Abstract
The majority of lung cancers are caused by long term exposure to the several classes of carcinogens present in tobacco smoke. While a significant fraction of lung cancers in never smokers may also be attributable to tobacco, many such cancers arise in the absence of detectable tobacco exposure, and may follow a very different cellular and molecular pathway of malignant transformation. Recent studies summarized here suggest that lung cancers arising in never smokers have a distinct natural history, profile of oncogenic mutations, and response to targeted therapy. The majority of molecular analyses of lung cancer have focused on genetic profiling of pathways responsible for metabolism of primary tobacco carcinogens. Limited research has been conducted evaluating familial aggregation and genetic linkage of lung cancer, particularly among never smokers in whom such associations might be expected to be strongest. Data emerging over the past several years demonstrates that lung cancers in never smokers are much more likely to carry activating mutations of the Epidermal Growth Factor Receptor (EGFR), a key oncogenic factor and direct therapeutic target of several newer anti-cancer drugs. EGFR mutant lung cancers may represent a distinct class of lung cancers, enriched in the never smoking population, and less clearly linked to direct tobacco carcinogenesis. These insights followed initial testing and demonstration of efficacy of EGFR-targeted drugs. Focused analysis of molecular carcinogenesis in lung cancers in never smokers is needed, and may provide additional biologic insight with therapeutic implications for lung cancers in both ever smokers and never smokers.
NATURAL HISTORY AND PROGNOSIS
The preceding papers in this issue of CCR Focus present an overview, and a description of clinical epidemiology and risk factors associated with lung cancer in never smokers (1, 2). This article is intended to summarize the current status of molecular profiling of lung cancer in never smokers, to indicate how profiles differ between lung cancer in ever smokers and never smokers, and to review the therapeutic implications of these molecular characteristics. To place the therapeutic implications in context, this section will first summarize recent studies of differential clinical outcomes in lung cancer patients by ever smoker vs. never smoker status, irrespective of therapies targeting particular molecular determinants.
Four recent retrospective analyses have compared the characteristics and treatment outcomes of never smokers and smokers with lung cancer across stages of disease and regardless of modality of treatment (3–6). All of these series report on data obtained prior to widespread use of EGFR inhibitors or other targeted therapies. Together these studies suggest that lung cancer in never smokers has peak incidence at a younger age than in smokers, is more likely to arise in women, and is more likely to be of adenocarcinoma histology. Furthermore, these studies demonstrate a survival advantage for never smokers compared to former and current smokers. These data are summarized in Table 1.
Table 1.
Results of studies on survival from lung cancer among never smokers
| Study | Design/population | Exposure Group | Reference Group | Risk Estimate (95% CI) |
Adjustment Variables |
|
|---|---|---|---|---|---|---|
| Tan et al. (2003) (3) | Cohort: 326 women with newly diagnosed lung cancer (187 never smokers) 180 deaths in cohort Study location: Singapore Study years: 1996–2000 |
Ever smoker | vs. | Never smoker | 1.3 (1.0–1.8) | age, stage at diagnosis, histology |
| Nordquist et al. (2004) (4) | Cohort: 654 men and women in treatment for lung cancer (132 never smokers) Number of deaths not stated Study location: USA (FL) Study years: 1985–2000 |
Current smoker | vs. | Never smoker | 1.33 (1.04–1.69) | gender, stage at diagnosis |
| Toh et al. (2006) (5) | Cohort: 975 men and women with newly diagnosed non-small cell lung cancer (286 never smokers) 805 deaths in cohort Study location: Singapore Study years: 1999–2005 |
Current smoker | vs. | Never smoker | 1.30 (1.04–1.62) | age, gender, ECOG status, stage at diagnosis, comorbidities, weight loss, treatment |
| Janjigian et al. (2008) (6) | Cohort: 2010 men and women with stage IIIb/IV non-small cell lung cancer (331 never smokers) Study location: USA (NY) |
Ever smoker | Vs. | Never smoker | 1.36 (1.16–1.60) | Age, gender, ECOG status, weight loss |
Abbreviations: CI -- confidence interval; ECOG --Eastern Cooperative Oncology Group
Age of onset
Two studies from Singapore considering lung cancer across histologic types suggest that cancer in never smokers occurs at a younger age of onset (3, 5) (p < 0.001). These data are further supported by an epidemiologic study in a Caucasian population (7). Etzel et al. report a higher proportion of never smokers (23.9%) among 230 cases of early onset lung cancer (≤ 50 years of age) than among 426 cases diagnosed at ≥ 70 years of age (17.6%) (p < 0.001). However, a fourth study limited only to patients with adenocarcinoma (4) reports the opposite finding: median age of onset 63.5 years for never smokers vs. 59.4 years for smokers (p = 0.0005).
Gender
Data from both Asia and the U.S. consistently report a higher proportion of women among never smokers with lung cancer relative to smokers with lung cancer. The study by Toh et al. found among a predominantly ethnic Chinese population in Singapore that over 68% of the never smokers with lung cancer were women, compared with 12% of current- and 13% of former smokers (p < 0.001) (5). Nordquist et al. reported that women comprised 78% of the never smoker cohort, compared with 54% of the smokers in their series limited to patients with adenocarcinoma (p < 0.0001) (4).
Histology
The Singapore series of Toh et al. was the only analysis that specifically focused on the distribution of histologic types among lung cancers arising in never, current, and former smokers (8). In this series adenocarcinomas comprised 69.9% of lung cancers in never smokers, versus 39.9% in current and 47.3% in former smokers (p < 0.001). Conversely, squamous cell carcinoma comprised 5.9% of lung cancer in never smokers, versus 35.7% in current and 28.0% in former smokers.
Outcome
Four of the retrospective series include multivariate analysis evaluating outcome in never smokers vs. ever-smokers with lung cancer across stages. These studies consistently report a consistent hazard ratio of approximately 1.3, favoring never smokers (Table 1).
Post-surgical outcome in never smokers
Two studies evaluated outcome among patients with surgically resected stage I non-small cell lung cancer (9, 10). Fujisawa reported 10-year overall and disease-specific survival of 84.9% and 88.2%, respectively, among individuals with 0 pack-years (N = 118), compared with 77.3% and 77.3% for smokers with 1 – 29 pack-years (N=39), and 36.7% and 64.7% for smokers with ≥ 30 pack-years (N = 212). P value for overall survival was < 0.001 comparing 0 to ≥ 1 pack-years, and comparing < 30 to ≥ 30 pack-years. Patients with a history of smoking had higher rates of death both from recurrent disease (p = 0.0004) and from nonmalignant causes (p = 0.026). However, survival was not significantly associated with smoking in a multivariate analysis including age, T stage, pleural invasion and gender. The definition of the 0 pack-year cohort in this study is not provided; it is unclear to what extent this cohort represents lifelong never smokers. Similarly, Yoshino et al. evaluated outcome in 428 patients with resected stage I lung adenocarcinomas (10). Never smokers (N = 193) had improved post-operative survival over 5 years relative to ever-smokers (p = 0.0001). In this study, ever-smoking status remained an independent prognostic factor in multivariate analysis including gender, histologic subtype, and pathologic sub-stage.
Evaluating surgically resectable non-small cell lung cancer more broadly, Nia et al. performed a retrospective post-surgical analysis on outcome of 311 patients with resected stage I – IIIB disease operated on by a single surgeon (11). The study excluded patients with perioperative death (death within 30 days of surgery, or during the same hospitalization as surgery). Populations analyzed included active smokers (54.3%), recent quitters (11.3%), former smokers (11.3%) and never smokers (8%; N = 25). Current active smokers were considered as the reference group. Post-surgical survival was significantly better in never smokers than active smokers (relative risk = 0.45; 95% CI 0.21 – 0.97; p = 0.042). However, similarly improved outcome was seen among former smokers (relative risk 0.54, 95% CI 0.35 – 0.84; p = 0.006) and recent quitters (relative risk 0.34; 95% CI 0.16 – 0.71; p = 0.004), suggesting that a primary difference in survival was attributable to smoking status at the time of surgery.
Outcome in the absence of active treatment
No studies to date have been designed specifically to compare the outcome of never smokers vs. ever smokers with advanced lung cancer in the absence of active therapy. However, two phase III randomized placebo-controlled trials of single agent EGFR inhibitor therapy in advanced recurrent disease – the ISEL study with gefitinib (Iressa) (12) and the BR.21 study with erlotinib (13) – collected data from the placebo control arms which are available for retrospective analysis. Never smokers in the ISEL study treated with placebo had a median survival of 6.1 months and a 1-year survival of 29% versus 4.9 months and an 18% 1-year survival for ever smokers. In the BR.21 study, the median survivals in the placebo group for never smokers and ever smokers were 5.6 months and 4.6 months, respectively. Although data from the placebo control arms in both studies suggests a trend toward improved outcome in never smokers, in neither case do these differences reach statistical significance, in contrast to the outcomes on the investigational arms of these studies (see below).
Chemotherapy treatment outcome in never smokers
Never smokers with lung cancer typically have not been considered as a separate clinical subgroup in trials of cytotoxic chemotherapies. Landmark trials used to set standard first-line (14) or second-line treatments (i.e. docetaxel and pemetrexed) (15, 16) for patients with advanced/metastatic NSCLC did not report the smoking history of patients studied. Even the more definitive study that demonstrated that the angiogenesis inhibitor, bevacizumab, confers a survival advantage when added to standard doublet chemotherapy in the first-line over chemotherapy alone (17), did not report data on smoking histories. Thus, there is little prospective data concerning response or survival rates in never smokers treated with conventional systemic therapies.
There are two retrospective studies on the outcomes of never smokers who received standard cytotoxic chemotherapy, and they present conflicting results. In one study, Tsao and colleagues studied response, progression-free survival, and overall survival in 873 evaluable patients with Stage III or IV NSCLC who received first-line chemotherapy at a single US institution from 1993 to 2002 (18). Never smokers (N = 137) had higher response rates than former or current smokers (19% vs. 8% vs. 12% respectively, p = 0.004), and improved overall survival (p < 0.0001). Never smoking status remained an independent predictor in multivariate analysis including adjustment for age, gender, stage, and performance status, with a hazard ratio of 1.47 for former smokers (p = 0.003) and a hazard ratio of 1.55 for current smokers (p = 0.0004). These data contrast with those found by Toh and colleagues, who studied 143 patients with Stage IIIB/IV NSCLC who received first-line chemotherapy at a single Singaporean institution from 1999 to 2002 (8). 41% (59) of patients were “never” smokers, while 59% (84) were smokers. They found no significant differences between never smokers and ever smokers in response, duration of response, or survival, even after adjusting for known prognostic factors.
At least four factors could account for the observed differences between the two studies. First, the investigators studied different ethnic populations (US vs. Singapore). Second, there were fewer patients in the Singaporean trial, which may have been underpowered to detect differences in outcome between never and ever smokers. Third, different types of chemotherapy were given to the US and Singaporean patients. In the US study, 78% of patients received platinum-based regimens, while in the Singapore study, only 59% of patients received platinum-based agents. Lastly, definitions of the populations studied slightly differed in the two analyses. The Singapore study defined never smokers as individuals “who had never smoked or smoked too little in the past to be regarded as an ex-smoker,” while the US study defined never smokers as individuals who smoked < 100 total lifetime cigarettes.
EGFR, KRAS, AND TREATMENT WITH EGFR INHIBITORS
EGFR mutations
Interest in defining the characteristics of lung cancer in never smokers over the past several years has been driven in large part from a striking observation in clinical trials of the EGFR small molecule tyrosine kinase inhibitors gefitinib and erlotinib: lung cancer patients with minimal or no history of tobacco use demonstrated markedly better clinical outcome when treated with these agents. Many retrospective studies of patients (from the US, Europe, and East Asia) have shown that compared to ever smokers, never smokers demonstrate preferential clinical benefit from treatment with an EGFR inhibitor as upfront or salvage treatment, including statistically significant higher responses rates, longer times to progression, and/or longer median overall survivals (Table 2). Additional intriguing clinical observations included that responses were higher in patients of East Asian ancestry, in women, and in tumors with adenocarcinoma histology (19).
Table 2.
