Abstract
The Burkholderia cepacia complex (Bcc) is a group of genetically related environmental bacteria that can cause chronic opportunistic infections in patients with cystic fibrosis (CF) and other underlying diseases. These infections are difficult to treat due to the inherent resistance of the bacteria to antibiotics. Bacteria can spread between CF patients through social contact and sometimes cause cepacia syndrome, a fatal pneumonia accompanied by septicemia. Burkholderia cenocepacia has been the focus of attention because initially it was the most common Bcc species isolated from patients with CF in North America and Europe. Today, B. cenocepacia, along with Burkholderia multivorans, is the most prevalent Bcc species in patients with CF. Given the progress that has been made in our understanding of B. cenocepacia over the past decade, we thought that it was an appropriate time to review our knowledge of the pathogenesis of B. cenocepacia, paying particular attention to the characterization of virulence determinants and the new tools that have been developed to study them. A common theme emerging from these studies is that B. cenocepacia establishes chronic infections in immunocompromised patients, which depend more on determinants mediating host niche adaptation than those involved directly in host cells and tissue damage.
Burkholderia cenocepacia is a motile, rod-shaped, metabolically diverse Gram-negative betaproteobacterium (168, 169) that is widespread in the environment, particularly within the rhizosphere (8), and is also an opportunistic pathogen causing chronic lung infections in patients with cystic fibrosis (CF) as well as other immunocompromised patients (169). Using recA sequencing and multilocus sequence typing, B. cenocepacia isolates can be subdivided into four distinct lineages, IIIA, IIIB, IIIC, and IIID (168). To date the majority of clinical isolates belong to the IIIA, IIIB, and IIID lineages (4, 96, 110, 168). Isolates from the IIIB and IIIC lineages may be readily cultivated from the natural environment (8, 96, 127, 168). However, even in the absence of culturable IIIA and IIID lineage bacteria from soil, members of these lineages can be detected in soil using non-culture-based methods (127), suggesting that they may also be present in soil but in low abundance. B. cenocepacia is one of at least 17 phenotypically similar species known as the Burkholderia cepacia complex (Bcc) (168-171). Although almost all the Bcc species have been isolated from CF patients, B. cenocepacia was initially the species most commonly isolated from patients with CF (97, 153) and associated with epidemic spread between CF patients (96). For these reasons, B. cenocepacia was the main focus of research groups studying the molecular biology, pathogenesis, and antibiotic resistance of Bcc bacteria. However, in recent years, Burkholderia multivorans has overtaken B. cenocepacia as the most common Bcc isolate in American and United Kingdom CF patients (102, 133). Some B. multivorans strains are widely distributed and have been associated with outbreaks (9). This article reviews our current understanding of the virulence determinants of B. cenocepacia as well as the tools developed to study them. Since Bcc bacteria are resistant to many clinically useful antibiotics (1, 22, 57, 118, 163), the study of virulence determinants is important for identifying bacterial processes that could be targeted by novel antibiotics or alternative anti-infective therapies.
(A portion of this work appears in S.A.L.'s Ph.D. thesis.)
Burkholderia in the environment.
Burkholderia sp. live in diverse ecological niches, often in either beneficial or pathogenic relationships with other organisms (for a recent review, see the work of Compant et al. [36]). Burkholderia spp. have been described as plant pathogens (7, 21, 72), symbiotic rhizospheric or endophytic plant growth promoters (35, 130), endosymbionts of fungi (5, 56, 70) and insects (77, 144), and animal pathogens (31, 59). They can degrade pollutants (25, 30, 83, 84, 147), fix nitrogen and solubilize metals for use by their symbiotic partners (25, 73), produce compounds that protect their host-associated partners from pathogenic fungi, bacteria, protozoa, and nematodes (26, 111, 114), and even induce plant host defense mechanisms (37). B. cenocepacia can be associated with plants, including onions, sugarcane, maize, wheat, and legumes (8, 96, 112). Conceivably, such diverse biological interactions exert selective pressure, giving rise to highly adaptable bacteria. In turn, this ability to adapt to different conditions could contribute significantly to the antibiotic resistance and pathogenesis of Burkholderia spp., including B. cenocepacia. Indeed, many factors discussed below that are required for B. cenocepacia pathogenesis have more to do with adaptation for survival under changing conditions (e.g., metabolic pathways, host antimicrobial molecule resistance mechanisms, and regulatory proteins required for the control of bacterial stress responses), which is likely necessary for establishing chronic infections, than with mounting a potent acute infection.
Genetics of B. cenocepacia.
