domains, including three extracellular loops and the N-terminal domain, determine mu, delta and kappa selectivity. These domains differ strongly between receptors, and likely form a protein gate (Fig. 1B) that selects agonists or antagonists entering the binding pocket thereby contributing to mu, delta and kappa selectivity. 3D computer modeling of the opioid binding pocket was developed principally on the crystal structure of rhodopsin and led to models of opioid binding sites that were investigated and refined by site-directed mutagenesis (see Décaillot and Kieffer, 2004) for the delta receptor). Very recently, the structure of the beta2-adrenergic receptor, a more closely related GPCR to opioid receptors than rhodopsin has been determined (Cherezov et al., 2007; Rosenbaum et al., 2007). The crystal structure shows much in common with rhodopsin and supports the models predicted from biochemical and site-directed mutagenesis regarding critical amino acids required for ligand binding and activation.
Fig. 1. Receptor structure and signaling.
(A) Lateral view of a 3D model of the human delta opioid receptor (from Décaillot and Kieffer, 2004). Helices are indicated as ribbons, side chains of aminoacids implicated in binding (dark gray) or both binding and activation (light grey are shown as sticks. The opioid binding site forms a pocket penetrating half-way into the helical bundle, and is similar across mu, delta and kappa receptors. (B) Opioid receptors are coupled to inhibitory G proteins and form signaling complexes with many protein partners. Opioid receptor activation modifies ion channel activities (decreased neuronal excitability or neurotransmitter release), decreases cAMP levels via inhibition of adenylate (Ad.) cyclase and activates phosphorylation pathways that lead to transcriptional regulations. As for all GPCRs, signaling is highly regulated by receptor phosphorylation and trafficking via scaffolding proteins. Signaling and regulatory proteins that have been identified are indicated in the figure. A current hypothesis is that different agonist ligands confer different patterns of receptor signaling and trafficking in vivo.
2. 2. From an inactive to an active state
As for all GPCRs, opioid receptors convey extracellular signals within the cell by activating heterotrimeric G proteins via conformational modulation of cytoplasmic domains of the receptor that interact with the G-proteins. Agonist binding modifies helical packing of the receptor, and a rearrangement in the positioning of transmembrane domains 3, 6 and 7 has been proposed to drive the transition between inactive and active conformations of the receptor (Decaillot et al., 2003). This helical movement modifies the receptor’s intracellular structure, hence the receptor-G protein interaction. Intracellular loops of the receptor form a large part of the receptor-G protein interface. These intracellular receptor domains are almost identical across mu, delta and kappa receptors, consistent with the fact that all three receptors interact with inhibitory G proteins of the Go/Gi type. G protein subunits dissociate from the activated receptor and, in turn, modulate intracellular effectors and pathways Intracellular signaling, in turn, lead either to short-term inhibition of neuronal activity, or to long-term genomic effects (Figure 1B, and see below).
2. 3. Pharmacological diversity
Opioid receptor pharmacology is complex and the existence of multiple mu, delta and kappa receptor types was proposed since the early 70’s. Gene cloning led to characterize three receptor genes only, and the molecular basis for pharmacological diversity has long remained a matter of debate (see Befort and Kieffer, 1997; Zaki et al., 1996). Alternative splicing has been reported, but it has been difficult to establish the biological relevance of these alternative transcripts in vivo, and to correlate their existence with the multiple opioid receptor subtypes that were described earlier by the pharmacology. Today, it is admitted that the three mu, delta and kappa receptors, encoded by the Oprm1, Oprd1 and Oprk1 genes, are highly dynamic ligand-recognition proteins that may indeed account for the wide diversity of opioid receptor subtypes reported by the pharmacology.
2. 4. Receptor complexes and many cellular responses
There are several ways to explain pharmacological heterogeneity of opioid receptors (Evans, 2004) other than by alternative transcripts or posttranslational modifications (phosphorylation, palmitoylation or glycosylation). First, increasing evidence support the notion that mu, delta and kappa receptors may adopt multiple active conformations. For example, mutagenesis data suggest the existence of multiple binding modes for opioids within the binding pocket (first evidence in Befort et al., 1996). Also, signaling studies in cellular models show that receptor activation and subsequent regulations (including phosphorylation and receptor internalization) are strongly drug-dependent (first evidence in Arden et al., 1995; Keith et al., 1996)). Hence the ligand-receptor complex, rather than the receptor itself, determines the ultimate physiological cellular response. A second source of heterogeneity is the direct cellular environment of the receptor. Heterotrimeric G protein expression differs across cell types, and the number of possible G protein-associated signaling pathways has expanded dramatically. The variable combinations of G proteins subunits and the nature of associated signaling networks necessarily generate neuron-specific, or even neuron compartment-specific responses. Also, as was demonstrated for several other GPCRs, many regulatory proteins directly interact with the receptor intracellular domains and C-terminal tail and potentially influence opioid receptor pharmacology (see Fig. 1B and reviewed in Contet et al., 2004). A third potential receptor modulator is another receptor molecule. The possibility that GPCRs exist as dimeric or oligomeric complexes has gained evidence in the recent years (first evidence (Jordan and Devi, 1999). Co-expression data suggest that the physical association of opioid receptors either as homodimers or heterodimers, or even with other GPCRs creates novel receptor entities with unique pharmacological properties, which would increase opioid receptor heterogeneity (see (Devi, 2001; Levac et al., 2002). Whether receptor dimerization truly occurs and modulates pharmacology in vivo remains an important question, which is generally highly investigated in GPCR research (see (Pin et al., 2007)). In conclusion, molecular approaches have provided a novel view of opioid receptors and it is likely that the complexity of opioid responses will extend far beyond the previously reported pharmacological subtypes as in vivo molecular pharmacology evolves. We must now envisage opioid receptors as dynamic multicomponent units, rather than single protein entities, and that ligands establish and stabilize different receptor-containing protein complexes. This inevitably leads to proposing that different agonist ligands confer different patterns of signaling and receptor trafficking and introduces the potential for agonist-directed opioid receptor signaling.