Results from studies of treatment with EGFR inhibitors and outcome by smoking status
| Study | Design/population | Smoking Status (No. subjects) |
Response Rate (95% CI) |
Time to Progression (95% CI) |
Overall Survival | Hazard Ratio (95% CI) |
|---|---|---|---|---|---|---|
| Gefitinib Therapy | ||||||
| Cohen et al. (2003) (24) | Prospective, retrospective subgroup: 142 male and female patients with with non-small cell lung cancer Study location: USA Study years: not stated |
Ever smoked (n = 108) |
4.6% (1.5%–10.6%) |
|||
| Never smoked (n = 34) |
29.4% (14.6%–46.3%) |
|||||
| Miller et al. (2004) (19) | Retrospective: 139 male and female patients with with non-small cell lung cancer Study location: USA (NY) Study years: 1998–2002 |
Ever smoked (n = 103) |
8% | |||
| Never smoked (n = 36) |
36% | |||||
| Han et al. (2005) (144) | Retrospective: 90 male and female patients with with non-small cell lung cancer Study location: Korea Study years: 2001–2004 |
Ever smoked (n = 47) |
7.3% | |||
| Never smoked (n = 43) |
21.9% | |||||
| Kim et al. (2005) (145) | Retrospective: 80 male and female patients with with non-small cell lung cancer Study location: Korea Study years: not stated |
Ever smoked (n = 66) |
15.9% | 243 days (29–457 days) |
||
| Never smoked (n = 17) |
58.8% | 382 days (91–673 days) |
||||
| Konishi et al. (2005) (146) | Retrospective: 122 male and female patients with with non-small cell lung cancer Study location: Japan Study years: 2002–2004 |
Ever smoked (n = 74) |
13.5% | 1 | ||
| Never smoked (n = 48) |
41.7% | 0.39 | ||||
| Lee et al. (27) (168) | Prospective first-line: 36 male and female patients with adenocarcinomas (all never smokers) Study location: Korea Study years: 2003–2004 |
Never smoked (n=36) |
69% | 33 weeks (4–54 weeks) |
||
| Lim et al. (2005) (147) | Retrospective: 110 male and female patients with with non-small cell lung cancer Study location: Singapore Study years: 2001–2003 |
Ever smoked (n = 44) |
9% | 7.6 months | 1 | |
| Never smoked (n = 66) |
47% | 13.6 months | 0.61 (0.38–0.99) |
|||
| Mitsudomi et al. (2005) (148) | Retrospective: 59 male and female patients with with non-small cell lung cancer Study location: Japan Study years: 1999–2003 |
Ever smoked (n = 31) |
36% | 1 | ||
| Never smoked (n = 28) |
68% | 0.511 (0.092–2.854) |
||||
| Thatcher et al. (2005) (12) | Prospective: 1694 male and female patients with with non-small cell lung cancer Study location: Asia, Europe, Central and South America, Canada, Australia Study years: up to October 2004 (start date not stated) |
Ever smoked, gefitinib arm (n = 879) |
5.3% | same as placebo arm | same as placebo arm (not stated) | 0.92 (0.79–1.06) |
| Never smoked, gefitinib arm (n = 250) |
18.1% | 5.6 months | 8.9 months | 0.67 (0.49–0.92) |
||
| Ever smoked, placebo arm (n = 437) |
same as gefitinib arm | same as gefitinib arm (not stated) | 1 | |||
| Never smoked, placebo arm (n = 125) |
2.8 months | 6.1 months | 1 | |||
| Shih et al. (2006) (149) | Retrospective: 62 male and female patients with with non-small cell lung cancer Study location: Taiwan Study years: 2001–2004 |
Ever smoked (n = 21) |
10% | 2.4 (1.1–4.9) |
||
| Never smoked (n = 41) |
51% | 1 | ||||
| Veronese et al. (2005) (150) | Retrospective: 112 male and female patients with with non-small cell lung cancer Study location: USA (PA) Study years: 2001–2003 |
Ever smoked (n = 93) |
||||
| Never smoked (n = 18) |
||||||
| Erlotinib Therapy: | ||||||
| Herbst et al.(2005) (31) | Prospective: 1059 male and female patients with with non-small cell lung cancer Study location: USA Study years: 2001–2002 |
Ever smoked, erlotinib arm (n = 467) |
current smokers: 8.4 months former smokers: 10.0 months |
|||
| Never smoked, erlotinib arm (n = 72) |
6.0 months | 22.5 months | 0.49 (0.28–0.85) |
|||
| Ever smoked, placebo arm (n = 495) |
current smokers: 9.1 months former smokers: 10.9 months |
|||||
| Never smoked, placebo arm (n = 44) |
4.3 months | 10.1 months | 1 | |||
Intensive molecular research by multiple groups seeking to explain these evident population differences ultimately led to three key publications in 2004, reporting that mutations affecting the ATP binding site of the tyrosine kinase domain of EGFR are strongly associated with response to EGFR tyrosine kinase inhibitors (20–22). Mutations in EGFR have become a primary focus of research in lung cancer. A number of EGFR point mutations and deletions affect the ATP binding site of the receptor and may lead to a constitutively active and ligand-independent receptor state. The most common activating mutations in the kinase domain of EGFR are exon 19 deletions that eliminate a leucine-arginine-glutamate-alanine (LREA) motif, and point mutations at position 858 in exon 21, resulting in substitution of arginine for leucine (L858R). Tumors with activating mutations in EGFR are highly dependent on continued EGFR signaling for proliferation and survival. Activating mutations in EGFR occur more commonly in never smokers, in women, in patients of East Asian ethnicity, and in adenocarcinomas.
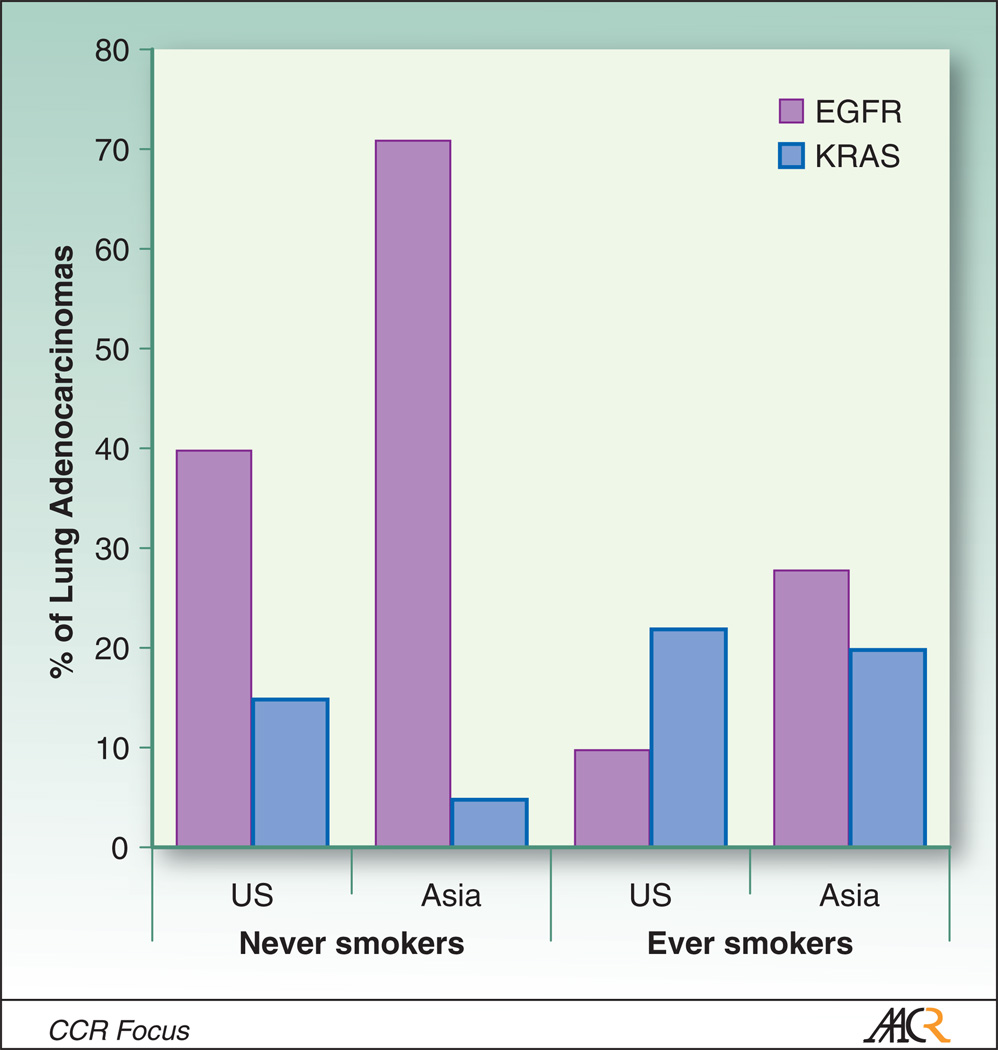
These data have been the subject of multiple recent retrospective studies; in one notable example, Shigematsu and Gazdar reported on analysis of more than 2,000 non-small cell lung cancers (23). EGFR mutations were found to be more common in adenocarcinoma (30%; 413 or 1380 cases) than in lung cancers of other histologies (2%; 16 of 993 cases), and more common in lung cancer from never smokers (45%) than from ever smokers (7%). There was a significant inverse correlation between smoking status and frequency of EGFR mutations (Figure 1).
Figure 1.
Differential frequencies of EGFR and KRAS mutations reported in lung adenocarcinomas in East Asia vs. United States, in never smokers and ever smokers. Activating mutations in both genes are found predominantly in adenocarcinomas, and occur in non-overlapping cohorts. US estimates of EGFR mutation frequencies are based on pooled data summarized in (41) and (22), and of KRAS mutation frequencies from (44). East Asian estimates for both genes are based on pooled data of (39) and (36).
EGFR targeted therapy
Several prospective studies have validated that never smokers favorably benefit from EGFR inhibitor monotherapy (either gefitinib or erlotinib) as compared to ever smokers. Among these, two were prospective clinical trials but retrospectively analyzed (24) for smoking association (i.e. IDEAL-1 (gefitinib) (25) and IDEAL-2 (gefitinib) (26)), and two were prospective randomized placebo-controlled trials for 2nd line therapy with pre-planned subgroup analyses (i.e. ISEL (gefitinib) (12) and BR.21 (erlotinib) (13)) (Table 2). In the ISEL study with gefitinib, never smokers had a response rate of 18.1% versus 5.3% for ever-smokers. The HR for survival for never smokers was 0.67 (p = 0.012). In the BR.21 study with erlotinib, the response rate for never smokers was 24.7% versus 3.9% in ever smokers, and the HR for survival was 0.4 (p = 0.02). These studies demonstrate that for patients with advanced NSCLC previously treated with chemotherapy, EGFR inhibitor monotherapy leads to greater benefit, in terms of improved response rate and prolonged survival, in never smokers compared to ever smokers. A treatment effect is further supported by the lack of a statistical difference in survival seen between never smokers and ever smokers in the placebo arms of the respective trials.
EGFR inhibitors may also be beneficial to never smokers in the first-line setting. In a prospective trial of gefitinib as first-line treatment in 36 never smokers, a 69% response rate was observed, with a median time-to-progression of 8 months (27). The median overall survival had not been reached at the time of publication. Recently, results were reported from a phase III randomized, open-label, first-line study of gefitinib vs. carboplatin and paclitaxel in “never” or “light” East Asian smokers with advanced NSCLC (i.e. the IRESSA Pan Asia Study or “IPASS”). Patients with EGFR mutations experienced longer progression-free survival with gefitinib, and those without mutations had longer progression-free survival with chemotherapy (EGFR mutation positive, HR 0.48; 95% CI 0.36 – 0.64; p < 0.001 [favors gefitinib], EGFR mutation negative, HR 2.85; 95% CI 2.05 – 3.98; p < 0.001 [favors chemotherapy] (28). Final results regarding overall survival have not yet been reported.
Taken together these several studies clearly demonstrate that never smokers benefit from treatment with EGFR inhibitors relative to ever smokers, and that this association is in large part attributable to the markedly higher rates of activating EGFR mutations in never smokers. Despite this important and consistent result, a standard treatment algorithm for never smokers with lung cancer remains to be established. Furthermore, although the addition of EGFR inhibitor to chemotherapy does not confer a survival advantage in unselected populations of NSCLC in four major studies ((29) (INTACT-2), (30) (INTACT-1), (31) (TRIBUTE), (32) (TALENT)), a retrospective analysis of never smokers in one of the studies (TRIBUTE (31)) showed that never smokers treated with the combination of erlotinib plus chemotherapy lived longer than those who received chemotherapy alone (22.5 vs. 10.1 months, p = 0.01). To follow up on this observation, the Cancer and Leukemia Group B (CALGB) is conducting a trial for untreated never smokers, randomizing patients to erlotinib alone or erlotinib plus concurrent chemotherapy. Because of the limited numbers of never smokers, completion of this trial is not expected for several years.
