Abstract
Glioblastoma multiforme is the most common and lethal primary brain tumor. Glioma progression depends on the rapid proliferation of tumor cells accompanied by an acute immunosuppressive environment, facilitated mainly by tumor infiltration of regulatory T cells (Tregs). In this study, we characterize the role of fibronectin, a high-molecular weight extracellular matrix glycoprotein secreted by tumor cells, in controlling glioma progression and in mediating immunosuppression. Fibronectin binds to membrane-spanning integrin receptors and plays an important role in cell signaling, in defining cellular shape, in mobility, and in regulating the cell cycle. We found that inhibition of fibronectin expression in glioma cells, using short hairpin RNA-mediated silencing of gene expression, delayed cell proliferation in vitro. This delayed growth is explained, in part, by the observed reduced expression of integrin β1 fibronectin receptor, which was restored by the inhibition of proteosomal activity. In our analysis of the downstream signaling targets of integrin β1, we demonstrated reduced phosphorylation of Src kinase and STAT-3. We also observed reduced survivin expression that induced a three-fold increased accumulation of fibronectin-knockdown cells in the G2/M phase. In an experimental animal model, the fibronectin knockdown tumors had a mean survival advantage of 23 days over wild-type tumors. Moreover, brain samples of animals bearing fibronectin-knockdown tumors showed delayed Treg recruitment. Collectively, we propose that fibronectin is a key mediator of glioma progression because its inhibition delays both tumor progression and immunosuppression.
Introduction
Glioblastoma multiforme is the most common and lethal primary tumor of the central nervous system in adults, representing more than 50% of the tumors in this category with a survival of 1 to 2 years [1,2]. Glioblastoma development is a bifaceted mechanism that involves rapid multiplication of tumor cells along with their ability to evade the immune response due to tumor infiltration of regulatory T cells (CD4+CD25+Foxp3+, Tregs) [2–6]. Several molecular targets have been identified which contribute to glioma progression but none of the described markers have been identified to be responsible for both tumor growth and immune suppression. Fibronectin secreted by tumor cells has been reported to promote brain tumor progression by binding integrin receptors on tumor cells, although the pathway by which it acts still remains unclear. In this study, we explain this mechanism by stable silencing of fibronectin production by GL261 glioma cells using a specific short hairpin RNA (shRNA). Silencing of fibronectin affects tumor cell proliferation, which was attributed to the loss of the integrin β1 fibronectin receptors. Activation of Src kinase and STAT-3 was reduced in fibronectin knocked-down GL261 cells. This delayed tumor growth by inhibiting survivin expression and Treg infiltration in brain tumors.
Fibronectin is a high-molecular weight extracellular matrix glycoprotein secreted by tumor cells [7]. Fibronectin can exist in either soluble plasma form or insoluble cellular form as a part of extracellular matrix. It is a ligand for the integrin β1 receptors [8,9]. Fibronectin plays an important role in cell signaling pertinent to tumor progression and is important for the proliferation of different types of cancers, including glioblastoma [10–12]. Binding of integrins with its ligand triggers downstream signal transduction cascades, of which activation of Src kinase has been deemed to be important for tumor development [13,14]. Src kinase is a tyrosine kinase that can be activated by phosphorylation at Tyr416. STAT3, a member of the JAK/STAT family, is one of the several downstream molecular targets of activated Src kinase [15,16]. STAT3 is considered to be the hub of several signal transduction pathways in brain tumor development (reviewed by Brantley and Benveniste [17]). Recent studies by several brain tumor research laboratories have identified activated STAT3 to be an important signal transduction regulatory molecule in glioma growth and tumor infiltration of Tregs [18,19].
We investigated the role played by fibronectin in GL261 mouse glioma cells. Fibronectin expression in GL261 was knocked down with shRNA for fibronectin gene. We show delayed growth of fibronectin-silenced GL261 tumors cells in tissue culture conditions. Fibronectin knockdown resulted in reduced expression of its receptor, the integrin β1 and consequential inhibition of Src kinase, STAT3 activity, and survivin expression in GL261 cells. Fibronectin-silenced tumors exhibited survival advantage in experimental animals over those implanted with wild-type GL261 tumor cells. Glioma Treg recruitment was also slower in the animals that were implanted with the shRNA-treated cells. These results indicate that fibronectin plays a major role in brain tumor development by signaling through integrin receptor-mediated signaling pathways.
Materials and Methods
Animals
Six- to eight-week-old male C57.BL6 (B6) mice were purchased from Jackson Laboratories (Bar Harbor, ME). All mice were housed in a specific pathogen-free facility at the University of Chicago, and experiments were conducted according to federal and institutional guidelines and with the approval of the University of Chicago Institutional Animal Care and Use Committee.
Reagents
Tissue culture reagents were obtained from Cellgro/Mediatech (Manassas, VA) and plasticware from BD Biosciences (San Jose, CA). shRNA, polymerase chain reaction (PCR), and quantitative PCR (qPCR) oligos were purchased from Integrated DNA Technologies (Coralville, IA). pSilencer 4.1-CMV puro vector was purchased from Ambion/Applied Biotechnologies (Austin, TX). Western blot reagents and iScript complementary DNA (cDNA) synthesis kit were purchased from Bio-Rad Laboratories (Hercules, CA). The primary and HRP-conjugated secondary antibodies for Western blots were obtained from Cell Signaling Technologies (Boston, MA), Santa Cruz Biotechnology (Santa Cruz, CA) and Jackson ImmunoResearch Laboratories (Westgrove, PA). The RNA isolation kit was obtained from Qiagen (Valencia, CA) and SYBR Green RT-PCRMasterMix from Invitrogen (Carlsbad, CA). Fluorescent-labeled antibodies for flow cytometry or microscopy were purchased from either BD Biosciences (San Jose, CA) or eBioscience (San Diego, CA). PP2 Src kinase inhibitor was purchased from EMD Bioscience (San Diego, CA). The stock solutions of 10 mM PP2 were prepared in DMSO. All other reagents were purchased from Thermo Fisher Scientific (Pittsburgh, PA), if otherwise mentioned.
shRNA Cloning and Characterization
The mouse cDNA sequence was analyzed for potential 21nts small interfering RNA (siRNA) target sequences (Dharmacon Design center; www.dharmacon.com). Five oligonucleotides and one scrambled siRNA were first analyzed for suppression of the fibronectin (Table 1), and siRNA 1 was chosen for maximal activity. The oligonucleotide was redesigned to have a hairpin loop in the middle and BamH1 and EcoR1 sites at the end and cloned in pSilencer 4.1-CMV puro vector. The following forward (Fwd) and reverse (Rev) oligonucleotides were used for fibronectin: Fwd 5′-GATCCGAACAAAGACAGAGACAATTTCAAGAGAATTGTCTCTGTCTTTGTTCAGA-3′ and Rev 5′-AGCTTCTGAACAAAGACAGAGACAATTCTCTTGAAATTGTCTCTGTCTTTGTTCG-3′. The oligonucleotides for scrambled shRNA were Fwd 5′-GATCCGAAACAGAACGAAGAACATTTCAAGAGATGTTCTTCGTTCTGTTTCAGA-3′ and Rev 5′-AGCTTCTGAAACAGAACGAAGAACATCTCTTGAAATGTTCTTCGTTCTGTTTCG-3′. Colonies isolated were sequenced to verify the presence of the insert and the recombinant plasmid was named FnKD. Scrambled shRNA was used as vector control (VC) in these experiments. GL261 cells at 80% confluency were transfected with either FnKD or VC plasmids using Lipofectamine (Invitrogen) as per the manufacturer's protocol. Forty-eight hours after transfection, cells were subjected to puromycin treatment at 1.25 µg/ml to select for stable transfectants, and selection was continued until the control untransfected cells were killed. These cells (GL261-FnKD) were then tested for fibronectin knockdown by checking for both messenger RNA (mRNA) and protein levels and were compared with the GL261-VC cells. For subsequent experiments, transfected cells were cultured in the complete medium that comprised Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum and antibiotics (complete medium).
