Abstract
Cross-sectional seroepidemiological studies of populations naturally exposed to Plasmodium falciparum suggest an association between protection from malaria and circulating antibodies to the carboxyl terminus of merozoite surface protein 1 (MSP1). Questions remain regarding the significance of cell-mediated immunity to MSP1 in conferring protection and inducing immunologic memory. Vaccine constructs have been based on the 42-kDa recombinant MSP1 protein (MSP142), which includes the 19-kDa (MSP119) and 33-kDa (MSP133) fragments containing the major B- and T-cell epitopes, respectively. To evaluate T-cell responses to the MSP133 fragment, two libraries of overlapping 18-mer peptides from the 3D7 and FVO MSP133 regions were used to screen a cohort of asymptomatic Kenyan adults. Gamma interferon (IFN-γ) measured by enzyme-linked immunospot assay (ELISPOT) at multiple time points assessed the magnitude and stability of these responses. The percentage of individuals with IFN-γ responses to single MSP133 peptides ranged from nil to 24%, were clustered among a subset of peptides, and were not consistently recalled over time. In comparison to peptide responses, IFN-γ ELISPOT responses to recombinant MSP142 were more prevalent, more frequently elicited by the 3D7 as opposed to the FVO allele, and more stable over time. The prevailing MSP133 genotype infection was 3D7, with few mixed infections and no sole FVO infections. This study demonstrates that immunity against MSP133 after cumulative natural infections consists of low-magnitude and difficult-to-detect IFN-γ responses. Although immunity against MSP1 alone will not confer protection against malaria, demonstrating a relative and sustained increase in T-cell immunity to MSP1 after vaccination would be a reasonable measurement of vaccine responsiveness.
Infection with Plasmodium spp., the protozoan parasites responsible for malaria, results in an estimated 350 to 500 million infections per year with an ensuing 1 to 2 million deaths, the majority of which are caused by Plasmodium falciparum (45). Adults who have maintained lifelong residence in areas where malaria transmission is stable and of high intensity gradually develop naturally acquired immunity, an age-related phenomenon marked by a reduced frequency and severity of clinical malaria and high-density blood stage parasitemia relative to that of infants and children. Maintaining naturally acquired immunity is thought to require continuing exposure to malaria blood stage antigens mediated through asymptomatic low-density blood stage infection. A malaria vaccine that would accelerate the development of naturally acquired immunity during infancy and childhood has been a high priority for many years. The recent success of vector interventions, such as insecticide-treated bed nets, in decreasing transmission of P. falciparum in sub-Saharan Africa suggests that a blood stage vaccine would also be desirable in order to maintain the strength and duration of naturally acquired immunity among adults (17).
Merozoite surface protein 1 (MSP1) is one of several candidates that have been considered for a blood stage vaccine. The primary ∼195-kDa MSP1 protein is expressed during schizogony and is processed initially to form a 42-kDa carboxyl-terminal fragment (MSP142) that remains attached to the merozoite surface. A second proteolytic cleavage results in the shedding of a 33-kDa fragment (MSP133) with formation of a 19-kDa carboxyl-terminal fragment (MSP119) anchored to the merozoite surface during invasion of the red blood cell (RBC) (4, 5). The 19-kDa fragment of MSP1 contains the major B-cell epitopes (18), the recognition of which is enhanced by T-helper epitopes in the 33-kDa fragment (1). Antibodies to MSP119 that interfere with RBC invasion by merozoites are one of several possible mechanisms by which naturally acquired immunity and experimental MSP1 vaccines may mediate protection against blood stage infection (27, 34). Studies of primates and mice immunized with MSP1 and more limited observations of residents of areas in which P. falciparum is endemic who have naturally acquired immunity and malaria-naïve human volunteers vaccinated with MSP142 suggest that T-cell responses to MSP1 are driven primarily by epitopes within the shed MSP133 fragment and that gamma interferon (IFN-γ) is important for optimal protective immunity (20, 23, 44, 46).
A challenge to developing malaria vaccines that enhance protective immunity against blood stage parasitemia has been antigenic polymorphism that arises through selection by the human immune response; i.e., inclusion of several blood stage alleles might be required in order to mitigate selection of alleles not included in the vaccine. The P. falciparum msp1 gene has been divided into 17 blocks based on conserved, semiconserved, and variant DNA sequences (30). The C-terminal MSP142 protein is encoded by blocks 15 to 17; MSP133 is encoded by block 15 and 16. MSP142 alleles expressed by most clinical isolates and laboratory cultures of P. falciparum exist as either the MAD20 (3D7) or FVO (K1 or Wellcome) allele of MSP119 and MSP133 (although genetic recombination between the two [and other] alleles encoded by block 17 and block 15 to 16 also occurs). Whereas most studies from sub-Saharan Africa and other areas where malaria is endemic have characterized msp1 block 17 haplotypes and cross-reactivity of IgG antibodies to the corresponding MSP119 polypeptides (14, 43), block 15 to 16 polymorphisms underlying the variation in MSP133 and its impact on naturally acquired T-cell immunity are less well understood. The objectives of the current study were to evaluate MSP133 polymorphism in an area of western Kenya in which malaria is holoendemic and the corresponding allele specificity of IFN-γ responses by adults with naturally acquired immunity to malaria.
MATERIALS AND METHODS
Study site and subjects.
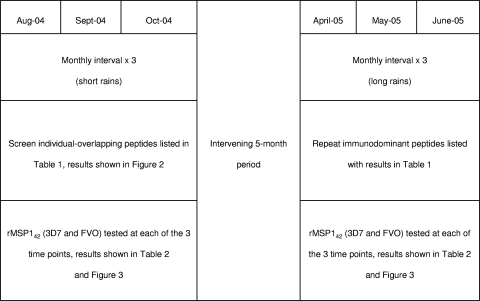
The studies were carried out in Kanyawegi, Nyanza Province, Kenya, within the auspices of a collaborative effort between Case Western Reserve University and the Kenya Medical Research Institute. The estimated entomologic inoculation rate when the study was conducted from 2004 to 2005 was ∼300 bites per year (2). Peaks in rainfall and malaria transmission occurred in September 2004 and May 2005. Twenty-four healthy asymptomatic adult volunteers (7 females, median age of 34; 17 males, median age of 44) were enrolled. Three 20-ml venous blood samples were obtained at 3-week intervals in August, September, and October 2004 and 6 months later in April, early May, and late May 2005 (the last time point is referred to as June for convenience) (Fig. 1). Data collected at the time of blood sampling included recent and current clinical features consistent with malaria morbidity (e.g., fever with malaise, decreased physical activity), blood stage infection status, and demography (age, gender, and family/household members and place of residence). Thick and thin blood smears were prepared, fixed in 100% methanol, stained with 5% Giemsa solution, and examined by light microscopy for P. falciparum-infected erythrocytes. The density of parasitemia was expressed as the number of asexual P. falciparum isolates per μl blood assuming a leukocyte count of 8,000 per μl. All participants were asymptomatic and reported taking no antimalarial drugs during the 11-month observation period.
FIG. 1.
Sample collection. Timeline, season, intervals, and MSP1 recombinant and peptide testing time points are depicted, as well as references to relevant tables and figures presented herein.
Ethical approval.
Approval for the study was obtained from the Kenya Medical Research Institute National Ethical Review Committee and the Institutional Review Board for Human Studies at University Hospitals of Cleveland, Case Western Reserve University. Written informed consent was obtained from the study participants.
PCR amplification of the region of Pfmsp1 encoding the MSP133 polypeptide.
To estimate the extent of antigenic polymorphism that would be needed to be considered in synthesizing peptides for use in T-cell studies, we used archival blood samples collected in 2003 from 10 adults and 10 children living in the study area. Ethical approval for collection and use of these samples was the same as described above. DNA was extracted from 200 μl blood using spin kits (QIAmp DNA blood kit; Qiagen Inc., Valencia, CA) according to the manufacturer's directions. Block 15 to 16 of msp1 was amplified by PCR in two ∼500-bp amplicons, grossly representing blocks 15 and 16 with primers selected using the Primer 3 program (http://frodo.wi.mit.edu/primer3/). PCR primers were chosen to distinguish between the 3D7 and FVO dimorphic variants, allowing for the detection of mixed or polyclonal infections. 3D7 primers for amplicon “M15” are 5′CCATTTTTGGAGAATCCGAAG 3′ (amino acid residues [aa] 1329 to 1336) and 5′ TTCGTCTGTTTTTGCTGGTG 3′ (aa 1504 to 1510), while “M16” primers are 5′ AAGGTTTTAGCGAAATATAAGGATGA 3′ (aa 1475 to 1484) and 5′ TTCTTCTCTTTCATCTAAATGTCTGAA 3′ (aa 1661 to 1669). FVO primers are amplicon “FVO15” (5′ TTGGAGAATCCGAAGAAGATT 3′ [aa 1332 to 1338] and 5′ TTTCACCTTGTTTGTCGTTGA 3′ [aa 1489 to 1515]) and “FVO16” (5′ TTCAATAGATACGGATATAAATTTTGC 3′ [aa 1556 to 1564] and 5′ ACATTCTTCTCTTTCATCTAAATGTCT 3′ [aa 1650 to 1658]). The PCR was conducted in 21-μl reaction volumes using 10.8 μl sterile water, 1 μl 2.5 mmol deoxynucleoside triphosphates (dNTPs), 2 μl 10× PCR buffer, 2 μl 25 mmol MgCl2, and 0.2 μl PE AmpliTaq Gold polymerase (5 U/μl) and 2 μl forward and reverse primers (2.5 μM). The cycling conditions included initial denaturation at 94°C for 4 min, 45 cycles of denaturation for 1 min 45 s at 94°C, and extension for 1 min at 63°C. This was followed by reannealing for 1 min at 72°C and a final extension for 10 min at 72°C. Three μl of PCR product was run on 1% agarose gel electrophoresis with development of bands in 1× TAE buffer and CyberGold with visualization on GelScanner. Only samples with bands in both blocks 15 and 16 for a particular allele are reported as positive. The identical protocol was used for determining block 15 and 16 sequence polymorphism on blood samples from the adults participating in the IFN-γ study described here.
