Abstract
Hepatocellular carcinoma (HCC) is a common cancer and hepatitis B virus (HBV) is a major etiological agent. Convincing epidemiological and experimental evidence also links HCC to aflatoxin, a naturally occurring mycotoxin that produces a signature p53-249ser mutation. Recently, we have reported that tumor-derived HBx variants encoded by HBV exhibited attenuated transactivation and pro-apoptotic functions, but retained their ability to block p53-mediated apoptosis. These results indicate that mutations in HBx may contribute to the development of HCC. In this study, we determined whether tumor-derived HBx mutants along, or in cooperation with p53-249ser, could alter cell proliferation and chromosome stability of normal human hepatocytes. To test this hypothesis, we established a telomerase immortalized normal human hepatocycte line HHT4 that exhibited a near diploid karyotype and expressed many hepatocyte-specific genes. We found that over-expression one of the tumor-derived HBx mutants, CT, significantly increased colony forming efficiency (CFE) while its corresponding wild-type allele CNT significantly decreased CFE in HHT4 cells. p53-249ser rescued CNT-mediated inhibition of colony formation. While HHT4 cells lacked an anchorage independent growth capability as they did not form any colonies in soft agar, the CT-expressing HHT4 cells could form colonies, which could be significantly enhanced by p53-249ser. Induction of aneuploidy could be observed in HHT4 cells expressing CT but additional recurring chromosome abnormalities could only be detected in cells coexpressing CT and p53-249ser. Our results are consistent with the hypothesis that certain mutations in HBx and p53 at codon 249 may cooperate in contributing to liver carcinogenesis.
Keywords: p53, HBx, cell proliferation, anchorage-independent growth
INTRODUCTION
Primary liver cancer is one of the most common and deadly cancers in the world. Based on epidemiological data, chronic HBV infection is a major etiological factor associated with the development of hepatocellular carcinoma (HCC), the major type of primary liver cancers (1). Epidemiological studies also indicate that dietary aflatoxin B1 (AFB1) intake contributes to the development of HCC, which is associated with a high frequency of a p53 mutation at codon 249 (2;3). Convincing evidence indicate that both HBV and AFB1 have a significant synergistic effect in HCC development (4). However, the molecular basis of the association of HBV infection with liver carcinogenesis remains unclear. Recent studies by many groups including ours indicate that HBx, a 17-Kd protein encoded by HBV, may act as an oncogene during liver carcinogenesis (5). In HBx transgenic mice, liver tumors were observed with a higher frequency compared to those in wild-type mice (6;7). In in vitro studies, HBx can induce transformation in NIH3T3 cells in cooperation with oncogenes such as ras (8), promote abnormal cell proliferation and abnormal mitosis (9–14). Functional studies also show that HBx binds directly to p53 and inhibits p53-mediated apoptosis (15). Expression of HBx has been shown in both pre-malignant and HCC samples (16). Recently, the availability of several naturally occurring tumor-derived HBx mutants provides a good tool to investigate the role of HBx in liver carcinogenesis (17–19). These tumor-derived HBx mutants mainly contain several point mutations distributed in different domains of HBx. These tumor-derived HBx mutants retain the ability to bind to p53 and block p53-mediated apoptosis, as compared with their corresponding wild-type variants. However, they have attenuated activity of NFκB and loss of inhibition of colony formation when overexpressed compared to wild-type HBx (17;20). These results support the hypothesis that HBx mutants may provide an advantage for tumor cells to grow in the host environment during liver carcinogenesis.
Previous in vitro studies related to HBV-associated liver carcinogenesis were mainly involved in HCC-derived cell lines or non-liver cell lines (8;21). This is largely due to the unavailability of a good normal liver-derived cell culture system. Data obtained in non-liver cells are often difficult to be interpreted because HBV carcinogenicity is hepatotropic. Normal human liver-derived cell lines were established previously in our laboratory by using a SV40 T immortalization protocol and these cell lines have been utilized in studies of carcinogen metabolism, mutagenesis and neoplastic transformation (22;23). However, similar to tumor-derived cell lines, T antigen immortalized cells are genetically unstable and aneuploid, and lose certain phenotypic traits including the loss of most cytochrome P450 (CYP) gene expression. Recent studies indicate that the human telomerase reverse transcriptase gene (hTERT) can immortalize human epithelial cells efficiently and that the immortalized cells remain mostly diploid and demonstrate a differentiated phenotype (24;25).
In this study, we established an hTERT-immortalized human hepatocyte cell line, HHT4. Using this cell model, we addressed the following questions: (1) whether tumor-derived HBx mutants induce neoplastic transformation of hTERT-immortalized human hepatocytes HHT4; and (2) whether tumor-derived HBx mutants and p53-249ser cooperate in the neoplastic transformation of HHT4 cells. Our results indicate that tumor-derived HBx mutants, but not their corresponding wild-type variants, can enhance cell proliferation and chromosome instability by cooperating with mutant p53.
MATERIALS AND METHODS
Plasmids and retrovirus production
The pCL-hTERT construct was produced by sub-cloning the human telomerase reverse transcriptase (hTERT) cDNA as an EcoR I fragment from pGRN145 (Geron Corporation, Menlo Park, CA) into the pCLXSN retroviral vector (26). pCLXSN is a derivative of the pLXSN retroviral vector containing a human cytomegalovirus immediate early promoter upstream of the multiple cloning site which allows more robust, long-term expression of the gene of interest. Various tumor-derived HBx mutant fragments (CT, CNT, MT, MNT) were amplified by polymerase-chain reaction (PCR) as previously described (20). These fragments were inserted into the HindIII and ClaI sites of a retroviral expression vector, pLHC (Invitrogen). The p53-249ser mutant fragment was cloned into the BamHI and SnaBI sites of a retroviral vector pBABE (kindly provided by Dr. Soreano). To produce hTERT-encoded retrovirus, 80% confluent Phoenix-A producer cells were transfected with pCL-hTERT construct using LipofectAMINE PLUS Reagent (Invitrogen, Carlsbad, CA) in serum-free DMEM medium. The medium was replaced 24 hours after transfection with either fresh LCM+10% chemically-denatured serum (CDS) (for primary hepatocytes) or DMEM (for all other cell types). Virus was collected as a raw supernatant 24–48 hours later, centrifuged, and filtered through 0.45-μm, low protein-binding filters. For HBx and p53 mutant retrovirus production, amphopack 293 cells were transfected with pLHC-CT, pLHC-CNT, pLHC-MT, pLHC-MNT, or pLHC-p53-249ser and viral supernatant collected 48 hours later. The titer of viral supernatant were quantified and stored at −80 °C for infection of HHT4 cells. The NIH 3T3 cell colony formation assay was used to determine viral titter.
Liver cell immortalization, culture, characterization and transfection
Freshly isolated normal human liver epithelial cells (hepatocytes enriched population) were obtained from organ donors who died of trauma at the University of Pittsburgh Liver Transplantation Center or from surgical resection at the University of Maryland. Normal human liver epithelial cells were obtained by a collagenase/dispase perfusion technique described previously (27) and plated onto flasks that were coated with BFN mix (10 μg/ml of bovine plasma fibronectin, Calbiochem; 0.1 mg/ml of BSA; collagen I (Vitrogen™, Cohesion, Palo Alto, CA); in LHC basal medium, (Invitrogen, Carlsbad, CA) at a cell density of 1×106 cells per 100mm plate. Cells were incubated overnight in HBM/HCM media (Clonetics, San Diego, CA) plus 10% fetal bovine serum.