Table 1.
List of siRNA Oligonucleotides Tested to Inhibit Fibronectin Expression in GL261 Mouse Glioma Cells.
| siRNA | Oligo | Exon |
| siRNA | 1 5′-GAACAAAGACAGAGACAA-3′ | 36 |
| siRNA 2 | 5′-GAACAAACACTAACGTAAA-3′ | 45/46 |
| siRNA 3 | 5′-ACTATTAGCTGGAGAACAA-3′ | 36 |
| siRNA 4 | 5′-GAACAAGACCAGAAGTATT-3′ | 8 |
| siRNA 5 | 5′-GGAACTGACTGATGTGAAA-3 | 19 |
| Scrambled | 5′-GAAACAGAACGAAGAACA-3′ |
PCR and qPCR
Silencing of the fibronectin gene was assessed by performing reverse transcription-PCR using the cDNA prepared from total RNA of GL261-FnKD and GL261-VC cells, respectively. RNA was isolated using the Qiagen RNeasy kit (Qiagen). cDNA was prepared from isolated RNA using the iScript cDNA synthesis kit (Bio-Rad). Primers used for mouse fibronectin were Fwd 5′-AATCACAGTAGTTGCGGCAGGAGA-3′ and Rev 5′-TCTGTCCCAGGCAGGAGATTTGTT-3′.
mRNA transcript levels of selected genes in GL261-VC and GL261-FnKD cells were tested by qPCR. Expression of mouse itgb1 (Fwd 5′-TGTGACCCATTGCAAGGAGAAGGA-3′ and Rev 5′-AATTGGGATGATGTCGGGACCAGT-3′) and survivin (birc5) (Fwd 5′-TTGAGGCCCTAGGTTCAATTCCCA-3′ and Rev 5′-AGCTGCTCAATTGACTGACGGGTA-3′) was studied using cDNA prepared from total RNA of GL261-VC and GL261-FnKD cells. qPCR was performed using Bio-RadOpticon 2 real-time PCRmonitor with SYBR GreenER qPCR Master Mix (Invitrogen). All reactions were performed in triplicate. Expression of each target gene was normalized to gapdh (Fwd 5′-TCAACAGCAACTCCCACTCTTCCA-3′ and Rev 5′-ACCCTGTTGCTGTAGCCGTATTCA-3′). Fold expression was calculated using the 2-ΔΔCT method [20].
Western Blots
Inhibition of fibronectin protein expression was assessed by Western blot using the cell lysates from GL261-FnKD and GL261-VC cells. Briefly, 1 x 106 cells of each type were lysed using M-PER mammalian protein extraction reagent with 1x Halt protease inhibitor cocktail and phosphatase inhibitor (Thermo Fisher Scientific). One hundred micrograms of protein from the cell lysates was denatured in the presence of β-mercaptoethanol and SDS at 100°C and loaded into each well of a Tris-HCl Precast Ready Gel for SDS-PAGE (Bio-Rad) and separated by electrophoresis. Separated peptides were then transferred on to a polyvinylidene difluoride membrane by semidry transfer blot (Bio-Rad). Transferred membranes were blocked with 5% nonfat milk in TBST (100 mM Tris and 150 mM NaCl [pH 7.4] with Tween-20) for 1 hour at room temperature and incubated overnight at 4°C with antibodies against mouse fibronectin. Antibody-treated membranes were washed with 1x TBST and reincubated with HRP-conjugated secondary antibody for 1 hour at room temperature. Immunoreactive bands were developed using the Immun-StarWestern C chemiluminescence reagent (Bio-Rad) and visualized by chemiluminescent camera in a Bio-Rad gel documentation system. Lysates from GL261-FnKD and GL261-VC cells were also used to detect the expression of phosphorylated Src kinase (phospho-Src [Tyr416]) and phosphorylated STAT3 (phospho-STAT3 [Tyr705]) and survivin protein levels using respective mouse-specific antibodies. The expression of total Src and STAT3 was used to measure the loading control for phospho-Src (Tyr416) and phospho-STAT3 (Tyr705), respectively. The expression of β-actin was used as a loading control for survivin expression.
In Vitro Labeling of Cultured Cells with Bromodeoxyuridine
GL261-FnKD and GL261-VC cells (50,000 cells per well) were plated in each well of a six-well tissue culture plate overnight in complete medium. On the next day, the medium was replaced with complete medium containing 10 µM of bromodeoxyuridine (BrdU; BD Biosciences). The cells were harvested at 24, 72, and 108 hours after the addition of BrdU and were stained according to the manufacturer's protocol. In brief, the cells were fixed and permeabilized, followed by permeabilization of the nuclear membrane, and finally refixed. The DNA was fragmented using DNAse I, and BrdU incorporation was detected by staining using a fluorescein isothiocyanate (FITC)-conjugated anti-BrdU monoclonal antibody (1:50). Mouse IgG1κ-FITC was used as the isotype control. Cells that were not treated with BrdU were tested as the experimental control. The cells were then analyzed by flow cytometry using BDFACSCalibur (BDBiosciences) and FlowJo software (Tree Star Inc, Ashland, OR). The experiment was conducted in triplicate wells and performed thrice. Similar experiments were performed in which cells were cultured in either serum-free medium or serum-free medium supplemented with 25 µg/ml of purified soluble bovine fibronectin (Sigma, St Louis, MO).
Flow Cytometry
The levels of integrin β1 on GL261-VC and GL261-FnKD cells were analyzed by flow cytometry. Briefly, cells were grown in 25-cm2 tissue culture flasks up to 50% confluency. The cells were harvested using a cell scraper to prevent trypsin-induced ablation of the integrins and was performed on ice to prevent internalization of the same. Harvested cells were fixed immediately with 1% paraformaldehyde and stained with anti-mouse integrin β1 antibodies conjugated with phycoerythrin (PE; 1:200). Hamster IgG-PE was used as antibody control. After incubation, the cells were washed twice with FACS buffer (PBS +1% BSA +0.02% NaN3), and the expression of integrin β1 was analyzed by flow cytometry.
MG132 Treatment of GL261 Cells
Fifty thousand GL261-VC or GL261-FnKD cells were plated in each well of a 24-well plate and cultured in complete medium under normal tissue culture conditions. After 24 hours, the medium was replaced with fresh medium, which was supplemented with 5 µM MG132 and incubated for 1, 2, and 5 hours under normal conditions. After incubation, the cells were harvested using cell scrapers on ice. Levels of integrin β1 were assessed by method as described above.
Cell Cycle Assay
To elucidate the delay in cellular proliferation, we performed a cell cycle analysis. Twenty thousand GL261-VC and GL261-FnKD cells were plated in each well of a 24-well plate and cultured in complete medium under regular tissue culture conditions for 48 hours. After incubation, the cells were washed twice with ice-cold PBS and fixed in ice-cold methanol. Fixed cells were rehydrated back with two washes of PBS and treated with RNase (100 µg/ml; Sigma) for 15 minutes at 37°C. The reaction was stopped by diluting it with three volumes of PBS. Propidium iodide (50 µg/ml; eBioscience) was added to the RNase-treated cells, and propidium iodide dilution was analyzed immediately by flow cytometry.
Src Kinase Inhibition and Survivin Expression
The role of Src kinase activity in survivin expression was tested in GL261 cells using PP2 Src kinase inhibitor. Briefly, 2 x 105 cells were plated in each well of a six-well tissue culture plate and were cultured exactly for 12 hours in complete medium supplemented with PP2 (0–15 µM). DMSO was used as vehicle control. After incubation, the cells were lysed, and Western blot was performed as described earlier using antimouse survivin antibody.
Intracranial Tumor Implantation
Male B6 mice were anesthetized with an intraperitoneal injection of ketamine hydrochloride (25 mg/ml)/xylazine (2.5 mg/ml) cocktail. A midline cranial incision and a right-sided burr hole were made 2 mm lateral to the sagittal sinus and approximately 2 mm superior to the lambda. The animals were positioned in stereotactic frame, and a Hamilton needle was then inserted into the burr hole and advanced 3 mm. Intracranial penetration was followed by the injection of 4 x 105 cells in 2.5 µl of sterile PBS. One group of five animals was injected with the GL261-VC cells, whereas a second group with the same number of animals received GL261-FnKD cells. All animals were kept under observation to assess survival.