Sequencing.
PCR products were purified using the QIAquick PCR purification kit (Qiagen, Inc., Valencia, CA) and sent to MWG Biotech (High Point, NC) for sequencing. Sequence alignments were performed using consed programs (16) comparing field isolates to 3D7 (3D7, GenBank sequence no. Z35327.1) and FVO (FVO, GenBank sequence no. X03371.1) vaccine strain sequences. P. falciparum infection status had already been independently determined by nested PCR of the highly conserved multicopy small ribosomal subunit (39).
Peptide synthesis.
Peptides (18-mer) of approximately 70% immunologic grade purity spanning the length of the 33-kDa region of both 3D7 (M) and FVO (K) variants were synthesized by SIGMA GENOSYS (The Woodlands, TX). The peptides overlapped by nine amino acid residues, creating a total of 30 peptides (M28 to M57) corresponding to the 3D7 allele and 28 peptides (K26 to K52) corresponding to the FVO allele (Table 1). Peptides were lyophilized and, immediately before use, solubilized in dimethyl sulfoxide (1%, wt/vol) and then diluted in sterile phosphate-buffered saline (PBS) for a working 10× concentration of 0.1 mg/ml.
TABLE 1.
Amino acid sequences of overlapping 18-mer peptides of the 33-kDa region of MSP1
| Peptide | Sequence | Position |
|---|---|---|
| MAD 20 allele | ||
| M28 | AISVTMDNILSGFENEYD | 1349-1367 |
| M29 | LSGFENEYDVIYLKPLAG | 1359-1376 |
| M30 | VIYLKPLAGVYRSLKKQI | 1368-1385 |
| M31 | VYRSLKKQIEKNIFTFNL | 1377-1394 |
| M32 | EKNIFTFNLNLNDILNSR | 1386-1403 |
| M33 | NLNDILNSRLKKRKYFLD | 1395-1412 |
| M34 | LKKRKYFLDVLESDLMQF | 1404-1421 |
| M35 | VLESDLMQFKHISSNEYI | 1413-1430 |
| M36 | KHISSNEYIIEDSFKLLN | 1422-1439 |
| M37 | IEDSFKLLNSEQKNTLLK | 1431-1448 |
| M38 | SEQKNTLLKSYKYIKESV | 1440-1457 |
| M39 | SYKYIKESVENDIKFAQE | 1449-1466 |
| M40 | ENDIKFAQEGISYYEKVL | 1458-1475 |
| M41 | GISYYEKVLAKYKDDLE | 1467-1483 |
| M42 | AKYKDDLESIKKVIKEE | 1476-1492 |
| M43 | SIKKVIKEEKEKFPSSPP | 1484-1501 |
| M44 | KEKFPSSPPTTPPSPAKT | 1493-1510 |
| M45 | TTPPSPAKTDEQKKESKF | 1502-1519 |
| M46 | DEQKKESKFLPFLTNIET | 1511-1528 |
| M47 | LPFLTNIETLYNNLVNKI | 1520-1537 |
| M48 | LYNNLVNKIDDYLINLKA | 1529-1546 |
| M49 | DDYLINLKAKINDCNVEK | 1538-1555 |
| M50 | KINDCNVEKDEAHVKITK | 1547-1564 |
| M51 | DEAHVKITKLSDLKAIDD | 1556-1573 |
| M52 | LSDLKAIDDKIDLFKNPY | 1565-1582 |
| M53 | KIDLFKNPYDFEAIKKLI | 1574-1591 |
| M54 | DFEAIKKLINDDTKKDML | 1583-1600 |
| M55 | NDDTKKDMLGKLLSTGLV | 1592-1609 |
| M56 | GKLLSTGLVQNFPNTIIS | 1601-1619 |
| M57 | QNFPNTIISKLIEGKFQD | 1611-1628 |
| K1 allele | ||
| K26 | AVTPSVIDNILSKIENEY | 1349-1366 |
| K27 | ILSKIENEYEVLYLKPLA | 1358-1375 |
| K28 | EVLYLKPLAGVYRSLKKQ | 1367-1384 |
| K29 | GVYRSLKKQLENNVMTFN | 1376-1393 |
| K30 | LENNVMTFNVNVKDILNS | 1385-1402 |
| K31 | VNVKDILNSRFNKRENFK | 1394-1411 |
| K32 | RFNKRENFKNVLESDLIP | 1403-1420 |
| K33 | NVLESDLIPYKDLTSSNY | 1412-1429 |
| K34 | YKDLTSSNYVVKDPYKFL | 1421-1438 |
| K35 | VVKDPYKFLNKEKRDKFL | 1430-1447 |
| K36 | NKEKRDKFLSSYNYIKDS | 1439-1456 |
| K37 | SSYNYIKDSIDTDINFAN | 1448-1465 |
| K38 | IDTDINFANDVLGYYKIL | 1457-1474 |
| K39 | DVLGYYKILSEKYKSDLD | 1466-1483 |
| K40 | SEKYKSDLDSIKKYINDK | 1475-1492 |
| K41 | SIKKYINDKQGENEKYLP | 1484-1501 |
| K42 | QGENEKLPFLNNIETLY | 1493-1510 |
| K43 | FLNNIETLYKTVNDKIDL | 1502-1529 |
| K44 | KTVNDKIDLFVIHLEAKV | 1511-1528 |
| K45 | FVIHLEAKVLNYTYEKSN | 1520-1537 |
| K46 | LNYTYEKSNVEVKIKELN | 1529-1546 |
| K47 | VEVKIKELNYLKTIQDKL | 1538-1555 |
| K48 | YLKTIQDKLADFKKNNNF | 1547-1564 |
| K49 | ADFKKNNNFVGIADLSTD | 1556-1573 |
| K50 | VGIADLSTDYNHNNLLTK | 1565-1582 |
| K51 | YNHNNLLTKFLSTGMVFE | 1574-1591 |
| K52 | FLSTGMVFENLAKTVLSN | 1583-1600 |
| K53 | NLAKTVLSNLLDGNLQG | 1592-1608 |
IFN-γ ELISPOT.
Blood was collected in sodium heparin Vacutainer tubes. Peripheral blood mononuclear cells (PBMCs) were separated from whole blood by Ficoll-Hypaque gradient centrifugation and suspended in complete culture medium (RPMI containing 10% human type AB heat-inactivated serum, 50 mg/ml gentamicin, 200 mM l-glutamine, 1 M HEPES). Enzyme-linked immunospot assay (ELISPOT) 96-well flat-bottomed plates were coated with 100 μl IFN-γ antibody Endogen M-700A (5 μg/ml; Endogen, Woburn, MA) and incubated overnight at 4°C. The plates were washed three times with sterile 1% PBS, blocked for 2 h with 100 μl solution of 10% fetal bovine serum in PBS, and washed again with 1% PBS. Then 0.5 ×106 PBMCs were added (100 μl) per well except for the positive control phytohemagglutinin (PHA), which had 0.1 × 106 cells. Single PBMC culture conditions for each individual included 10 μg/ml of each 18-mer MSP1 peptide, 5 μg/ml recombinant 3D7 and FVO MSP142, 10 μl PBS (negative-control well), and 1 μg/ml PHA (positive-control well). After incubation of plates for 3.5 days at 37°C with humidity and 5% CO2, plates were washed with PBS and then PBS/Tween. Secondary antibody Endogen M-701B biotin was added at 100 μl/well (0.75 μg/ml). After 90 min incubation at 37°C, plates were washed with PBS/Tween followed by the addition of 100 μl streptavidin substrate (Dako P0397). After 2 h of incubation at room temperature, plates were developed over 15 to 20 min by the addition of 1% 3-amino-9-ethylcarbazole (AEC) in acetate buffer and 0.015% H2O2 was added. Spot-forming units (SFU) were counted on-site by stereoscope examination and later validated by Immunospot Satellite analyzer (Cellular Technology Ltd., Shaker Heights, OH).