Cells were switched to HBM/HCM media containing pCL-hTERT virus in the presence of 4 mg/ml of polybrene (Sigma, St. Louis, MO) for five hours. CDS was added at the final concentration of 10%. Cells were incubated overnight and fresh HBM/HCM media plus 10% CDS with or without 35 μg/ml of G418 were added. Fresh media were changed every 3–4 days. G418 was only used for the first two weeks of cell culture. Cells without any virus showed minimum signs of cell proliferation and died, usually within four weeks. In viral-infected cells, healthy colonies with epithelial cell morphology were visible in about two weeks. After four weeks of culture, healthy colonies were isolated using cloning cylinders into plated into 12-well plates. Cells were maintained in culture with less than 80% confluency, and were passaged every week with 1:4 split ratio; using EPET solution (Invitrogen, Carlsbad, CA) followed by SBTI solution (Invitrogen, Carlsbad, CA). From two batches of hepatocytes, two independent clones (i.e., HHT3 and HHT4) were successfully expanded and continuously passaged. These cells were cultured routinely in HBM/HCM plus 3% CDS and further characterized.
cDNA microarrays and telomerase activity analyses
Total RNA samples were isolated from log-phase growing HHT3 and HHT4 cells using Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. The detailed microarray platform, hybridization, quality control, data acquisition and data filtering were performed essentially as previously described (28). Telomerase activity assays were performed on the cells using a real-time PCR based assay (29). This assay utilizes the TRAPeze ® XL Telomerase detection kit (Intergen, Purchase, NY) modified for fluorescence detection from a probe as the telomerase is performs its telomere lengthening function with a Sequence Detection System 7700 instrument (Applied Biosystems, Foster City, CA ). Each assay was performed using 1.5μg of total cellular protein, in triplicate, with a heat-inactivated sample used as a negative control. NCI-H1299 cells were used as the quantitation standard for the assay. All values were reported as the cell equivalent activity of NCI-H1299 cells. The detection limit of the assay was 5 NCI-H1299 cell equivalents.
Colony formation and soft agar assays
HHT4 cells were infected with retroviruses containing HBx mutants (MOI=1) and selected by hygromycin (50ug/ml). Drug-resistant colonies appearing 3 weeks later were fixed, stained and counted. Cells from five clonally derived CT lines (10,000 cells per 10cm plate) were mixed with 0.5% agar and seeded on 0.8% agar at bottom layer. Colony formation was monitored under the microscope. Three weeks later, the plates were fixed, stained and colonies were counted. To recover colonies from soft agar cultures, colonies were randomly chosen and picked up under the microscope using 200μl pipette tips. They were then incubated with 50μl E-PET for 5 minutes and cultured with HCM with 10% CDS in a 37 °C incubator supplemented with 5% CO2. Each cell line was tested in triplicate.
Western blotting and RT-PCR
Proteins from total cell extracts were fractioned on 16% SDS-PAGE gels, followed by transfer and blocking in 5% non-fat dry milk. Blots were probed with anti-M2 antibody, followed by incubation with horseradish peroxidase-conjugated secondary antibody. Antibody-antigen complexes were detected by enhanced chemiluminescence (ECL) (GE Healthcare, Piscataway, NJ) according to the manufacturer’s protocol.
Metaphase preparation and conventional G-banding analysis
Metaphase preparations were obtained for cytogenetic studies using standard chromosome harvest techniques for both the HHT3 and HHT4 cell lines (30). Chromosomal G-banding was performed using a trypsin pretreatment followed by Giemsa staining protocol (30). For each cell line, 20 metaphases were analyzed. Chromosomes were identified and classified according to the nomenclature proposed by the International System for Human Cytogenetic Nomenclature (ISCN) (31).
Spectral karyotyping (SKY) and FISH analyses
SKY was performed as previously described (32). In brief, twenty-four human chromosome-specific DNA libraries were generated by bivariate, high-resolution flow sorting and amplified using degenerate oligonucleotide primed PCR. DNA labeling was performed by directly incorporating haptenized or fluorochrome-conjugated dUTPs. The differentially labeled probe sets were combined and precipitated in the presence of an excess of unlabeled human Cot-1 DNA (Invitrogen, Carlsbad, CA). Hybridization and post-hybridization washes were completed as previously described (32). Spectral images were acquired with a SD200 SpectraCube system (Applied Spectral Imaging, Vista, CA) coupled via a c-mount adapter to a Leica DMRBE microscope followed by spectrabased classification. FISH analysis using whole chromosome painting probes for chromosomes 1 and 16, and subtelomeric probes for 1p and 16q was performed in separate experiments, using commercially available probes (Vysis, Inc.) according to standard laboratory protocols.
RESULTS
Establishment of hTERT-immortalized human hepatocyte-derived cell lines
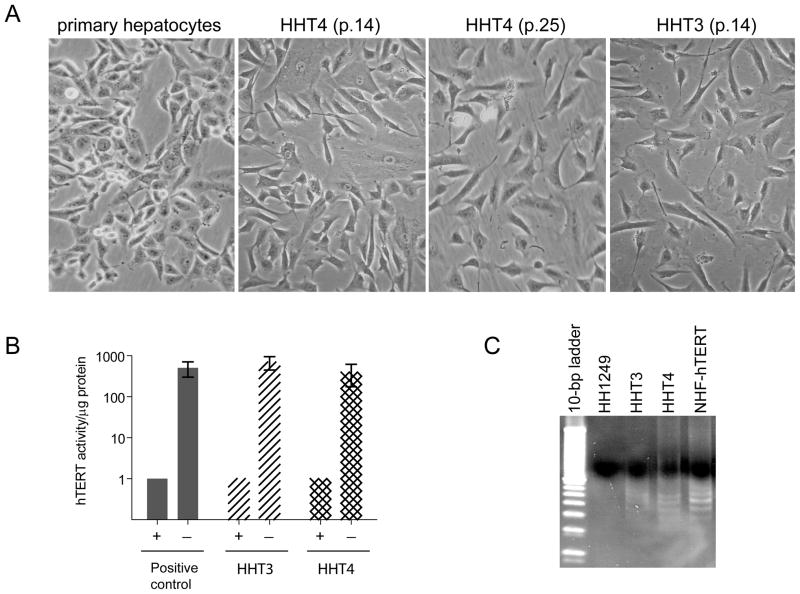
We attempted to establish immortalized human liver cell lines from freshly-isolated primary normal hepatocytes by expressing a human telomerase reverse transcriptase gene (hTERT) as this gene could effectively immortalize normal human fibroblasts and epithelial cells. A modified retroviral vector (pCLXSN) (26) was used as it contained a human cytomegalovirus immediate early promoter (CMV) upstream of the multiple cloning site which allows more robust, long-term expression of hTERT. Two batches of donor hepatocytes were used for infection and selection under G418. Among many colonies that initially survived G418 selection, only two independent clones were capable of expanding into mass cultures (data not shown). These clones were designated as HHT3 and HHT4, respectively. These cells were passaged for more than 12 months in a serum-free LCM medium. They displayed epithelial morphology reassembly of primary cultured hepatocytes (Fig 1A), expressed telomerase activity as determined by the TRAP assay (Fig 1B) and retained some of the hepatocytes markers, i.e., immuopositivity for cytokeratin 14 and 18 but were unable to grow in soft agar and were not tumorigenic in athymic nude mice (data not shown).
Figure 1.