2. 5. Ligand-directed signaling: implications for drug design
One dramatic case of agonist-selective signaling is observed with drugs acting at serotonin receptors. NIDA-supported research has elegantly shown that hallucinogenic drugs such as LSD, mediate their behaviors via 5HT2A receptors by activating different downstream signaling pathways from non-hallucinogenic 5HT2A agonists such as lisuride (Gonzalez-Maeso et al., 2007). The ability for drugs activating the same receptor yet eliciting different behavioral responses offers an important concept for refining pharmaceuticals and in the case of opioids perhaps separating signaling pathways leading to respiratory depression or addiction from those inhibiting pain. An important clinical feature of opioid drugs beside their acute actions is the development of opioid tolerance and dependence. Tolerance is initiated by receptor activation followed by cascade of adaptive responses initially involving the receptor itself (perhaps receptor phosphorylation and interaction with proteins such as arrestins, in some cases triggering trafficking changes), then adaptive changes of signaling pathways and cellular homeostatic mechanisms, and finally circuitry modulated by altered activity of the receptor-containing cells. Opponent processes resulting from continued opioid drugs are the signature of dependence and opioid withdrawal with increased sensitivity to pain, diarrhea, dysphoria, and agitation, reveal adaptive compensatory responses to drug treatment that opposing the acute effects of the drug. Opponent processes may also contribute tolerance since they begin to oppose the acute effects of the drug. Recent data suggests that different opioid drugs may utilize different desensitization mechanisms for receptor signaling although which pathways would be clinically beneficial for reducing tolerance and withdrawal is still not known (Kelly et al., 2008). Therefore a remaining challenge is to determine if behaviorally relevant signaling pathways can be targeted by different opioid drugs. Rationale design of opioid compounds that activate a specific subset of mu, delta or kappa receptor-associated signaling pathways is a possible strategy to develop novel drugs of high therapeutic value and low adverse activities, but this still remains a distant goal.
Finally, ligand-directed signaling may also influence endogenous opioid physiology. The existence of about thirty endogenous opioid peptides derived by alternative proteolytic cleavage of three precursor proteins has been puzzling for many years (Evans et al., 1988). In addition to differential stability to extracellular proteases and differential selectivity towards opioid receptors, individual opioid peptides may show subtle differences regarding signaling and desensitization characteristics in vivo.
3. From receptor genes to behavior
3. 1. Opioid receptor knockout mice
Since a decade, mice lacking mu, delta or kappa receptors, as well as preproenkephalin, preprodynorphin or β–endorphin, have been created by gene targeting –so-called knockout mice (see Kieffer and Gavériaux-Ruff, 2002). Mice lacking a single opioid receptor type, or even the triple receptor knockout mice, are viable and fertile and show no obvious developmental deficit, indicating that the opioid system is not essential for survival, at least under home cage conditions. These mutant mice have been extensively analyzed either for spontaneous behaviors, or in response to opioid and non-opioid drugs. Usually confirming, and further extending the pharmacology, the genetic approach has clarified the specific contribution of each opioid receptor in opioid-controlled physiology and behaviors. The comparative analysis of mu, delta and kappa knockout mice has definitely highlighted very distinct activity patterns for each receptor in vivo. Similar comparisons are currently underway for opioid peptide knockout mice. Main conclusions from receptor gene knockout are as follows:
3. 2. Addictive behaviors and emotional responses
Mu receptors represent the primary molecular target for morphine in vivo and mediate both beneficial and adverse effects of the most broadly used opiate (Matthes et al., 1996). Mu receptors also mediate rewarding properties of non-opioid drugs of abuse including cannabinoids (Ghozland et al., 2002), alcohol (Roberts et al., 2000) and nicotine (Berrendero et al., 2002), or even natural reinforcers such as social interactions (Moles et al., 2004). Mu receptors therefore represent a key molecular trigger for reward, and most likely contribute to the initiation of addictive behaviors (Contet et al., 2004). Kappa receptors mediate dysphoric activities of both kappa opioids (Simonin et al., 1998) and cannabinoids (Ghozland et al., 2002) and therefore oppose mu receptors in regulating the hedonic tone, as previously proposed (Spanagel et al., 1992). Recent studies have implicated pro-dynorphin derived opioid peptide activation of kappa receptors as modulating stress-induced relapse and suggest the potential of kappa antagonists as a therapeutic target for relapse (Land et al., 2008). Delta receptors are less directly involved in hedonic control. Very distinct from mu and kappa receptors, delta receptors regulate emotional responses and show anxiolytic and antidepressant activity (Filliol et al., 2000). This specific function of delta receptors is now confirmed by recent pharmacological studies (see (Jutkiewicz, 2006; Saitoh et al., 2004)) using SNC80, the only commercially available highly selective delta compound. Further analysis of delta knockout mice and the development of more selective compounds will likely reveal other activities of delta receptors, that will be of potential interest in the field of psychiatric disorders.
3. 3. Pain
Mu, delta and kappa receptor-deficient mice all exhibit enhanced pain sensitivity. This confirms that the three receptors, activated by endogenous opioid peptides, tonically inhibit nociceptive responses. Phenotypes of mutant mice differ across pain assays (Martin et al., 2003). In models of physiological or acute pain, mu receptors modulate mechanical, chemical and supraspinally-controlled thermal nociception, while kappa receptors modulate spinally-mediated thermal nociception and visceral pain. Again delta receptors differ from mu and kappa receptors in that there is no obvious regulation of acute pain. In contrast, there is strong evidence for a role of delta receptors in reducing hyperalgesia in situations of inflammatory (Gaveriaux-Ruff et al., in press) and neuropathic (Nadal et al., 2006) pain. These data combined with many pharmacological studies clearly demonstrate a specific role for each receptor in regulating the broad diversity of pain modalities (see Dickenson T. H and L., 2005).
3. 4. Perspectives: the neuroanatomical aspects
Opioid receptors are broadly expressed throughout the nervous system. In order to understand the basis of opioid receptor-controlled behaviors, it is critical to localize receptors that operate within neural circuits. Both transgenic and viral approaches are being developed to create mouse models with regionally-targeted opioid receptor knockout (conditional knockout) or knock-down (small interference RNA). These approaches will lead to identify specific opioid receptor populations responsible for pain control, either in peripheral or central nociceptive pathways, and reveal the specific opioid receptor populations regulating emotional responses, as well as drug reward, craving and relapse within the complex addiction circuitry (see Baler and Volkow, 2006; Nestler, 2005; O'Brien and Gardner, 2005; Koob and Kreek, 2007).