Burkholderia species have some of the largest, most complex bacterial genomes described to date (93, 105). They are high in percent G+C content, characterized by a multireplicon structure, and possess numerous gene duplications, insertion sequences, and mobile elements. These elements are thought to contribute to the plasticity of Burkholderia genomes and their ability to acquire a wide range of metabolic pathways (93). Burkholderia genomes can also mutate rapidly when the organisms are subject to in vitro stress conditions or during infections (46, 125, 128). The genome sequence of B. cenocepacia strain J2315 was published in 2009 (65), although the Wellcome Trust Sanger Institute made initial sequencing and annotation of the genome available to researchers since 2000. J2315 is a member of the electrophoretic type 12 lineage of strains that caused transmissible infections in CF patients in Canada, the United Kingdom, and Europe (58, 103, 168); strain J2315 is also a member of the IIIA phylogenetic lineage of B. cenocepacia (65, 168). The genome has over 8 Mbp and consists of three circular chromosomes and a plasmid (65). Most studies of B. cenocepacia conducted utilize strain J2315 or another CF clinical isolate, strain K56-2, which is clonally related to J2315 by pulsed-field gel electrophoresis (104) and is a double-locus variant by multilocus sequence typing (65). In this case, the available genome sequence of J2315 is applied to K56-2; however, there are some differences between these two strains, including differences in lipopolysaccharide (LPS) structure (120), pigment production (75), and interactions with eukaryotic cells (142). Strain K56-2 has often been used because it has lower resistance to antimicrobial agents than J2315, which facilitates genetic selection and makes it more amenable to genetic manipulation. The U.S. Department of Energy Joint Genome Institute has sequenced the genomes of three other B. cenocepacia strains from lineage IIIB. They include one isolate from the blood of a CF patient with cepacia syndrome and two rhizosphere isolates. These strains may be useful for genomic comparisons to IIIA lineage strains. The Burkholderia Genome Database (http://www.burkholderia.com) provides users with an excellent resource for annotations of genomes and comparative genome analysis and includes all of the sequenced B. cenocepacia genomes as well as genomes from other Burkholderia spp. (177). For the genes described below that contribute to virulence in B. cenocepacia, we have listed in Table 1 the systematic gene numbers from B. cenocepacia strain J2315.
TABLE 1.
B. cenocepacia strain J2315 systematic gene names for virulence determinants described in this review
| Gene or function | Systematic gene name(s) in J2315 |
|---|---|
| rpoE | BCAL0998 and BCAL2872 (two copies) |
| rpoN | BCAL0813 |
| htrA | BCAL2829 |
| rpoN-EBP | BCAL1536 |
| cepRI | BCAM1868/BCAM1870 |
| aidA | BCAS0293 |
| cciRI | BCAM0239a/BCAM0240 |
| cepR2 | BCAM0188 |
| BDSFa synthase (rpfF) | BCAM0581 |
| shvR | BCAS0225 |
| atsR | BCAM0379 |
| Type 3 secretion system | BCAM2045 to BCAM2057 |
| Type 4 secretion systems | pBCA020 to pBCA059 (virulence related) BCAM0324 to BCAM0340 (plasmid mobilization) |
| Type 6 secretion system | BCAL0337 to BCAL0351 |
| zmpA | BCAS0409 |
| zmpB | BCAM2307 |
| katA | BCAM2107 |
| katB | BCAL3299 |
| sodC | BCAL2643 |
| hppD | BCAL0207 |
| Ornibactin synthesis | BCAL1688 to BCAL1702 |
| LPS core oligosaccharideb | |
| hldA | BCAL2945 |
| waaC | BCAL3112 |
| Ara4N biosynthesis | BCAL1929 to BCAL1935 |
| Flagellumb | |
| fliC | BCAL0114 |
| fliJ | BCAL0521 |
| Cable pili | BCAM2756 to BCAM2762 |
| acp | BCAL0995 and BCAL2875 (two copies) |
| mgtC | BCAM1867 |
| Phenylacetic acid catabolismb | |
| paaE | BCAL0212 |
| paaA | BCAL0216 |
| amiI | BCAM0265 |
| opcI | BCAM0267 |
| pbr | Not present |
BDSF, cis-2-dodecenoic acid, diffusible, nonhomoserine lactone signal molecule.
Numerous genes located in multiple clusters are required for these determinants; those listed have been mutated for characterization of virulence determinants.
Genetic tools for use with B. cenocepacia.