Establishment of hTERT-immortalized human hepatocyte-derived cell lines. (A) Morphological characteristics of primary cultured human hepatocytes and their hTERT-immortalized derivatives HHT3 and HHT4 at different passages. (B) Telomerase activities of HHT3 and HHT4 cells determined by the telomeric repeat amplification protocol (TRAP) assay. (C) Representative amplification products, as analyzed by electrophoresis in a non-denaturing 10% polyacrylamide gel, from (B) are shown. The primary human hepatocytes (HH1249) are included as a negative control, and hTERT-immortalized NHF cells (NHF-hTERT) as a positive control.
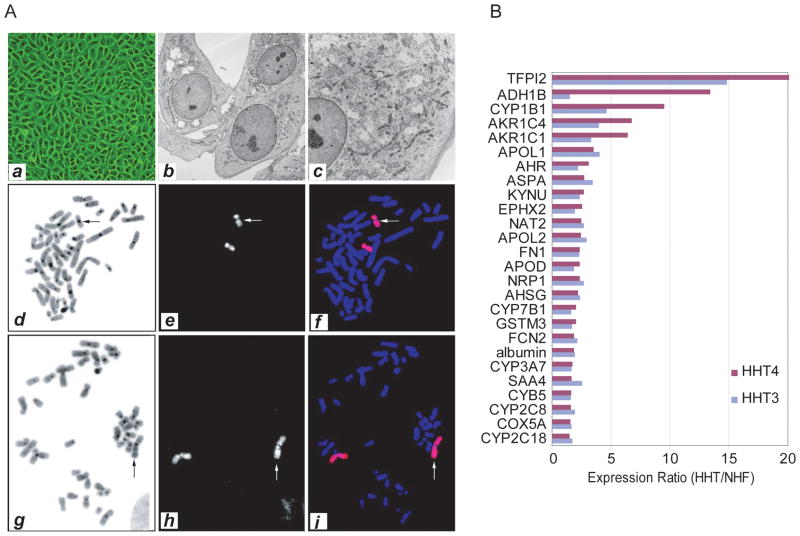
Further characterization of HHT3 and HHT4 cells revealed that these cells appeared to have normal cellular features. For example, they showed the same feature of contact inhibition as immortalized normal human fibroblasts (Fig 2A). Electron microscope analysis indicated that HHT4 cells shared features found in primary hepatocytes such as abundant glycogen granules in the cytoplasm (Fig 2A, b-c). Both HHT3 and HHT4 were diploid/near diploid as determined by G-band metaphase analysis. In 20 metaphases of HHT3 cells examined, we observed a non-mosaic abnormal male karyotype with extra material of unknown origin attached to 16q. Twenty four color fluorescence in situ hybridization (FISH) analysis by spectral karyotyping (SKY) showed that the additional material on the abnormal chromosome 16 was derived from chromosome 16 (data not shown). Subsequent FISH analysis with chromosome 16 whole chromosome painting probe confirmed the above SKY finding. FISH analysis with a 16q subtelomere probe showed no signal on the abnormal 16q. Information from these FISH studies as well as the G-band analysis permitted us to reinterpret the abnormal chromosome 16 to have an inverted duplication of the long arm from band q22 to q23 with accompanying loss of 16q24 (Fig 2A, d-f). All 20 metaphases of HHT4 cells examined had the following clonal chromosomal abnormalities: a structural rearrangement of 1p, loss of chromosome 6 (monosomy 6) and an apparent terminal deletion of 9p. SKY analysis of HHT4 cells showed that the rearranged 1p consisted entirely of chromosome 1 material. FISH analysis with whole chromosome 1 painting probes confirmed the SKY results, and FISH analysis with a 1p subtelomere probe showed absence of signal on the structurally abnormal chromosome 1. These tests suggested that the abnormality on chromosome 1 is an inverted duplication of the short arm from band p13 to p34.3 with accompanying loss of the material from p35 to the p terminus (Fig 2A, g-i). In summary, we concluded that HHT3 had a karyotype of 46, XY, der(16)dup(16)(q23 q22)del(16)(q24) and HHT4 had a karyotype of 45, XY, der(1)dup(1)(p34.3 p13)del(1)(p35), −6, del(9)(p21). HHT3 and HHT4 had few apparent structural and/or numerical chromosomal abnormalities which may represent a random event during immortalization with hTERT.
Figure 2.
Characterization of HHT3 and HHT4 cells. (A) Representative phase-contrast cell image (a) and electron micrographs (b, c) of HHT4 cells are shown. Representative images of a mitotic HHT3 cell (d-f) and a mitotic HHT4 cell (g-i) analyzed by fluorescence in situ hybridization (FISH) with a whole chromosome 16 painting probe or whole chromosome 1 painting probe, respectively. (B) Expression profiles of hepatocyte-specific genes in HHT3 and HHT4 cells analyzed by cDNA microarray. The expression ratios of HHT3 or HHT4 cells over hTERT-immortalized normal human fibroblasts (NHF) are shown. Hepatocyte-specific genes were selected based on a SAGE transcript profile of freshly isolated and cultured normal primary human hepatocytes (33).
To further determine whether HHT cells express hepatocyte-specific genes, we performed cDNA microarray analysis by comparing HHT3 and HHT4 to an hTERT-immortalized normal human fibroblast cell line (hTERT-NHF). A total of 201 genes had a ≥2-fold increased expression in HHT3 or HHT4 cells as compared to hTERT-NHF (data not shown). Among them, 28 (14%) (Fig 2B) were known genes specific for hepatocytes (33). For example, the most abundantly expressed gene was tissue factor pathway inhibitor 2 (TFPI2) followed by alcohol dehydrogenase 1B (ADH1B), apolipoprotein L (APOL), glutathione S-transferase (GST), aldo-keto reductase (AKR) family, P450 cytochrome genes, including CYP7B1, CYP1B1, CYP3A7, CYPB5, CYP2CB and CYP2C18, and epithelial-specific markers such as fibronectin (Fig 2B). Thus, there is an enrichment of hepatocyte specific gene expression associated with HHT3 and HHT4 cells.