4. From opioid receptor genes to downstream target genes
Exposure to opioids induces genetic reprogramming of neuronal function. The analysis of responses to chronic morphine in about thirty knockout mouse lines has revealed that many genes contribute to the development of morphine dependence in vivo (reviewed in Contet et al., 2004). Differential gene expression experiments, including gene profiling or subtractive approaches, have identified several gene families whose transcription is regulated following treatment with drugs of abuse (Pollock, 2002). Psychostimulants have been mainly used, while morphine studies are still limited (Ammon et al., 2003; Jacobs et al., 2003; McClung and Nestler, 2008).
Morphine essentially activates mu receptors in vivo. Hence the development of morphine tolerance and dependence –and other adaptations to chronic morphine- results from excessive mu receptor activation throughout the nervous system. Exposure to other drugs of abuse, also likely triggers continual mu opioid receptor activation through endogenous mechanisms. The genetic consequences of repeated mu receptor stimulation may be a key strategy towards understanding the molecular bases of drug abuse, and a recent study has focused on mu opioid receptor signaling-associated events. In this study, a genome-wide investigation identified a specific set of morphine-induced gene regulations, which occurred in wild-type mice but not in mu receptor knockout mice. These regulations were studied in the central extended amygdala (Befort et al, in press) which interfaces brain reward and stress systems, and in the lateral hypothalamus (Befort et al., in press), an area critical for reward and motivation. These sets of genes, many of which have not been associated to mu receptor stimulation as yet, provide unique molecular repertoires towards understanding mu receptor-mediated neural plasticity, and perhaps discover novel mechanisms of drug craving and relapse.
5. From receptor genes to receptor imaging in vivo
5. 1. Non invasive imaging approaches in addiction
While PET and MRI techniques offer unique access to visualization of structure and function in the living brain (see (Volkow et al., 2004)), their spatial and temporal resolution are limited. Because receptor subcellular localization and traffic in neurons is critical to understand receptor function, there is tremendous interest in developing non-invasive imaging approaches that achieve subcellular resolution, and allow direct visualization of receptor movements in live neurons. The green fluorescent protein (GFP) from the jellyfish Aequora victoria (Tsien, 1998) and GFP variants have become reporters of choice to study dynamic biological processes in living cells, and mouse engineering has opened the way to functional imaging in mammals (Hadjantonakis et al., 2003). Driven by selected promoters in transgenic mouse strains, the fluorescent reporter has revealed the localization, shape, movement and growth of specific cell populations in neural regeneration and plasticity (Trachtenberg et al., 2002). In a small number of reports, gene targeting in mouse has been used to create fluorescent versions of well-characterized proteins in vivo and explore their distribution and dynamics within a native environment. Recently, a mouse line expressing a GFP-tagged delta receptor in place of the native delta receptor (delta-eGFP knock-in mice) has been created. In these animals the receptor is expressed at levels comparable to wild-type, and is functionally coupled to G proteins. This is the first example of knock-in mice expressing a fully functional fluorescent G protein coupled receptor (Scherrer et al., 2006).