Genetic manipulation of Burkholderia strains is challenging, and this initially hampered rapid progress in the field. However, a series of constantly improving mutagenesis systems specifically tailored for B. cenocepacia were developed over the last decade. Most of these systems employed a strategy in which nonreplicative plasmids are integrated into specific locations in the chromosome via homologous recombination, resulting in polar (51), nonpolar (53), or conditional (122) mutations. Recently, five systems for the creation of unmarked gene deletions have been published for Burkholderia spp., including B. cenocepacia (12, 33, 52, 61, 98). These systems allow the creation of multiple deletions within a strain and can also be used to integrate DNA into heterologous locations in the chromosome for cis complementation experiments and other applications. Other genetic tools for the study of B. cenocepacia include transposon mutagenesis (28, 68), microarray technology (47), subtractive hybridization (16), and high-throughput sequencing of RNA (RNA-seq) for quantification of transcriptional responses (179). Together, these tools facilitate much more rapid and refined manipulation of B. cenocepacia today than was possible a decade ago.
In vivo and ex vivo models of infection.
In vivo and ex vivo models have been used to study the ability of B. cenocepacia to cause disease. The most widely used animal model for studies of members of the Bcc is the rat agar bead model of chronic lung infection that was originally developed for Pseudomonas aeruginosa infections (29). In this model, bacteria are embedded in agar beads, which are then inserted into rat lungs via the trachea, where they establish a chronic but nonlethal infection (29). Similar experiments using Bcc bacteria in agar beads have also been conducted in mice (159). A mouse model of chronic granulomatous disease, another condition in which patients are susceptible to opportunistic Bcc infections (176), has also been established for the study of Bcc virulence (148). Three simpler, less expensive animal models, the nematode Caenorhabditis elegans (82), Galleria mellonella moth larvae (146), and zebra fish (44) models, have also been used for the study of Bcc pathogenicity. As members of the Bcc are also plant pathogens, plant models developed for measuring the virulence of Bcc bacteria include the alfalfa seedling (15) and onion tissue (71) models.
The ability of B. cenocepacia to infect and survive in macrophages, both with and without a functional cystic fibrosis transmembrane conductance regulator (CFTR) protein (86, 88), epithelial cells (140, 145), dendritic cells (101), neutrophils (23, 24, 141), and amoeba (87), has been investigated; and these models have also been used for the study of putative virulence factors (reviewed in reference 143). A common theme that emerges from these studies is that after phagocytosis, the internalized B. cenocepacia cells delay the maturation of bacterium-containing vacuoles. In macrophages, the maturation delay results in reduced acidification of the lumen of bacterium-containing vacuoles and inhibition of the assembly of the NADPH oxidase complex on the vacuolar membrane, and these phenotypes are greatly exaggerated in the absence of a functional CFTR protein (74, 86, 88, 137). The B. cenocepacia organinisms in infected epithelial cells also interfere with the normal endocytic pathway, but intracellular bacteria eventually enter the endoplasmic reticulum, where they replicate. In dendritic cells, engulfed B. cenocepacia cells can alter cell function and induce necrosis (101). Studies of tissue culture systems have been hindered by the inability to complete traditional gentamicin protection assays due to bacterial resistance to gentamicin. However, new strains that are gentamicin sensitive but otherwise behave as the wild type during macrophage infections were recently constructed (60) and should facilitate these types of experiments.
VIRULENCE DETERMINANTS OF B. CENOCEPACIA
Several virulence determinants that may play a role in the ability of B. cenocepacia to cause disease have been proposed (Fig. 1). A signature tagged mutagenesis study of B. cenocepacia using the rat agar bead model of chronic lung infection identified over 100 genes that were required by B. cenocepacia for survival in this model (68). All three chromosomes and the plasmid possessed genes required for B. cenocepacia survival. These genes encoded proteins with diverse predicted functions, including regulatory, transport, and metabolism proteins, proteins involved in cell surface biogenesis, and conserved proteins of unknown function (68).
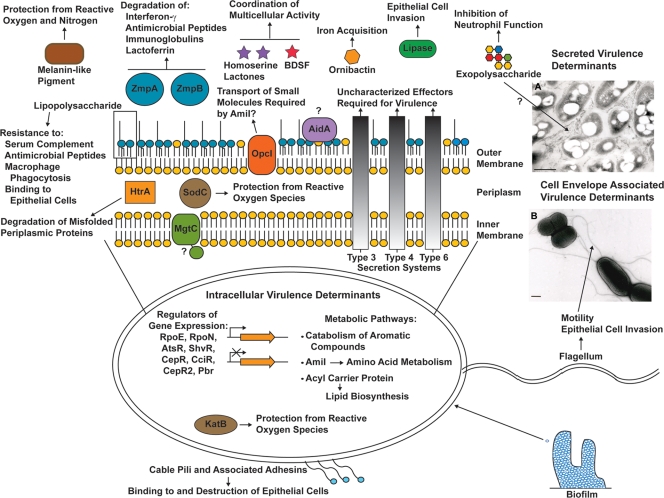
FIG. 1.