Statistics.
Statistical analyses were conducted using SAS version 8.2 (SAS, Cary, NC). An IFN-γ ELISPOT was considered positive if the proportion of SFU in the stimulated well was significantly greater than that in the unstimulated background well using Fisher's exact test (P < 0.05). If an individual did not have IFN-γ responses to PHA, his or her samples were eliminated from the analysis. The 3D7 and FVO MSP142 alleles were tested by IFN-γ ELISPOT using PBMCs from malaria-naïve North American adults, and no responders were detected (data not shown). Response rates of 15% among the study population were used as an initial indicator of moderate population recognition for each peptide, and response rate was thus deemed the threshold criterion in order to include that peptide for repeat testing. Associations between IFN-γ responses and other variables of interest were evaluated with Fisher's exact test. Temporal stability of responses was evaluated using kappa statistics (to assess agreement of responses over time) and McNemar's test (to assess the homogeneity of an individual's responses across time points).
RESULTS
MSP133 polymorphism in P. falciparum isolates from western Kenya.
In order to ascertain genetic diversity in naturally circulating MSP133 allelic sequences, DNA was isolated from 20 blood smear-positive samples previously collected from children and adults residing in the Kenya study site where the current study was performed. The region of msp1 encoding both allelic variants of MSP133 was amplified by PCR, and genetic sequences were aligned with the corresponding vaccine strains of 3D7 and FVO. No synonymous or nonsynonymous polymorphisms were found between the natural circulating strains and the vaccine strains (data not shown). Eighteen-mer peptides thus were constructed with amino acid sequences based on the referent 3D7 and FVO alleles (Table 1).
The prevalence of blood smears positive for asexual-stage P. falciparum from the 24 asymptomatic adults participating in the current study ranged from 22 to 42% (density, 80 to 13,000 parasites/μl) from the three samplings in 2004 and from 25 to 62% (density, 80 to 7,280 parasites/μl) in 2005. Block 15 to 16 msp1 genotyping for each variant was performed on all available samples. Single infections with the 3D7 allele predominated over three time points in 2004 (23 to 42%), with no single FVO infections and 5 to 9% mixed 3D7/FVO infections (Table 2 ). Similar percentages were observed in the three monthly collections in 2005 (29 to 48% single 3D7 infections, no single FVO infections, and nil to 9% mixed infections; Table 2). Of the 133 samples with results available for both blood smear and PCR, there was 85% concordance between the two methods: 9 of 133 (7%) positive blood smears were PCR negative, and 11 of 133 (8%) negative blood smears were PCR positive. Blood smear positivity and block 15 to 16 msp1 genotype detected for a single individual varied both within each 3-month period and between the 2004 and 2005 transmission seasons. Treatment guidelines of the Kenya Ministry of Health at the time this study was performed were to administer antimalaria drugs to adults with malaria-positive blood smears only when accompanied by symptoms of malaria-attributable illness, such as fever. None of the participants in this study had recent or ongoing signs or symptoms consistent with clinical malaria.
TABLE 2.
P. falciparum density by blood smear, 33-kDa MSP1 genotype infection, and corresponding IFN-γ ELISPOT responses
| Individual | 2004 resulta |
2005 result |
||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| August |
September |
October |
April |
May |
June |
|||||||||||||||||||||||||
| Density | MSP1 K1 infection | IFN-γ response to MSP1 (K1) | MSP1 MAD20 infection | IFN-γ response to MSP1 (MAD20) | Density | MSP1 K1 infection | IFN-γ response to MSP1 (K1) | MSP1 MAD20 infection | IFN-γ response to MSP1 (MAD20) | Density | MSP1 K1 infection | IFN-γ response to MSP1 (K1) | MSP1 MAD20 infection | IFN-γ response to MSP1 (MAD20) | Density | MSP1 K1 infection | IFN-γ response to MSP1 (K1) | MSP1 MAD20 infection | IFN-γ response to MSP1 (MAD20) | Density | MSP1 K1 infection | IFN-γ response to MSP1 (K1) | MSP1 MAD20 infection | IFN-γ response to MSP1 (MAD20) | Density | MSP1 K1 infection | IFN-γ response to MSP1 (K1) | MSP1 MAD20 infection | IFN-γ response to MSP1 (MAD20) | |
| 1 | 280 | − | + | + | + | 480 | − | ND | + | ND | 0 | − | − | − | + | 80 | − | + | + | + | 0 | − | − | − | + | 400 | − | − | + | + |
| 2 | 0 | − | + | − | + | 80 | − | − | − | − | 80 | − | ND | − | ND | 0 | − | + | − | + | 0 | − | ND | − | ND | 0 | − | − | − | + |
| 3 | 80 | − | − | + | − | 0 | − | − | − | − | − | − | − | + | 80 | − | + | + | + | 0 | − | − | − | − | 0 | − | − | − | + | |
| 4 | 0 | − | − | − | + | 0 | − | − | − | − | 400 | − | − | + | − | 80 | − | − | − | − | 80 | − | ND | + | ND | 0 | − | − | − | + |
| 5 | 0 | − | − | + | + | 0 | − | − | + | − | 400 | + | − | + | − | 0 | − | − | − | + | 80 | − | − | + | − | 80 | − | − | − | + |
| 6 | 0 | − | + | − | + | 0 | − | − | − | + | ND | ND | ND | ND | ND | 80 | − | − | + | + | 0 | − | + | − | + | 0 | − | − | − | + |
| 7 | 920 | + | − | + | − | 0 | − | − | − | − | 0 | − | − | − | − | 1,040 | − | − | + | − | 0 | − | − | − | − | 0 | − | ND | − | ND |
| 8 | 0 | − | + | − | + | 0 | − | − | − | + | 80 | − | + | − | + | 80 | − | − | − | + | 0 | − | + | − | + | 80 | − | − | + | + |
| 9 | 0 | − | − | + | + | 0 | − | − | − | + | 0 | − | ND | − | ND | 80 | − | − | + | − | 80 | − | − | + | − | 200 | − | − | + | − |
| 10 | 0 | − | − | − | + | 0 | − | ND | + | ND | 0 | − | − | + | − | 0 | − | − | − | + | 0 | − | − | − | + | 80 | − | − | − | + |
| 11 | 0 | − | ND | + | ND | 1,400 | − | ND | + | ND | 0 | − | ND | − | ND | 80 | − | − | − | + | 0 | − | − | − | − | 0 | − | + | − | + |
| 12 | 0 | − | − | − | − | 0 | − | + | − | − | 280 | − | − | + | + | 0 | − | − | − | + | 0 | − | − | − | − | 160 | − | − | + | − |
| 13 | 0 | − | ND | − | ND | 80 | − | ND | + | ND | 200 | − | ND | + | ND | 80 | − | − | + | − | 7,280 | − | − | + | − | ND | ND | ND | ND | ND |
| 14 | 80 | − | − | − | + | 0 | − | + | − | + | 80 | − | + | + | + | 0 | − | − | − | + | 0 | − | − | − | − | 0 | − | − | − | − |
| 15 | 0 | − | − | − | − | 0 | − | − | + | − | 80 | − | − | − | + | 0 | − | − | − | + | 0 | − | − | − | + | 80 | − | − | − | + |
| 16 | 0 | − | − | − | + | ND | ND | ND | ND | ND | 0 | − | ND | − | ND | |||||||||||||||
| 17 | 440 | − | − | + | − | 0 | − | + | + | + | 0 | − | − | − | − | 80 | + | − | + | − | 0 | − | − | − | − | 520 | − | − | + | − |
| 18 | 0 | − | + | − | + | 0 | − | + | − | + | 120 | − | + | − | + | 0 | − | + | − | + | 0 | − | ND | − | ND | 320 | − | ND | + | ND |
| 19 | 320 | − | − | + | − | 0 | − | − | + | − | 0 | − | ND | − | ND | 0 | − | − | − | − | 240 | − | ND | + | ND | 200 | − | − | + | − |
| 20 | 280 | − | − | + | − | 0 | + | ND | + | ND | 0 | − | ND | − | ND | 160 | − | − | + | + | 0 | − | − | − | + | 480 | − | − | + | − |
| 21 | 0 | − | − | − | + | 0 | − | + | + | + | 0 | − | ND | − | ND | 0 | − | − | − | + | 80 | − | − | + | − | 0 | − | + | − | + |
| 22 | 80 | − | ND | + | ND | 0 | − | ND | − | ND | 0 | − | ND | − | ND | 80 | − | − | + | − | ND | ND | ND | ND | ND | 80 | − | − | + | − |
| 23 | 3,400 | − | + | + | + | 0 | − | − | − | + | 0 | − | ND | − | ND | |||||||||||||||
| 24 | 240 | + | ND | + | ND | 13,000 | + | − | + | − | 0 | − | ND | − | ND | 240 | + | ND | + | ND | 0 | − | − | − | − | 400 | − | − | + | − |
| No. positive/total no. | 10/24 | 2/24 | 6/20 | 12/24 | 13/20 | 5/23 | 2/23 | 5/17 | 11/23 | 8/17 | 9/22 | 1/23 | 3/12 | 6/23 | 7/12 | 13/22 | 2/22 | 4/21 | 10/22 | 14/21 | 5/21 | 0/21 | 2/17 | 6/21 | 6/17 | 13/21 | 0/21 | 2/19 | 10/21 | 11/19 |
| Frequency | 42% | 8% | 30% | 50% | 65% | 22% | 8% | 29% | 46% | 47% | 41% | 4% | 25% | 25% | 58% | 59% | 9% | 19% | 45% | 67% | 25% | 0% | 12% | 29% | 35% | 62% | 0% | 10% | 48% | 58% |
Density is expressed as parasites/ml. +, positive response; −, negative response. ND, no data.