5. 2. From the gene to a trackable receptor in vivo
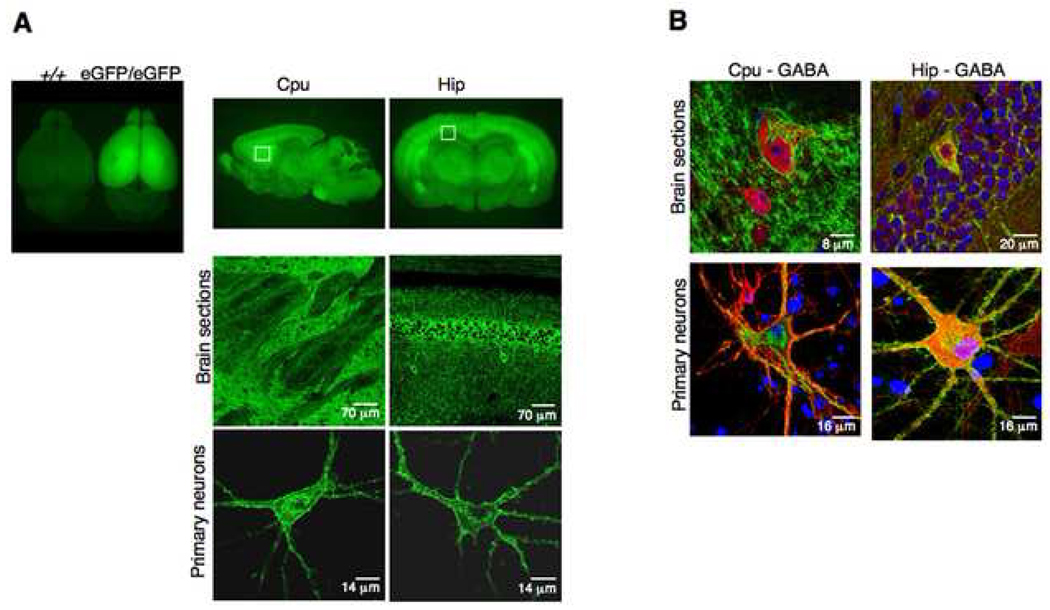
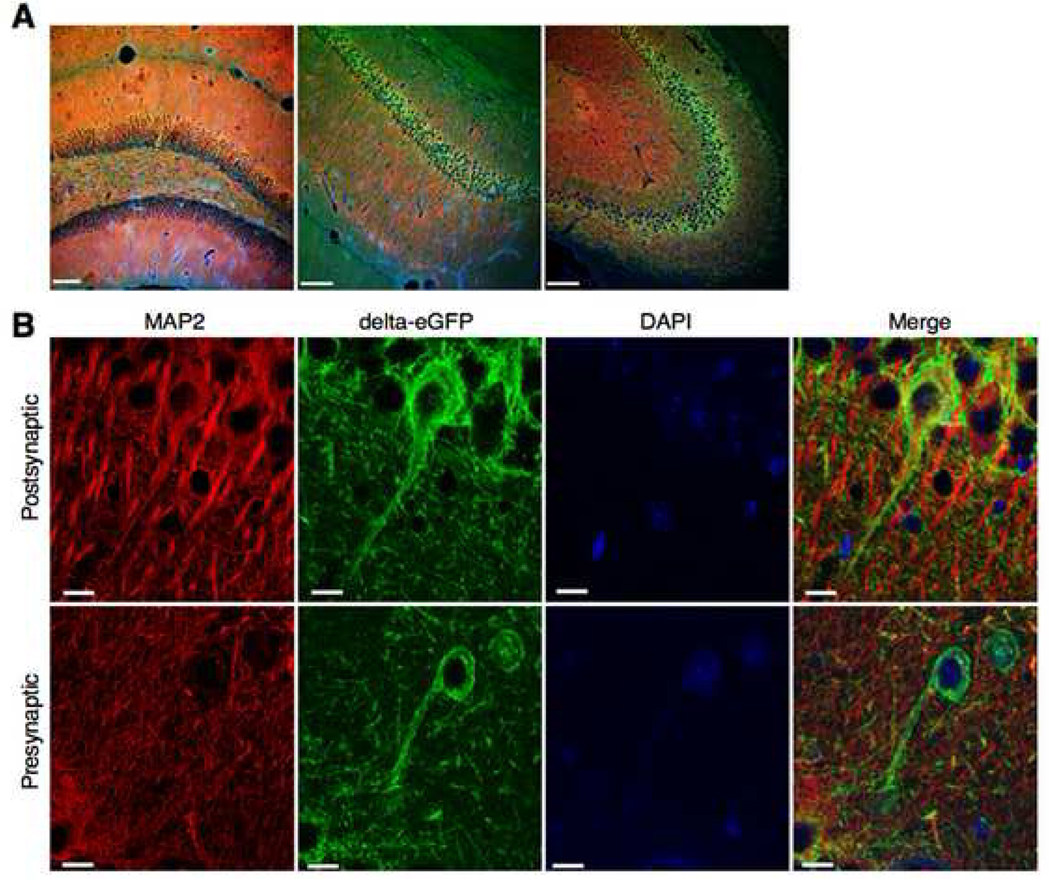
The mutant delta-eGFP mice prove to be an extraordinary tool to study receptor biology. The receptor neuroanatomy is visible throughout the nervous system (Fig. 2 and 3). Confocal imaging of brain section reveals distinct architectures of the fluorescent signal across brain areas, which reflect the natural distribution of delta receptors in brain circuits. Fluorescent cell bodies and processes are easily detectable in hippocampal interneurons while strong homogenous and diffuse fluorescence is observed in the basolateral amygdala. Striatal sections show fluorescent cell bodies embedded in dense fluorescence likely arising from high dendritic receptor expression (Fig. 2A). Immunohistochemistry using standard neuronal markers allows to readily identify phenotypic characteristics of delta receptor-expressing neurons (Fig. 2B) and further, allows to precisely localize receptor localization in distinct neuronal compartment. As an example, preliminary data using MAP-2 labeling show the existence of both presynaptic and postsynaptic receptors in hippocampal neurons (Fig. 3, Massotte and Kieffer, unpublished). Detailed functional mapping of fluorescent delta receptors will greatly help our understanding of delta receptor function in vivo.
Fig. 2. Receptor imaging in delta-eGFP knock-in mice.
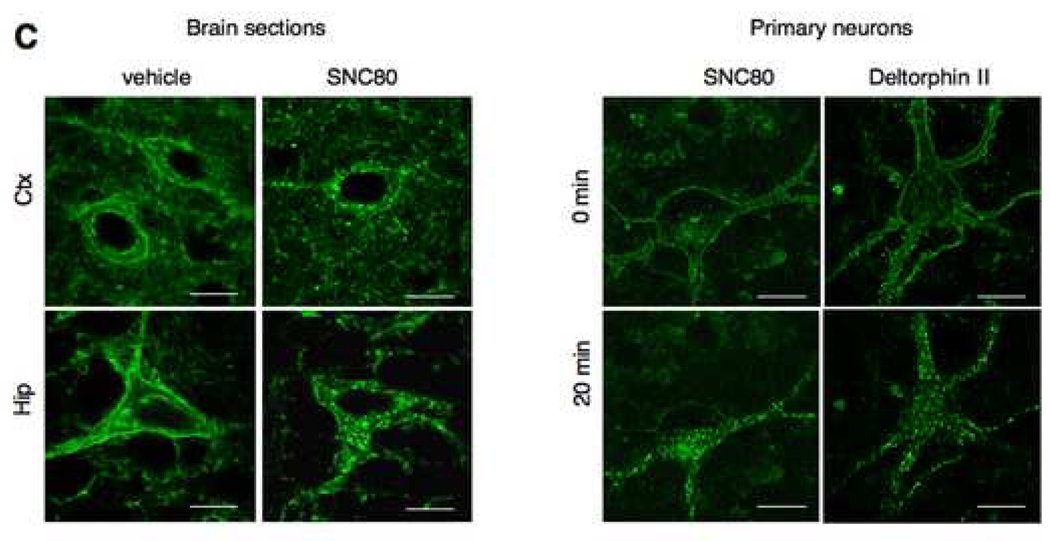
Images are adapted from Scherrer et al. (Scherrer et al., 2006) (A) Epifluorescence macroscopy shows the general anatomical distribution of delta receptors. Top left, whole brain from wild-type (+/+) and knock-in (eGFP/eGFP) mice; top midde, coronal section at the level of the caudate putamen (Cpu); top right, sagittal section at the level of the hippocampus (Hip). Confocal microscopy of regions delimited by insets reveals receptor distribution with a cellular resolution (middle panels). Confocal microscopy of primary neurons from caudate putamen and hippocampus highlights the subcellular distribution of delta receptors (bottom panels). (B) Immunostaining with GAD antibodies (red) identifies GABAergic neurons expressing fluorescent delta receptors in neurons from the Cpu (Cpu-GABA) and Hip (Hip-GABA) in either brain sections (top) or primary cultures (bottom). Nuclei are stained in blue. (C) Delta receptor internalization in vivo, upon exposure to delta agonists. Left panels show prominent surface labeling in cortical (Ctx) and hippocampal (Hip) neurons in brain sections of vehicle-treated animals (left) and the typical punctate pattern of internalized receptors in brain sections of SNC80-treated animals (right); Scale bars 8 µm. Right panels shows delta-eGFP labeling in striatal primary neurons before (0 min) and 20 minutes after exposure to both a non-peptidic (SNC80) and a peptidic (deltorphin II) delta agonist; scale bars 12 µm. In those preparations, the receptor internalization process can be observed in real time, using time-lapse confocal microscopy (see Scherrer et al., 2006).
Fig. 3. Delta opioid receptors are localized both pre- and post-synaptically in the hippocampus (D. Massotte and B. Kieffer, unpublished).