Similar experiments were performed in which two groups of animals were implanted with GL261-VC and GL261-FnKD cells, respectively, and three animals per group were killed on the 7th, 14th, and 21st day post implantation (PI). Brain tissues around the tumor injection site were harvested from each animal for further studies.
Histology
Twenty-one days after tumor implantation, mouse brains were harvested, flash frozen, sectioned at a thickness of 8 µm, and thaw-mounted (37°C for 5–10 minutes) onto precleaned SuperFrost slides (Thermo Fisher Scientific). The slides were stained with hematoxylin and eosin to visualize the brain tumors and were dehydrated sequentially as follows: alcoholic formalin (30 seconds), H2O (10 dips), hematoxylin (filtered Gill 3; 2 minutes), H2O x 2 (10 dips), bluing solution (60 seconds), H2O (10 dips), alcoholic eosin (10 dips), 95% EtOH x 2 (10 dips), 100% EtOH x 2 (10 dips), and xylene x 3 (10 dips). Sections were covered with Permount, coverslipped, and viewed using a Zeiss Axioskop microscope (Carl Zeiss, Hamburg, Germany). Images were captured using Improvision's OpenLab software (PerkinElmer, Waltham, MA) microscope attached to a Zeiss Axiocam CCD camera and image capturing system (Carl Zeiss).
Survivin Expression in Tumor-Implanted Brain
To analyze whether survivin played any role in tumor growth in vivo, qPCR for survivin mRNA transcript levels was performed with cDNA prepared from the brain tissues that were harvested from the injection site of GL261-VC and GL261-FnKD tumor-implanted animals at 7, 14, and 21 days after tumor implantation. qPCR was performed as described earlier. The expression of survivin (birc5) was normalized to gapdh. Fold expression was calculated using the 2-ΔΔCT method [20].
Analysis of Treg Infiltration in Tumor-Implanted Brains
The effects of fibronectin silencing on Treg were studied in tumor-implanted brains harvested at 7, 14, and 21 days PI. Brains were harvested, and cells were resuspended in staining buffer (1x Hank's balanced salt solution, 1% bovine serum albumin, 0.02% sodium azide) after red blood cell was lysed using ACK lysing buffer. Brain cells were stained with anti-CD4-FITC monoclonal antibody (1:200) for surface CD4 expression, and intranuclear expression of FoxP3 was assessed by staining with anti-FoxP3-APC monoclonal antibody (1:100) after permeabilizing and fixing the cells with FoxP3 staining kit (eBioscience). Respective isotype controls were used for each antibody. Treg infiltration was then analyzed by flow cytometry.
Statistical Analysis
GraphPad Prism software package forWindows (version 4.0; GraphPad Software, Inc, La Jolla, CA) was used to run analysis of variance, two-tailed unequal-variance t test, and Student's t test, and SD and SEM of the data were presented. A value of P < .05 was considered significant.
Results
Stable Knockdown of Fibronectin Expression in GL261 Glioma Cells
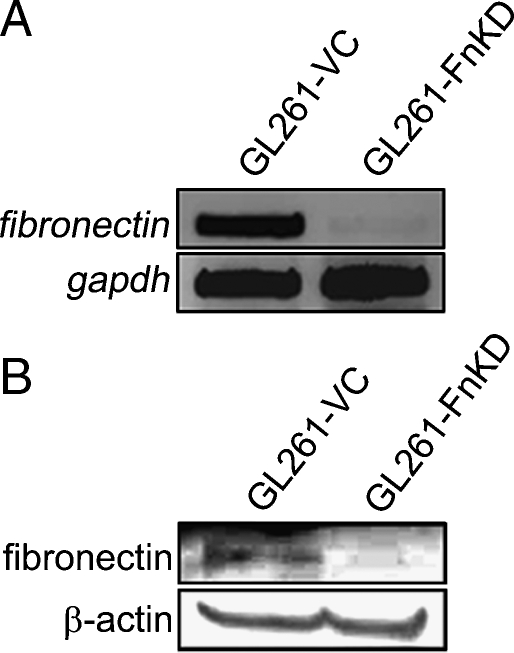
GL261 mouse glioma cells were transfected with a plasmid encoding mouse fibronectin-silencing shRNA (FnKD) or scrambled shRNA (VC). The fibronectin shRNA was chosen from a set of five siRNA oligonucleotides that were tested for their efficiency in inhibiting fibronectin expression (Table 1). Transfected cells were selected by puromycin treatment and a positive clone was selected when the nontransfected cells were killed by the antibiotic treatment. Reverse transcription-PCR for fibronectin expression in the GL261-FnKD cells did not show any amplified product when compared with those of the GL261-VC cells (Figure 1A). Similar results were observed when Western blots were performed with the lysates from GL261-VC and GL261-FnKD cells and probed with the anti-fibronectin antibody. GL261-VC lysates showed the fibronectin band, whereas it was absent in the GL261-FnKD lysates (Figure 1B). These results confirm that fibronectin expression is stably silenced in the GL261-FnKD cells.
Figure 1.
Fibronectin knockdown using shRNA. shRNA was designed to silence the mouse fibronectin gene and was cloned in an shRNA-expressing plasmid and transfected in GL261 mouse glioma cell line (GL261-FnKD). Cells transfected with scrambled plasmid were used as vector control (GL261-VC). (A) Expression of fibronectin gene in the GL261-VC and GL261-FnKD as observed by PCR using cDNA prepared from stably transfected cells. (B) Western blot band for fibronectin protein in cell lysates of GL261-VC and GL261-FnKD cell lysates. β-Actin protein was used as the loading control.
Fibronectin Knockdown Delays Glioma Proliferation
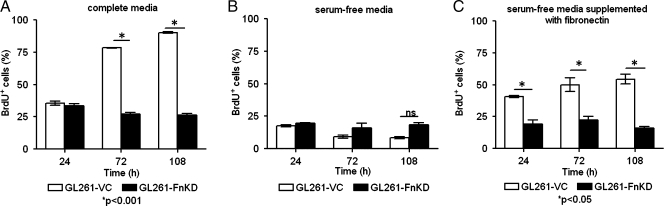
Glioma progression depends on the exponential growth of tumor cells. Under regular culture conditions, the GL261-FnKD cells showed very slow growth in comparison to the GL261-VC or the GL261-wild type cells, which warranted a BrdU uptake assay. BrdU uses nucleotide substitution to replace thymidine with uridine in the DNA structure of dividing cells both in vitro and in vivo. Equal numbers of GL261-VC and GL261-FnKD cells were cultured with 10 µM BrdU in complete medium for 24, 72, and 108 hours and BrdU uptake was analyzed by flow cytometry after fixing and staining the cells with anti-BrdU antibody. Although there was no difference in BrdU uptake between the two groups at 24 hours, a rapid increase in BrdU accumulation was observed in GL261-VC cells after 72 hours and was further increased at 108 hours. In contrast, the GL261-FnKD cells failed to significantly incorporate BrdU even at 108 hours (Figure 2A).
Figure 2.
BrdU uptake assay. BrdU uptake is often used as a marker for cell proliferation because it is rapidly taken up by dividing cells, and accumulates in these cells since it is notmetabolized. (A) GL261-VC or GL261-FnKD cells were incubated with BrdU in complete medium for different time points. (B) Cells incubated with BrdU in serum-free media. (C) Cells incubated with BrdU in serum-free medium, which was supplemented with 25 µg/ml of purified fibronectin. BrdU uptake was analyzed as described in Materials and Methods. Percentage values of BrdU-positive cells plotted in a bar graph showing BrdU-positive GL261-VC cells (white bars) versus GL261-FnKD cells (black bars) at similar time points. Error bars represent SD within triplicate observations.