Screening of overlapping peptides from the MSP133 3D7 and FVO alleles.
Due to the limited number of PBMCs available from 20-ml blood volumes and the large number of peptides to be screened as T-cell epitopes, we designed a sampling strategy which allowed individual testing of each of the 30 3D7 and 28 FVO MSP1 peptides listed in Table 1. Screening was performed in the first, second, and third sampling periods in August, September, and October 2004. This procedure produced a list of immunodominant peptides that could be prioritized for subsequent repeat measurements to evaluate the stability of responses over two distinct high-transmission seasons in 2004 and 2005, as well as the relationship of T-cell IFN-γ responses to previous and coexisting P. falciparum infection. Figure 1 summarizes the study design and explains which conditions were tested and retested at each time point. The results for the first three sampling periods are described below.
3D7 MSP133 peptides tested in August 2004.
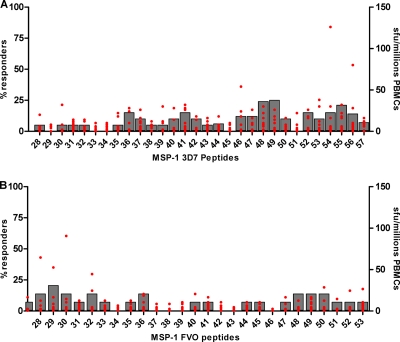
The first 26 of 30 overlapping peptides from the 3D7 allele were tested. The proportion of donors with PBMCs responding to individual peptides ranged from nil to 24% (Fig. 2 A). Responses to five peptides were observed for ≥15% individuals, all of which were encoded in msp1 block 16 (M36 [3 of 20], M41 [3 of 20], M48 [4 of 20], M49 [3 of 20], and M52 [3 of 20]). The mean SFU per million PBMCs for each of the peptide epitopes is also shown in Fig. 2A. As a basis of comparison, IFN-γ ELISPOT responses to the 3D7 MSP142 protein are included for each time point (Table 2). We observed that 65% (13 of 20) of individuals responded to the recombinant protein, with a mean IFN-γ SFU value of 63 per million PBMCs. A lower proportion of PBMC donors (30% [6 of 20]) responded to FVO MSP142 protein, with a mean of 20 IFN-γ SFU per million PBMCs.
FIG. 2.
Frequency of IFN-γ ELISPOT responders to overlapping MSP1 33-kDa peptides from Table 1, for 3D7-specific peptides (A) and MSP1 (33-kDa) FVO-specific peptides (B) screened in August to October 2004, illustrated with bars and corresponding to the left margin legend. Magnitude of IFN-γ ELISPOT-forming units (sfu) per million PBMCs is represented by red spots and range referenced on the right margin legend.
FVO MSP133 peptides tested in September 2004.
Twenty-eight overlapping peptides from the FVO allele of MSP133 were used to assess PBMC IFN-γ responses (Fig. 2B). PBMCs from only 17 of 23 individuals had evaluable IFN-γ responses, and results from 6 participants were excluded due to high endogenous background IFN-γ expression (due to bacterial contamination confined to a single plate). Of the PBMCs from the remaining 11 study participants with interpretable results, only a single peptide stimulated an IFN-γ response by ≥15% of individuals: K29 (block 15). The mean magnitude of the response to the K29 MSP1 FVO peptide was 10 SFU per million PBMCs. With respect to responses to recombinant MSP142, 8 of 17 (47%) donors' PBMCs responded to the 3D7 allele and 5 of 17 (29%) to the FVO allele (Table 2). The mean IFN-γ SFU values for the 3D7 and FVO proteins were 90 and 34 per million PBMCs, respectively.
Testing the remaining 3D7 and FVO MSP133 peptides in October 2004.
Of the 24 study participants, 16 agreed to a third consecutive monthly venous blood donation in 2004. Both alleles of the MSP142 protein and the remaining six untested 3D7 and FVO peptides (M54, M55, M56, M57, K52, and K53) were screened. The proportions of responders to the 3D7 and FVO MSP142 proteins were similar to previous measurements in August and September (Table 2) (for the 3D7 allele, 7 of 12 [58%], 108 IFN-γ SFU per million PBMCs [mean]; for the FVO allele, 3 of 12 [25%], 52 IFN-γ SFU per million PBMCs [mean]). From this remaining subset, the peptides stimulating IFN-γ responses by ≥15% individuals included M54 (3 of 14), M55 (4 of 14), M56 (3 of 14), and M57 (3 of 14).
Association between Pfmsp1 infection genotype and IFN-γ responses to allelic variants of MSP133 peptides.
Two approaches were taken to determine the relationship between the MSP133-infecting genotype and allele-specific IFN-γ responses. First, the correlation between the coexisting genotype and allele-specific IFN-γ response was evaluated at each time point. We hypothesized that if an individual demonstrated an allele-specific IFN-γ response, he or she would be less likely to be infected with the cognate genotype. Second, it was determined whether an infecting genotype induced an allele-specific IFN-γ response that was detectable at the ensuing time point 1 month later. No significant association between infecting genotype and coexisting PBMC IFN-γ responses to the corresponding 3D7 or FVO peptides or MSP142 proteins was detected by the chi-square test statistic. Furthermore, there was no significant association between an IFN-γ response to a particular allele and the infecting genotype detected 1 month earlier (Table 2). Kappa statistics for temporal stability were low (<0.6), though McNemar's test failed to detect significant heterogeneity across time.
Immunogenicity and stability of MSP133 peptide IFN-γ responses across transmission seasons.
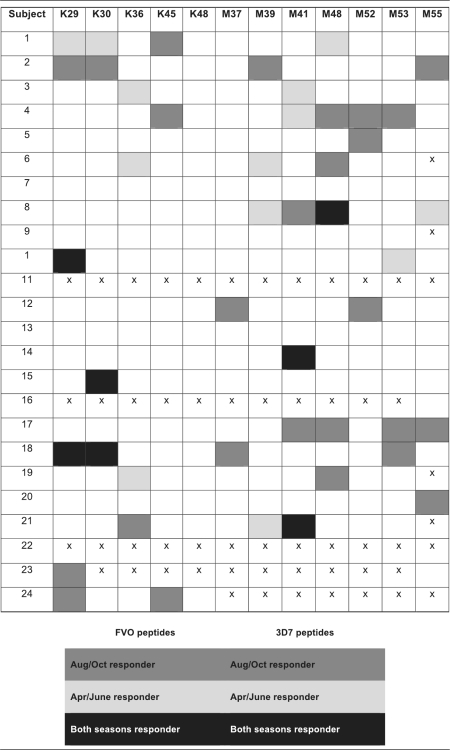
In April, May, and June 2005, IFN-γ ELISPOT recall responses were determined for both the 3D7 and FVO MSP142 recombinant proteins (Table 2) and 12 immunodominant peptides selected by screening performed in 2004 (M37, M39, M41, M48, M52, M53, M55, K29, K30, K36, K45, and K48; Table 3). Due to limitations in the ELISPOT 12-by-8-well-plate format, when high IFN-γ responses were clustered within overlapping peptides (i.e., M52 to M57) we selected representative peptides (i.e., M52, M53, M55) with fewer overlapping amino acids from this region. The proportion of responders to one or more of these peptides ranged from nil to 16% across the three sampling times in 2005. Only one peptide generated recall responses by ≥15% of the participants—M41 (3 of 19 in June). Table 3 displays repeat peptide-specific recall responses for each individual.
TABLE 3.
Stability of INF-γ ELISPOT responses to individual 3D7 and FVO MSP1 (33-kDa) peptides
Peptides generating the highest number of responders in 2005 were K29, M39, and M41. The individuals responding to these peptides differed from those responding to the same peptides in 2004. When data collected during the two seasons were combined, peptides eliciting the highest number of responders were K29 (five total responders: three responders in 2004 only, one responder in 2005 only, and one responder in both seasons) and M41 (seven total responders: three in 2004 and four in 2005). Three peptides elicited responses in both 2004 and 2005: K29, K30, and M48. However, this was observed for only two individuals. One individual responded to K29 and K30 in September 2004, October 2004, and April 2005, and one individual responded to M48 in August 2004, October 2004, April 2005, and May 2005. Of note, if an individual responded to one peptide, he or she tended to respond to other peptides of the same allele at that time point. Overall, few individuals retained consistent responses to single peptides in both 2004 and 2005 (Table 3).