The T790M mutation in EGFR, associated with acquired resistance to gefitinib and erlotinib, has been reported as a rare somatic germline mutation associated with genetic susceptibility to lung cancer in a family with multiple lung adenocarcinomas and bronchoalveolar cancers (33). The smoking status of only one family member was reported; he had a history of smoking. In contrast to multiple prior reports, recent data suggests that this alteration in the catalytic domain of EGFR might result in increased kinase activity and confer a proliferative advantage to cells relative to wildtype EGFR (34).
KRAS mutations
Oncogenic mutations in KRAS are found in approximately 20% of lung adenocarcinomas, and are rare in other histologic subtypes. A meta-analysis evaluating the prognostic significance of KRAS mutations in lung adenocarcinoma concluded that KRAS mutation is a negative prognostic factor, with an overall hazard ratio for death of 1.35 (1.16 – 1.56) (35). KRAS and EGFR mutations, although both found primarily in adenocarcinomas, are essentially never found together in the same tumor, suggesting that these mutations may serve as alternative mechanisms for activating an overlapping set of oncogenic pathways (36–41). The EGFR cell surface receptor activates multiple downstream signaling pathways controlling cell proliferation and cell survival, including the Ras-Raf-ERK and PI3K-Akt pathways. A constitutively active mutant KRAS appears to obviate the need for upstream signaling from EGFR in activating the ERK pathway, and in many cells, PI3K-Akt as well.
Several investigators specifically evaluated whether KRAS mutations in lung adenocarcinomas correlate with smoking status. Of these studies, one (39) clearly demonstrated an association with tobacco exposure. This study, focused on an Asian population reported that KRAS mutations were associated with ever smoking status (p = 0.003), male sex (p = 0.009), and poor histologic differentiation (p = 0.037). Two smaller studies of American and European populations failed to find a significant association between smoking status and KRAS mutations, although in both, a trend toward lower KRAS mutational frequency in never smokers was observed (42, 43). A recently published larger study evaluating KRAS mutational frequency in 482 lung adenocarcinomas in an American population also failed to demonstrate a statistically significant association between KRAS mutation and either smoking status or gender, although KRAS transversion mutations were strongly associated with smoking status (p < 0.0001) (44). It is unclear whether the evident discrepancy between the Riely and Tam studies represents a true difference in lung adenocarcinoma biology between Asian and European/American populations (Figure 1). An association between KRAS mutation and smoking status was confirmed in a recent large scale gene sequencing study of adenocarcinomas (discussed below) (45).
Alternative kinase mutations: EGFR family members, STK11, and EML4-ALK
In addition to activating mutations in EGFR, similar activating mutations in related family members HER2 and HER4 have been found in a smaller fraction (approximately 2%) of lung adenocarcinomas. Gene amplification of HER2 and HER3 have been reported to confer increased sensitivity to EGFR tyrosine kinase inhibitors in lung cancer cell lines (46, 47). Interestingly, EGFR family member mutations, like EGFR and KRAS mutations, appear to be mutually exclusive, presumably representing mechanisms for activation of overlapping or redundant downstream oncogenic pathways (48). HER2 mutation is found at higher frequency among lung cancers of never smokers (p = 0.02) (38). Associations of HER3 mutations with smoking status have not been definitively reported.
STK11 (LBK1) encodes a serine-threonine kinase implicated in cell proliferation and cell survival pathways, and has been found to be mutated in about 11% of non-small cell lung cancers (49). STK11 mutations are more frequent in lung cancers from ever smokers than never smokers (p = 0.007). STK11 mutations are more commonly found in KRAS mutant tumors (p = 0.042) and are very rare in EGFR mutant tumors (p = 0.002). The biological basis for these associations has not been defined.
A recently identified translocation results in an in-frame fusion between the EML4 and ALK genes (EML4-ALK) resulting in a fusion protein with preservation of the ALK kinase domain. Among 266 resected non-small cell lung cancers in an East Asian population, the EML4-ALK fusion gene was found in about 5% of cases (50). EML4-ALK was associated with younger age of cancer onset (p = 0.018) and with never smoking status (p = 0.009). EML4-ALK, EGFR, and KRAS mutations were all mutually exclusive, suggesting that EML4-ALK may be an important oncogenic factor, and a potential therapeutic target, in EGFR wildtype, KRAS wildtype, lung cancer in never smokers.
GENETIC EPIDEMIOLOGY
Familial aggregation
Many lines of evidence suggest that individual susceptibility to lung carcinogens, including SHS, plays a role in the development of lung cancer. Increased susceptibility may have a genetic component, as suggested by consistent reports of familial aggregation of lung cancer (51). A number of studies of familial clustering of lung cancer have been undertaken, most of which report an approximately 1.5 to 2-fold increased risk associated with having a first-degree relative with lung cancer (51). Fewer studies have been conducted in never smokers. The first study of familial risk of lung cancer was conducted more than 40 years ago by Tokuhata et al. (52). This study found that nonsmokers with lung cancer were 40% more likely than nonsmoking controls to report a first-degree relative with lung cancer. Women were more likely than men to report such a family history. Some studies have shown no increased lung cancer risk in never smokers associated with family history of lung cancer in any first-degree relative (53–57), but results vary based on type of relative affected, number of relatives affected, and age or sex of affected relative. While Brownson et al. showed little increase in risk with one family member affected with lung cancer, a 2.6-fold increase (95% CI 1.1–6.2) in risk of lung cancer was seen for never smokers with a family history of five or more first-degree relatives affected (58). Wu et al. focused specifically on familial risk in never smoking women and found that risk of adenocarcinoma of the lung was increased more than three-fold with a family history of lung cancer in a mother (OR 3.24; 95% CI 1.1–9.9) or sister (OR 3.59; 95% CI 1.3–9.8) (54). Young age at diagnosis often suggests an underlying genetic contribution to risk and several studies have shown that family history of an early onset lung cancer is associated with increased risk of lung cancer among never smoking relatives. Schwartz et al. (56) reported a 6-fold increased risk of lung cancer among relatives of never smokers with lung cancer diagnosed between ages 40 and 59 (95% CI 1.1–33.4), while Kreuzer et al. (53) found a non-significant 3-fold increase in risk of lung cancer in female never smokers under age 46 with a family history (OR 3.28; 95% CI 0.71–15.1). A large cohort study in Japan reported that family history of lung cancer in either a parent or sibling was associated with a 2.5-fold increase in lung cancer risk among never smokers (RR = 1.27–4.84) (59). A recent meta-analysis, including 11 studies, evaluated risk associated with family history of lung cancer among never smokers and reported that family history contributed to risk (RR = 1.51; 95% CI 1.11–2.06) (51). In 6 studies with information on number of relatives affected, lung cancer risk in never smokers was increased 57% (95% CI 1.34–1.84) when one relative was affected and 2.5-fold when two or more relatives were affected (95% CI 1.72–3.70).
Several limitations are relevant to the interpretation of these studies, the most notable of which are incomplete or often no adjustment for family structure and smoking among relatives, and lack of validation of family histories. Also, when risk among never smokers is of particular interest, sample sizes tend to be small and analyses that detail smoking status of all affected relatives are lacking. Despite these caveats, there is strong evidence of a familial contribution to risk of developing lung cancer among never smokers.
Linkage studies
The findings of family aggregation suggest the possibility of one or more susceptibility genes for lung cancer. Family linkage studies have been used successfully to identify highly penetrant, low frequency susceptibility genes for diseases. To date, there is only one ongoing lung cancer family linkage study. This national study, being conducted by the Genetic Epidemiology of Lung Cancer Consortium, is not limited to lung cancer among never smokers, and high risk families often include smoking and never smoking affected members. The first findings from this study were reported by Bailey-Wilson et al. and include linkage of lung cancer to a region on chromosome 6q23–25 (146cM to 164cM) (60). It was also shown that lung cancer risk among putative carriers of the linkage region was increased even in never smokers. The search for a lung cancer gene within this region is ongoing. Only about 1% of lung cancer patients have such extensive family histories (with three or more affected relatives), but 10–15% have at least one first-degree relative with the disease.
Candidate gene association studies
While linkage studies are typically used to detect highly penetrant genes that are rare, alternative approaches such as association studies are used to detect susceptibility genes that are more common, i.e., with minor allele frequencies of 5% or more. Such studies in lung cancer have primarily focused on polymorphisms in genes coding for enzymes involved in phase I and phase II metabolism of carcinogens in tobacco smoke and DNA damage repair. This body of work was recently reviewed and few consistent results have been identified using this approach even in large populations primarily comprised of smokers (61). The work in never smokers is limited to a few studies of modest sample size. These studies focus on the same set of candidate genes evaluated in smokers. The rationale for studying polymorphisms in the tobacco metabolism pathway in non-smokers is that a significant subset of non-smokers have exposure to SHS, suggesting that some of the underlying mechanisms of lung carcinogenesis may be the same as seen in smokers (62). Conversely, a larger fraction of cancers in never smokers may have no clear relationship to tobacco carcinogens, and inclusion of such cases in analysis makes negative findings somewhat difficult to interpret. It has been suggested that a genetic contribution to risk might be more evident when environmental exposures are low, as in the case of SHS exposure and in never smokers. These populations have generally not been a primary focus of candidate gene analyses: data summarized below is primarily derived from subset analysis of larger studies.
Metabolism of polycyclic aromatic hydrocarbons (PAHs), tobacco-specific nitrosamines, and aromatic amines in cigarette smoke occurs via two classes of enzymes: phase I (oxidation/reduction/hydrolysis) and phase II (conjugation) enzymes. The following phase I and II enzymes have been studied in at least 100 never smokers with lung cancer: CYP1A1, CYP1B1, NAD(P)H quinone oxidoreductase 1 (NQO1), N-acetyltransferase 2 (NAT2) and the glutathione S-transferases (GSTM1, P1 and T1).
CYP1A1 and CYP1B1 are active in the metabolism of PAHs found in tobacco smoke. Two CYP1A1 polymorphisms are the most frequently studied, a T3801C substitution resulting in a MspI restriction site and a A2455G substitution resulting in a Ile462Val change in exon 7. A CYP1B1 Leu432Val polymorphism has also been studied. Glutathione S-transferases occur in a number of classes and act to conjugate electrophilic compounds with glutathione. GSTM1 and GSTT1 occur in null forms resulting in enzyme deficiencies, while an Ile105Val polymorphism in GSTP1 is the most frequently studied. NQO1 can act in both carcinogen activation and detoxification and the Pro187Ser polymorphism has been evaluated. Individuals can carry a fast/rapid or slow acetylator phenotype that can be defined by a series of SNPs in NAT2.
Because never smokers make up a small fraction of all lung cancer cases, some of the larger studies of never smokers have come from consortia. A pooled analysis of 11 primarily European studies by the Genetic Susceptibility to Environmental Carcinogenesis (GSEC) consortium included 130 never smoking cases and 925 never smoking controls (63). Never smokers carrying at least one CYP1A1 Val allele at Ile462Val were at 2-fold (95% CI 1.36–3.13) increased risk of developing lung cancer. The association was stronger in women than in men. A follow-up pooled analysis by GSEC, now including 14 studies and 302 never smoking cases and 1631 never smoking controls, showed a three-fold increased risk associated with the CYP1A1 Val allele of Ile462Val among never smokers (95% CI 1.51–5.91), but no effect of the MspI polymorphism or the GSTM1 null genotype (64, 65). In evaluating the results of these 14 studies independently, few of the studies showed positive findings and only one of the studies included more than 100 never smoking cases. Only three studies provided data on SHS exposure.
Raimondi et al. (66) evaluated the role of GSTT1 in lung cancer in a large pooled and meta-analysis based on 34 studies, which included data on more than 4,000 never smokers. No association was found between the GSTT1 null genotype and lung cancer risk among never smokers in these analyses.
In addition to the pooled analyses, there are few studies of never smokers that include at least 100 cases. Only these larger studies of over 100 never smoking lung cancer cases are discussed. None of the polymorphisms in GSTM1, GSTT1 and GSTP1 studied by Wenzlaff et al. (67) were associated with risk of lung cancer among never smokers. However, in individuals with 20 or more years of household SHS exposure, carrying the GSTM1 null genotype was associated with a 2.3-fold increased risk (95% CI 1.05–5.13). Risk was further increased in these individuals if they also carried the GSTP1 Val allele at Ile105Val (OR 4.56; 95% CI 1.21–17.21). Similar findings of increased risk among GSTM1 never smokers exposed to SHS were noted in two other studies. In one, GSTM1 null women exposed to 40 or more pack-years of SHS exposure through their husbands were at 2.3-fold (95% CI 1.13–4.57) increased risk of lung cancer (68). In a case-only study, an OR of 2.3 (95% CI 1.15–4.51) was noted in the subset of never smoking women with lung cancer who were both GSTM1 null and exposed to SHS (69). No significant associations for GSTM1 or GSTT1 and lung cancer risk were reported by Malatas et al. (70).