Similar experiments were performed in which serum-free Dulbecco's modified Eagle medium was used in place of complete medium to investigate the role of serum-associated soluble plasma fibronectin in glioma proliferation. Levels of BrdU incorporation were reduced in both GL261-VC and GL261-FnKD in the absence of serum at all of the time points studied (Figure 2B).
Purified soluble fibronectin was supplemented to the serum-free medium in a third set of experiments. Incorporation of BrdU by GL261-VC cells was recovered to an intermediary level in the presence of soluble fibronectin. However, GL261-FnKD cells showed no signs of increase in proliferation (Figure 2C).
Expression of β1 Integrin Is Reduced in GL261-FnKD Cells
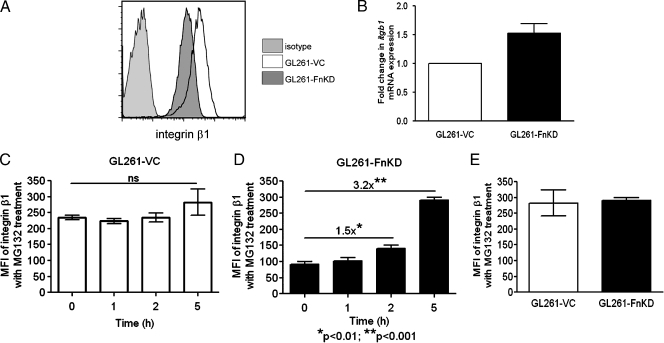
A major receptor for fibronectin on tumor cells is the membrane-spanning integrin β1 [9]. Failure of GL261-FnKD cells to respond to soluble plasma fibronectin or purified soluble fibronectin prompted us to check the levels of integrin β1 in these cells. GL261-VC and GL261-FnKD cells were stained with PE-conjugated anti-β1 antibody. Flow cytometric analysis revealed reduced mean fluorescence intensity (MFI) of integrin β1 expression in GL261-FnKD cells when compared with those of GL261-VC cells (Figure 3A). However, no significant difference in mRNA transcript levels of itgb1 gene expression was observed between the GL261-VC and GL261-FnKD cells (Figure 3B).
Figure 3.
β1 Integrin expression in glioma cells. (A) Flow cytometry profile of cell surface expression of β1 integrin on GL261-VC (open histogram) and GL261-FnKD cells (dark gray histogram). Hamster IgG was used as antibody control (light gray histogram). (B) Fold change in itgb1 mRNA transcript levels in GL261-FnKD cells (black bar) with respect to GL261-VC (white bar)measured by quantitative PCR. (C) MFI of cell surface β1 integrin expression calculated from flow cytometric analysis of GL261-VC cells treated with MG132. (D) MFI of cell surface β1 integrin expression calculated from flow cytometric analysis of GL261-FnKD cells treated with MG132. A 1.5-fold increase in MFI was observed after 2 hours of treatment, and a 3.2-fold increase was observed after 5 hours. (E) Bar diagram comparing MFI of cells surface β1 expression between GL261-VC (white bar) and GL261-FnKD (black bar) cells treated with MG132 for 5 hours. Error bars for all figures represent SD within triplicate observations.
MG132 Treatment Facilitates Integrin β1 Expression in GL261-FnKD Cells
We tested the effects of MG132, a potent inhibitor of proteosomal activity, on the reversal of integrin β1 protein expression in GL261-FnKD cells. Both GL261-VC and GL261-FnKD cells were incubated for 5 hours in the presence of 5 µM MG132. Integrin β1 expression in GL261-VC cells was not affected by the treatment (Figure 3C). However, in GL261-FnKD cells, there was a 1.5-fold increase in integrin β1 expression after 2 hours, which increased up to 3.2-fold when the cells were incubated for 5 hours with MG132 (Figure 3D). Five hours of treatment with MG132 increased the MFI of integrin β1 expression in GL261-FnKD cells to the same levels of GL261-VC cells (Figure 3E).
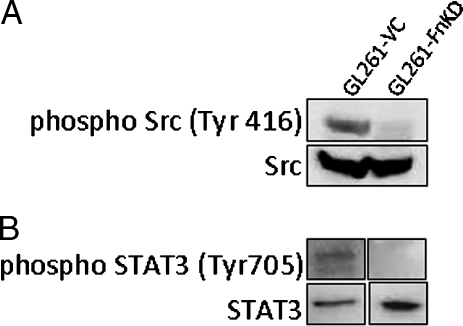
Src Kinase and STAT-3 Activity Are Downregulated in Fibronectin Knockdown GL261 Cells
Integrins signal through Src kinase and play an important role in tumor progression [13,14,21]. We wanted to test the status of Src kinase expression in GL261-FnKD cells. Western blot results showed complete abrogation of phospho-Src (Tyr 416) in GL261-FnKD cells. Clean bands of phospho-Src (Tyr416) were, however, observed in GL261-VC cell lysates (Figure 4A). Because Src kinase is the first major molecule in the downstream signaling cascade of integrin receptors, its loss further indicated that integrin β1 was downregulated in GL261-FnKD cells.
Figure 4.
Reduced Src kinase and STAT3 activity in GL261-FnKD cells. (A) Western blots for phospho-Src (Tyr416) in GL261-VC and GL261-FnKD cell lysates. Expression of total Src levels was measured as loading control. (B) Phospho-STAT3 (Tyr705) in GL261-VC and GL261-FnKD cells. Total STAT3 expression was used as a control.
STAT-3 is a downstream target for Src kinase [15,16,22,23]. It is also an important oncogene promoting transcription control, cell cycle progression and cellular transformation [24,25] and is a target for cancer target drug discovery [26]. STAT3 is also an important marker for glioma progression and is under intense investigation [19,27–31]. The status of STAT3 activation in GL261-FnKD cells was analyzed by Western blot. Results demonstrated reduced levels of phospho-STAT3 (Tyr705) in GL261-FnKD cells with respect to GL261-VC cells (Figure 4B).
G2/M Block in the GL261-FnKD Cell Cycle Pathway
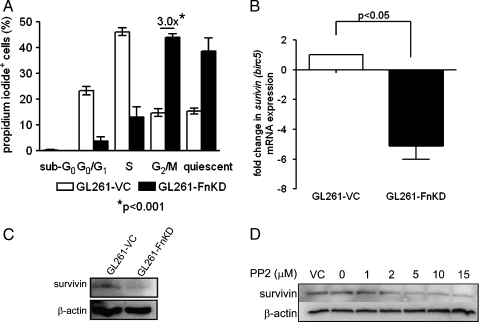
Delay in GL261-FnKD cell growth required us to look at the cell cycle status of these cells. GL261-VC and GL261-FnKD cells were cultured for 48 hours in complete medium, and cell cycle analysis of these cells was performed according to Materials and Methods. The results showed a three-fold increase in accumulation of GL261-FnKD cells in the G2/M phase in comparison to the GL261-VC cells. The GL261-VC cells, however, were mostly in the G0/G1 and S phases (Figure 5A).
Figure 5.
Survivin expression in GL261-FnKD cells. (A) Cell cycle assay with propidium iodide incorporation for 48 hours in culture. White bars represent GL261-VC cells, and black bars represent GL261-FnKD cells. Three-fold higher levels of propidium iodide-positive GL261-FnKD cells were observed in the G2/M phase. Error bars represent SD within six replicate observations. (B) Fold change in the mRNA transcript level of survivin (birc5) between GL261-VC (white bars) and GL261-FnKD (black bars) measured by quantitative PCR. Error bars represent SD between triplicate qPCR observations. (C) Immunoblot for survivin expression in GL261-VC and GL261-FnKD cell lysates. β-Actin was used as a loading control. (D) Western blot for survivin protein expression in GL261 cells after treatment with PP2 (0–15 µM). DMSO was used a vehicle control (VC). β-Actin was used as loading control.