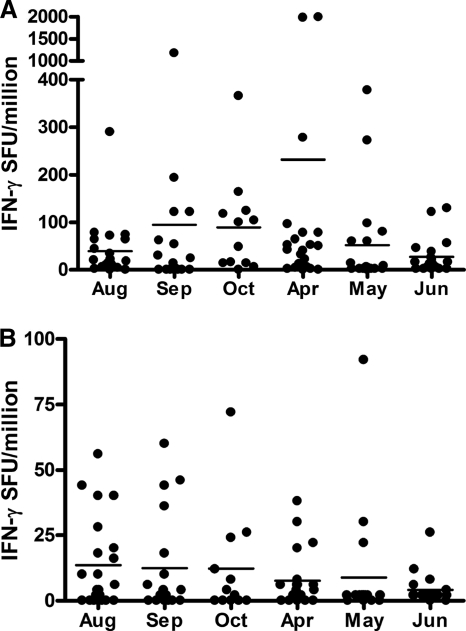
Frequency and magnitude of IFN-γ responses to rMSP142 allelic variants.
The stability and magnitude of IFN-γ responses to the 3D7 and FVO MSP142 recombinant proteins were ascertained for the six time points over the course of this 11-month observational study (Fig. 3 A and B). Overall, 58% (14 of 24) of individuals had IFN-γ responses to 3D7 MSP142 at ≥2 time points. For the FVO variant, 29% (7 of 24) of study participants demonstrated responses at multiple time points. For individual study participants with IFN-γ responses to 3D7 MSP142 in August, September, and October 2004, 90% (eight of nine) individuals had at least one response, but only three maintained a response across the entire 3-month period (three of nine). For participants with FVO MSP142 ELISPOTs successfully performed in 2004, 60% (6 of 10) individuals had at least one response, but only one PBMC donor maintained a response across each of the 3 months. Of the original 24 individuals, 19 had ELISPOTs completed in April, May, and June 2005. For the 3D7 MSP142, six individuals had reproducible responses in these 3 months. For the FVO MSP142, no study participants had responses at all three monthly time points and only one individual responded twice.
FIG. 3.
Magnitude of IFN-γ ELISPOT responses to allelic variants of MSP1 42-kDa recombinant protein, 3D7 (A) and FVO (B), during six time periods. ELISPOT responses are reported as spot-forming units (SFU) per million PBMCs.
When responses between the two 3-month study periods in 2004 and 2005 were compared, the majority of individuals with responses in 2004 also had responses in 2005 (87% [13 of 15]) to the 3D7 and FVO allele (70% [7 of 10]). Of the 11 individuals with IFN-γ responses to 3D7 MSP142 at ≥3 time points, 64% also had IFN-γ responses to the FVO variant at multiple time points. On the other hand, three other individuals had responses to 3D7 MSP142 at ≥3 time points but no response to the FVO MSP142 at any time. However, the overall percentage of responders to MSP142 in 2005 was not dissimilar to that observed 5 months earlier: 63% (April), 35% (May), and 58% (June) to the 3D7 allele and 18% (April), 12% (May), and 11% (June) to the FVO allele. The magnitudes of IFN-γ recall responses to FVO MSP142 were consistently lower than those to 3D7 MSP142, and the mean magnitude of responses to each allelic variant did not vary over time even though the responses may have been generated by different sets of individuals (Fig. 3A and B).
DISCUSSION
The majority of clinical signs and complications of malaria result from the erythrocytic phase of malaria infection, and individuals with high-density parasitemia have a greater likelihood of experiencing malaria-related illness when results are adjusted for age (29). Thus, a vaccine that confers protective immunity directed against the merozoite stage of Plasmodium falciparum would have great benefit in preventing or mitigating the clinical burden of disease, particularly among infants and children who have not yet developed naturally acquired immunity and adults who experience waning immunity due to reduced exposure to malaria (15, 36). The merozoite protein and vaccine antigen MSP1 displays extensive polymorphisms between the two main naturally circulating alleles, 3D7 and FVO. Understanding the influence of circulating alleles on the stability or “boosting” of an allele-specific immune response in naturally exposed populations will help guide interpretation of immunogenicity outcome measurements in vaccine trials. In our study based on the DNA sequence of the region of msp1 encoding MSP133, we found that both 3D7 and FVO alleles of P. falciparum MSP1 are responsible for infections in western Kenya, with the majority being a single infection with 3D7. This is supported by other MSP1 genomic typing done in Kisumu, Kenya (C. Ockenhouse, personal communication). Due to technical limitations in sequencing the entire length of MSP142 we were not able to ascertain if other allelic variants of MSP1, i.e., CAMP/FUP (33-kDa 3D7 plus 19-kDa FVO) were present. It is also possible that detection of the FVO MSP133 allele was underestimated as a result of inadequate sensitivity of our PCR assay to detect low-level FVO parasitemia in the face of a coinfection with high-level parasitemia of the 3D7 allele. Using another PCR-based method to detect P. falciparum allelic coinfections within this community in 2003, we found the FVO MSP119 variant to be dominant (11), suggesting that the CAMP/FUP strain may be the dominant infection.
The genetic sequences of the block 15 to 16 msp1, suggesting (MSP133) isolated from 20 random samples from Kenyan adults and children demonstrated genetic sequences identical to those of the referent 3D7 and FVO vaccine strains, with no synonymous or nonsynonymous polymorphisms. This information provides optimism for time-consuming MSP1 vaccine development, since it suggests that although the 3D7 and FVO alleles differ, recent isolates do not reveal novel polymorphisms. Similar results were found in a study conducted in Thailand (22), where sequencing of the region of msp1 encoding MSP133 from 19 individuals demonstrated only four nucleotide differences, all resulting in nonsynonymous changes. Taken together, these results suggest that MSP1 vaccines should not be hampered by emerging allelic variants within the 33-kDa region of individual MSP1 strains. In comparison, the extent of circulating polymorphisms of the MSP119 has been well documented from a vaccine trial site in Mali (43). In that study, among 14 haplotypes identified, the prevalence of the 3D7 MSP119 allele was only 16%, with the CAMP/FUP and FVO alleles at 36 and 46%, respectively.
Previous work has supported the importance of MSP119 in inducing humoral immune responses and of MSP133 in providing necessary T-cell responses (8, 13, 19, 44). Udhayakumar et al. (44) examined T-cell proliferation by residents of western Kenya to various 17-mer peptides from the 33-kDa region chosen by amphiphilic score. The peptide with the highest proliferation was PL147 (block 16). We used a peptide identical to PL147 (designated M41) that also generated one of the higher proportions of responders, reinforcing this peptide's immunodominance. An additional peptide from block 15, K29, was repeatedly immunogenic in our study. A recent study on the coast of Kenya used our overlapping MSP1 peptides to evaluate IFN-γ responses by cord blood T cells, reflecting prenatal exposure to malarial antigens (28). Consistent with the findings presented here, the peptides generating IFN-γ responses by the greatest number of newborns' cord blood mononuclear cells were M41, M42, and M43, as well as K29, K30, and K31. The M41 peptide lies in a variable region with 10 of 18 amino acids being different between the FVO and 3D7 alleles, while the K29 peptide is in a semiconserved region with four amino acid differences between the two alleles, supporting the data from Udhayakumar et al. that cellular immune responses to 33 kDa are greatest in regions differing between these two allelic forms of MSP1.
A study by Wipasa et al. (46) used pools of overlapping peptides from the MSP133 region of Plasmodium yoelii to examine the immune responses in mice. They found that two peptide pools induced responses by T cells that, when transferred to naïve immunodeficient (SCID) mice prior to challenge with blood stage P. yoelii, generated partial protection against parasitemia and death. Our study, while not using peptide pools, found the number of participants with IFN-γ responses to MSP142 recombinant proteins to be much higher than the number of responders to individual peptides. These findings suggest that multiple epitopes in the C terminus of MSP1 generate effective T-cell immune responses. With the unavailability of recombinant MSP133 for use in the assays performed here, we were unable to determine if the increased IFN-γ responses to MSP142 were due to additional T-cell epitopes in MSP119. However, in the study done by Udhayakumar et al. (44) the peptides derived from MSP119 elicited weaker T-cell proliferation responses than peptides from MSP133. Also, in a study by Lee et al. (24), fewer than 10% of Kenyan adults demonstrated IFN-γ responses to individual 20-mer peptides from MSP119. Lastly, in a study of five malaria-naïve adults vaccinated with the 3D7 allele of MSP142, interleukin-5 (IL-5) levels from PBMCs stimulated with homologous MSP133 or MSP142 were both higher than in cells stimulated with MSP119 (20). Six additional volunteers were vaccinated with MSP142 FVO, and similarly IL-5 production with MSP142 was higher than with MSP119. The aggregate of evidence from these studies would support our conclusion that the dominant T-cell epitopes lie in the 33-kDa fragment of MSP1, although we did not rule out contribution by N-terminal T-cell epitopes.