Neither Kiyohara et al. in a study of Japanese women, Wenzlaff et al. (71) in a population-based study in the U.S., nor Bennett et al. (72) in a case-only analysis found lung cancer risk associated with the CYP1A1 polymorphisms, even when taking into account SHS exposure. In one of these studies, polymorphisms in the gene coding for CYP1B1 were also evaluated. Carrying at least one Val allele in CYP1B1 Leu432Val was associated with an almost 3-fold (95% CI 1.63–5.07) increase in risk of lung cancer in Caucasian never smokers. This association was most significant in never smokers with SHS exposure.
Neither Chao et al. (73) nor Bock et al. (74) report significant associations of the NQO1 Pro187Ser polymorphism with risk of lung cancer in never smokers. In one study, however, the subset of individuals age 50 or greater carrying the T allele risk was at approximately 50% decreased risk of lung cancer (95% CI 0.27–0.87) (74). The acetylation phenotype was evaluated for its contribution to lung cancer risk in a Taiwanese population of never smokers (75). Several SNPs in NAT2 were genotyped allowing classification of never smokers as either rapid acetylators or slow acetylators. Rapid acetylators were at 2.5-fold increased risk of developing lung cancer (95% CI 1.40–4.23).
In addition to work focused on phase I and II enzyme polymorphisms, studies have looked at the role of SNPs in genes coding for DNA repair enzymes. These studies also suffer from limited sample sizes, particularly of never-smokers. In a case-control study nested in a large cohort study, Matullo et al. evaluated 16 DNA repair polymorphisms and found no association with risk of lung cancer in never and former smokers (76). Likewise, no significant lung cancer risk has been found to be associated with genotype at polymorphisms in OGG1 (77, 78) among never smokers. Although prior studies have failed to find evident associations between polymorphisms in XRCC1 and lung cancer risk in never smokers (77, 79), one recent case-control study of Chinese never-smoking women reported significant associations with two of five polymorphisms examined (80). One of these two is a silent base change, of questionable biological significance, and these have not been confirmed in other similar (and larger) data sets. O6-alkylguanine DNA aklyltransferse (AGT) repairs DNA adducts caused by the metabolism of N-nitroso compounds in cigarette smoke. Three SNPs in AGT have been studied in never smokers (81). The 143Val/178Arg variant alleles were associated with a 2-fold (95% CI 1.03–4.07) increased risk of lung cancer in this study. A recent pooled analysis of 14 studies of sequence variants in 12 DNA repair enzymes, including data from 8,454 cases and 9,344 controls (878 never smoking cases and 3326 never smoking controls), revealed only four sequence variants weakly associated with lung cancer risk, none with evident specificity for never smoking status (82).
Genome wide association studies
A number of large-scale genomic analyses of lung cancers, including genome-wide association studies characterizing genetic contributors to lung cancer risk in case-control series, and genomic and epigenetic studies characterizing tumor-specific somatic genomic alterations in histologic subtypes of lung cancer, have been recently presented.
High density arrays of single nucleotide polymorphism loci (SNPs) can be screened to produce detailed profiles of germline alterations associated with increased cancer risk. Remarkably, three recent genome wide association studies, each involving analysis of over 300,000 SNPs in several thousand cases and controls, independently converged on the 15q24–25.1 chromosomal locus as a polymorphic site, genomic amplification of which was associated with lung cancer risk (83–85). This region contains genes for nicotinic acetylcholine receptor subunits, suggesting a possible association with nicotine dependence. A subsequent analysis taking into account data from all three studies and additional never smoking cases suggests that the 15q24–25.1 locus polymorphism is not associated with lung cancer in never smokers, supporting that its primary influence on lung cancer risk may be through an influence on tobacco addiction (86). To date, genome-wide association studies of lung cancer have not defined genetic factors contributing to lung cancer risk in never smokers.
Broad-based screens for both tumor-specific (somatic) epigenetic and genetic alterations have recently been performed. Weir et al. conducted a large-scale SNP analysis of adenocarcinomas, the most common type of lung cancer in never smokers (87). Although this dataset of 371 patients included 82 never smokers, no correlations between amplifications or deletions and smoking status were significant after correction for multiple hypothesis testing. Shames et al. published an epigenetic genome-wide screen, evaluating promoter methylation profiles in 107 primary lung cancers (88). Unfortunately this dataset included only 9 “never smokers or non-smokers,” precluding any assessment of epigenetic alterations specific to lung cancer in the never smoker. Ding et al. recently applied high throughput gene sequencing to 188 primary lung adenocarcinomas including 20 never smokers, sequencing 623 genes suggested to play a role in cancer biology (45). The investigators identified a total of 26 genes with sufficiently high frequencies of mutation to implicate them in lung carcinogenesis. The average number of mutations in smokers was significantly higher than in never smokers (p = 0.021), and the authors were able to confirm the previously noted associations between never-smoking status and EGFR mutation (p = 0.0046), and smoking status and both KRAS mutation (p = 0.021) and STK11 mutation (p = 0.044).
Summary: genetic epidemiology
The goal of profiling the genetic characteristics of an individual at high risk for lung cancer is to better understand the underlying biology of lung carcinogenesis in an effort to direct treatment and prevention strategies. This is particularly important to advance the state of knowledge of lung carcinogenesis among never smokers. Familial aggregation of lung cancer has been fairly well characterized and accounts for an approximate 50% increase in risk of lung cancer among never smokers. Family linkage studies, with the goal of identifying a lung cancer susceptibility gene, have included families with both smoking and never smoking members and have yet to identify such a gene. While hundreds of candidate gene studies have been conducted in smokers, the specific genes that are associated with alterations in risk remain poorly understood. Fewer studies have focused on never smokers and these studies have substantial limitations. The studies discussed represent the largest studies conducted, however many are underpowered to detect moderate risks especially when allele frequencies are low and lack information on SHS exposure. Gene-gene or gene-environment interactions have not been studied and require extremely large sample sizes. False positive results are a potential problem and more likely when initially small data sets are stratified by age, race, gender and histologic type. Larger studies from consortia have the capacity to pool findings across studies to increase sample size and power. These consortia should focus on genetic susceptibility in never smokers, but have their own limitations including population heterogeneity due to significant differences in allelic frequencies between races and ethnicities, differing case and exposure definitions, and differing genotyping methods.
A limited number of candidate SNPs has been studied that represent only some of the variation within a gene, may not be functional, and are unlikely to be acting alone. These analyses have primarily focused on genes encoding carcinogen metabolic enzymes relevant to tobacco carcinogens. Newer approaches that select candidate genes within pathways and genotype at multiple markers within a gene have begun to be applied to lung cancer generally, and to lung cancer in never smokers. Given the complexity of the genome and pathways involved in carcinogenesis, it is likely that there are relevant pathways yet to be fully characterized. The use of new technology to genotype thousands of candidate SNPs per sample is allowing for more complete coverage of the variation within candidate genes in multiple pathways. In addition to candidate gene studies, whole genome association studies offer a powerful approach when relative risks are modest and where environmental factors play a role. Genome-wide studies in lung cancer have included limited numbers of never smokers. Genomic analyses of tumor-specific alterations, including copy number and mutational studies, have confirmed previously identified mutations associated with never smoking status, but to date have not defined novel mutations in the etiology of lung cancer of never smokers.
BIOMARKERS OF EXPOSURE
Within the population of never smokers with lung cancer, a critical distinction may exist between cancers related to SHS, and cancers unrelated to tobacco exposure. Limited methods exist for quantitation of SHS exposure by both intensity of exposure and duration. Previous studies have focused on current smokers compared to either ex-smokers or never smokers. Studies of never smokers stratified by exposure to passive smoke, indoor pollution, and other risk factors are limited. A comprehensive review of exposure and cancer-related biomarkers of tobacco smoke has been recently published (89) (Supplementary Table 1).
TP53 mutations as markers of exposure
There are several molecular differences in lung cancers from smokers as compared to never smokers (Table 3). One of the more promising genes, considering biomarkers of exposure, is the TP53 tumor suppressor gene which is located in chromosome 17p13.1. It encodes a multifunctional p53 phosphoprotein, important in apoptosis, cell cycle regulation, senescence as well as DNA-repair; for reviews see e.g., (90–92).
Table 3.
Key biomarkers of differences in lung cancer between ever smokers and never smokers
| Biomarker | Never smokers | Reference |
|---|---|---|
| TP53 mutation spectrum | Lower frequency of G:C to T:A mutations Lower frequency of mutations Lower frequency of mutations at hotspots |
Hainaut et al. (102) Vahakangas et al. (43) Toyooka et al. (98) |
| KRAS mutations | Lower frequency Lower frequency of transversions |
Kosaka et al. (36); Tam et al. (39) Riely et al. (44) |
| STK11 (LBK1) mutations | Lower frequency | Koivunen et al. (49) |
| EGFR mutations | Higher frequency | Mitsudomi et al. (148); Shigematsu et al. (23,41) |
| HER2 mutations | Higher frequency | Shigematsu et al. (38) |
| EML4-ALK transloations | Higher frequency | Wong et al. (50) |
| N-tyr expression | Higher level | Le Calvez et al. (111) |
Most (if not all) human tumors lack function of p53 either through a gene deletion or mutation or through an indirect or posttranslational mechanism, such as increased expression of negative regulators of p53, like the overexpression of Jab1 (93). In some cases mutations of the TP53 gene, of which 73.7% are missense mutations, show a mutation spectrum implicating carcinogen-specificity (Supplementary Table 2) and may serve as environmental and clinical biomarkers (94–96). There are over 2900 TP53 mutations from lung cancer (in 38.6% of the tumors analyzed) listed in the latest release of IARC-based database (1; see also (97)). Information about smoking/nonsmoking can be found in over 1000 cases, but about passive smoke exposure in very few cases. Never smoker is not a designation in this database.
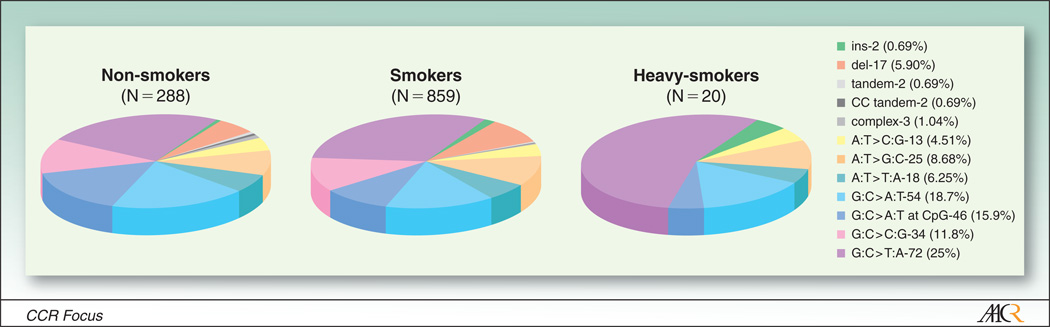
While both TP53 mutation frequency and the number of hotspot mutations are higher in smokers than in never smokers (98), major differences in the mutation spectra are less clear when all smokers are compared to all nonsmokers in the IARC database (Figure 2). Deletions and insertions (10% in smokers, 5% in nonsmokers) and G:C to T:A mutations (31% in smokers, 26% in nonsmokers) show some difference. The extent to which TP53 mutations observed in nonsmokers reflect exposure to SHS cannot be defined currently because of sparse data related to quantitation of exposure. G:C to T:A transversions, typically induced by the cigarette smoke carcinogen benzo(a)pyrene (BP) in experimental systems (99), are very common at CpG sites in smoking-related lung cancer, and their frequency increases with increased smoking (100–102). One report (103) suggests a higher TP53 mutation frequency in smokers with asbestos exposure compared to smokers without asbestos. Experimental studies by Pfeifer and coworkers (104–109) support a PAH-specific fingerprint in TP53 mutation spectrum in smokers (102).
Figure 2.
TP53 mutation spectra in cancers in lung cancer by smoking status. Source: IARC Database version R13, November 2008 release.