Expression of tumor-derived HBx and p53-249ser mutants in HHT4 cells
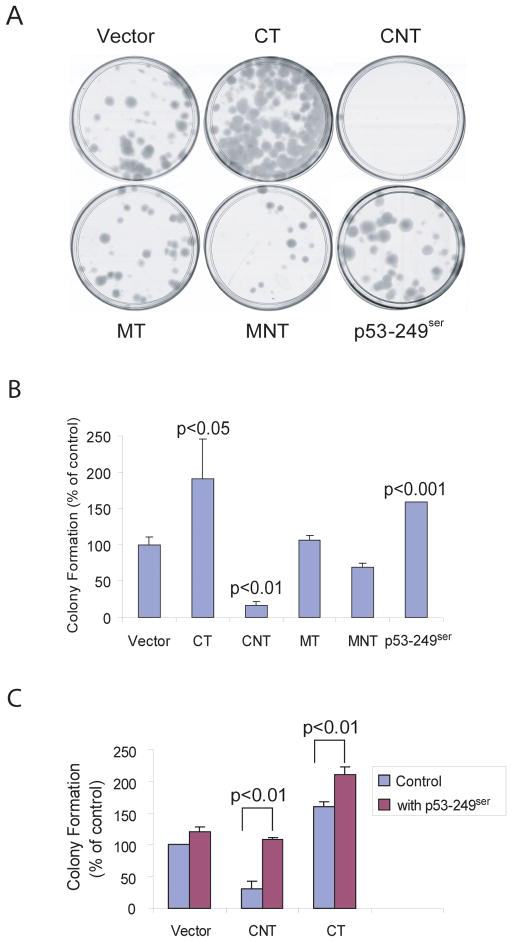
To improve success in obtaining clones that stably express HBx recombinant DNA, we constructed retroviral vectors encoding various tumor-derived HBx mutants and their corresponding variants from the same individuals, which were previously described as CT, CNT, MT and MNT, respectively (20). Specifically, CNT encodes a wild-type HBx gene isolated from non tumor tissue in patient C and CT is the corresponding HBx mutant isolated from tumor tissue of the same patient. Similarly, MNT is a HBx gene isolated from non-tumor tissue in patient M and MT is the corresponding HBx mutant isolated from tumor tissue of the same patient. We also constructed a retroviral vector encoding a p53-249ser gene, a signature mutant allele frequently occurring in HCC from areas where AFB1 food contamination and HBV chronic infection were prevalent (4). HHT4 cells were infected individually with equal amounts of retroviruses (MOI=1) encoding various mutants or a control virus (vector) followed by a selection with hygromycin. Drug-resistant colonies appearing 3 weeks later were fixed, stained and counted. Reproducibly, one tumor-derived HBx mutant CT significantly increased colony-forming efficiency, while it’s variant CNT, which was derived from the same patient, significantly decreased colony-forming efficiency when compared to a vector control virus (Fig 3A-B). While the MT mutant had a minimum activity, the corresponding MNT also appeared to inhibit HHT4 cell colony formation although the difference was not significant. Consistent with previous published reports (34;35), p53-249ser significantly induced HHT4 cell colony formation (Fig 3A-B). These results indicate that a mutation in HBx can enhance colony formation in immortalized HHT4 cells. Interestingly, the CNT-mediated growth inhibitory activity in HHT4 cells could be reversed by expression of p53-249ser (Fig 3C). Co-expression of p53-249ser further enhanced CT-mediated colony-forming efficiency (Fig 3C). However, when we monitored cell cycle progression of HHT4 cells expressing HBx mutants and/or p53-249ser mutant as determined by bromodeoxyurindine (BrdU) pulse labeling followed by flow cytometric analysis, we found minimum cell cycle perturbation or change in sub-G1 population indicative of apoptosis (data not shown). It appears there is no strong effect of HBx mutants or p53-249ser mutant on cell cycle transition or apoptosis. Nevertheless, the p53-249ser mutant appeared to act cooperatively with the tumor-derived HBx mutant CT in promoting HHT4 cell colony formation.
Figure 3.
Effect of p53-249ser mutant or various tumor-derived HBx mutants on HHT4 cell growths. (A) Representative cultured plates with HHT4 cell colonies expressing various recombinant genes. (B) Colony formation efficiency of HHT4 cells expressing CT, CNT, MT, MNT or p53-259ser mutants. (C) Effect of p53-249ser on colony formation of HHT4 cells expressing CNT or CT.
In vitro cellular transformation of HHT4 cells by CT and p53-249ser
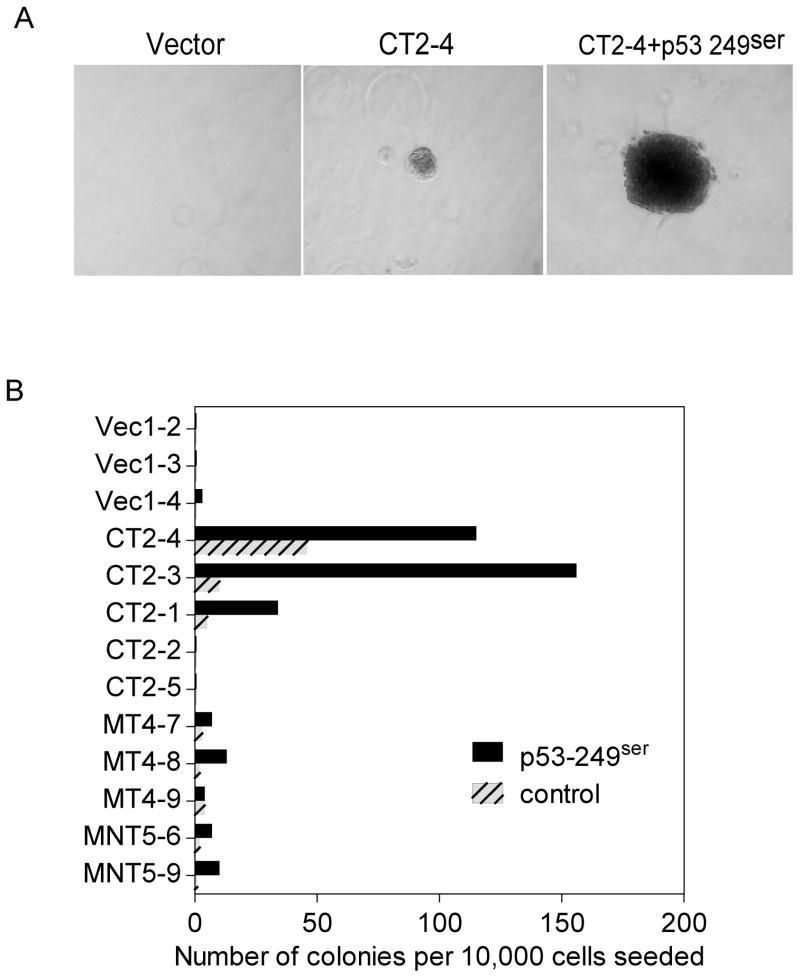
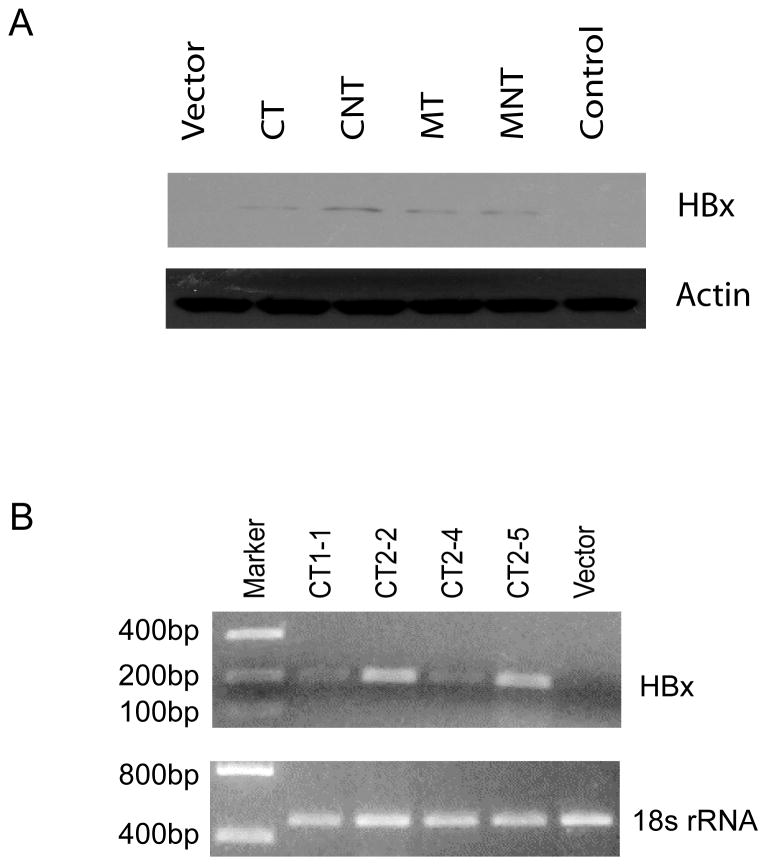
An ability to grow in soft agar is one of the hallmarks of in vitro cellular transformation of normal human cells. The soft agar growth potential of HHT4 cells expressing various HBx mutants was monitored. Colonies arising from the vector control, CT, MT, or MNT-infected HHT4 cells were isolated and attempts were made to expand them into mass cultures. No viable surviving colonies could be isolated from CNT-expressing cells thus were not included for further studies. Similarly, no viable survival colonies from p53-249ser virus alone infected cultures could be expanded into a mass culture. Thus, we were unable to assess clonally derived HHT4 cells stably expressing only p53-249ser mutant. The clonally derived cells expressing CT, MT or MNT were then infected with a control retrovirus or p53-249ser virus, and subsequently mixed with 0.5% agar and seeded on 0.8% agar at bottom layer. After 3 weeks, the plates were fixed, stained and the number of viable colonies was counted. Representative colonies growing on soft agar expressing CT alone or co-expressed with p53-249ser are shown in Fig 4A, and quantitative histograms are shown in Fig 4B. While no colony growth was observed in the parental cell line or 3 clones derived from the vector-infected HHT4 cells, expression of the CT, MT and MNT mutants resulted in an induction of soft agar colony formation with a much more pronounced effect by the CT mutant. About 2 in 10,000 seeded MT or MNT-expressing cells could form colonies in soft agar. In contrast, 3 of 5 clones of CT-expressing cells formed colonies in soft agar efficiently. Interestingly, infection of these cells with p53-249ser appeared to improve their ability to grow in soft agar, with more pronounced effects observed in CT-expressing cells but minimum effects in MT or MNT-expressing cells (Fig 4B). Expression of p53-249ser alone had a minimum effect on soft agar growth. Expression of HBx in HHT4 cells infected with retroviral vectors encoding various HBx variants was verified by western blotting analysis (Fig 5A). In addition, expression of HBx in CT stably transduced clones, i.e., CT1-1, CT2-2, CT2-4 and CT2-5, was verified by RT-PCR analysis (Fig 5B).