Over the 11-month observation period, IFN-γ responses to 3D7 recombinant MSP142 tended to be more prevalent, robust, and stable than those to FVO protein at all monthly time points and over two malaria seasons. This may reflect increased circulating prevalence of the 3D7 variant of MSP133 (which would include the 3D7 MSP142 variant plus the CAMP/FUP variant), the repeated exposure of adults in a region where malaria is holoendemic, or perhaps an increased immunogenicity of the 3D7 MSP133. We were not able to demonstrate any temporal relationship between IFN-γ responses and the prevalence of parasitemia in the same or preceding month as when the ELISPOTs were performed. As this study involved only 24 adults who were asymptomatic, it is unclear whether the IFN-γ responses were sufficient to be protective against malaria-related illness or partially responsible for the observed clearance of blood stage infections (in the absence of antimalarial drugs).
PBMC IFN-γ responses to the MSP133 18-mer peptides did not appear to be stable, due to the relatively low frequency of responders and low magnitude of IFN-γ responses measured by ELISPOT. There were only two individuals who consistently responded to a single peptide, suggesting that overcoming HLA haplotype restriction might improve responses; however, HLA matching a vaccine to individuals would be logistically impossible and prohibitively expensive due to the extreme HLA heterogeneity in African populations (25). In this study, we were not able to determine the HLA class I and II genotypes of the study participants, results which would have allowed us to evaluate the influence of HLA restriction on the stability of IFN-γ-specific peptide responses. However, from our previously published studies we know that there is a high degree of HLA diversity with few dominant alleles and only rare instances of homozygosity (7). Unlike other pathogens such as viruses and bacteria, eukaryotic parasites with complex life cycles and larger genomes display many more antigens and allelic variants of those antigens to the host immune system. This may thus diminish the impact of a single “immunodominant” T-cell epitope commonly used in recall assays, as there may be many such epitopes within and between epitope-variant malaria proteins. Accordingly, determining HLA genotype has less importance for studies using recombinant malaria proteins for vaccine or immunologic profiling studies. In addition, in this study we used overlapping peptides that should be able to be recognized by different HLA types, thereby supporting one of the main points of our study showing the lack of reproducible responses over time.
Other longitudinal studies examining cellular responses to the carboxyl-terminal region of the FVO allele (i.e., Wellcome strain) by individuals who had been mainly infected with parasites with 3D7 (i.e., MAD20) alleles were conducted in the Gambia by Riley et al. (37, 38). Consistent with our findings, lymphoproliferative and IFN-γ responses to MSP1 in adults fluctuated over time and were not associated with parasitemia or malaria transmission season. Their studies of children (3 to 8 years of age), however, showed that lymphoproliferative but not IFN-γ responses (by enzyme-linked immunosorbent assay [ELISA]) before a malaria season were significantly associated with asymptomatic malaria infections, although IFN-γ production did increase with age. Another study of children age 2 to 11 years in Uganda found only 9 of 117 had IFN-γ responses to MSP1. All of the responders were older than 4 years, with no relationship with prior episodes of parasitemia (40). It thus appears that cellular immune responses to MSP133 increase with age and cumulative exposure, similar to humoral immunity to MSP119 (6). Studies examining antibody levels in adults, including one evaluating the same adult population in this paper, showed that titers remain high and stable over time (6, 10, 38). The IFN-γ responses seen here may possibly reflect low-frequency memory T cells involved in protection in conjunction with antibodies. Longitudinal studies of effector and memory T-cell responses and cross-reactivity/cross-protection conferred by sequential allelic infections are ongoing to address the clinical relevance of IFN-γ-mediated responses.
As the 3D7 MSP142 protein antigen is a leading blood stage vaccine candidate, several vaccine trials testing it have been completed (33, 35, 41). In a phase 1 trial determining the safety and immunogenicity of MSP142 formulated in AS02A in malaria-naïve adults, moderate increases in IFN-γ ELISPOT responses were seen in most volunteers after the first but not the second or third immunization (33). Preimmunization IFN-γ responses in malaria-experienced populations are important for establishing baseline values for subsequent vaccine trials, although as we have shown in this study that such responses vary greatly within and between individuals. Similar to our previous cross-sectional studies of pre-erythrocytic-stage malaria peptides (31, 32), while an individual's responses varied between time points, the overall proportions of responders to each allele were similar across each season. Given the large proportion of adults responding to the 3D7 allele in this study, outcome measurements for an MSP1 vaccine tested in areas in which the disease is endemic may have to incorporate a fold increase over baseline and prolonged duration of immune response in order to distinguish between a vaccine-attributable affect and normal variation as the result of natural exposure.
It has long been postulated that malaria may interfere with the generation of IFN-γ-producing memory T cells; thus, repeated natural infections may pose additional hurdles for vaccines in areas in which malaria is endemic. Evidence of this inherent difficulty was shown with another blood stage antigen, apical membrane antigen 1 (AMA1), when tested as a vaccine formulation with the adjuvant system AS02A in malaria-exposed adults in Mali (26). Fifty-five percent of volunteers at baseline had measurable IFN-γ ELISPOT responses, and while there was an increase in IFN-γ production after three vaccinations, there was no statistical difference in outcomes between volunteers who received the AMA1 candidate vaccine and those who received the rabies control vaccine. This phase 1 study did not evaluate efficacy, but it is hoped that future efficacy trials conducted in areas of endemicity will have cellular assay endpoints to help elucidate the role of IFN-γ.
Humoral immune responses to MSP1 will play an essential role in vaccine-induced protection to disease, and with MSP1, antibodies generated to the conformational epitopes of the 19-kDa region are important in inhibition of erythrocyte invasion (3, 21). However, as has been shown in Aotus monkey trials, better immunogenicity and homologous protection were achieved with immunization with recombinant MSP142 than with recombinant MSP119 (9, 42), underscoring the importance of the 33-kDa region. Selecting the appropriate allele(s) of MSP142 to go forward as a vaccine candidate will be dependent upon the role the MSP133 plays in immunity. Our study has shown that the 3D7 MSP133 allele, the most prevalent circulating variant in western Kenya, generates a higher and more consistent IFN-γ response in asymptomatic adults with naturally acquired immunity to malaria. Such IFN-γ production has been shown to be protective in children in Papua New Guinea, with the underlying mechanism postulated to be increased cellular immunity and/or induction of antibody-dependent cell-mediated inhibition (ADCI) (12). Alternatively, the MSP133 variant may, through T-cell help, be the determinant of effective MSP119 humoral responses. It is possible that the 3D7 allele in this regard may not be as desirable, as in the phase 2b vaccine trial of 400 Kenyan children where the 3D7 allele of MSP142 did not demonstrate any protection (35). The ability of a vaccine to induce cross-reactive cellular responses to the multiple alleles of P. falciparum encountered in the field may also be a determining factor in its success. In the study of malaria-naïve adults vaccinated with either the 3D7 or FVO allele of MSP142, there was little IFN-γ produced by PBMCs when they were stimulated with the heterologous allele compared to when they were stimulated with the homologous allele (20). A single blood stage antigen will not likely be sufficient as a stand-alone vaccine; thus, planned studies examining the nature and development of immune responses to MSP1 in Kenyan infants and children, and the involvement of these responses in protection, will be crucial to the development of a successful multistage, multiantigen vaccine candidate that can benefit children as well as adults, particularly as increased global efforts to reduce malaria transmission may reduce the strength and duration of naturally acquired immunity.
Acknowledgments
This work was supported by National Institutes of Health, 1R01-AI43906 (J.W.K. and A.M.M.), 5R01-CA102667 (R.R.), FIC-1D43TW006576 (K.C. and P.O.S.), and T32-AI007427 (M.D.S.).
This work was performed with the permission of the director of the Kenya Medical Research Institute. We thank Jeffrey Bailey and Christopher Yohn for technical assistance with sequencing and alignments. We also thank Fredrick Opinya, John Ogone, and John Oyombe for study participant recruitment and coordinating longitudinal field activities and data collection and Wanda DePasquale for manuscript preparation. We are grateful to the study participants for their contribution.
We declare no competing financial interests.
Editor: J. F. Urban, Jr.
Footnotes
Published ahead of print on 9 August 2010.