Representation of the localization and known functions of B. cenocepacia virulence determinants. All of the established and proposed virulence determinants are discussed in the appropriate subsections of this review. BDSF denotes cis-2-dodecenoic acid, diffusible, nonhomoserine lactone signal molecule. Virulence determinants with proposed or unknown functions are denoted with question marks. (Inset A) Bacteria fixed in the presence of alcian blue and stained with uranyl acetate and lead citrate to visualize the extracellular matrix produced by bacterial cells from the virulent rough colony morphology phenotype described by Bernier et al. (14). The exact composition of this extracellular matrix is unknown but is proposed to consist of exopolysaccharide and possibly lipopolysaccharide and proteins (14). (Inset B) Bacterial cells negatively stained with uranyl acetate to visualize flagella. Bars, 0.5 μm. (Electron micrographs courtesy of Maria Soledad Saldías, reproduced with permission.)
Putative virulence determinants were also identified through comparative transcriptomic studies using either microarray or high-throughput sequencing of RNA from closely related soil and clinical isolates of B. cenocepacia from the IIIB lineage, which were grown under conditions mimicking soil and CF sputum (179, 180). Similar microarray experiments were conducted with the IIIA lineage strain J2315 (47). These studies revealed hundreds of genes that were differentially regulated under conditions mimicking CF sputum. However, there was little convergence between the data obtained for CF isolates from the two different lineages, with only nine genes being differentially regulated in a similar fashion in the two studies (47, 179). This could be due to differences in experimental procedures or the result of important differences between strains of B. cenocepacia. The latter possibility is conceivable, since several virulence properties, described in more detail below, are strain specific. Therefore, we have listed in Table 2 virulence determinants of B. cenocepacia that have been characterized in multiple infection models and in Table 3 those that have been tested in only single infection models.
TABLE 2.
Virulence determinants tested in multiple infection models
| Virulence determinant | Mutant phenotype(s) |
References | |||
|---|---|---|---|---|---|
| Vertebrate models | Invertebrate models | Plant models | Eukaryotic cell culture models | ||
| Ornibactin siderophore | 3-log decrease in bacterial burden, rat agar bead model | Decreased virulence in C. elegans model, avirulent in G. mellonella model | Full virulence in alfalfa seedling model | No data | 150, 164 |
| LPS core oligosaccharide | Avirulent in rat agar bead model | Decreased virulence in C. elegans and G. mellonella models | Full virulence in alfalfa seedling model | Does not survive in murine macrophages | 99, 121, 164 |
| CepRI quorum-sensing system | Less lung tissue inflammation in rat agar bead model, decreased virulence after intranasal infection in Cftr−/− mice | Decreased virulence in C. elegans model (strain independent), decreased virulence in G. mellonella model (strain dependent) | Decreased virulence in alfalfa seedling model (strain dependent) | No data | 82, 151, 164 |
| CciRI quorum-sensing system | Less lung tissue inflammation, rat agar bead model | Full virulence in C. elegans and G. mellonella models | Full virulence in alfalfa seedling model | No data | 10, 164 |
| HtrA protease | Avirulent in rat agar bead model | Full virulence in C. elegans and G. mellonella models | Full virulence in alfalfa seedling model | No data | 51, 164 |
| AidA | Virulent in rat agar bead model | Decreased virulence in C. elegans model, full virulence in G. mellonella model | Virulent in alfalfa seedling model | No data | 66, 164 |
| BDSFa | Decreased virulence in zebra fish | Decreased virulence in G. mellonella model | No data | Does not inhibit C. albicans germ tube formation | 19, 44, 136 |
| Type 3 secretion system | 3-log decrease in bacterial burden in rat agar bead model | Decreased virulence in C. elegans model, full virulence in G. mellonella model | Full virulence in alfalfa seedling model | Virulent in murine macrophages | 86, 159, 164 |
| Type 4 secretion system | No data | No data | Does not cause tissue destruction in onions | Decreased virulence in airway epithelial and macrophage cells | 48, 138 |
| Type 6 secretion system | Decreased virulence in rat agar bead model | No data | No data | Decreased survival in the presence of amoeba | 6, 68 |
| ZmpA protease | 3- to 4-log decrease in bacterial burden, rat agar bead model (strain dependent) | Full virulence in C. elegans and G. mellonella models | Virulent in alfalfa seedling model with decreased bacterial burden | No data | 15, 40, 81, 164 |
| ZmpB protease | Less lung tissue inflammation in rat agar bead model | Full virulence in C. elegans and G. mellonella models | Full virulence in alfalfa seedling model | No data | 81, 164 |
| ShvR regulator | Less lung tissue inflammation in rat agar bead model | Full virulence in C. elegans and G. mellonella models | Avirulent in alfalfa seedling model | No data | 14, 164 |
| OpcI outer membrane porin | Less lung tissue inflammation in rat agar bead model | Full virulence in C. elegans and G. mellonella models | Full virulence in alfalfa seedling model | No data | 10, 164 |
| MgtC | Decreased virulence in rat agar bead model | No data | No data | Does not survive in murine macrophages | 68, 107 |
| Phenylacetic acid catabolic pathway | Decreased virulence in rat agar bead model | Decreased virulence in C. elegans model | No data | No data | 68, 90 |
| Cable pili | No data | Decreased virulence in G. mellonella model | No data | Does not kill airway epithelial cells | 32, 136 |
| Flagellum | Avirulent in mouse agar bead model | Decreased virulence in G. mellonella model | No data | Reduced invasion of epithelial cells | 136, 160, 165 |
BDSF, cis-2-dodecenoic acid, diffusible, nonhomoserine lactone signal molecule.