Figure 4.
Anchorage-independent growth of HHT4 cells stably expressing CT, MT or MNT with or without expression of p53-249ser mutant. (A) Representative colonies of a CT stably expressing HHT4 clone CT2-4 in soft agar or coexpressing p53-249ser. (B) Six CT-transduced clones, 3 MT-transduced clones and 2 MNT-transduced clones from HHT4 cells alone with 3 vector control clones were evaluated for colony formation in soft agar.
Figure 5.
Expression of HBx in HHT4-derived clones stably transducing various HBx mutants analyzed by western blot (A) or qRT-PCR (B).
It appeared that G-banding and 24-color FISH analyses including two additional p53-249ser-expressing CT2-4 derivative clones independently isolated from soft agar culture, i.e., CT2-4-1, CT2-4-3, indicated that additional recurring chromosome abnormalities were acquired after an introduction of HBx and p53 mutants (Table 1). While additional persistent abnormal chromosome abnormality was not visible in CT2-4 cells as compared to parental HHT4 cells, translocation involved in chromosome 15 (M2) and deletion in chromosome 9 (M3) were frequent events in 3 CT2-4 derivatives expressing p53-249ser (Table 1). An induction of aneuploidy could be observed in HHT4 cells expressing CT and p53-249ser mutants. We also performed karyotype analyses in HHT4 cells stably expressing only p53-249ser as a control. Since single colony-derived cells could not be expanded into mass culture, we combined all survival colonies infected with p53-249ser mutant virus for karyotype analyses. Interestingly, we found that p53-249ser expressing cells had recurrent loss of chromosomes 4 and 13. These changes appear unique to p53-249ser since they were absent in CT2-4 derivatives (Table 1).
Table 1.
Karyotypes of HHT4 cells expressing CT and p53-249ser
| Cells | der 1 | −4 | −6 | del 9 | −13 | del 15 |
|---|---|---|---|---|---|---|
| HHT4 | x | – | x | x | – | – |
| CT2-4 | x | – | x | – | – | – |
| CT2-4-1 | x | – | x | – | – | x |
| CT2-4-2 | x | – | x | – | – | x |
| CT2-4-3 | x | – | x | x | – | – |
| HHT4 + p53-249ser | x | x | x | x | x | – |
Karyotype analysis of HHT4 cells expressing CT and p53-249ser
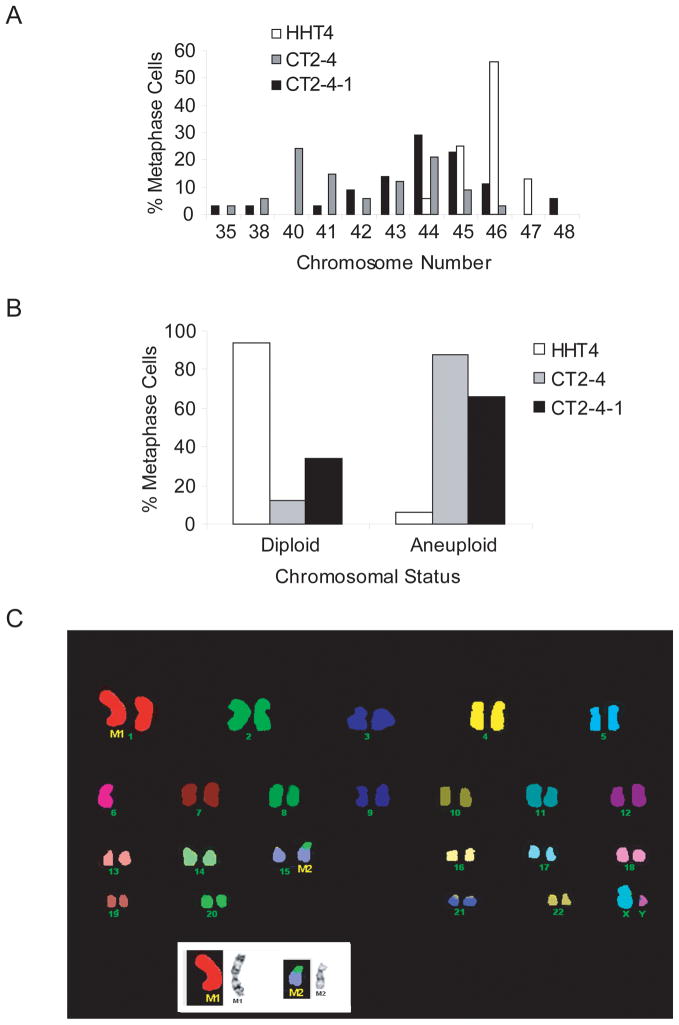
G-band karyotype analysis was performed on CT2-4 as well as CT2-4-1, CT2-4-2, and CT2-4-3, which are derivative clones of CT2-4 expressing p53-249ser that were recovered from soft agar culture. While HHT4 cells had a mean chromosome count of 45, CT2-4 and CT2-4-2 cells had a mean chromosome count of 44 and 43, respectively, with an increase in the percent of aneuploid cells, as determined by metaphase counting analysis (Fig 6A-B). Twenty-four color FISH assisted in further characterization of structural abnormalities (Fig 6C). All cell lines were lacking a normal copy of chromosome 1 which was replaced by a complex structurally abnormal chromosome 1 with duplication and deletion of material. Furthermore, all cell lines were lacking chromosome 6. CT2-4-3 was identical to the parental HHT4 cell line. CT2-4-1 and CT2-4-2 had an additional anomaly comprised of a rearrangement between chromosome 15 and 20. Therefore, additional recurring chromosomal abnormalities were acquired in CT2-4-1 and CT2-4-2 after the introduction of a p53 mutant to CT2-4 (Table 1). The presence of p53-249ser mutant in CT-expressing cells was confirmed using a diagnostic restriction enzyme digestion approach based on RT-PCR and HaeIII digestion as previously described (data not shown) (36).