Reduced Survivin Expression in GL261-FnKD Cells
Survivin is a member of inhibitor of apoptosis protein family. It is known to facilitate mitosis and drive cells into the S phase. Overexpression of survivin has often been shown to result in accelerated S phase shift and vice versa [32]. Survivin has also been reported to be one of the most important proliferation markers of glioma [33–35]. qPCR analysis of survivin (birc5) mRNA transcript level demonstrated five-fold lower level in GL261-FnKD cells when compared with that of GL261-VC cells (Figure 5B). Western blot for survivin protein expression mirrored the qPCR observation. Reduced expression of survivin protein was observed in GL261-FnKD cells (Figure 5C).
Survivin Expression in GL261 Cells Is Regulated by Src Kinase Activity
To draw a link between the integrin β1 signaling and survivin activity, we tested the role of Src kinase activation in survivin expression in glioma. Survivin has also been shown to be a downstream target of Src kinase/STAT-3 pathway, which further warranted this investigation [36,37]. GL261-VC cells were treated for 12 hours with the increasing concentrations (0–15 µM) of PP2 to block Src kinase activity. Survivin protein expression was analyzed by Western blot and showed decreasing survivin expression with increasing concentrations of PP2 (Figure 5D).
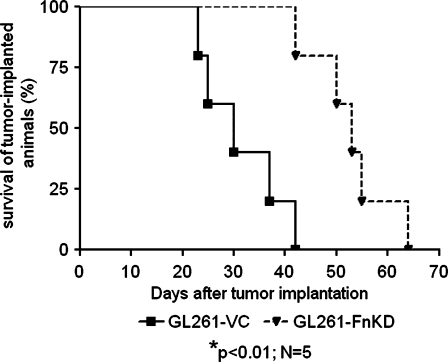
Increased Survival of GL261-FnKD Tumor-Implanted Animals
Intracranial implantation of either GL261-VC or GL261-FnKD was performed in two groups of five B6 mice. The first casualty of the GL261-VC-implanted group was recorded on the 23rd day PI, and all animals in this group succumbed to the tumor by the 42nd day PI. However, the first death in GL261-FnKD-implanted animals was on 42nd day PI, and final death was recorded on the 64th day PI. Kaplan-Meier survival curve demonstrated a 23-day survival advantage of GL261-FnKD-implanted animals versus GL261-VC (Figure 6).
Figure 6.
Fibronectin knockdown in glioma cells prolongs survival time of tumor implanted animals. Kaplan-Meier survival curve for two groups of B6 mice implanted intracranially with GL261-VC (solid line) and GL261-FnKD (broken line) tumors, respectively. Five animals were included in each group. Mean survival time of GL261-VC-implanted animals was 30 days, whereas it was 53 days for GL261-FnKD-implanted group.
Lower Survivin Expression and Delay in Tumor Development In Vivo
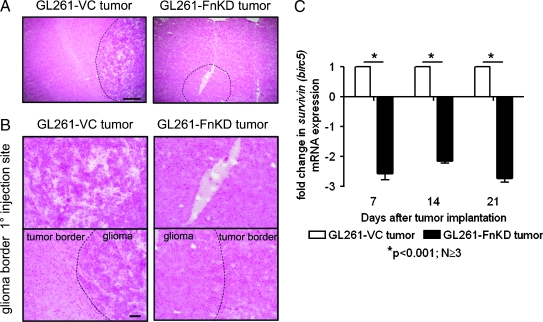
Histopathology of tumor development was performed by staining brain sections with hematoxylin and eosin. Implantation of GL261-VC cells led to reproducible tumor growth by the 21st day PI, which is evident by the buildup of basophilic stained cells around the injection site. Although injected cells were observed in GL261-FnKD-implanted brains, typical tumor formation was never observed at identical time point (Figure 7, A and B).
Figure 7.
Tumor development and survivin transcript levels in experimental animals. (A) Hematoxylin-eosin (HE) staining of tumor-implanted brain sections at 21st day PI. Harvested brains were sectioned through the site of tumor implant, and tumor distribution was observed under microscope. Scale bars indicate 500 µm. (B) Upper panel shows higher-magnification image of the primary injection site in the HE-stained sections of GL261-VC (left) and GL261-FnKD (right) tumor implanted brain. Lower panel shows the tissue distribution at the glioma border in the tumor-implanted HE-stained brain sections. Scale bars indicate 100 µm. (C) Fold change in mRNA transcript level of survivin (birc5) in GL261-VC- (white bars) and GL261-FnKD-implanted (black bars) brains at 7, 14, and 21 days PI. Error bars represent SEM of observations obtained from three animals per treatment group.
Previous experiments showed reduced survivin expression in GL261-FnKD cells grown in vitro (Figure 5, B and C). We observed similar pattern of reduced survivin (birc5) mRNA transcripts levels in GL261-FnKD tumor-implanted brains when compared with those of GL261-VC-implanted brains. Tumor-implanted animals were killed on days 7, 14, and 21 PI. The right posterior section of the brain that was injected with tumor cells was harvested from animals to prepare cDNA from total RNA. qPCR results showed roughly 2- to 2.5-fold decrease in the transcription of survivin (birc5) in GL261-FnKD-implanted brains at all the time points observed when compared with GL261-VC (Figure 7C).
Delay in Treg Recruitment in GL261-FnKD-Implanted Brains
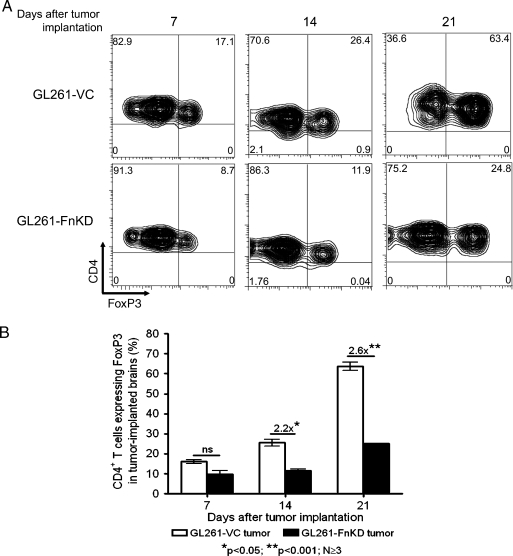
Delayed in vivo tumor growth was further evidenced by a lag in Treg recruitment in the GL261-FnKD implanted brains (Figure 8). The expression of FoxP3+-expressing CD4+ T cells in brain tumors was analyzed after the 7th, 14th, and 21st day PI. Although on the 7th day, the difference in Treg recruitment was not significant, the level of FoxP3 expression by CD4+ T cells in GL261-FnKD-implanted brains at 14th day was almost 2.2-fold lower than that in GL261-VC tumors. After 21 days, the difference was increased to almost 2.6-fold with 63.4% of CD4+ T cells expressing FoxP3 in GL261-VC tumors, whereas 24.8% CD4+ T cells expressed FoxP3 in GL261-FnKD tumors.
Figure 8.
Treg infiltration is affected in GL261-FnKD-implanted brains. (A) Flow cytometric measurement of FoxP3+ CD4+ T cells' infiltration in tumor-implanted brains at 7th, 14th and 21st day PI. CD4 is distributed along the y axis, whereas FoxP3 is along the x axis of the plots. These observations are derived from total CD4+ T cells derived from each tumor-implanted brain. Quadrants were drawn based on the isotype staining for each antibody. Numbers in each quadrant represent respective distribution frequency. Treg infiltration in GL261-VC tumor-implanted brains (top panel) is compared with GL261-FnKD-implanted brains (lower panel). This figure represents one of three animals per group at each time point. (B) Bar diagram representation of CD4+ T cells that express FoxP3 in tumor-implanted brains. FoxP3 expression was increased by 2.2-fold in GL261-VC tumor-implanted brains (white bars) over GL261-FnKD tumor-implanted brains (black bars) by 14 days PI and increased to 2.6-fold at 21 days. Error bars indicate SEM of three or more replicate animals per group at their respective time points.