REFERENCES
- 1.Ahlborg, N., I. T. Ling, W. Howard, A. A. Holder, and E. M. Riley. 2002. Protective immune responses to the 42-kilodalton (kDa) region of Plasmodium yoelii merozoite surface protein 1 are induced by the C-terminal 19-kDa region but not by the adjacent 33-kDa region. Infect. Immun. 70:820-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beier, J. C., C. N. Oster, F. K. Onyango, J. D. Bales, J. A. Sherwood, P. V. Perkins, D. K. Chumo, D. V. Koech, R. E. Whitmire, and C. R. Roberts. 1994. Plasmodium falciparum incidence relative to entomologic inoculation rates at a site proposed for testing malaria vaccines in western Kenya. Am. J. Trop. Med. Hyg. 50:529-536. [DOI] [PubMed] [Google Scholar]
- 3.Bergmann-Leitner, E. S., E. H. Duncan, and E. Angov. 2009. MSP-1p42-specific antibodies affect growth and development of intra-erythrocytic parasites of Plasmodium falciparum. Malar. J. 8:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackman, M. J., H. G. Heidrich, S. Donachie, J. S. McBride, and A. A. Holder. 1990. A single fragment of a malaria merozoite surface protein remains on the parasite during red cell invasion and is the target of invasion-inhibiting antibodies. J. Exp. Med. 172:379-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackman, M. J., H. Whittle, and A. A. Holder. 1991. Processing of the Plasmodium falciparum major merozoite surface protein-1: identification of a 33-kilodalton secondary processing product which is shed prior to erythrocyte invasion. Mol. Biochem. Parasitol. 49:35-44. [DOI] [PubMed] [Google Scholar]
- 6.Branch, O. H., A. J. Oloo, B. L. Nahlen, D. Kaslow, and A. A. Lal. 2000. Anti-merozoite surface protein-1 19-kDa IgG in mother-infant pairs naturally exposed to Plasmodium falciparum: subclass analysis with age, exposure to asexual parasitemia, and protection against malaria. V. The Asembo Bay Cohort Project. J. Infect. Dis. 181:1746-1752. [DOI] [PubMed] [Google Scholar]
- 7.Cao, K., A. M. Moormann, K. E. Lyke, C. Masaberg, O. P. Sumba, O. K. Doumbo, D. Koech, A. Lancaster, M. Nelson, D. Meyer, R. Single, R. J. Hartzman, C. V. Plowe, J. Kazura, D. L. Mann, M. B. Sztein, G. Thomson, and M. A. Fernandez-Vina. 2004. Differentiation between African populations is evidenced by the diversity of alleles and haplotypes of HLA class I loci. Tissue Antigens 63:293-325. [DOI] [PubMed] [Google Scholar]
- 8.Daly, T. M., and C. A. Long. 1995. Humoral response to a carboxyl-terminal region of the merozoite surface protein-1 plays a predominant role in controlling blood-stage infection in rodent malaria. J. Immunol. 155:236-243. [PubMed] [Google Scholar]
- 9.Darko, C. A., E. Angov, W. E. Collins, E. S. Bergmann-Leitner, A. S. Girouard, S. L. Hitt, J. S. McBride, C. L. Diggs, A. A. Holder, C. A. Long, J. W. Barnwell, and J. A. Lyon. 2005. The clinical-grade 42-kilodalton fragment of merozoite surface protein 1 of Plasmodium falciparum strain FVO expressed in Escherichia coli protects Aotus nancymai against challenge with homologous erythrocytic-stage parasites. Infect. Immun. 73:287-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dent, A. E., K. Chelimo, P. O. Sumba, M. D. Spring, B. S. Crabb, A. M. Moormann, D. J. Tisch, and J. W. Kazura. 2009. Temporal stability of naturally acquired immunity to merozoite surface protein-1 in Kenyan adults. Malar. J. 8:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dent, A. E., C. T. Yohn, P. A. Zimmerman, J. Vulule, J. W. Kazura, and A. M. Moormann. 2007. A polymerase chain reaction/ligase detection reaction fluorescent microsphere assay to determine Plasmodium falciparum MSP-119 haplotypes. Am. J. Trop. Med. Hyg. 77:250-255. [PMC free article] [PubMed] [Google Scholar]
- 12.D'Ombrain, M. C., L. J. Robinson, D. I. Stanisic, J. Taraika, N. Bernard, P. Michon, I. Mueller, and L. Schofield. 2008. Association of early interferon-gamma production with immunity to clinical malaria: a longitudinal study among Papua New Guinean children. Clin. Infect. Dis. 47:1380-1387. [DOI] [PubMed] [Google Scholar]
- 13.Egan, A. F., P. Burghaus, P. Druilhe, A. A. Holder, and E. M. Riley. 1999. Human antibodies to the 19kDa C-terminal fragment of Plasmodium falciparum merozoite surface protein 1 inhibit parasite growth in vitro. Parasite Immunol. 21:133-139. [DOI] [PubMed] [Google Scholar]
- 14.Egan, A. F., J. A. Chappel, P. A. Burghaus, J. S. Morris, J. S. McBride, A. A. Holder, D. C. Kaslow, and E. M. Riley. 1995. Serum antibodies from malaria-exposed people recognize conserved epitopes formed by the two epidermal growth factor motifs of MSP1(19), the carboxy-terminal fragment of the major merozoite surface protein of Plasmodium falciparum. Infect. Immun. 63:456-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Good, M. F. 2001. Towards a blood-stage vaccine for malaria: are we following all the leads? Nat. Rev. Immunol. 1:117-125. [DOI] [PubMed] [Google Scholar]
- 16.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 17.Greenwood, B., and G. Targett. 2009. Do we still need a malaria vaccine? Parasite Immunol. 31:582-586. [DOI] [PubMed] [Google Scholar]
- 18.Hirunpetcharat, C., J. H. Tian, D. C. Kaslow, N. van Rooijen, S. Kumar, J. A. Berzofsky, L. H. Miller, and M. F. Good. 1997. Complete protective immunity induced in mice by immunization with the 19-kilodalton carboxyl-terminal fragment of the merozoite surface protein-1 (MSP1[19]) of Plasmodium yoelii expressed in Saccharomyces cerevisiae: correlation of protection with antigen-specific antibody titer, but not with effector CD4+ T cells. J. Immunol. 159:3400-3411. [PubMed] [Google Scholar]
- 19.Hirunpetcharat, C., P. Vukovic, X. Q. Liu, D. C. Kaslow, L. H. Miller, and M. F. Good. 1999. Absolute requirement for an active immune response involving B cells and Th cells in immunity to Plasmodium yoelii passively acquired with antibodies to the 19-kDa carboxyl-terminal fragment of merozoite surface protein-1. J. Immunol. 162:7309-7314. [PubMed] [Google Scholar]
- 20.Huaman, M. C., L. B. Martin, E. Malkin, D. L. Narum, L. H. Miller, S. Mahanty, and C. A. Long. 2008. Ex vivo cytokine and memory T cell responses to the 42-kDa fragment of Plasmodium falciparum merozoite surface protein-1 in vaccinated volunteers. J. Immunol. 180:1451-1461. [DOI] [PubMed] [Google Scholar]
- 21.John, C. C., R. A. O'Donnell, P. O. Sumba, A. M. Moormann, T. F. de Koning-Ward, C. L. King, J. W. Kazura, and B. S. Crabb. 2004. Evidence that invasion-inhibitory antibodies specific for the 19-kDa fragment of merozoite surface protein-1 (MSP-1 19) can play a protective role against blood-stage Plasmodium falciparum infection in individuals in a malaria endemic area of Africa. J. Immunol. 173:666-672. [DOI] [PubMed] [Google Scholar]
- 22.Jongwutiwes, S., K. Tanabe, and H. Kanbara. 1993. Sequence conservation in the C-terminal part of the precursor to the major merozoite surface proteins (MSP1) of Plasmodium falciparum from field isolates. Mol. Biochem. Parasitol. 59:95-100. [DOI] [PubMed] [Google Scholar]
- 23.Kumar, S., F. Villinger, M. Oakley, J. C. Aguiar, T. R. Jones, R. C. Hedstrom, K. Gowda, J. Chute, A. Stowers, D. C. Kaslow, E. K. Thomas, J. Tine, D. Klinman, S. L. Hoffman, and W. W. Weiss. 2002. A DNA vaccine encoding the 42 kDa C-terminus of merozoite surface protein 1 of Plasmodium falciparum induces antibody, interferon-gamma and cytotoxic T cell responses in rhesus monkeys: immuno-stimulatory effects of granulocyte macrophage-colony stimulating factor. Immunol. Lett. 81:13-24. [DOI] [PubMed] [Google Scholar]