A definitive demonstration of a specific TP53 mutation spectrum related to smoking requires the comparison of the mutation spectra from smokers and never smokers. Both possible gender (98, 110) and geographical differences (96, 101) have to be taken on account in such studies. The few existing studies of never smokers support the hypothesis of smoking-related TP53 mutations with a higher frequency in smokers than in never smokers (43, 111). Toyooka and coworkers (98) analyzed in detail the R6 version of IARC database and found that the G:C to T:A difference between smokers and nonsmokers can be entirely accounted for by the difference in lung cancers in women, with a 13 % frequency in never smokers compared to 36% in smokers; lung cancers in men demonstrated no difference. These results were interpreted as suggestive of higher sensitivity of women to tobacco carcinogens.
Even fewer papers have evaluated the effect of former smoking or exposure to SHS on TP53 mutations. Lung cancers from passive (112) and former smokers (43) have a higher prevalence of TP53 mutations than cases with no history of tobacco exposure, and lower prevalence than that of current smokers (111). That a different TP53 mutation frequency and spectrum between smokers and nonsmokers has also been described in colorectal (113) and in bladder cancer (114) gives support to the idea that TP53 mutations may be useful as a biomarker of smoking etiology of cancer. An ongoing challenge is to design studies incorporating reliable data on exposure to SHS.
An interesting correlation has recently emerged between analysis of p53 pathway and EGFR pathway mutations in lung cancer in never smokers. The factor p14ARF complexes with and inhibits Mdm2, the primary regulator of p53 stability. p14ARF deficiency leads to secondary p53 instability and decreased p53 function. Mounawar et al. analyzed a series of cases in the IARC database for mutations in factors including EGFR, p53, and p14ARF (115). Downregulation of p14ARF expression was more frequently observed in tumors of never smokers (62.5%) than ever smokers (35%; p = 0.0008). Among never smokers, 11 of 16 EGFR mutant tumors were also TP53 mutant, while only 2 of 17 EGFR wildtype tumors had TP53 mutations (p = 0.0008). All EGFR mutant tumors with wildtype TP53 demonstrated suppression of p14ARF expression. Taken together these data suggest a functional interaction between loss of p53 activity and dysregulation of EGFR in lung cancer in never smokers.
As discussed in the accompanying CCR Focus paper, the high incidence of lung cancer among non-smoking Chinese women has been closely associated with the use of smoky coal (116–119) with a dose-dependent increase in the risk (120, 121). Smoky coal emissions are very rich in PAH-compounds (43% of organic emissions) (122). Consequently, there is a higher number of BP-DNA adducts in the bronchoalveolar-lavage cells, peripheral blood mononuclear cells, and placentas of smoky coal exposed women than in non-exposed controls (123). Supporting the benzo(a)pyrene-specific mutation spectrum in smokers, DeMarini and coworkers (124) found the TP53 mutations in the lung cancer tissue of these women clustering in codons 153–158, with most of the mutations being G to T, and with all G to T transversions found on the nontranscribed strand. A clear difference between lung cancers associated with coal smoke vs. tobacco smoke was the TP53 codon 154, a hotspot for PAH adducts (104, 106) but not associated with lung cancers from smokers (102). In general, both the frequency and type of mutations (primarily GC to TA transversions) induced by smoky coal, cigarette smoke condensate, and benzo(a)pyrene in bacterial strains are similar to those found in lung cancers from non-smoking women exposed to smoky coal (122).
Taylor and coworkers (125) originally reported the intriguing finding of a TP53 hotspot in codon 249 in radon-exposed uranium miners from Colorado (Table 4). Among the 52 studied lung cancers, 16 contained an AGG – ATG transversion leading to an amino acid change (arg to met), distinct from the aflatoxin-related mutation in liver cancers at the same locus (AGG - AGT, arg to ser, (126, 127)). However, other studies of uranium miners with radon exposure (128–132), or of other environmentally exposed populations (43, 133, 134) have found very few or no codon 249 AGG-ATG mutations in lung cancers. The predominant type of mutations induced by ionizing radiation in mammalian cells are large deletions or translocations (127). An alternative hypothesis about mycotoxins contributing to the peculiar codon 249 mutation found by Taylor and coworkers (125) has been proposed (135). Although the radon-induced mutation spectrum in lung cancer may be different from the mutation spectrum in cigarette smoke-induced lung cancer from people not exposed to radon, no definitive mutation or set of mutations typical of radon has been defined.
Table 4.
TP53 mutations in populations exposed to radon
| Population | Exposure (mean WLM) |
Number of cases |
No cases (%) with TP53 mutations |
Codon 249 AGG-ATG |
Reference |
|---|---|---|---|---|---|
| Uranium miners: | |||||
| New Mexico | 111 | 19 | 7 mutations in 5 cases (26%) | 0 | Vahakangas et al. (128) |
| Colorado | 1382 | 52 | 16 (31%) | Taylor et al. (125) | |
| Colorado | 1270 | 23 | NS | 0 | McDonald et al. (129) |
| Germany | 1011 | 50 | NS | 0 | Bartsch et al. (130); Hollstein et al. (131) |
| Germany | NK | 29 | NS | 2 (7%) | Yang et al. (132) |
| Environmental exposure: | |||||
| England | 9.2 | 17 | NS | 0 | Lo et al. (133) |
| Sweden | 0.25 0.5 2 |
139 90 14 |
29 (20.9%) 23 (25.6%) 6 (42.9%) |
0 0 0 |
Yngveson et al. (151) |
| Missouri, USA Women |
NK | Non-smokers120 Ex-smokers 11 |
22 (18%) 6 (54%) |
0 0 |
Vahakangas et al. (43) |
WLM, working level months (1WLM corresponds to 170 hours in a place where air contains 130 000 MeV of α-radiation in 1 liter of air; 1Bq/m3 is equivalent to 0.005 WLM)
NK, not known, NS, not studied
Other biomarkers of exposure
Highly specific and sensitive analytical assays of metabolites of nicotine and tobacco-specific N-nitrosamines have been developed that can measure these metabolites in biofluids from infants and adults exposed to SHS. Of these, the most commonly used as a biomarker of exposure is cotinine, which has been extensively applied to quantitate exposure in non-smokers (136, 137). This analytical approach is limited to recent exposure and does not measure lifetime exposure to tobacco smoke.
Carcinogen-DNA or -protein adducts are also measures of tobacco smoke exposure. Blood hemoglobin adducts with 4-aminobiphenyl, acrylamide, ethylene oxide and acrylonitrile are higher in current smokers than in never smokers (138–141), but are not specific for tobacco smokers. DNA adducts have also been measured by the sensitive and non-specific 32P-postlabeling assay but this method also has problems with quantitation (89).
Detection of mutagens in urine using S. typhimurium strains has been widely used and have shown the mutagenicity to be generally dependent on tobacco smoking (142). However, confounding factors such as diet affect urinary mutagenicity. The specific chemicals in tobacco smoke that cause the DNA damage and subsequent mutations are unknown.
Chromosomal assays, e.g. frequencies of sister chromatid exchanges in blood lymphocytes are generally increased in smokers (142) but are not specific for chemicals in tobacco smoke.
We agree with the conclusion of Hatsukami et al (89) (p. 604), that there exists “…no comprehensive set of biomarkers of carcinogen exposure or biological effects as a predictive measure of the total carcinogenicity related to exposure to tobacco or tobacco smoke.” Definition and application of highly sensitive and specific methods for quantitating both acute and chronic SHS exposure are clearly primary research needs in relationship to lung cancer in never smokers.
SUMMARY AND CONCLUSIONS
The emergence of targeted therapies with a differential benefit in clinically and molecularly defined categories of lung cancer emphasizes the need for close interaction between epidemiologists, laboratory scientists, and clinicians to promote better treatment for distinct categories of patients with this disease. Lung cancers can be divided into subgroups based on both differential natural history and differential response to therapeutic response among different patient groups. A critical dichotomy in lung cancer appears to be that between ever smokers and never smokers. These categories have both prognostic and therapeutic implications. The etiologic differences between lung cancer in ever and never smokers, in terms of underlying genetic risk factors and differential pathways of molecular carcinogenesis, are beginning to be defined, but have only recently become a sharp focus of investigation.
Although lung cancer in never smokers would rank independently among the ten most common causes of cancer mortality, there has been a relative paucity of attention to this important patient population. Despite a few published reports in the field, familial aggregation and genetic linkage studies in lung cancer have generally not focused on never smoking cohorts as a separate entity. Similarly, candidate gene association studies in lung cancer have primarily focused on genes relevant to pathways involved in tobacco carcinogen metabolism, which may be of less immediate relevance to genetic contributors to lung cancer risk in the absence of tobacco smoke. Genome-wide analyses of lung cancer in never smokers specifically may be of great interest in defining genetic factors augmenting lung cancer risk. Risk factors identified in never smokers, in which the overwhelming effects of tobacco-induced carcinogenesis are minimized, are likely to be distinct from those currently identified in relation to tobacco metabolic pathways, and may help define lung cancer risk both in the presence and absence of tobacco exposure.
Key tumor suppressor genes and proto-oncogenes with mutational profiles that differ between lung cancers in never smokers and ever smokers include TP53, KRAS, and EGFR. TP53 mutations that show a dose-dependent increase in frequency with tobacco smoke exposure include G to T transversions at hotspots directly influenced by known tobacco carcinogens. However, there is sufficient overlap between the TP53 mutational spectra of lung cancers in smokers and never smokers to preclude differentiating tumors solely on the basis of TP53 genotype. Mutations in KRAS and in EGFR are mutually exclusive, and appear to represent alternative oncogenic pathways to transformation, both resulting in activation of the downstream MAPK pathway as well as other pathways regulating proliferation and survival. EGFR mutations are relatively common in lung cancer in never smokers, but rare in lung cancers in smokers. Small molecule inhibitors of the EGFR tyrosine kinase such as erlotinib and gefitinib demonstrate very high response rates and offer significant clinical benefit against tumors with activating mutations of EGFR. In contrast, tumors with constitutively activating mutations of KRAS (predominantly active or former smokers) demonstrate essentially no objective responses to these drugs.
The biased distribution of KRAS and EGFR mutations in ever and never smokers and the differential clinical benefit from EGFR-directed therapy in these patient subsets have spurred significant interest in the issue of lung cancer in never smokers. However, KRAS and EGFR mutant tumors together represent a minority of non-small cell lung cancers in both ever and never smoking patients.
Molecular pathways of lung carcinogenesis are emerging and need additional definition regarding pathways particularly relevant to the never smoker. Evidence in hand suggests that many of the molecular pathways in lung cancer in never smokers are different from those typically implicated in smokers. Supportive evidence includes the observed differences in histology of lung cancers (higher frequency of adenocarcinomas in never smokers than in smokers), differences in mutational spectra found in the tumors, and differences in response to therapy. Recent estimates based on pooled data from 17 published reports, totaling over 26,000 cases, suggests that the ratio of adenocarcinoma to squamous cell carcinoma is approximately 0.4 in lung cancers in smokers, compared to 3.4 in never smokers (143).
Mutational spectra in several genes differ in lung cancer of never smokers compared to smokers. Most of the current literature has focused on TP53, EGFR and KRAS mutations. Other critical genes/pathways will need to be defined. Defining the many other genetic and molecular differences that distinguish tobacco-driven tumorigenesis from the development of lung cancers in never-smokers is of critical importance in identifying alternative risk factors for lung cancer, of particular but not exclusive relevance to never smokers. In addition, the lessons of EGFR-targeted therapies teach us that defining these differences at a molecular level may identify important therapeutic strategies for targeting key oncogenic events in the thousands of patients with this disease.
Supplementary Material
Acknowledgements
Supported by the Flight Attendant Medical Research Institute (FAMRI).