Discussion
Fibronectin in brain tumors was first described by Sherbet et al. [11] and has been primarily associated with cell migration upon binding with integrin receptors [12]. The expression of fibronectin and other extracellular matrix components by glioma is directly linked to the expression of integrins on cell surface. Ritchie et al. [38] showed that in glioma cells with induced and ectopic expression of nuclear factor κB, there was an increase in the expression of both fibronectin and integrins. More recently, fibronectin has been reported to maintain extracellular matrix rigidity to promote structural rigidity, motility, and proliferation of established glioma cell lines in vitro [39]. However, the functional mechanism of fibronectin-mediated regulation of glioma progression has remained unknown.
Fibronectin expression in GL261 glioma cells was silenced with the introduction of shRNA targeted toward the mouse fibronectin gene in the cells (Figure 1). Inhibition of fibronectin expression in GL261 glioma cells (GL261-FnKD) delayed cell proliferation in vitro in comparison to the GL261 cells that were treated with scrambled shRNA (GL261-VC). When GL261-FnKD cells were implanted in the brain of experimental animals, a delay in tumor formation was observed (Figure 7A), and these animals exhibited a 23-day survival advantage over the GL261-VC-implanted ones (Figure 6). Our observation is the first report linking fibronectin with in vivo tumor growth and survival of experimental animals.
We performed a BrdU uptake assay to monitor the delay in cell growth. BrdU uptake is often used as a marker for cell proliferation because it is rapidly assimilated by dividing cells [40,41]. The results showed that the slow-growing GL261-FnKD cells accumulated lower amounts of BrdU over time, whereas the GL261-VC cells incorporated increasing amounts of BrdU at comparable time points (Figure 2A). It is important to note here that GL261-FnKD cells showed lesser incorporation of BrdU in the presence of serum, which is a rich source of soluble plasma fibronectin. We performed experiments to show the importance of soluble fibronectin in glioma cellular proliferation. In one such experiment, GL261-VC failed to accumulate any BrdU in serum-free condition (Figure 2B). This observation was partially recovered by supplementing the serum-free medium with purified soluble fibronectin. These experiments were done in the presence of 25 µg/ml of purified fibronectin because it was the optimal concentration found in bovine serum [42]. The levels of BrdU incorporated by GL261-VC cells increased partially in response to the purified fibronectin (Figure 2C). These observations indicated that soluble fibronectin was important for glioma cellular proliferation. However, fibronectin-silenced GL261-FnKD failed to respond again, even in the presence of purified soluble fibronectin.
Extracellular fibronectin is a ligand for integrin receptors on tumor cell surface [8,43]. Integrin β1 has been shown to be a major cell surface receptor for glioma and to play important roles in glioma progression [9]. The inability of GL261-FnKD cells to respond to soluble fibronectin warranted further investigation of the status of the integrin β1 fibronectin receptors in these cells. We measured integrin β1 expression by qPCR for its mRNA transcript levels followed by flow cytometry to assess the cellular protein expression. We observed a decrease in the MFI of integrin β1 expression in the GL261-FnKD cells in comparison to the GL261-VC cells (Figure 3A). This phenomenon explained the lack of response by GL261-FnKD to plasma-associated and purified soluble fibronectin. However, we could not detect any difference between the itgb1 mRNA transcript levels between the GL261-VC and GL261-FnKD cells (Figure 3B). This indicated that silencing of fibronectin expression caused a posttranscriptional inhibition of integrin β1 fibronectin receptor. We tested this hypothesis by inhibiting proteosomal degradation of integrin β1 expression by treating cells with MG132, an irreversible proteosomal inhibitor. The inhibitor did not have any effect on the integrin β1 expression in the GL261-VC cells (Figure 3C). However, the MFI of integrin expression in GL261-FnKD cells increased by 3.2-fold after incubating the cells for 5 hours with the inhibitor (Figure 3D). In fact, similar levels of integrins were expressed in both GL261-VC and GL261-FnKD cells after 5 hours of treatment with MG132 (Figure 3E). These results indicated that shRNA-mediated inhibition of fibronectin expression in GL261 glioma cells induced a posttranscriptional degradation of the integrin β1 fibronectin receptors by activation of proteosomal enzymes. It is important to note here that integrin β1 has been shown to be required for diffuse brain invasion of glioma cells in an experimental rat glioma model [44]. Our results also correlate with this observation, and the lack of integrin β1 expression might contribute to the delay in tumor growth and increased survival of GL261-FnKD-implanted animals (Figures 6 and 7, A and B). However, we believe that in addition to glioma invasion, the delayed proliferation of glioma cells in these animals played an even more important role in increased survival. We further investigated the role of integrin β1 in glioma cell proliferation.
Degradation of integrin β1 in GL261-FnKD cells was predicted to affect integrin-induced proliferation signals. Our hypothesis was confirmed when the activated phospho-Src (Tyr416), a downstream target of integrin signaling [13,14], was almost obliterated in GL261-FnKD cells (Figure 4A). Next, we tested the activation of STAT3 in GL261-FnKD cells because of two reasons: first, Src kinase is one of several regulators of STAT3 function [16,22], and second, activated STAT3 is a vital marker for glioma progression [19,29–31]. Our hypothesis of reduced STAT3 activity in GL261-FnKD cells was confirmed by the absence of the activated phospho-STAT3 (Tyr705) in these cells (Figure 4B). We performed a cell cycle assay to understand the relation between degradation of integrin receptors and delay in proliferation of the GL261-FnKD cells. Analysis of the data acquired from these experiments revealed three-fold more accumulation of the slow growing GL261-FnKD cells in the G2/M phase. In comparison, the GL261-VC cells were mostly in the S phase indicating their active proliferation (Figure 5A). This observation suggested a defect in proliferation regulators of GL261-FnKD cells.
Survivin or baculoviral inhibitor of apoptosis repeat containing 5 (birc5) is a member of the inhibitor of apoptosis family [45]. Survivin is observed uniquely in tumor and developmental cells, which undergo either inappropriate or programmed cell growth [46]. Survivin participates in cell cycle entry and progression by binding with mitotic spindle fibers and by facilitating the cells to roll into the S phase and controlling apoptosis by inhibiting caspase activation [32,47]. Survivin has been identified as an important oncogene for glioblastoma promoting tumor proliferation, and inhibition of survivin in U251 glioma cells has been reported to exhibit decreased cell growth and cell cycle arrest [33,34]. Moreover, survivin has also been shown to be a downstream target of Src kinase and STAT3 activity in different types of cancer [36,37,48,49]. As such, we sought to study the status of survivin in GL261-FnKD cells over other proliferation markers. We observed decreased survivin expression, both at the mRNA transcript and protein levels (Figure 5, B and C). These results corroborated with the earlier reports that noted the importance of survivin in glioma progression [33,35]. Our observation also agreed with the recent report published by Mellai et al. [34], in which the authors observed that survivin expression in glioma cases positively correlated only with tumor proliferation, whereas it failed to show any correlation with apoptosis. We further demonstrated that survivin expression in glioma was directly related to the Src/STAT3 pathway by treating GL261 cells with the PP2 Src kinase inhibitor. Western blot with the PP2-treated cell lysates showed decreased levels of survivin protein (Figure 5D). This result matched with the recently reported observation of Src kinase-dependent survivin expression in osteosarcoma [37]. We further tested the expression of survivin in animal models implanted with tumors. A two-fold decrease in survivin mRNA transcript levels was observed in murine brains bearing GL261-FnKD tumors when compared with those of GL261-VC-implanted brains during the first 3 weeks after tumor implantation (Figure 7C). These observations confirmed that fibronectin silencing aborted integrin signaling in GL261 cells and failed to initiate Src kinase and STAT3 activity, thus aggressively reducing survivin expression.