Figure 6.
Aneuploidy in hTERT-immortalized normal human hepatocytes expressing a tumor-derived HBx mutant and p53-249ser hot-spot mutant. (A, B) Percent of metaphase cells derived from HHT4, CT2-4 (CT) and CT2-4-1 (CT+p53-249ser) cells with abnormal chromosome numbers. (C) Spectral karyotyping of a representative metaphase from CT-2-4-1 cells.
DISCUSSION
In this study, we have shown that by using a newly established telomerase-immortalized human hepatocyte-derived cell line (HHT4), the tumor-derived HBx mutant CT significantly increased colony formation of HHT4 cells, while the non-tumor-derived variant (CNT) derived from the same carrier patient significantly decreased colony formation. Coexpression of p53-249ser rescued CNT-induced colony formation inhibition. Expression of p53-249ser enhanced soft agar growth in HHT4 cells expressing CT, while cells expressing CT alone grew poorly in soft agar. The cloned CT cells coexpressing p53-249ser were aneuploid and showed additional abnormal changes in chromosome 15. However, HHT4 cells expressing p53-249ser alone have lost one copy of chromosome 4 and one copy of chromosome 13 and these changes appear unique to p53 mutant alone (Table 1). Our results are consistent with the hypothesis that CT or p53-249ser mutant alone could induce random chromosome changes but a right combination of chromosome changes when acting together may provide a favorable condition and promote neoplastic transformation.
The newly established telomerase-immortalized human hepatocyte-derived cell line HHT4 has proven to be a good ex vivo model to study liver carcinogenesis. Previous studies related to HBV-associated liver carcinogenesis were mainly involved in liver tumor-derived cell lines or non-liver cell lines (8;17;20;21). These cell lines could not truly reflect the normal process in liver carcinogenesis. This is largely due to the unavailability of a good normal liver-derived cell culture system. We have shown that this newly established cell line HHT4 is near diploid, expresses hepatocyte-specific genes, does not grow in soft agar and is not tumorigenic in nude mice.
The observation of suppression of colony formation by wild-type HBx in HHT 4 cells is consistent with previous reports (17;20;21). It has been reported that overexpression of wild-type HBx can induce or enhance apoptosis in a p53-dependent or p53-independent manner (17;20). Furthermore, HBx expression can induce a late G1 cell cycle block prior to their counterselection by apoptosis (17).
In our study, we also found that one tumor-derived HBx mutant CT increased colony formation in HHT4 cells, which is different from previous studies. The tumor-derived HBx mutant CT contains point mutations in multiple domains of HBx. One possibility is due to the ex vivo model used in this study. As mentioned before, the HHT4 cell line may more accurately reflect the natural process of human liver carcinogenesis. Previous studies from our group and others have shown that these tumor-derived HBx mutants exhibit attenuated transactivation and pro-apoptotic functions, but retained their ability to block p53-mediated apoptosis (17;20;37). Our result further supports the idea that mutations in HBx may contribute to the development of hepatocellular carcinoma.
The observation of a cooperative effect of tumor-derived HBx mutants and p53 249ser mutant in both colony formation and growth in soft agar supports the following hypothesis. The mutation in HBx may provide a growth advantage to those liver cells with mutation in p53. Epidemiological studies as well as molecular studies have shown that there is a high correlation between HBx mutations and p53 mutation at codon 249 (38–41). However, these cells failed to produce any tumor when 2×106 cells of these clones were injected subcutaneously into either nude mice or NOD/SCID mice (data not shown). Taken together, these results indicate that HBx mutants and p53-249ser mutant are weak oncogenes but forced expression of these genes can improve cell viability and anchorage-independent growth. Additional steps may be needed for the tumorigenic conversion of HHT4 cells.
An earlier study by Ponchel and colleagues indicates that p53-249ser mutant increases cell survival and mitotic activity in a p53-deficient hepatoma cell line, Hep3B (35). Interestingly, when we monitored cell cycle progression of HHT4 cells expressing HBx mutants and/or p53-249ser mutant as determined by BrdU pulse labeling followed by flow cytometric analysis, we found minimum cell cycle perturbation or change in sub-G1 population as an indicator of apoptosis (data not shown). It appears there is no strong effect of HBx mutants or p53-249ser mutant on cell cycle transition or apoptosis in hTERT-immortalized normal hepatocyte derived cells. In contrast, single colonies derived from p53-249ser transduced HHT4 cells always fail to propagate into a mass culture, regardless of repeated attempts. The reason for failure to expand p53-249ser mutant expressing HHT4 cells is unclear, but it suggests that cell-cell contact may be critical in supporting cell growth in p53-249ser expressing HHT4 cells. It also suggests that the effect of HBx and/or p53-249ser mutant on cell growth is complex and that the carcinogenic activity of these genes must be subtle and it may take several cell divisions to reach full effect. We speculate that cell type differences (i.e., normal liver cells vs HCC cells) may contribute to the observed discrepancy.
It is interesting to note that hot-spot mutations in HBx at codons 130 and 131, corresponding to lysine to methionine and valine to threonine substitutions, respectively, are prevalent in HCC cases from Qidong, China, where p53-249ser mutation and the HBV genotype C are common (42). In contrast, the CT mutant, derived from an individual with HBV genotype A from an area where p53-249ser mutations are not common, contains multiple amino acid substitutions including those at codons 130 and 131 from lysine to methione and valine to threonine, respectively. The biological difference among these various mutants is unclear at the present time and thus it is cautionary when interpreting the data presented. Future studies are needed to explore the oncogenecity of these mutations.
In summary, our study indicates that certain tumor-derived HBx mutants, not their wild-type variants, may contribute to liver carcinogenesis by cooperating with a hot-spot p53 mutant, i.e., p53-249ser commonly found to be associated with chronic HBV carriers. The finding of a tumor-derived HBx mutant CT in promoting cellular transformation of normal human hepatocytes is important as it may provide a rationale to develop a diagnostic tool by detecting this allele for liver carcinogenesis. It will be interesting to determine molecular profiles of these tumor-derived HBx mutants in telomerase-immortalized human hepatocytes, similar to those published previously (43–46).