Confocal imaging of brain sections from delta-eGFP mice, labeled with a MAP2 antibody that labels somatodendritic, but not axonal, compartments. MAP immunostaining is shown in red, fluorescently labeled delta receptors are shown in green, and cell nuclei in blue (DAPI). (A) A general view at the level of the hippocampus. Dentate gyrus (left panel), CA1 region (central panel) and CA3 region (right panel). Scale bars 100 µm. (B) Top panels: the delta receptor is expressed in dendrites. In this neuron MAP2 and delta-eGFP are co-localized (merged image on the right), suggesting a postsynaptic localization of the receptor. Scale bar 10 µm. Bottom panels: the delta receptor is also expressed in axons. In this neuron, lack of MAP2 and delta-eGFP co-staining (merged image on the right) indicates a presynaptic receptor localization. Scale bar 10µm.
5. 3. The dynamics and implications of receptor trafficking in vivo
The eGFP fusion strategy allows the study of real-time receptor trafficking in live neurons. Primary neurons from delta-eGFP mice have been exposed to opioid ligands and rapid receptor internalization could be observed in real-time, upon exposure to several delta agonists (see Fig. 2C and Scherrer et al., 2006). Preliminary studies using these ex-vivo preparations indicate that delta-eGFP receptors are targeted to lysosomes (Tryoen-Toth and Kieffer, unpublished), definitely classifying delta receptors among slow-recycling/fast degrading GPCRs (Tanowitz and von Zastrow, 2003). In the future, real-time receptor trafficking studies will be expanded to slice preparations from delta-eGFP mice, and possibly in live animals while resolution of optic fiber technology improves.
The biological significance of GPCR internalization in vivo remains unknown, and whether receptor internalization negatively or positively regulates receptor function is highly debated. At present, most receptor trafficking studies after agonist administration have been performed in cellular models and their physiological relevance is limited. These in vitro systems may not reflect in vivo situations in terms of receptor density, protein content of receptor-expressing cells, or even receptor localization within subcellular compartments as is the case for neurons (Bernard et al., 2006). Additionally, data from cellular models provide no understanding of how receptor trafficking influences integrated responses in the living organism. Intracellular trafficking of native receptors in tissues have been rarely studied, due to limited availability of specific antibodies for the receptors (Sternini et al., 1996; Tappe-Theodor et al., 2007; Van Bockstaele and Commons, 2001 and see Bernard et al., 2006)). Delta-eGFP mice represent a unique tool to readily examine the subcellular localization of endogenously expressed receptors in neurons and correlate receptor trafficking with the behavioral effects of agonists in vivo. First data showed that treatment with the delta agonist SNC80 triggers massive receptor endocytosis throughout the nervous system (Fig. 2C), together with locomotor activation. Interestingly, mice with internalized receptors do not respond behaviorally to a second drug administration (Scherrer et al., 2006). This was a first indication that internalization may impact delta receptor signaling in vivo. Further studies using agonists with variable internalization potencies, and examining other -possibly therapeutically relevant- behavioral responses, will establish the extend to which receptor internalization controls receptor function and drug efficacy, or influences the development of in vivo tolerance.
Delta-eGFP mice also represent a particularly appealing tool to report for endogenous peptide release. Delta receptor internalization may be observable in the mouse, following activation of the endogenous opioid system, and this opens an entire field of investigation. The identification of behavioral situations and sites where opioid peptides stimulate delta receptors will be helpful to understand many of the opioid-controlled behaviors. Finally, the study of opioid receptor expression, distribution and dynamics (responsivity to agonists) in drug-dependent or post-dependent animals will be greatly facilitated in these mutant mice. Imaging data from this – or similar- mouse model in situations relevant to drug abuse may ultimately provide invaluable information at the neuronal level, which is currently lacking in PET or MRI addiction studies.
5. 4. A novel approach in GPCR research
GPCRs represent the largest and most versatile family of membrane receptors. Each member has a specific cellular life cycle (Tan et al., 2004) and is subjected to a particular set of signaling regulatory mechanisms (Pierce et al., 2002). Our eGFP-knock-in approach could be extended to other GPCRs, as for example to mu receptor as a prototypic fast-recycling receptor, or to orphan receptors for which in vivo pharmacology is still in its infancy. Also the development of fluorescently tagged receptors with distinct colors may be instrumental to address the issue of receptor heterodimerization in vivo.
6. Conclusion
6. 1. Targeting opioid receptors for therapeutics
One fundamental goal of opioid research remains the rational design of more effective opioid analgesics. In addition there is increasing interest in developing therapeutic drugs targeting delta receptors for emotional disorders and conditions of chronic pain, and kappa receptors in the treatment of drug abuse and stress. Drug design in the past has been focused on drug selectivity (mu, delta and kappa), intrinsic efficacy of opioid drugs at these receptors and drug metabolism. Based on the newfound knowledge of opioid receptor complexes, new pharmacology categories of opioid drugs are emerging. Properties such as ability to recruit specific kinases or arrestins, or selective signaling cascades and desensitization processes, are becoming discriminating criteria in drug development. Our understanding about ligand –receptor contacts and conformational states that modulate the formation of selective receptor complexes are still in their early stages, yet differential signaling and trafficking do manifest in altered cellular and behavioral responses. It will be important to determine optimal complex-forming profiles for drugs that infer maximal clinical efficacy and minimal deleterious side-effects for all opioid therapeutic uses. This new challenge for opioid pharmacology has implications for maintaining synthesis efforts. These will lead to develop new opioid drugs that trigger the formation of defined receptor complexes to achieve optimal therapeutic efficacy in the human nervous system.
6. 2. Understanding the physiology and plasticity of the opioid system in vivo
On-going studies have returned to analyzing opioid receptor and peptides operating in their physiological environment, using molecular and genetic tools as well as high resolution imaging techniques. There is a need to identify neural sites where opioid peptide and receptors operate within nociceptive, emotional and motivational circuits. Also, and in line with the search for drugs that activate specific receptor-signaling complexes, it will be important to characterize signaling pathways that are relevant to specific behavioral or physiological responses to opioids in vivo. Among these, opioid-associated pathways that underlie addictive behaviors will be of particular interest in drug abuse research. Additionally, novel insights into brain function will arise from understanding interactions of the opioid system with other neurotransmitter systems. For example, whether the opioid system interacts with the cannabinoid system or anti-opioid systems at molecular, cellular or circuit level remains an open question. Lastly, the elucidation of molecular and network adaptations to chronic opiates will undoubtedly shed light on general mechanisms of brain plasticity.