A major cause of brain tumor progression is increased Treg infiltration in the tumor [4,5]. We measured Treg infiltration in the tumor-implanted brains of experimental animals by flow cytometry (Figure 8). A gradual but definitive increase in Treg accumulation was observed in animals' brains implanted with GL261-VC tumors as early as 14th day PI. At the 21st day PI, Tregs accounted for 63.4% of all the CD4+ T cells in the brain. This observation correlated with proliferative increase in tumor growth and survival of the GL261-VC-implanted animals. In comparison, Treg infiltration in GL261-FnKD-implanted brains was slower and accumulated 2.6-fold less Tregs than the GL261-VC-implanted animals by the 21st day PI. These observations indicated a reduced immunosuppressive environment in fibronectin-silenced tumors. These observations also demonstrated that Treg infiltration was dependent on the tumor growth and corroborated with recent observations by Grauer et al. [50], in which the authors reported gradual accumulation of Tregs during tumor growth.
Collectively, we conclude that this study for the first time contributes toward understanding the role of fibronectin in glioma progression. It explains the mechanism that involves the maintenance of integrin β1 fibronectin receptors in glioma cells required by fibronectin to induce a Src kinase-dependent survivin activity that promotes brain tumor proliferation.
Acknowledgments
The authors thank Sherise D. Ferguson, University of Chicago, for her comments, suggestions, and critical review of this work.
Footnotes
This work was supported by the National Cancer Institute (R01-CA122930, R01-CA138587, and R21-CA135728), the National Institute of Neurological Disorders and Stroke (K08-NS046430), The Alliance for Cancer Gene Therapy Young Investigator Award, and the American Cancer Society (RSG-07-276-01-MGO).
References
- 1.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Choi BD, Archer GE, Mitchell DA, Heimberger AB, McLendon RE, Bigner DD, Sampson JH. EGFRvIII-targeted vaccination therapy of malignant glioma. Brain Pathol. 2009;19:713–723. doi: 10.1111/j.1750-3639.2009.00318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ksendzovsky A, Feinstein D, Zengou R, Sharp A, Polak P, Lichtor T, Glick RP. Investigation of immunosuppressive mechanisms in a mouse glioma model. J Neurooncol. 2009;93:107–114. doi: 10.1007/s11060-009-9884-6. [DOI] [PubMed] [Google Scholar]
- 4.El Andaloussi A, Lesniak MS. An increase in CD4+CD25+FOXP3+ regulatory T cells in tumor-infiltrating lymphocytes of human glioblastoma multiforme. Neuro Oncol. 2006;8:234–243. doi: 10.1215/15228517-2006-006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heimberger AB, Abou-Ghazal M, Reina-Ortiz C, Yang DS, Sun W, Qiao W, Hiraoka N, Fuller GN. Incidence and prognostic impact of FoxP3+ regulatory T cells in human gliomas. Clin Cancer Res. 2008;14:5166–5172. doi: 10.1158/1078-0432.CCR-08-0320. [DOI] [PubMed] [Google Scholar]
- 6.Sugihara AQ, Rolle CE, Lesniak MS. Regulatory T cells actively infiltrate metastatic brain tumors. Int J Oncol. 2009;34:1533–1540. doi: 10.3892/ijo_00000282. [DOI] [PubMed] [Google Scholar]
- 7.Pankov R, Yamada KM. Fibronectin at a glance. J Cell Sci. 2002;115:3861–3863. doi: 10.1242/jcs.00059. [DOI] [PubMed] [Google Scholar]
- 8.Pytela R, Pierschbacher MD, Ruoslahti E. Identification and isolation of a 140 kd cell surface glycoprotein with properties expected of a fibronectin receptor. Cell. 1985;40:191–198. doi: 10.1016/0092-8674(85)90322-8. [DOI] [PubMed] [Google Scholar]
- 9.Rooprai HK, Vanmeter T, Panou C, Schnull S, Trillo-Pazos G, Davies D, Pilkington GJ. The role of integrin receptors in aspects of glioma invasion in vitro. Int J Dev Neurosci. 1999;17:613–623. doi: 10.1016/s0736-5748(99)00051-9. [DOI] [PubMed] [Google Scholar]
- 10.Han S, Khuri FR, Roman J. Fibronectin stimulates non-small cell lung carcinoma cell growth through activation of Akt/mammalian target of rapamycin/S6 kinase and inactivation of LKB1/AMP-activated protein kinase signal pathways. Cancer Res. 2006;66:315–323. doi: 10.1158/0008-5472.CAN-05-2367. [DOI] [PubMed] [Google Scholar]
- 11.Sherbet GV, Tindle ME, Stidolph S. Fibronectin in human astrocytomas grown in tissue culture. Anticancer Res. 1982;2:251–254. [PubMed] [Google Scholar]
- 12.Ohnishi T, Hiraga S, Izumoto S, Matsumura H, Kanemura Y, Arita N, Hayakawa T. Role of fibronectin-stimulated tumor cell migration in glioma invasion in vivo: clinical significance of fibronectin and fibronectin receptor expressed in human glioma tissues. Clin Exp Metastasis. 1998;16:729–741. doi: 10.1023/a:1006532812408. [DOI] [PubMed] [Google Scholar]
- 13.Desgrosellier JS, Barnes LA, Shields DJ, Huang M, Lau SK, Prevost N, Tarin D, Shattil SJ, Cheresh DA. An integrin α(v)β(3)-c-Src oncogenic unit promotes anchorage-independence and tumor progression. Nat Med. 2009;15:1163–1169. doi: 10.1038/nm.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Putnam AJ, Schulz VV, Freiter EM, Bill HM, Miranti CK. Src, PKCα, and PKCδ are required for avβ3 integrin-mediated metastatic melanoma invasion. Cell Commun Signal. 2009;7:10. doi: 10.1186/1478-811X-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu CL, Meyer DJ, Campbell GS, Larner AC, Carter-Su C, Schwartz J, Jove R. Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science. 1995;269:81–83. doi: 10.1126/science.7541555. [DOI] [PubMed] [Google Scholar]
- 16.Niu G, Bowman T, Huang M, Shivers S, Reintgen D, Daud A, Chang A, Kraker A, Jove R, Yu H. Roles of activated Src and Stat3 signaling in melanoma tumor cell growth. Oncogene. 2002;21:7001–7010. doi: 10.1038/sj.onc.1205859. [DOI] [PubMed] [Google Scholar]
- 17.Brantley EC, Benveniste EN. Signal transducer and activator of transcription-3: a molecular hub for signaling pathways in gliomas. Mol Cancer Res. 2008;6:675–684. doi: 10.1158/1541-7786.MCR-07-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abou-Ghazal M, Yang DS, Qiao W, Reina-Ortiz C, Wei J, Kong LY, Fuller GN, Hiraoka N, Priebe W, Sawaya R, et al. The incidence, correlation with tumor-infiltrating inflammation, and prognosis of phosphorylated STAT3 expression in human gliomas. Clin Cancer Res. 2008;14:8228–8235. doi: 10.1158/1078-0432.CCR-08-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dasgupta A, Raychaudhuri B, Haqqi T, Prayson R, Van Meir EG, Vogelbaum M, Haque SJ. Stat3 activation is required for the growth of U87 cell-derived tumours in mice. Eur J Cancer. 2009;45:677–684. doi: 10.1016/j.ejca.2008.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Chien S. Analysis of integrin signaling by fluorescence resonance energy transfer. Methods Enzymol. 2007;426:177–201. doi: 10.1016/S0076-6879(07)26009-4. [DOI] [PubMed] [Google Scholar]
- 22.Cao X, Tay A, Guy GR, Tan YH. Activation and association of Stat3 with Src in v-Src-transformed cell lines. Mol Cell Biol. 1996;16:1595–1603. doi: 10.1128/mcb.16.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhattacharya S, Ray RM, Johnson LR. STAT3-mediated transcription of Bcl-2, Mcl-1 and c-IAP2 prevents apoptosis in polyamine-depleted cells. Biochem J. 2005;392:335–344. doi: 10.1042/BJ20050465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 25.Bromberg J, Darnell JE., Jr The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 2000;19:2468–2473. doi: 10.1038/sj.onc.1203476. [DOI] [PubMed] [Google Scholar]