Footnotes
http://www-p53.iarc.fr/index.html; R13, November 2008
References
- 1.Rudin CM, Avila-Tang E, Samet JM. Lung cancer in never smokers: a call to action. Clin Cancer Res. 2009;15 doi: 10.1158/1078-0432.CCR-09-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Samet JM, Avila-Tang E, Boffetta P, et al. Lung cancer in never smokers: clinical epidemiology and environmental risk factors. Clin Cancer Res. 2009;15 doi: 10.1158/1078-0432.CCR-09-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan YK, Wee TC, Koh WP, et al. Survival among Chinese women with lung cancer in Singapore: a comparison by stage, histology and smoking status. Lung Cancer. 2003;40:237–246. doi: 10.1016/s0169-5002(03)00038-2. [DOI] [PubMed] [Google Scholar]
- 4.Nordquist LT, Simon GR, Cantor A, Alberts WM, Bepler G. Improved survival in never-smokers vs current smokers with primary adenocarcinoma of the lung. Chest. 2004;126:347–351. doi: 10.1378/chest.126.2.347. [DOI] [PubMed] [Google Scholar]
- 5.Toh CK, Gao F, Lim WT, et al. Never-smokers with lung cancer: epidemiologic evidence of a distinct disease entity. J Clin Oncol. 2006;24:2245–2251. doi: 10.1200/JCO.2005.04.8033. [DOI] [PubMed] [Google Scholar]
- 6.Janjigaian YY, McDonnell K, Kris MG, et al. ASCO. Chicago, IL: The American Society of Clinical Oncology; 2008. Pack years of cigarette smoking as a predictor of survival in 2013 patients with stage IIIb/IV non-small cell lung cancer (NSCLC) p. 425s. May 30–June 3, 2008. [Google Scholar]
- 7.Etzel CJ, Lu M, Merriman K, Liu M, Vaporciyan A, Spitz MR. An epidemiologic study of early onset lung cancer. Lung Cancer. 2006;52:129–134. doi: 10.1016/j.lungcan.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 8.Toh CK, Lim WT. Lung cancer in never-smokers. J Clin Pathol. 2007;60:337–340. doi: 10.1136/jcp.2006.040576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujisawa T, Iizasa T, Saitoh Y, et al. Smoking before surgery predicts poor long-term survival in patients with stage I non-small-cell lung carcinomas. J Clin Oncol. 1999;17:2086–2091. doi: 10.1200/JCO.1999.17.7.2086. [DOI] [PubMed] [Google Scholar]
- 10.Yoshino I, Kawano D, Oba T, Yamazaki K, Kometani T, Maehara Y. Smoking status as a prognostic factor in patients with stage I pulmonary adenocarcinoma. Ann Thorac Surg. 2006;81:1189–1193. doi: 10.1016/j.athoracsur.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 11.Sardari Nia P, Weyler J, Colpaert C, Vermeulen P, Van Marck E, Van Schil P. Prognostic value of smoking status in operated non-small cell lung cancer. Lung Cancer. 2005;47:351–359. doi: 10.1016/j.lungcan.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 12.Thatcher N, Chang A, Parikh P, et al. Gefitinib plus best supportive care in previously treated patients with refractory advanced non-small-cell lung cancer: results from a randomised, placebo-controlled, multicentre study (Iressa Survival Evaluation in Lung Cancer) Lancet. 2005;366:1527–1537. doi: 10.1016/S0140-6736(05)67625-8. [DOI] [PubMed] [Google Scholar]
- 13.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 14.Schiller JH, Harrington D, Belani CP, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 15.Hanna N, Shepherd FA, Fossella FV, et al. Randomized phase III trial of pemetrexed versus docetaxel in patients with non-small-cell lung cancer previously treated with chemotherapy. J Clin Oncol. 2004;22:1589–1597. doi: 10.1200/JCO.2004.08.163. [DOI] [PubMed] [Google Scholar]
- 16.Shepherd FA, Dancey J, Ramlau R, et al. Prospective randomized trial of docetaxel versus best supportive care in patients with non-small-cell lung cancer previously treated with platinum-based chemotherapy. J Clin Oncol. 2000;18:2095–2103. doi: 10.1200/JCO.2000.18.10.2095. [DOI] [PubMed] [Google Scholar]
- 17.Sandler A, Gray R, Perry MC, et al. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006;355:2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 18.Tsao AS, Liu D, Lee JJ, Spitz M, Hong WK. Smoking affects treatment outcome in patients with advanced nonsmall cell lung cancer. Cancer. 2006;106:2428–2436. doi: 10.1002/cncr.21884. [DOI] [PubMed] [Google Scholar]
- 19.Miller VA, Kris MG, Shah N, et al. Bronchioloalveolar pathologic subtype and smoking history predict sensitivity to gefitinib in advanced non-small-cell lung cancer. J Clin Oncol. 2004;22:1103–1109. doi: 10.1200/JCO.2004.08.158. [DOI] [PubMed] [Google Scholar]
- 20.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 21.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 22.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shigematsu H, Gazdar AF. Somatic mutations of epidermal growth factor receptor signaling pathway in lung cancers. Int J Cancer. 2006;118:257–262. doi: 10.1002/ijc.21496. [DOI] [PubMed] [Google Scholar]
- 24.Cohen MH, Williams GA, Sridhara R, Chen G, Pazdur R. FDA drug approval summary: gefitinib (ZD1839) (Iressa) tablets. Oncologist. 2003;8:303–306. doi: 10.1634/theoncologist.8-4-303. [DOI] [PubMed] [Google Scholar]
- 25.Fukuoka M, Yano S, Giaccone G, et al. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (The IDEAL 1 Trial) [corrected] J Clin Oncol. 2003;21:2237–2246. doi: 10.1200/JCO.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 26.Kris MG, Natale RB, Herbst RS, et al. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: a randomized trial. Jama. 2003;290:2149–2158. doi: 10.1001/jama.290.16.2149. [DOI] [PubMed] [Google Scholar]
- 27.Lee DH, Han JY, Lee HG, et al. Gefitinib as a first-line therapy of advanced or metastatic adenocarcinoma of the lung in never-smokers. Clin Cancer Res. 2005;11:3032–3037. doi: 10.1158/1078-0432.CCR-04-2149. [DOI] [PubMed] [Google Scholar]
- 28.Mok T, Leong SS, Liu X, et al. Gefitinib vs Carboplatin/paclitaxel in Clinically Selected Chemonaive Patients with Advanced Non-small-Cell Lung Cancer (NSCLC) in Asia: Randomized, Open-label, Phase III Study. J Thorac Oncol. 2008;3:S302. [Google Scholar]
- 29.Giaccone G, Herbst RS, Manegold C, et al. Gefitinib in combination with gemcitabine and cisplatin in advanced non-small-cell lung cancer: a phase III trial--INTACT 1. J Clin Oncol. 2004;22:777–784. doi: 10.1200/JCO.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Herbst RS, Giaccone G, Schiller JH, et al. Gefitinib in combination with paclitaxel and carboplatin in advanced non-small-cell lung cancer: a phase III trial--INTACT 2. J Clin Oncol. 2004;22:785–794. doi: 10.1200/JCO.2004.07.215. [DOI] [PubMed] [Google Scholar]
- 31.Herbst RS, Prager D, Hermann R, et al. TRIBUTE: a phase III trial of erlotinib hydrochloride (OSI-774) combined with carboplatin and paclitaxel chemotherapy in advanced non-small-cell lung cancer. J Clin Oncol. 2005;23:5892–5899. doi: 10.1200/JCO.2005.02.840. [DOI] [PubMed] [Google Scholar]
- 32.Gatzemeier U, Pluzanska A, Szczesna A, Kaukel E, Roubec J, et al. Results of a phase III tiral of erlotinib (OSI-744) combined with cisplatin and gemcitabine (GC) chemotherapy in advanced non-small cell lung cancer (NSCLC) Proceedings of the American Society of Clinical Oncology. 2004;2004:7010. [Google Scholar]
- 33.Bell DW, Gore I, Okimoto RA, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet. 2005;37:1315–1316. doi: 10.1038/ng1671. [DOI] [PubMed] [Google Scholar]
- 34.Vikis H, Sato M, James M, et al. EGFR-T790M is a rare lung cancer susceptibility allele with enhanced kinase activity. Cancer Res. 2007;67:4665–4670. doi: 10.1158/0008-5472.CAN-07-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mascaux C, Iannino N, Martin B, et al. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. British journal of cancer. 2005;92:131–139. doi: 10.1038/sj.bjc.6602258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 37.Soung YH, Lee JW, Kim SY, et al. Mutational analysis of EGFR and K-RAS genes in lung adenocarcinomas. Virchows Arch. 2005;446:483–488. doi: 10.1007/s00428-005-1254-y. [DOI] [PubMed] [Google Scholar]
- 38.Shigematsu H, Takahashi T, Nomura M, et al. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res. 2005;65:1642–1646. doi: 10.1158/0008-5472.CAN-04-4235. [DOI] [PubMed] [Google Scholar]
- 39.Tam IY, Chung LP, Suen WS, et al. Distinct epidermal growth factor receptor and KRAS mutation patterns in non-small cell lung cancer patients with different tobacco exposure and clinicopathologic features. Clin Cancer Res. 2006;12:1647–1653. doi: 10.1158/1078-0432.CCR-05-1981. [DOI] [PubMed] [Google Scholar]
- 40.Marks JL, Broderick S, Zhou Q, et al. Prognostic and therapeutic implications of EGFR and KRAS mutations in resected lung adenocarcinoma. J Thorac Oncol. 2008;3:111–116. doi: 10.1097/JTO.0b013e318160c607. [DOI] [PubMed] [Google Scholar]
- 41.Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. Journal of the National Cancer Institute. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 42.Gealy R, Zhang L, Siegfried JM, Luketich JD, Keohavong P. Comparison of mutations in the p53 and K-ras genes in lung carcinomas from smoking and nonsmoking women. Cancer Epidemiol Biomarkers Prev. 1999;8:297–302. [PubMed] [Google Scholar]
- 43.Vahakangas KH, Bennett WP, Castren K, et al. p53 and K-ras mutations in lung cancers from former and never-smoking women. Cancer Res. 2001;61:4350–4356. [PubMed] [Google Scholar]
- 44.Riely GJ, Kris MG, Rosenbaum D, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res. 2008;14:5731–5734. doi: 10.1158/1078-0432.CCR-08-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cappuzzo F, Toschi L, Domenichini I, et al. HER3 genomic gain and sensitivity to gefitinib in advanced non-small-cell lung cancer patients. British journal of cancer. 2005;93:1334–1340. doi: 10.1038/sj.bjc.6602865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cappuzzo F, Varella-Garcia M, Shigematsu H, et al. Increased HER2 gene copy number is associated with response to gefitinib therapy in epidermal growth factor receptor-positive non-small-cell lung cancer patients. J Clin Oncol. 2005;23:5007–5018. doi: 10.1200/JCO.2005.09.111. [DOI] [PubMed] [Google Scholar]
- 48.Gandhi J, Zhang J, Xie Y, et al. Alterations in genes of the EGFR signaling pathway and their relationship to EGFR tyrosine kinase inhibitor sensitivity in lung cancer cell lines. PLoS ONE. 2009;4:e4576. doi: 10.1371/journal.pone.0004576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koivunen JP, Kim J, Lee J, et al. Mutations in the LKB1 tumour suppressor are frequently detected in tumours from Caucasian but not Asian lung cancer patients. British journal of cancer. 2008;99:245–252. doi: 10.1038/sj.bjc.6604469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong DW, Leung EL, So KK, et al. The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer. 2009;115:1723–1733. doi: 10.1002/cncr.24181. [DOI] [PubMed] [Google Scholar]
- 51.Matakidou A, Eisen T, Houlston RS. Systematic review of the relationship between family history and lung cancer risk. British journal of cancer. 2005;93:825–833. doi: 10.1038/sj.bjc.6602769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tokuhata GK, Lilienfeld AM. Familial aggregation of lung cancer in humans. Journal of the National Cancer Institute. 1963;30:289–312. [PubMed] [Google Scholar]