Acknowledgments
We thank Dr. Christian Brechot for advice, Maria Motz for technical assistance, and Ms. Karen MacPherson for bibliographic assistance. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- 1.Chen CJ, Yu MW, Liaw YF. Epidemiological characteristics and risk factors of hepatocellular carcinoma. J Gastroenterol Hepatol. 1997;12(9–10):S294–308. doi: 10.1111/j.1440-1746.1997.tb00513.x. [DOI] [PubMed] [Google Scholar]
- 2.Hsu IC, Metcalf RA, Sun T, Welsh JA, Wang NJ, Harris CC. Mutational hotspot in the p53 gene in human hepatocellular carcinomas. Nature. 1991;350:427–8. doi: 10.1038/350427a0. [DOI] [PubMed] [Google Scholar]
- 3.Viviani S, Carrieri P, Bah E, Hall AJ, Kirk GD, Mendy M, Montesano R, Plymoth A, Sam O, Vander SM, Whittle H, Hainaut P. 20 years into the Gambia Hepatitis Intervention Study: assessment of initial hypotheses and prospects for evaluation of protective effectiveness against liver cancer. Cancer Epidemiol Biomarkers Prev. 2008 Nov;17(11):3216–23. doi: 10.1158/1055-9965.EPI-08-0303. [DOI] [PubMed] [Google Scholar]
- 4.Hussain SP, Schwank J, Staib F, Wang XW, Harris CC. TP53 mutations and hepatocellular carcinoma: insights into the etiology and pathogenesis of liver cancer. Oncogene. 2007 Apr 2;26(15):2166–76. doi: 10.1038/sj.onc.1210279. [DOI] [PubMed] [Google Scholar]
- 5.Budhu A, Wang XW. The functional relevance of HBx subcellular localization and nuclear shuttling. In: Kobarg J, editor. The Pleiotropic Functions of the Viral Protein HBx in Hepatitis B Virus Infection and the Development of Liver Cancer. Kerala: Research Signpost; 2008. pp. 105–33. [Google Scholar]
- 6.Kim CM, Koike K, Saito I, Miyamura T, Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;351:317–20. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- 7.Yu DY, Moon HB, Son JK, Jeong S, Yu SL, Yoon H, Han YM, Lee CS, Park JS, Lee CH, Hyun BH, Murakami S, et al. Incidence of hepatocellular carcinoma in transgenic mice expressing the hepatitis B virus X-protein. J Hepatol. 1999 Jul;31:123–32. doi: 10.1016/s0168-8278(99)80172-x. [DOI] [PubMed] [Google Scholar]
- 8.Kim YC, Song KS, Yoon G, Nam MJ, Ryu WS. Activated ras oncogene collaborates with HBx gene of hepatitis B virus to transform cells by suppressing HBx-mediated apoptosis. Oncogene. 2001;20:16–23. doi: 10.1038/sj.onc.1203840. [DOI] [PubMed] [Google Scholar]
- 9.Hodgson AJ, Keasler VV, Slagle BL. Premature cell cycle entry induced by hepatitis B virus regulatory HBx protein during compensatory liver regeneration. Cancer Res. 2008 Dec 15;68(24):10341–8. doi: 10.1158/0008-5472.CAN-08-2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin-Lluesma S, Schaeffer C, Robert EI, van Breugel PC, Leupin O, Hantz O, Strubin M. Hepatitis B virus X protein affects S phase progression leading to chromosome segregation defects by binding to damaged DNA binding protein 1. Hepatology. 2008 Nov;48(5):1467–76. doi: 10.1002/hep.22542. [DOI] [PubMed] [Google Scholar]
- 11.Kim S, Park SY, Yong H, Famulski JK, Chae S, Lee JH, Kang CM, Saya H, Chan GK, Cho H. HBV X protein targets hBubR1, which induces dysregulation of the mitotic checkpoint. Oncogene. 2008 May 29;27(24):3457–64. doi: 10.1038/sj.onc.1210998. [DOI] [PubMed] [Google Scholar]
- 12.Wen Y, Golubkov VS, Strongin AY, Jiang W, Reed JC. Interaction of hepatitis B viral oncoprotein with cellular target HBXIP dysregulates centrosome dynamics and mitotic spindle formation. J Biol Chem. 2008 Feb 1;283(5):2793–803. doi: 10.1074/jbc.M708419200. [DOI] [PubMed] [Google Scholar]
- 13.Forgues M, Difilippantonio MJ, Linke SP, Ried T, Nagashima K, Feden J, Valerie K, Fukasawa K, Wang XW. Involvement of Crm1 in hepatitis B virus X protein-induced aberrant centriole replication and abnormal mitotic spindles. Mol Cell Biol. 2003 Aug;23(15):5282–92. doi: 10.1128/MCB.23.15.5282-5292.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang W, Budhu A, Forgues M, Wang XW. Temporal and spatial control of nucleophosmin by the Ran-Crm1 complex in centrosome duplication. Nat Cell Biol. 2005 Aug;7(8):823–30. doi: 10.1038/ncb1282. [DOI] [PubMed] [Google Scholar]
- 15.Wang XW, Hussain SP, Huo TI, Wu CG, Forgues M, Hofseth LJ, Brechot C, Harris CC. Molecular pathogenesis of human hepatocellular carcinoma. Toxicology. 2002;181–182:43–7. doi: 10.1016/s0300-483x(02)00253-6. [DOI] [PubMed] [Google Scholar]
- 16.Iavarone M, Trabut JB, Delpuech O, Carnot F, Colombo M, Kremsdorf D, Brechot C, Thiers V. Characterisation of hepatitis B virus X protein mutants in tumour and non-tumour liver cells using laser capture microdissection. J Hepatol. 2003 Aug;39(2):253–61. doi: 10.1016/s0168-8278(03)00217-4. [DOI] [PubMed] [Google Scholar]
- 17.Sirma H, Giannini C, Poussin K, Paterlini P, Kremsdorf D, Brechot C. Hepatitis B virus X mutants, present in hepatocellular carcinoma tissue abrogate both the antiproliferative and transactivation effects of HBx. Oncogene. 1999 Aug 26;18:4848–59. doi: 10.1038/sj.onc.1202867. [DOI] [PubMed] [Google Scholar]
- 18.Chen WN, Oon CJ, Leong AL, Koh S, Teng SW. Expression of integrated hepatitis B virus X variants in human hepatocellular carcinomas and its significance. Biochem Biophys Res Commun. 2000 Oct 5;276(3):885–92. doi: 10.1006/bbrc.2000.3562. [DOI] [PubMed] [Google Scholar]
- 19.Ma NF, Lau SH, Hu L, Xie D, Wu J, Yang J, Wang Y, Wu MC, Fung J, Bai X, Tzang CH, Fu L, et al. COOH-terminal truncated HBV X protein plays key role in hepatocarcinogenesis. Clin Cancer Res. 2008 Aug 15;14(16):5061–8. doi: 10.1158/1078-0432.CCR-07-5082. [DOI] [PubMed] [Google Scholar]
- 20.Huo TI, Wang XW, Forgues M, Wu CG, Spillare EA, Giannini C, Brechot C, Harris CC. Hepatitis B virus x mutants derived from human hepatocellular carcinoma retain the ability to abrogate p53-induced apoptosis. Oncogene. 2001;20:3620–8. doi: 10.1038/sj.onc.1204495. [DOI] [PubMed] [Google Scholar]
- 21.Wang JC, Hsu SL, Hwang GY. Inhibition of tumorigenicity of the hepatitis B virus X gene in Chang liver cell line. Virus Res. 2004 Jun 15;102(2):133–9. doi: 10.1016/j.virusres.2004.01.023. [DOI] [PubMed] [Google Scholar]
- 22.Pfeifer AMA, Cole KE, Smoot DT, Weston A, Groopman JD, Shields PG, Vignaud J-M, Juillerat M, Lipsky MM, Trump BF, Lechner JF, Harris CC. SV40 T-antigen immortalized normal human liver epithelial cells express hepatocyte characteristics and metabolize chemical carcinogens. Proc Natl Acad Sci U S A. 1993;90:5123–7. doi: 10.1073/pnas.90.11.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mace K, Offord EA, Harris CC, Pfeifer AM. Development of in vitro models for cellular and molecular studies in toxicology and chemoprevention. Arch Toxicol Suppl. 1998;20:227–36. doi: 10.1007/978-3-642-46856-8_20. [DOI] [PubMed] [Google Scholar]