- 26.Costantino L, Barlocco D. STAT 3 as a target for cancer drug discovery. Curr Med Chem. 2008;15:834–843. doi: 10.2174/092986708783955464. [DOI] [PubMed] [Google Scholar]
- 27.Rahaman SO, Harbor PC, Chernova O, Barnett GH, Vogelbaum MA, Haque SJ. Inhibition of constitutively active Stat3 suppresses proliferation and induces apoptosis in glioblastoma multiforme cells. Oncogene. 2002;21:8404–8413. doi: 10.1038/sj.onc.1206047. [DOI] [PubMed] [Google Scholar]
- 28.Thomas CY, Chouinard M, Cox M, Parsons S, Stallings-Mann M, Garcia R, Jove R, Wharen R. Spontaneous activation and signaling by overexpressed epidermal growth factor receptors in glioblastoma cells. Int J Cancer. 2003;104:19–27. doi: 10.1002/ijc.10880. [DOI] [PubMed] [Google Scholar]
- 29.Iwamaru A, Szymanski S, Iwado E, Aoki H, Yokoyama T, Fokt I, Hess K, Conrad C, Madden T, Sawaya R, et al. A novel inhibitor of the STAT3 pathway induces apoptosis in malignant glioma cells both in vitro and in vivo. Oncogene. 2007;26:2435–2444. doi: 10.1038/sj.onc.1210031. [DOI] [PubMed] [Google Scholar]
- 30.Brantley EC, Nabors LB, Gillespie GY, Choi YH, Palmer CA, Harrison K, Roarty K, Benveniste EN. Loss of protein inhibitors of activated STAT-3 expression in glioblastoma multiforme tumors: implications for STAT-3 activation and gene expression. Clin Cancer Res. 2008;14:4694–4704. doi: 10.1158/1078-0432.CCR-08-0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hussain SF, Kong LY, Jordan J, Conrad C, Madden T, Fokt I, Priebe W, Heimberger AB. A novel small molecule inhibitor of signal transducers and activators of transcription 3 reverses immune tolerance in malignant glioma patients. Cancer Res. 2007;67:9630–9636. doi: 10.1158/0008-5472.CAN-07-1243. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki A, Hayashida M, Ito T, Kawano H, Nakano T, Miura M, Akahane K, Shiraki K. Survivin initiates cell cycle entry by the competitive interaction with Cdk4/p16(INK4a) and Cdk2/cyclin E complex activation. Oncogene. 2000;19:3225–3234. doi: 10.1038/sj.onc.1203665. [DOI] [PubMed] [Google Scholar]
- 33.Zhen HN, Li LW, Zhang W, Fei Z, Shi CH, Yang TT, Bai WT, Zhang X. Short hairpin RNA targeting survivin inhibits growth and angiogenesis of glioma U251 cells. Int J Oncol. 2007;31:1111–1117. [PubMed] [Google Scholar]
- 34.Mellai M, Caldera V, Patrucco A, Annovazzi L, Schiffer D. Survivin expression in glioblastomas correlates with proliferation, but not with apoptosis. Anticancer Res. 2008;28:109–118. [PubMed] [Google Scholar]
- 35.Zhen HN, Zhang X, Hu PZ, Yang TT, Fei Z, Zhang JN, Fu LA, He XS, Ma FC, Wang XL. Survivin expression and its relation with proliferation, apoptosis, and angiogenesis in brain gliomas. Cancer. 2005;104:2775–2783. doi: 10.1002/cncr.21490. [DOI] [PubMed] [Google Scholar]
- 36.Cai L, Zhang G, Tong X, You Q, An Y, Wang Y, Guo L, Wang T, Zhu D, Zheng J. Growth inhibition of human ovarian cancer cells by blocking STAT3 activation with small interfering RNA. Eur J Obstet Gynecol Reprod Biol. 2010;148:73–80. doi: 10.1016/j.ejogrb.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 37.Fossey SL, Liao AT, McCleese JK, Bear MD, Lin J, Li PK, Kisseberth WC, London CA. Characterization of STAT3 activation and expression in canine and human osteosarcoma. BMC Cancer. 2009;9:81. doi: 10.1186/1471-2407-9-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ritchie CK, Giordano A, Khalili K. Integrin involvement in glioblastoma multiforme: possible regulation by NF-κB. J Cell Physiol. 2000;184:214–221. doi: 10.1002/1097-4652(200008)184:2<214::AID-JCP9>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 39.Ulrich TA, de Juan Pardo EM, Kumar S. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res. 2009;69:4167–4174. doi: 10.1158/0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Preto A, Goncalves J, Rebocho AP, Figueiredo J, Meireles AM, Rocha AS, Vasconcelos HM, Seca H, Seruca R, Soares P, et al. Proliferation and survival molecules implicated in the inhibition of BRAF pathway in thyroid cancer cells harbouring different genetic mutations. BMC Cancer. 2009;9:387. doi: 10.1186/1471-2407-9-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jung KM, Bae IH, Kim BH, Kim WK, Chung JH, Park YH, Lim KM. Comparison of flow cytometry and immunohistochemistry in non-radioisotopic murine lymph node assay using bromodeoxyuridine. Toxicol Lett. 2010;192:229–237. doi: 10.1016/j.toxlet.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 42.Hayman EG, Ruoslahti E. Distribution of fetal bovine serum fibronectin and endogenous rat cell fibronectin in extracellular matrix. J Cell Biol. 1979;83:255–259. doi: 10.1083/jcb.83.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harima A, Nakaseko C, Yokota A, Kitagawa M, Morimoto C, Harigaya K, Saito Y. Fibronectin promotes cell proliferation of human pre-B cell line via its interactions with VLA-4 and VLA-5. Hematology. 2008;13:236–243. doi: 10.1179/102453308X348315. [DOI] [PubMed] [Google Scholar]
- 44.Paulus W, Baur I, Beutler AS, Reeves SA. Diffuse brain invasion of glioma cells requires β1 integrins. Lab Invest. 1996;75:819–826. [PubMed] [Google Scholar]
- 45.Granziero L, Ghia P, Circosta P, Gottardi D, Strola G, Geuna M, Montagna L, Piccoli P, Chilosi M, Caligaris-Cappio F. Survivin is expressed on CD40 stimulation and interfaces proliferation and apoptosis in B-cell chronic lymphocytic leukemia. Blood. 2001;97:2777–2783. doi: 10.1182/blood.v97.9.2777. [DOI] [PubMed] [Google Scholar]
- 46.Sah NK, Khan Z, Khan GJ, Bisen PS. Structural, functional and therapeutic biology of survivin. Cancer Lett. 2006;244:164–171. doi: 10.1016/j.canlet.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki A, Ito T, Kawano H, Hayashida M, Hayasaki Y, Tsutomi Y, Akahane K, Nakano T, Miura M, Shiraki K. Survivin initiates procaspase 3/p21 complex formation as a result of interaction with Cdk4 to resist Fas-mediated cell death. Oncogene. 2000;19:1346–1353. doi: 10.1038/sj.onc.1203429. [DOI] [PubMed] [Google Scholar]
- 48.Black KL, Chen K, Becker DP, Merrill JE. Inflammatory leukocytes associated with increased immunosuppression by glioblastoma. J Neurosurg. 1992;77:120–126. doi: 10.3171/jns.1992.77.1.0120. [DOI] [PubMed] [Google Scholar]
- 49.Zhou J, Bi C, Janakakumara JV, Liu SC, Chng WJ, Tay KG, Poon LF, Xie Z, Palaniyandi S, Yu H, et al. Enhanced activation of STAT pathways and overexpression of survivin confer resistance to FLT3 inhibitors and could be therapeutic targets in AML. Blood. 2009;113:4052–4062. doi: 10.1182/blood-2008-05-156422. [DOI] [PubMed] [Google Scholar]
- 50.Grauer OM, Nierkens S, Bennink E, Toonen LW, Boon L, Wesseling P, Sutmuller RP, Adema GJ. CD4+FoxP3+ regulatory T cells gradually accumulate in gliomas during tumor growth and efficiently suppress antiglioma immune responses in vivo. Int J Cancer. 2007;121:95–105. doi: 10.1002/ijc.22607. [DOI] [PubMed] [Google Scholar]