- 24.Lee, E. A., D. R. Palmer, K. L. Flanagan, W. H. Reece, K. Odhiambo, K. Marsh, M. Pinder, M. B. Gravenor, W. A. Keitel, K. E. Kester, C. Diggs, D. Kaslow, V. Apostolopoulos, W. R. Ballou, A. V. Hill, U. Krzych, and M. Plebanski. 2002. Induction of T helper type 1 and 2 responses to 19-kilodalton merozoite surface protein 1 in vaccinated healthy volunteers and adults naturally exposed to malaria. Infect. Immun. 70:1417-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo, M., J. Embree, S. Ramdahin, J. Ndinya-Achola, S. Njenga, J. B. Bwayo, S. Pan, X. Mao, M. Cheang, T. Stuart, R. C. Brunham, and F. A. Plummer. 2002. HLA-A and HLA-B in Kenya, Africa: allele frequencies and identification of HLA-B*1567 and HLA-B*4426. Tissue Antigens 59:370-380. [DOI] [PubMed] [Google Scholar]
- 26.Lyke, K. E., M. Daou, I. Diarra, A. Kone, B. Kouriba, M. A. Thera, S. Dutta, D. E. Lanar, D. G. Heppner, Jr., O. K. Doumbo, C. V. Plowe, and M. B. Sztein. 2009. Cell-mediated immunity elicited by the blood stage malaria vaccine apical membrane antigen 1 in Malian adults: results of a phase I randomized trial. Vaccine 27:2171-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyon, J. A., E. Angov, M. P. Fay, J. S. Sullivan, A. S. Girourd, S. J. Robinson, E. S. Bergmann-Leitner, E. H. Duncan, C. A. Darko, W. E. Collins, C. A. Long, and J. W. Barnwell. 2008. Protection induced by Plasmodium falciparum MSP1(42) is strain-specific, antigen and adjuvant dependent, and correlates with antibody responses. PLoS One 3:e2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malhotra, I., A. N. Wamachi, P. L. Mungai, E. Mzungu, D. Koech, E. Muchiri, A. M. Moormann, and C. L. King. 2008. Fine specificity of neonatal lymphocytes to an abundant malaria blood-stage antigen: epitope mapping of Plasmodium falciparum MSP1(33). J. Immunol. 180:3383-3390. [DOI] [PubMed] [Google Scholar]
- 29.Miller, L. H., D. I. Baruch, K. Marsh, and O. K. Doumbo. 2002. The pathogenic basis of malaria. Nature 415:673-679. [DOI] [PubMed] [Google Scholar]
- 30.Miller, L. H., T. Roberts, M. Shahabuddin, and T. F. McCutchan. 1993. Analysis of sequence diversity in the Plasmodium falciparum merozoite surface protein-1 (MSP-1). Mol. Biochem. Parasitol. 59:1-14. [DOI] [PubMed] [Google Scholar]
- 31.Moormann, A. M., C. C. John, P. O. Sumba, D. Tisch, P. Embury, and J. W. Kazura. 2006. Stability of interferon-gamma and interleukin-10 responses to Plasmodium falciparum liver stage antigen-1 and thrombospondin-related adhesive protein in residents of a malaria holoendemic area. Am. J. Trop. Med. Hyg. 74:585-590. [PubMed] [Google Scholar]
- 32.Moormann, A. M., P. O. Sumba, D. J. Tisch, P. Embury, C. H. King, J. W. Kazura, and C. C. John. 2009. Stability of interferon-gamma and interleukin-10 responses to Plasmodium falciparum liver stage antigen 1 and thrombospondin-related adhesive protein immunodominant epitopes in a highland population from Western Kenya. Am. J. Trop. Med. Hyg. 81:489-495. [PMC free article] [PubMed] [Google Scholar]
- 33.Ockenhouse, C. F., E. Angov, K. E. Kester, C. Diggs, L. Soisson, J. F. Cummings, A. V. Stewart, D. R. Palmer, B. Mahajan, U. Krzych, N. Tornieporth, M. Delchambre, M. Vanhandenhove, O. Ofori-Anyinam, J. Cohen, J. A. Lyon, and D. G. Heppner. 2006. Phase I safety and immunogenicity trial of FMP1/AS02A, a Plasmodium falciparum MSP-1 asexual blood stage vaccine. Vaccine 24:3009-3017. [DOI] [PubMed] [Google Scholar]
- 34.O'Donnell, R. A., T. F. de Koning-Ward, R. A. Burt, M. Bockarie, J. C. Reeder, A. F. Cowman, and B. S. Crabb. 2001. Antibodies against merozoite surface protein (MSP)-1(19) are a major component of the invasion-inhibitory response in individuals immune to malaria. J. Exp. Med. 193:1403-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogutu, B. R., O. J. Apollo, D. McKinney, W. Okoth, J. Siangla, F. Dubovsky, K. Tucker, J. N. Waitumbi, C. Diggs, J. Wittes, E. Malkin, A. Leach, L. A. Soisson, J. B. Milman, L. Otieno, C. A. Holland, M. Polhemus, S. A. Remich, C. F. Ockenhouse, J. Cohen, W. R. Ballou, S. K. Martin, E. Angov, V. A. Stewart, J. A. Lyon, D. G. Heppner, and M. R. Withers. 2009. Blood stage malaria vaccine eliciting high antigen-specific antibody concentrations confers no protection to young children in Western Kenya. PLoS One 4:e4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richie, T. L., and A. Saul. 2002. Progress and challenges for malaria vaccines. Nature 415:694-701. [DOI] [PubMed] [Google Scholar]
- 37.Riley, E. M., S. J. Allen, J. G. Wheeler, M. J. Blackman, S. Bennett, B. Takacs, H. J. Schonfeld, A. A. Holder, and B. M. Greenwood. 1992. Naturally acquired cellular and humoral immune responses to the major merozoite surface antigen (PfMSP1) of Plasmodium falciparum are associated with reduced malaria morbidity. Parasite Immunol. 14:321-337. [DOI] [PubMed] [Google Scholar]
- 38.Riley, E. M., S. Morris-Jones, M. J. Blackman, B. M. Greenwood, and A. A. Holder. 1993. A longitudinal study of naturally acquired cellular and humoral immune responses to a merozoite surface protein (MSP1) of Plasmodium falciparum in an area of seasonal malaria transmission. Parasite Immunol. 15:513-524. [DOI] [PubMed] [Google Scholar]
- 39.Snounou, G., and B. Singh. 2002. Nested PCR analysis of Plasmodium parasites. Methods Mol. Med. 72:189-203. [DOI] [PubMed] [Google Scholar]
- 40.Ssewanyana, I., C. Pietras, C. A. Baker, F. Nghania, N. G. Jones, P. J. Rosenthal, G. Dorsey, and H. Cao. 2008. Pattern of malaria-specific T-cell responses in a cohort of Ugandan children. J. Trop. Pediatr. 54:6-13. [DOI] [PubMed] [Google Scholar]
- 41.Stoute, J. A., J. Gombe, M. R. Withers, J. Siangla, D. McKinney, M. Onyango, J. F. Cummings, J. Milman, K. Tucker, L. Soisson, V. A. Stewart, J. A. Lyon, E. Angov, A. Leach, J. Cohen, K. E. Kester, C. F. Ockenhouse, C. A. Holland, C. L. Diggs, J. Wittes, and D. G. Heppner, Jr. 2007. Phase 1 randomized double-blind safety and immunogenicity trial of Plasmodium falciparum malaria merozoite surface protein FMP1 vaccine, adjuvanted with AS02A, in adults in western Kenya. Vaccine 25:176-184. [DOI] [PubMed] [Google Scholar]
- 42.Stowers, A. W., V. Cioce, R. L. Shimp, M. Lawson, G. Hui, O. Muratova, D. C. Kaslow, R. Robinson, C. A. Long, and L. H. Miller. 2001. Efficacy of two alternate vaccines based on Plasmodium falciparum merozoite surface protein 1 in an Aotus challenge trial. Infect. Immun. 69:1536-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takala, S. L., D. Coulibaly, M. A. Thera, A. Dicko, D. L. Smith, A. B. Guindo, A. K. Kone, K. Traore, A. Ouattara, A. A. Djimde, P. S. Sehdev, K. E. Lyke, D. A. Diallo, O. K. Doumbo, and C. V. Plowe. 2007. Dynamics of polymorphism in a malaria vaccine antigen at a vaccine-testing site in Mali. PLoS Med. 4:e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Udhayakumar, V., D. Anyona, S. Kariuki, Y. P. Shi, P. B. Bloland, O. H. Branch, W. Weiss, B. L. Nahlen, D. C. Kaslow, and A. A. Lal. 1995. Identification of T and B cell epitopes recognized by humans in the C-terminal 42-kDa domain of the Plasmodium falciparum merozoite surface protein (MSP)-1. J. Immunol. 154:6022-6030. [PubMed] [Google Scholar]
- 45.WHO. 2005. The world malaria report 2005, prepared by Roll Back Malaria (RBM) Department, United Nations Children's Fund (UNICEF) and WHO. World Health Organization, Geneva, Switzerland. http://rbm.who.int/wmr2005/.
- 46.Wipasa, J., C. Hirunpetcharat, Y. Mahakunkijcharoen, H. Xu, S. Elliott, and M. F. Good. 2002. Identification of T cell epitopes on the 33-kDa fragment of Plasmodium yoelii merozoite surface protein 1 and their antibody-independent protective role in immunity to blood stage malaria. J. Immunol. 169:944-951. [DOI] [PubMed] [Google Scholar]