- 24.Shay JW, Wright WE. Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis. 2005 May;26(5):867–74. doi: 10.1093/carcin/bgh296. [DOI] [PubMed] [Google Scholar]
- 25.Masutomi K, Yu EY, Khurts S, Ben Porath I, Currier JL, Metz GB, Brooks MW, Kaneko S, Murakami S, DeCaprio JA, Weinberg RA, Stewart SA, et al. Telomerase maintains telomere structure in normal human cells. Cell. 2003 Jul 25;114(2):241–53. doi: 10.1016/s0092-8674(03)00550-6. [DOI] [PubMed] [Google Scholar]
- 26.Naviaux RK, Costanzi E, Haas M, Verma IM. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J Virol. 1996 Aug;70:5701–5. doi: 10.1128/jvi.70.8.5701-5705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu IC, Lipsky MM, Cole KE, Su CH, Trump BF. Isolation and culture of hepatocytes from human liver of immediate autopsy. In Vitro. 1985;21:154–60. doi: 10.1007/BF02621352. [DOI] [PubMed] [Google Scholar]
- 28.Ye QH, Qin LX, Forgues M, He P, Kim JW, Peng AC, Simon R, Li Y, Robles AI, Chen Y, Ma ZC, Wu ZQ, et al. Predicting hepatitis B virus-positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning. Nat Med. 2003 Apr;9(4):416–23. doi: 10.1038/nm843. [DOI] [PubMed] [Google Scholar]
- 29.Elmore LW, Forsythe HL, Ferreira-Gonzalez A, Garrett CT, Clark GM, Holt SE. Real-time quantitative analysis of telomerase activity in breast tumor specimens using a highly specific and sensitive fluorescent-based assay. Diagn Mol Pathol. 2002 Sep;11(3):177–85. doi: 10.1097/00019606-200209000-00009. [DOI] [PubMed] [Google Scholar]
- 30.The AGT Cytogenetics Laboratory Manual. 3. Philadelphia: Lippincott-Raven; 1997. [Google Scholar]
- 31.An International System for Human Cytogenetic Nomenclature. Basel: S. Karger; 1995. [Google Scholar]
- 32.Haddad BR, Schrock E, Meck J, Cowan J, Young H, Ferguson-Smith MA, du MS, Ried T. Identification of de novo chromosomal markers and derivatives by spectral karyotyping. Hum Genet. 1998 Nov;103(5):619–25. doi: 10.1007/s004390050878. [DOI] [PubMed] [Google Scholar]
- 33.Wu CG, Forgues M, Siddique S, Farnsworth J, Valerie K, Wang XW. SAGE transcript profiles of normal primary human hepatocytes expressing oncogenic hepatitis B virus X protein. FASEB J. 2002;16(12):1665–7. doi: 10.1096/fj.02-0074fje. [DOI] [PubMed] [Google Scholar]
- 34.Coursen JD, Bennett WP, Khan MA, Forrester K, Pietenpol JA, Harris CC. Differential effects of p53 mutants on the growth of human bronchial epithelial cells. Mol Carcinog. 1997 Jul;19:191–203. [PubMed] [Google Scholar]
- 35.Ponchel F, Puisieux A, Tabone E, Michot JP, Froschl G, Morel AP, Frebourg T, Fontaniere B, Oberhammer F, Ozturk M. Hepatocarcinoma-specific mutant p53-249ser induces mitotic activity but has no effect on transforming growth factor beta 1- mediated apoptosis. Cancer Res. 1994;54:2064–8. [PubMed] [Google Scholar]
- 36.Hussain SP, Kennedy CH, Amstad P, Lui H, Lechner JF, Harris CC. Radon and lung carcinogenesis: mutability of p53 codons 249 and 250 to 238Pu alpha-particles in human bronchial epithelial cells. Carcinogenesis. 1997;18:121–5. doi: 10.1093/carcin/18.1.121. [DOI] [PubMed] [Google Scholar]
- 37.Kwun HJ, Jang KL. Natural variants of hepatitis B virus X protein have differential effects on the expression of cyclin-dependent kinase inhibitor p21 gene. Nucleic Acids Res. 2004;32(7):2202–13. doi: 10.1093/nar/gkh553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sylla A, Diallo MS, Castegnaro J, Wild CP. Interactions between hepatitis B virus infection and exposure to aflatoxins in the development of hepatocellular carcinoma: a molecular epidemiological approach. Mutat Res. 1999 Jul 16;428(1–2):187–96. doi: 10.1016/s1383-5742(99)00046-0. [DOI] [PubMed] [Google Scholar]
- 39.Oguey D, Dumenco LL, Pierce RH, Fausto N. Analysis of the tumorigenicity of the X gene of hepatitis B virus in a nontransformed hepatocyte cell line and the effects of cotransfection with a murine p53 mutant equivalent to human codon 249. Hepatology. 1996 Nov;24(5):1024–33. doi: 10.1002/hep.510240508. [DOI] [PubMed] [Google Scholar]
- 40.Sohn S, Jaitovitch-Groisman I, Benlimame N, Galipeau J, Batist G, Alaoui-Jamali MA. Retroviral expression of the hepatitis B virus x gene promotes liver cell susceptibility to carcinogen-induced site specific mutagenesis. Mutat Res. 2000 Jun 30;460(1):17–28. doi: 10.1016/s0921-8777(00)00010-0. [DOI] [PubMed] [Google Scholar]
- 41.Kuang SY, Lekawanvijit S, Maneekarn N, Thongsawat S, Brodovicz K, Nelson K, Groopman JD. Hepatitis B 1762T/1764A mutations, hepatitis C infection, and codon 249 p53 mutations in hepatocellular carcinomas from Thailand. Cancer Epidemiol Biomarkers Prev. 2005 Feb;14(2):380–4. doi: 10.1158/1055-9965.EPI-04-0380. [DOI] [PubMed] [Google Scholar]
- 42.Hsia CC, Kleiner DE, Jr, Axiotis CA, Di Bisceglie A, Nomura AM, Stemmermann GN, Tabor E. Mutations of p53 gene in hepatocellular carcinoma: roles of hepatitis B virus and aflatoxin contamination in the diet. J Natl Cancer Inst. 1992;84:1638–41. doi: 10.1093/jnci/84.21.1638. [DOI] [PubMed] [Google Scholar]
- 43.Tarn C, Bilodeau ML, Hullinger RL, Andrisani OM. Differential immediate early gene expression in conditional hepatitis B virus pX-transforming versus nontransforming hepatocyte cell lines. J Biol Chem. 1999 Jan 22;274(4):2327–36. doi: 10.1074/jbc.274.4.2327. [DOI] [PubMed] [Google Scholar]
- 44.Minami M, Daimon Y, Mori K, Takashima H, Nakajima T, Itoh Y, Okanoue T. Hepatitis B virus-related insertional mutagenesis in chronic hepatitis B patients as an early drastic genetic change leading to hepatocarcinogenesis. Oncogene. 2005 Jun 23;24(27):4340–8. doi: 10.1038/sj.onc.1208628. [DOI] [PubMed] [Google Scholar]
- 45.Hann HW, Lee J, Bussard A, Liu C, Jin YR, Guha K, Clayton MM, Ardlie K, Pellini MJ, Feitelson MA. Preneoplastic markers of hepatitis B virus-associated hepatocellular carcinoma. Cancer Res. 2004 Oct 15;64(20):7329–35. doi: 10.1158/0008-5472.CAN-04-1095. [DOI] [PubMed] [Google Scholar]
- 46.Kim JW, Wang XW. Gene expression profiling of preneoplastic liver disease and liver cancer: a new era for improved early detection and treatment of these deadly diseases? Carcinogenesis. 2003 Mar;24(3):363–9. doi: 10.1093/carcin/24.3.363. [DOI] [PubMed] [Google Scholar]