- 53.Kreuzer M, Heinrich J, Kreienbrock L, Rosario AS, Gerken M, Wichmann HE. Risk factors for lung cancer among nonsmoking women. Int J Cancer. 2002;100:706–713. doi: 10.1002/ijc.10549. [DOI] [PubMed] [Google Scholar]
- 54.Wu AH, Fontham ET, Reynolds P, et al. Family history of cancer and risk of lung cancer among lifetime nonsmoking women in the United States. Am J Epidemiol. 1996;143:535–542. doi: 10.1093/oxfordjournals.aje.a008783. [DOI] [PubMed] [Google Scholar]
- 55.Mayne ST, Buenconsejo J, Janerich DT. Familial cancer history and lung cancer risk in United States nonsmoking men and women. Cancer Epidemiol Biomarkers Prev. 1999;8:1065–1069. [PubMed] [Google Scholar]
- 56.Schwartz AG, Yang P, Swanson GM. Familial risk of lung cancer among nonsmokers and their relatives. Am J Epidemiol. 1996;144:554–562. doi: 10.1093/oxfordjournals.aje.a008965. [DOI] [PubMed] [Google Scholar]
- 57.Gorlova OY, Zhang Y, Schabath MB, et al. Never smokers and lung cancer risk: a case-control study of epidemiological factors. Int J Cancer. 2006;118:1798–1804. doi: 10.1002/ijc.21561. [DOI] [PubMed] [Google Scholar]
- 58.Brownson RC, Alavanja MC, Caporaso N, Berger E, Chang JC. Family history of cancer and risk of lung cancer in lifetime non-smokers and long-term ex-smokers. Int J Epidemiol. 1997;26:256–263. doi: 10.1093/ije/26.2.256. [DOI] [PubMed] [Google Scholar]
- 59.Nitadori J, Inoue M, Iwasaki M, et al. Association between lung cancer incidence and family history of lung cancer: data from a large-scale population-based cohort study, the JPHC study. Chest. 2006;130:968–975. doi: 10.1378/chest.130.4.968. [DOI] [PubMed] [Google Scholar]
- 60.Bailey-Wilson JE, Amos CI, Pinney SM, et al. A major lung cancer susceptibility locus maps to chromosome 6q23–25. Am J Hum Genet. 2004;75:460–474. doi: 10.1086/423857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwartz AG, Prysak GM, Bock CH, Cote ML. The molecular epidemiology of lung cancer. Carcinogenesis. 2007;28:507–518. doi: 10.1093/carcin/bgl253. [DOI] [PubMed] [Google Scholar]
- 62.Besaratinia A, Pfeifer GP. Second-hand smoke and human lung cancer. The lancet oncology. 2008;9:657–666. doi: 10.1016/S1470-2045(08)70172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Le Marchand L, Guo C, Benhamou S, et al. Pooled analysis of the CYP1A1 exon 7 polymorphism and lung cancer (United States) Cancer Causes Control. 2003;14:339–346. doi: 10.1023/a:1023956201228. [DOI] [PubMed] [Google Scholar]
- 64.Hung RJ, Boffetta P, Brockmoller J, et al. CYP1A1 and GSTM1 genetic polymorphisms and lung cancer risk in Caucasian non-smokers: a pooled analysis. Carcinogenesis. 2003;24:875–882. doi: 10.1093/carcin/bgg026. [DOI] [PubMed] [Google Scholar]
- 65.Benhamou S, Lee WJ, Alexandrie AK, et al. Meta- and pooled analyses of the effects of glutathione S-transferase M1 polymorphisms and smoking on lung cancer risk. Carcinogenesis. 2002;23:1343–1350. doi: 10.1093/carcin/23.8.1343. [DOI] [PubMed] [Google Scholar]
- 66.Raimondi S, Paracchini V, Autrup H, et al. Meta- and pooled analysis of GSTT1 and lung cancer: a HuGE-GSEC review. Am J Epidemiol. 2006;164:1027–1042. doi: 10.1093/aje/kwj321. [DOI] [PubMed] [Google Scholar]
- 67.Wenzlaff AS, Cote ML, Bock CH, Land SJ, Schwartz AG. GSTM1, GSTT1 and GSTP1 polymorphisms, environmental tobacco smoke exposure and risk of lung cancer among never smokers: a population-based study. Carcinogenesis. 2005;26:395–401. doi: 10.1093/carcin/bgh326. [DOI] [PubMed] [Google Scholar]
- 68.Kiyohara C, Wakai K, Mikami H, Sido K, Ando M, Ohno Y. Risk modification by CYP1A1 and GSTM1 polymorphisms in the association of environmental tobacco smoke and lung cancer: a case-control study in Japanese nonsmoking women. Int J Cancer. 2003;107:139–144. doi: 10.1002/ijc.11355. [DOI] [PubMed] [Google Scholar]
- 69.Bonner MR, Bennett WP, Xiong W, et al. Radon, secondhand smoke, glutathione-S-transferase M1 and lung cancer among women. Int J Cancer. 2006;119:1462–1467. doi: 10.1002/ijc.22002. [DOI] [PubMed] [Google Scholar]
- 70.Malats N, Camus-Radon AM, Nyberg F, et al. Lung cancer risk in nonsmokers and GSTM1 and GSTT1 genetic polymorphism. Cancer Epidemiol Biomarkers Prev. 2000;9:827–833. [PubMed] [Google Scholar]
- 71.Wenzlaff AS, Cote ML, Bock CH, et al. CYP1A1 and CYP1B1 polymorphisms and risk of lung cancer among never smokers: a population-based study. Carcinogenesis. 2005;26:2207–2212. doi: 10.1093/carcin/bgi191. [DOI] [PubMed] [Google Scholar]
- 72.Bennett WP, Alavanja MC, Blomeke B, et al. Environmental tobacco smoke, genetic susceptibility, and risk of lung cancer in never-smoking women. Journal of the National Cancer Institute. 1999;91:2009–2014. doi: 10.1093/jnci/91.23.2009. [DOI] [PubMed] [Google Scholar]
- 73.Chao C, Zhang ZF, Berthiller J, Boffetta P, Hashibe M. NAD(P)H:quinone oxidoreductase 1 (NQO1) Pro187Ser polymorphism and the risk of lung, bladder, and colorectal cancers: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2006;15:979–987. doi: 10.1158/1055-9965.EPI-05-0899. [DOI] [PubMed] [Google Scholar]
- 74.Bock CH, Wenzlaff AS, Cote ML, Land SJ, Schwartz AG. NQO1 T allele associated with decreased risk of later age at diagnosis lung cancer among never smokers: results from a population-based study. Carcinogenesis. 2005;26:381–386. doi: 10.1093/carcin/bgh314. [DOI] [PubMed] [Google Scholar]
- 75.Chiou HL, Wu MF, Chien WP, et al. NAT2 fast acetylator genotype is associated with an increased risk of lung cancer among never-smoking women in Taiwan. Cancer Lett. 2005;223:93–101. doi: 10.1016/j.canlet.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 76.Matullo G, Dunning AM, Guarrera S, et al. DNA repair polymorphisms and cancer risk in non-smokers in a cohort study. Carcinogenesis. 2006;27:997–1007. doi: 10.1093/carcin/bgi280. [DOI] [PubMed] [Google Scholar]
- 77.Hung RJ, Brennan P, Canzian F, et al. Large-scale investigation of base excision repair genetic polymorphisms and lung cancer risk in a multicenter study. Journal of the National Cancer Institute. 2005;97:567–576. doi: 10.1093/jnci/dji101. [DOI] [PubMed] [Google Scholar]
- 78.Kohno T, Kunitoh H, Toyama K, et al. Association of the OGG1-Ser326Cys polymorphism with lung adenocarcinoma risk. Cancer Sci. 2006;97:724–728. doi: 10.1111/j.1349-7006.2006.00240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hao B, Miao X, Li Y, et al. A novel T-77C polymorphism in DNA repair gene XRCC1 contributes to diminished promoter activity and increased risk of non-small cell lung cancer. Oncogene. 2006;25:3613–3620. doi: 10.1038/sj.onc.1209355. [DOI] [PubMed] [Google Scholar]
- 80.Yin J, Vogel U, Ma Y, Qi R, Wang H. Association of DNA repair gene XRCC1 and lung cancer susceptibility among nonsmoking Chinese women. Cancer Genet Cytogenet. 2009;188:26–31. doi: 10.1016/j.cancergencyto.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 81.Cohet C, Borel S, Nyberg F, et al. Exon 5 polymorphisms in the O6-alkylguanine DNA alkyltransferase gene and lung cancer risk in non-smokers exposed to second-hand smoke. Cancer Epidemiol Biomarkers Prev. 2004;13:320–323. doi: 10.1158/1055-9965.epi-03-0120. [DOI] [PubMed] [Google Scholar]
- 82.Hung RJ, Christiani DC, Risch A, et al. International Lung Cancer Consortium: pooled analysis of sequence variants in DNA repair and cell cycle pathways. Cancer Epidemiol Biomarkers Prev. 2008;17:3081–3089. doi: 10.1158/1055-9965.EPI-08-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hung RJ, McKay JD, Gaborieau V, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 85.Thorgeirsson TE, Geller F, Sulem P, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Spitz MR, Amos CI, Dong Q, Lin J, Wu X. The CHRNA5-A3 region on chromosome 15q24–25.1 is a risk factor both for nicotine dependence and for lung cancer. J Natl Cancer Inst. 2008;100:1552–1556. doi: 10.1093/jnci/djn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shames DS, Girard L, Gao B, et al. A genome-wide screen for promoter methylation in lung cancer identifies novel methylation markers for multiple malignancies. PLoS medicine. 2006;3:e486. doi: 10.1371/journal.pmed.0030486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hatsukami DK, Benowitz NL, Rennard SI, Oncken C, Hecht SS. Biomarkers to assess the utility of potential reduced exposure tobacco products. Nicotine Tob Res. 2006;8:600–622. doi: 10.1080/14622200600858166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 91.Yee KS, Vousden KH. Complicating the complexity of p53. Carcinogenesis. 2005;26:1317–1322. doi: 10.1093/carcin/bgi122. [DOI] [PubMed] [Google Scholar]
- 92.Sengupta S, Harris CC. p53: traffic cop at the crossroads of DNA repair and recombination. Nat Rev Mol Cell Biol. 2005;6:44–55. doi: 10.1038/nrm1546. [DOI] [PubMed] [Google Scholar]
- 93.Lee EW, Oh W, Song J. Jab1 as a mediator of nuclear export and cytoplasmic degradation of p53. Mol Cells. 2006;22:133–140. [PubMed] [Google Scholar]
- 94.Harris CC. Structure and function of the p53 tumor suppressor gene: clues for rational cancer therapeutic strategies. Journal of the National Cancer Institute. 1996;88:1442–1455. doi: 10.1093/jnci/88.20.1442. [DOI] [PubMed] [Google Scholar]
- 95.Hainaut P, Hollstein M. p53 and human cancer: the first ten thousand mutations. Adv Cancer Res. 2000;77:81–137. doi: 10.1016/s0065-230x(08)60785-x. [DOI] [PubMed] [Google Scholar]
- 96.de Moura Gallo CV, Azevedo ESMG, de Moraes E, Olivier M, Hainaut P. TP53 mutations as biomarkers for cancer epidemiology in Latin America: current knowledge and perspectives. Mutat Res. 2005;589:192–207. doi: 10.1016/j.mrrev.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 97.Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum Mutat. 2002;19:607–614. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- 98.Toyooka S, Tsuda T, Gazdar AF. The TP53 gene, tobacco exposure, and lung cancer. Hum Mutat. 2003;21:229–239. doi: 10.1002/humu.10177. [DOI] [PubMed] [Google Scholar]
- 99.Hainaut P, Vahakangas K. p53 as a sensor of carcinogenic exposures: mechanisms of p53 protein induction and lessons from p53 gene mutations. Pathol Biol (Paris) 1997;45:833–844. [PubMed] [Google Scholar]
- 100.Hernandez-Boussard TM, Hainaut P. A specific spectrum of p53 mutations in lung cancer from smokers: review of mutations compiled in the IARC p53 database. Environ Health Perspect. 1998;106:385–391. doi: 10.1289/ehp.98106385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bennett WP, Hussain SP, Vahakangas KH, Khan MA, Shields PG, Harris CC. Molecular epidemiology of human cancer risk: gene-environment interactions and p53 mutation spectrum in human lung cancer. J Pathol. 1999;187:8–18. doi: 10.1002/(SICI)1096-9896(199901)187:1<8::AID-PATH232>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 102.Hainaut P, Pfeifer GP. Patterns of p53 G-->T transversions in lung cancers reflect the primary mutagenic signature of DNA-damage by tobacco smoke. Carcinogenesis. 2001;22:367–374. doi: 10.1093/carcin/22.3.367. [DOI] [PubMed] [Google Scholar]
- 103.Wang X, Christiani DC, Wiencke JK, et al. Mutations in the p53 gene in lung cancer are associated with cigarette smoking and asbestos exposure. Cancer Epidemiol Biomarkers Prev. 1995;4:543–548. [PubMed] [Google Scholar]