Acknowledgements
We thank G. Scherrer for creating the mutant delta-eGFP knock-in mice and are grateful to D. Massotte for providing preliminary images showing pre- and postsynaptic localization of the delta receptor (Fig. 3). BK also wishes to thank C. Gavériaux-Ruff, K. Befort, D. Filliol and J. Becker for their invaluable contributions to the field. We are grateful to NIDA for supporting the Center for Opioid Receptors and Drugs of Abuse (# DA 005010). Funding was also from CNRS, INSERM and University Strasbourg. We also thank the Shirley and Stefan Hatos Neuroscience Research Foundation for supporting our research programs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Brigitte L. Kieffer, Email: briki@igbmc.u-strasbg.fr.
Christopher J. Evans, Email: cevans@ucla.edu.
References
- Akil H, Watson SJ, Young E, Lewis ME, Khachaturian H, Walker JJ. Endogenous opioids: Biology and function. Ann. Rev. Neurosci. 1984;7:223–255. doi: 10.1146/annurev.ne.07.030184.001255. [DOI] [PubMed] [Google Scholar]
- Ammon S, Mayer P, Riechert U, Tischmeyer H, Hollt V. Microarray analysis of genes expressed in the frontal cortex of rats chronically treated with morphine and after naloxone precipitated withdrawal. Brain Res Mol Brain Res. 2003;112:113–125. doi: 10.1016/s0169-328x(03)00057-3. [DOI] [PubMed] [Google Scholar]
- Arden JR, Segredo V, Wang Z, Lameh J, Sadée W. Phosphorylation and agonist specific intracellular trafficking of an epitope-tagged m-opioid receptor expressed in HEK 293 cells. J. Neurochem. 1995;65:1636–1645. doi: 10.1046/j.1471-4159.1995.65041636.x. [DOI] [PubMed] [Google Scholar]
- Baler RD, Volkow ND. Drug addiction: the neurobiology of disrupted self-control. Trends Mol Med. 2006;12:559–566. doi: 10.1016/j.molmed.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Befort K, Filliol D, Darcq E, Ghate A, Matifas A, Lardenois A, Muller J, Thibault C, Dembele D, Poch O, Kieffer BL. Gene altered expression in lateral hypothalamus upon mu opioid receptor activation by morphine Annals of the New York Academy of Sciences. in press. [DOI] [PubMed] [Google Scholar]
- Befort K, Filliol D, Ghate A, Darcq E, Matifas A, Muller J, Lardenois A, Thibault C, Dembele D, Le Merrer J, Becker JAJ, Poch O, Kieffer BL. Mu-opioid receptor activation induces transcriptional plasticity in the central extended amygdala. Eur J Neurosci. doi: 10.1111/j.1460-9568.2008.06273.x. in press. [DOI] [PubMed] [Google Scholar]
- Befort K, Kieffer BL. Structure-activity relationships in the delta opioid receptor. In: Budd K, Hamann W, editors. Pain reviews. London: Arnold; 1997. pp. 100–121. [Google Scholar]
- Befort K, Tabbara L, Kling D, Maigret B, Kieffer BL. Role of transmembrane residues of the d-opioid receptor in ligand recognition. J. Biol. Chem. 1996;271:10161–10168. doi: 10.1074/jbc.271.17.10161. [DOI] [PubMed] [Google Scholar]
- Bernard V, Decossas M, Liste I, Bloch B. Intraneuronal trafficking of G-protein-coupled receptors in vivo. Trends Neurosci. 2006;29:140–147. doi: 10.1016/j.tins.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Berrendero F, Kieffer BL, Maldonado R. Attenuation of nicotine-induced antinociception, rewarding effects and dependence in mu-opioid receptor knockout mice. J. Neurosci. 2002;22:10935–10940. doi: 10.1523/JNEUROSCI.22-24-10935.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar RJ. Endogenous opiates and behavior: 2006. Peptides. 2007;28:2435–2513. doi: 10.1016/j.peptides.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contet C, Kieffer BL, Befort K. Mu opioid receptor: a gateway to drug addiction. Curr Op. Neurobiol. 2004;14:1–9. doi: 10.1016/j.conb.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Décaillot F, Kieffer BL. In vitro and in vivo mutagenesis: insights into delta receptor structure and function. 2004:41–60. In: ?, (Ed), ? ?, ? [Google Scholar]
- Décaillot FM, Befort K, Filliol D, Yue S, Walker P, Kieffer BL. Opioid receptor random mutagenesis reveals a mechanism for G protein-coupled receptor activation. Nat Struct Biol. 2003;10:629–636. doi: 10.1038/nsb950. [DOI] [PubMed] [Google Scholar]
- Devi LA. Heterodimerization of G-protein-coupled receptors: pharmacology, signaling and trafficking. Trends Pharmacol Sci. 2001;22:532–537. doi: 10.1016/s0165-6147(00)01799-5. [DOI] [PubMed] [Google Scholar]
- Dickenson THLKB. p. Chap 27. In: Koltzenburg Ma., editor. Opiates-Basic. London, UK: Wall & Melzack’s Textbook of Pain 5th Elsevier; 2005. [Google Scholar]
- Evans CJ. Secrets of The Opium Poppy Revealed. Neuropharmacology. 2004;47 Supp. 1:293–299. doi: 10.1016/j.neuropharm.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Evans CJ, Hammond DL, Frederickson RCA. The opioid peptides. In: Pasternak GW, editor. The Opiate Receptors. Clifton, N.J.: Humana Press; 1988. pp. 23–71. [Google Scholar]
- Evans CJ, Keith DE, Morrison H, Magendzo K, Edwards RH. Cloning of a delta opioid receptor by functional expression. Science. 1992;258:1952–1955. doi: 10.1126/science.1335167. [DOI] [PubMed] [Google Scholar]
- Filliol D, Ghozland S, Chluba J, Martin M, Matthes HW, Simonin F, Befort K, Gaveriaux-Ruff C, Dierich A, LeMeur M, Valverde O, Maldonado R, Kieffer BL. Mice deficient for delta- and mu-opioid receptors exhibit opposing alterations of emotional responses. Nat Genet. 2000;25:195–200. doi: 10.1038/76061. [DOI] [PubMed] [Google Scholar]