- 104.Denissenko MF, Pao A, Tang M, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science. 1996;274:430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- 105.Denissenko MF, Chen JX, Tang MS, Pfeifer GP. Cytosine methylation determines hot spots of DNA damage in the human P53 gene. Proc Natl Acad Sci U S A. 1997;94:3893–3898. doi: 10.1073/pnas.94.8.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Smith LE, Denissenko MF, Bennett WP, et al. Targeting of lung cancer mutational hotspots by polycyclic aromatic hydrocarbons. Journal of the National Cancer Institute. 2000;92:803–811. doi: 10.1093/jnci/92.10.803. [DOI] [PubMed] [Google Scholar]
- 107.Tang MS, Zheng JB, Denissenko MF, Pfeifer GP, Zheng Y. Use of UvrABC nuclease to quantify benzo[a]pyrene diol epoxide-DNA adduct formation at methylated versus unmethylated CpG sites in the p53 gene. Carcinogenesis. 1999;20:1085–1089. doi: 10.1093/carcin/20.6.1085. [DOI] [PubMed] [Google Scholar]
- 108.Pfeifer GP, Denissenko MF. Formation and repair of DNA lesions in the p53 gene: relation to cancer mutations? Environ Mol Mutagen. 1998;31:197–205. doi: 10.1002/(sici)1098-2280(1998)31:3<197::aid-em1>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 109.Denissenko MF, Pao A, Pfeifer GP, Tang M. Slow repair of bulky DNA adducts along the nontranscribed strand of the human p53 gene may explain the strand bias of transversion mutations in cancers. Oncogene. 1998;16:1241–1247. doi: 10.1038/sj.onc.1201647. [DOI] [PubMed] [Google Scholar]
- 110.Tseng JE, Rodriguez M, Ro J, Liu D, Hong WK, Mao L. Gender differences in p53 mutational status in small cell lung cancer. Cancer Res. 1999;59:5666–5670. [PubMed] [Google Scholar]
- 111.Le Calvez F, Mukeria A, Hunt JD, et al. TP53 and KRAS mutation load and types in lung cancers in relation to tobacco smoke: distinct patterns in never, former, and current smokers. Cancer Res. 2005;65:5076–5083. doi: 10.1158/0008-5472.CAN-05-0551. [DOI] [PubMed] [Google Scholar]
- 112.Husgafvel-Pursiainen K, Boffetta P, Kannio A, et al. p53 mutations and exposure to environmental tobacco smoke in a multicenter study on lung cancer. Cancer Res. 2000;60:2906–2911. [PubMed] [Google Scholar]
- 113.Miyaki M, Iijima T, Ishii R, et al. Increased frequency of p53 mutation in sporadic colorectal cancer from cigarette smokers. Jpn J Clin Oncol. 2002;32:196–201. doi: 10.1093/jjco/hyf047. [DOI] [PubMed] [Google Scholar]
- 114.Wallerand H, Bakkar AA, de Medina SG, et al. Mutations in TP53, but not FGFR3, in urothelial cell carcinoma of the bladder are influenced by smoking: contribution of exogenous versus endogenous carcinogens. Carcinogenesis. 2005;26:177–184. doi: 10.1093/carcin/bgh275. [DOI] [PubMed] [Google Scholar]
- 115.Mounawar M, Mukeria A, Le Calvez F, et al. Patterns of EGFR, HER2, TP53, and KRAS mutations of p14arf expression in non-small cell lung cancers in relation to smoking history. Cancer Res. 2007;67:5667–5672. doi: 10.1158/0008-5472.CAN-06-4229. [DOI] [PubMed] [Google Scholar]
- 116.Lan Q, Chen W, Chen H, He XZ. Risk factors for lung cancer in non-smokers in Xuanwei County of China. Biomed Environ Sci. 1993;6:112–118. [PubMed] [Google Scholar]
- 117.Mumford JL, He XZ, Chapman RS, et al. Lung cancer and indoor air pollution in Xuan Wei, China. Science. 1987;235:217–220. doi: 10.1126/science.3798109. [DOI] [PubMed] [Google Scholar]
- 118.Chapman RS, Mumford JL, Harris DB, He ZZ, Jiang WZ, Yang RD. The epidemiology of lung cancer in Xuan Wei, China: current progress, issues, and research strategies. Arch Environ Health. 1988;43:180–185. doi: 10.1080/00039896.1988.9935850. [DOI] [PubMed] [Google Scholar]
- 119.Band PR, Spinelli JJ, Gallagher RP, et al. Identification of occupational cancer risks using a population-based cancer registry. Recent Results Cancer Res. 1990;120:106–121. doi: 10.1007/978-3-642-84068-5_8. [DOI] [PubMed] [Google Scholar]
- 120.Lan Q, He X, Costa DJ, et al. Indoor coal combustion emissions, GSTM1 and GSTT1 genotypes, and lung cancer risk: a case-control study in Xuan Wei, China. Cancer Epidemiol Biomarkers Prev. 2000;9:605–608. [PubMed] [Google Scholar]
- 121.Lan Q, Feng Z, Tian D, et al. p53 gene expression in relation to indoor exposure to unvented coal smoke in Xuan Wei, China. J Occup Environ Med. 2001;43:226–230. doi: 10.1097/00043764-200103000-00010. [DOI] [PubMed] [Google Scholar]
- 122.Granville CA, Hanley NM, Mumford JL, DeMarini DM. Mutation spectra of smoky coal combustion emissions in Salmonella reflect the TP53 and KRAS mutations in lung tumors from smoky coal-exposed individuals. Mutat Res. 2003;525:77–83. doi: 10.1016/s0027-5107(02)00314-7. [DOI] [PubMed] [Google Scholar]
- 123.Gallagher J, Mumford J, Li X, Shank T, Manchester D, Lewtas J. DNA adduct profiles and levels in placenta, blood and lung in relation to cigarette smoking and smoky coal emissions. IARC Sci Publ. 1993:283–292. [PubMed] [Google Scholar]
- 124.DeMarini DM, Landi S, Tian D, et al. Lung tumor KRAS and TP53 mutations in nonsmokers reflect exposure to PAH-rich coal combustion emissions. Cancer Res. 2001;61:6679–6681. [PubMed] [Google Scholar]
- 125.Taylor JA, Watson MA, Devereux TR, Michels RY, Saccomanno G, Anderson M. p53 mutation hotspot in radon-associated lung cancer. Lancet. 1994;343:86–87. [PubMed] [Google Scholar]
- 126.Hsu IC, Metcalf RA, Sun T, Welsh JA, Wang NJ, Harris CC. Mutational hotspot in the p53 gene in human hepatocellular carcinomas. Nature. 1991;350:427–428. doi: 10.1038/350427a0. [DOI] [PubMed] [Google Scholar]
- 127.Bressac B, Kew M, Wands J, Ozturk M. Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature. 1991;350:429–431. doi: 10.1038/350429a0. [DOI] [PubMed] [Google Scholar]
- 128.Vahakangas KH, Samet JM, Metcalf RA, et al. Mutations of p53 and ras genes in radon-associated lung cancer from uranium miners. Lancet. 1992;339:576–580. doi: 10.1016/0140-6736(92)90866-2. [DOI] [PubMed] [Google Scholar]
- 129.McDonald JW, Taylor JA, Watson MA, Saccomanno G, Devereux TR. p53 and K-ras in radon-associated lung adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 1995;4:791–793. [PubMed] [Google Scholar]
- 130.Bartsch H, Hollstein M, Mustonen R, et al. Screening for putative radon-specific p53 mutation hotspot in German uranium miners. Lancet. 1995;346:121. doi: 10.1016/s0140-6736(95)92144-3. [DOI] [PubMed] [Google Scholar]
- 131.Hollstein M, Bartsch H, Wesch H, et al. p53 gene mutation analysis in tumors of patients exposed to alpha-particles. Carcinogenesis. 1997;18:511–516. doi: 10.1093/carcin/18.3.511. [DOI] [PubMed] [Google Scholar]
- 132.Yang Q, Wesch H, Mueller KM, Bartsch H, Wegener K, Hollstein M. Analysis of radon-associated squamous cell carcinomas of the lung for a p53 gene hotspot mutation. British journal of cancer. 2000;82:763–766. doi: 10.1054/bjoc.1999.0995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lo YM, Darby S, Noakes L, et al. Screening for codon 249 p53 mutation in lung cancer associated with domestic radon exposure. Lancet. 1995;345:60. doi: 10.1016/s0140-6736(95)91183-9. [DOI] [PubMed] [Google Scholar]
- 134.Hei TK, Bedford J, Waldren CA. 53 mutation hotspot in radon-associated lung cancer. Lancet. 1994;343:1158–1159. doi: 10.1016/s0140-6736(94)90262-3. [DOI] [PubMed] [Google Scholar]
- 135.Venitt S, Biggs PJ. Radon, mycotoxins, p53, and uranium mining. Lancet. 1994;343:795. doi: 10.1016/s0140-6736(94)91872-4. [DOI] [PubMed] [Google Scholar]
- 136.Benowitz NL, Bernert JT, Caraballo RS, Holiday DB, Wang J. Optimal serum cotinine levels for distinguishing cigarette smokers and nonsmokers within different racial/ethnic groups in the United States between 1999 and 2004. Am J Epidemiol. 2009;169:236–248. doi: 10.1093/aje/kwn301. [DOI] [PubMed] [Google Scholar]
- 137.Seccareccia F, Zuccaro P, Pacifici R, et al. Serum cotinine as a marker of environmental tobacco smoke exposure in epidemiological studies: the experience of the MATISS project. Eur J Epidemiol. 2003;18:487–492. doi: 10.1023/a:1024672522802. [DOI] [PubMed] [Google Scholar]
- 138.Bergmark E. Hemoglobin adducts of acrylamide and acrylonitrile in laboratory workers, smokers and nonsmokers. Chem Res Toxicol. 1997;10:78–84. doi: 10.1021/tx960113p. [DOI] [PubMed] [Google Scholar]
- 139.Carmella SG, Chen M, Villalta PW, Gurney JG, Hatsukami DK, Hecht SS. Ethylation and methylation of hemoglobin in smokers and non-smokers. Carcinogenesis. 2002;23:1903–1910. doi: 10.1093/carcin/23.11.1903. [DOI] [PubMed] [Google Scholar]
- 140.Fennell TR, MacNeela JP, Morris RW, Watson M, Thompson CL, Bell DA. Hemoglobin adducts from acrylonitrile and ethylene oxide in cigarette smokers: effects of glutathione S-transferase T1-null and M1-null genotypes. Cancer Epidemiol Biomarkers Prev. 2000;9:705–712. [PubMed] [Google Scholar]
- 141.Airoldi L, Vineis P, Colombi A, et al. 4-Aminobiphenyl-hemoglobin adducts and risk of smoking-related disease in never smokers and former smokers in the European Prospective Investigation into Cancer and Nutrition prospective study. Cancer Epidemiol Biomarkers Prev. 2005;14:2118–2124. doi: 10.1158/1055-9965.EPI-05-0150. [DOI] [PubMed] [Google Scholar]
- 142.IARC. Tobacco smoke and involuntary smoking. France: International Agency for Research on Cancer; IARC Monograph 83. 2004
- 143.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers--a different disease. Nat Rev Cancer. 2007;7:778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 144.Han SW, Kim TY, Hwang PG, et al. Predictive and prognostic impact of epidermal growth factor receptor mutation in non-small-cell lung cancer patients treated with gefitinib. J Clin Oncol. 2005;23:2493–2501. doi: 10.1200/JCO.2005.01.388. [DOI] [PubMed] [Google Scholar]
- 145.Kim KS, Jeong JY, Kim YC, et al. Predictors of the response to gefitinib in refractory non-small cell lung cancer. Clin Cancer Res. 2005;11:2244–2251. doi: 10.1158/1078-0432.CCR-04-2081. [DOI] [PubMed] [Google Scholar]
- 146.Konishi J, Yamazaki K, Kinoshita I, et al. Analysis of the response and toxicity to gefitinib of non-small cell lung cancer. Anticancer Res. 2005;25:435–441. [PubMed] [Google Scholar]
- 147.Lim ST, Wong EH, Chuah KL, et al. Gefitinib is more effective in never-smokers with non-small-cell lung cancer: experience among Asian patients. British journal of cancer. 2005;93:23–28. doi: 10.1038/sj.bjc.6602652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mitsudomi T, Kosaka T, Endoh H, et al. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J Clin Oncol. 2005;23:2513–2520. doi: 10.1200/JCO.2005.00.992. [DOI] [PubMed] [Google Scholar]
- 149.Shih JY, Gow CH, Yu CJ, et al. Epidermal growth factor receptor mutations in needle biopsy/aspiration samples predict response to gefitinib therapy and survival of patients with advanced nonsmall cell lung cancer. Int J Cancer. 2006;118:963–969. doi: 10.1002/ijc.21458. [DOI] [PubMed] [Google Scholar]
- 150.Veronese ML, Algazy K, Bearn L, et al. Gefitinib in patients with advanced non-small cell lung cancer (NSCLC): the expanded access protocol experience at the University of Pennsylvania. Cancer Invest. 2005;23:296–302. doi: 10.1081/cnv-61528. [DOI] [PubMed] [Google Scholar]
- 151.Yngveson A, Williams C, Hjerpe A, Lundeberg J, Soderkvist P, Pershagen G. p53 Mutations in lung cancer associated with residential radon exposure. Cancer Epidemiol Biomarkers Prev. 1999;8:433–438. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.