- Gaveriaux-Ruff C, Karchewski L, Hever X, Matifas A, Kieffer BL. Inflammatory pain is enhanced in delta-opioid receptor knockout mice. Eur. J. Neurosci. doi: 10.1111/j.1460-9568.2008.06223.x. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghozland S, Matthes HW, Simonin F, Filliol D, Kieffer BL, Maldonado R. Motivational effects of cannabinoids are mediated by mu- and kappa-opioid receptors. J. Neurosci. 2002;22:1146–1154. doi: 10.1523/JNEUROSCI.22-03-01146.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Maeso J, Weisstaub NV, Zhou M, Chan P, Ivic L, Ang R, Lira A, Bradley-Moore M, Ge Y, Zhou Q, Sealfon SC, Gingrich JA. Hallucinogens recruit specific cortical 5-HT(2A) receptor-mediated signaling pathways to affect behavior. Neuron. 2007;53:439–452. doi: 10.1016/j.neuron.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Hadjantonakis AK, Dickinson ME, Fraser SE, Papaioannou VE. Technicolour transgenics: imaging tools for functional genomics in the mouse. Nat Rev Genet. 2003;4:613–625. doi: 10.1038/nrg1126. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Ide S, Han W, Hayashida M, Uhl GR, Sora I. How individual sensitivity to opiates can be predicted by gene analyses. Trends Pharmacol Sci. 2005;26:311–317. doi: 10.1016/j.tips.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Jacobs EH, Smit AB, de Vries TJ, Schoffelmeer AN. Neuroadaptive effects of active versus passive drug administration in addiction research. Trends Pharmacol Sci. 2003;24:566–573. doi: 10.1016/j.tips.2003.09.006. [DOI] [PubMed] [Google Scholar]
- Jordan BA, Devi LA. G-protein-coupled receptor heterodimerization modulates receptor function. Nature. 1999;399:697–700. doi: 10.1038/21441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jutkiewicz EM. The antidepressant -like effects of delta-opioid receptor agonists. Mol Interv. 2006;6:162–169. doi: 10.1124/mi.6.3.7. [DOI] [PubMed] [Google Scholar]
- Keith DE, Murray SR, Zaki PA, Chu PC, Lissin DV, Kang L, Evans CJ, Von Zastrow M. Morphine activates opioid receptors without causing their rapid internalization. J. Biol. Chem. 1996;277:19021–19024. doi: 10.1074/jbc.271.32.19021. [DOI] [PubMed] [Google Scholar]
- Kelly E, Bailey CP, Henderson G. Agonist-selective mechanisms of GPCR desensitization. Br J Pharmacol. 2008;153 Suppl 1:S379–S388. doi: 10.1038/sj.bjp.0707604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer BL. Recent advances in molecular recognition and signal transduction of active peptides: receptors for opioid peptides. Cell Mol Neurobiol. 1995;15:615–635. doi: 10.1007/BF02071128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer BL, Befort K, Gaveriaux-Ruff C, Hirth CG. The d-opioid receptor: isolation of a cDNA by expression cloning and pharmacological characterization. Proc. Natl. Acad. Sci. USA. 1992;89:12048–12052. doi: 10.1073/pnas.89.24.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer BL, Gavériaux-Ruff C. Exploring the opioid system by gene knockout. Prog Neurobiol. 2002;66:285–306. doi: 10.1016/s0301-0082(02)00008-4. [DOI] [PubMed] [Google Scholar]
- Koob G, Kreek MJ. Stress, dysregulation of drug reward pathways, and the transition to drug dependence. Am J Psychiatry. 2007;164:1149–1159. doi: 10.1176/appi.ajp.2007.05030503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaForge KS, Yuferov V, Kreek MJ. Opioid receptor and peptide gene polymorphisms: potential implications for addictions. Eur. J. Pharmacol. 2000;410:249–268. doi: 10.1016/s0014-2999(00)00819-0. [DOI] [PubMed] [Google Scholar]
- Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- Land BB, Bruchas MR, Lemos JC, Xu M, Melief EJ, Chavkin C. The dysphoric component of stress is encoded by activation of the dynorphin kappa-opioid system. J Neurosci. 2008;28:407–414. doi: 10.1523/JNEUROSCI.4458-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levac BA, O'Dowd BF, George SR. Oligomerization of opioid receptors: generation of novel signaling units. Curr Opin Pharmacol. 2002;2:76–81. doi: 10.1016/s1471-4892(02)00124-8. [DOI] [PubMed] [Google Scholar]
- Mansour A, Fox CA, Akil H, Watson SJ. Opioid-receptor mRNA expression in the rat CNS: anatomical and functional implications. Trends Neurosci. 1995;18:22–29. doi: 10.1016/0166-2236(95)93946-u. [DOI] [PubMed] [Google Scholar]
- Martin M, Matifas A, Maldonado R, Kieffer BL. Acute antinociceptive responses in single and combinatorial opioid receptor knockout mice: dictinct mu, delta and kappa tones. Eur J Neurosci. 2003;17:1–8. doi: 10.1046/j.1460-9568.2003.02482.x. [DOI] [PubMed] [Google Scholar]
- Martin WR, Eades CG, Thompson JA, Huppler RE, Gilbert PE. The effects of morphine and nalorphine-like drugs in the nondependent and morphine-dependent chronic spinal dog. Journal of Pharmacology and Experimental Therapeutics. 1976;197:517–532. [PubMed] [Google Scholar]
- Matthes HWD, Maldonado R, Simonin F, Valverde O, Slowe S, Kitchen I, Befort K, Dierich A, LeMeur M, Dollé P, Tzavara E, Hanoune J, Roques BP, Kieffer BL. Loss of morphine-induced analgesia, reward effect and withdrawal symptoms in mice lacking the µ-opioid receptor gene. Nature. 1996;383:819–823. doi: 10.1038/383819a0. [DOI] [PubMed] [Google Scholar]
- Mayer P, Hollt V. Pharmacogenetics of opioid receptors and addiction. Pharmacogenet Genomics. 2006;16:1–7. doi: 10.1097/01.fpc.0000182781.87932.0d. [DOI] [PubMed] [Google Scholar]
- McClung CA, Nestler EJ. Neuroplasticity mediated by altered gene expression. Neuropsychopharmacology. 2008;33:3–17. doi: 10.1038/sj.npp.1301544. [DOI] [PubMed] [Google Scholar]
- Moles A, Kieffer BL, D'Amato FR. Deficit in attachment behavior in mice lacking the mu-opioid receptor gene. Science. 2004;304:1983–1986. doi: 10.1126/science.1095943. [DOI] [PubMed] [Google Scholar]
- Nadal X, Banos JE, Kieffer BL, Maldonado R. Neuropathic pain is enhanced in delta-opioid receptor knockout mice. Eur J Neurosci. 2006;23:830–834. doi: 10.1111/j.1460-9568.2006.04569.x. [DOI] [PubMed] [Google Scholar]
- Nestler EJ. Is there a common molecular pathway for addiction? Nat Neurosci. 2005;8:1445–1449. doi: 10.1038/nn1578. [DOI] [PubMed] [Google Scholar]
- O'Brien CP, Gardner EL. Critical assessment of how to study addiction and its treatment: human and non-human animal models. Pharmacol Ther. 2005;108:18–58. doi: 10.1016/j.pharmthera.2005.06.018. [DOI] [PubMed] [Google Scholar]
- Pert CB, Snyder SH. Opiate receptor: demonstration in nervous tissue. Science. 1973;179:1011–1014. doi: 10.1126/science.179.4077.1011. [DOI] [PubMed] [Google Scholar]
- Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- Pin JP, Neubig R, Bouvier M, Devi L, Filizola M, Javitch JA, Lohse MJ, Milligan G, Palczewski K, Parmentier M, Spedding M. International Union of Basic and Clinical Pharmacology. LXVII. Recommendations for the recognition and nomenclature of G protein-coupled receptor heteromultimers. Pharmacol Rev. 2007;59:5–13. doi: 10.1124/pr.59.1.5. [DOI] [PubMed] [Google Scholar]
- Pollock JD. Gene expression profiling: methodological challenges, results, and prospects for addiction research. Chem Phys Lipids. 2002;121:241–256. doi: 10.1016/s0009-3084(02)00160-3. [DOI] [PubMed] [Google Scholar]
- Roberts A, Mcdonald JS, Heyser CJ, Kieffer BL, Matthes HWD, Koob GF, Gold LH. mu-opioid receptor knockout mice do not self-administer alcohol. J. Pharm. Exp. Ther. 2000;293:1002–1008. [PubMed] [Google Scholar]
- Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- Saitoh A, Kimura Y, Suzuki T, Kawai K, Nagase H, Kamei J. Potential anxiolytic and antidepressant-like activities of SNC80, a selective delta-opioid agonist, in behavioral models in rodents. J Pharmacol Sci. 2004;95:374–380. doi: 10.1254/jphs.fpj04014x. [DOI] [PubMed] [Google Scholar]
- Scherrer G, Tryoen-Toth P, Filliol D, Matifas A, Laustriat D, Cao YQ, Basbaum AI, Dierich A, Vonesh JL, Gaveriaux-Ruff C, Kieffer BL. Knockin mice expressing fluorescent delta-opioid receptors uncover G protein-coupled receptor dynamics in vivo. Proc Natl Acad Sci U S A. 2006;103:9691–9696. doi: 10.1073/pnas.0603359103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon EJ, Hiller JM, Edelman I. Stereospecific binding of the potent narcotic analgesic [3H] etorphine to rat brain homogenate. Proc. Natl. Acad. Sci. 1973;70:1947–1949. doi: 10.1073/pnas.70.7.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonin F, Valverde O, Smadja S, Slowe S, Kitchen I, Dierich A, Le Meur M, Roques BP, Maldonado R, Kieffer BL. Disruption of the k-opioid receptor gene in mice enhances sensitivity to chemical visceral pain, impairs pharmacological actions of the selective k-agonist U-50,488H and attenuates morphine withdrawal. EMBO J. 1998;17:886–897. doi: 10.1093/emboj/17.4.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanagel R, Herz A, Shippenberg TS. Opposing tonically active endogenous opioid systems modulate the mesolimbic dopaminergic pathway. Proc Natl Acad Sci U S A. 1992;89:2046–2050. doi: 10.1073/pnas.89.6.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternini C, Spann M, Anton B, Keith DE, Bunnett NW, Von Zastrow M, Evans CJ, Brecha NC. Agonist-selective endocytosis of m opioid receptor by neurons in vitro. Proc. Natl. Acad. Sci. USA. 1996;93:9241–9246. doi: 10.1073/pnas.93.17.9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CM, Brady AE, Nickols HH, Wang Q, Limbird LE. Membrane trafficking of G protein-coupled receptors. Annu Rev Pharmacol Toxicol. 2004;44:559–609. doi: 10.1146/annurev.pharmtox.44.101802.121558. [DOI] [PubMed] [Google Scholar]
- Tanowitz M, von Zastrow M. A novel endocytic recycling signal that distinguishes the membrane trafficking of naturally occurring opioid receptors. J Biol Chem. 2003;278:45978–45986. doi: 10.1074/jbc.M304504200. [DOI] [PubMed] [Google Scholar]
- Tappe-Theodor A, Agarwal N, Katona I, Rubino T, Martini L, Swiercz J, Mackie K, Monyer H, Parolaro D, Whistler J, Kuner T, Kuner R. A molecular basis of analgesic tolerance to cannabinoids. J Neurosci. 2007;27:4165–4177. doi: 10.1523/JNEUROSCI.5648-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenius L. Stereospecific interaction between narcotic analgesics and a synaptic plasma membrane fraction of rat cerebral cortex. Acta Pharmacol. Toxicol. 1973;32:317–319. doi: 10.1111/j.1600-0773.1973.tb01477.x. [DOI] [PubMed] [Google Scholar]
- Trachtenberg JT, Chen BE, Knott GW, Feng G, Sanes JR, Welker E, Svoboda K. Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature. 2002;420:788–794. doi: 10.1038/nature01273. [DOI] [PubMed] [Google Scholar]
- Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- Van Bockstaele EJ, Commons KG. Internalization of mu-opioid receptors produced by etorphine in the rat locus coeruleus. Neuroscience. 2001;108:467–477. doi: 10.1016/s0306-4522(01)00426-2. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Fowler JS, Wang GJ. The addicted human brain viewed in the light of imaging studies: brain circuits and treatment strategies. Neuropharmacology. 2004;47 Suppl 1:3–13. doi: 10.1016/j.neuropharm.2004.07.019. [DOI] [PubMed] [Google Scholar]
- Zaki PA, Bilsky EJ, Vanderah TW, Lai J, Evans CJ, Porreca F. Opioid receptor types and subtypes: the d receptor as a model. Annu. Rev. Pharmacol. Toxicol. 1996;36:379–401. doi: 10.1146/annurev.pa.36.040196.002115. [DOI] [PubMed] [Google Scholar]