TABLE 3.
Virulence determinants tested in only one infection model
| Virulence determinant | Mutant phenotype in selected virulence model | Reference |
|---|---|---|
| RpoE alternative sigma factor | Does not survive in murine macrophages | 53 |
| RpoN alternative sigma factor | Does not survive in murine macrophages | 141 |
| RpoN enhancer binding protein | Decreased virulence in rat agar bead model | 68 |
| Secreted lipase | Decreased epithelial cell invasion | 116 |
| KatB catalase/ peroxidase | Decreased survival in murine macrophages | —a |
| SodC superoxide dismutase | Does not survive in murine macrophages | 76 |
| Melanin-like pigment | Does not survive in murine macrophages | 75 |
| Pbr regulator | Decreased virulence in C. elegans | 131 |
| AmiI amidase | Decrease (1 log unit) in bacterial burden in rat agar bead infections | 10 |
| Acyl carrier protein | Decreased virulence in C. elegans | 152 |
—, K. E. Keith and M. A. Valvano, unpublished results.
Alternative sigma factors and related proteins.
Alternative sigma factors are regulatory proteins that activate transcription of particular gene subsets by binding at sites within promoter regions and interacting with the RNA polymerase complex, to allow the initiation of transcription (2). Two alternative sigma factors, RpoE and RpoN, are required for the ability of engulfed B. cenocepacia to delay phagolysosomal fusion in murine macrophages (53, 141).
RpoE is also required for the ability of B. cenocepacia to grow under conditions of high osmolarity and high temperature (53). In Gram-negative bacteria, RpoE mediates gene activation in response to extracytoplasmic stress (2), and individual components of the extracytoplasmic stress response are also likely to be important for the virulence of B. cenocepacia. These include HtrA-like periplasmic proteases, of which B. cenocepacia is predicted to encode four (51). Under conditions of extracytoplasmic stress, HtrA can act either as a chaperone to aid in the refolding of misfolded periplasmic proteins or as a protease to degrade misfolded proteins (154). Mutation of one of the HtrA proteases (BCAL2829) of B. cenocepacia resulted in a strain that is avirulent in the rat agar bead infection model and has increased sensitivity to osmotic and thermal stress (51).
RpoN is necessary for B. cenocepacia motility and biofilm formation (141). In other bacteria, RpoN activates the expression of genes necessary for a wide range of functions, and unlike the activation of other sigma factors, this activation also requires an enhancer binding protein (113). A mutant defective in 1 of the 20 predicted enhancer binding proteins (BCAL1536) of B. cenocepacia is attenuated in the rat agar bead infection model (68).
Quorum sensing.
Quorum sensing allows regulation of gene expression on the basis of the density of the bacterial population. Bacteria secrete compounds that accumulate outside the cell, and once sufficient cell densities are reached, the concentration of the diffusible compound reaches a threshold and the bacteria begin to alter gene expression (54). These systems represent a way for bacterial cells to communicate with one another and coordinate multicellular behavior.
The cepRI quorum-sensing system, which mediates production of N-octanoylhomoserine lactone, regulates numerous functions in B. cenocepacia. Initial studies showed that disruption of the system leads to increased siderophore production and decreased extracellular protease and lipase activities (94). Further studies showed that this system was required for motility (95), biofilm stability (162), virulence in the rat agar bead model of chronic lung infection (151), and killing of C. elegans (82). A random promoter library screen identified almost 90 B. cenocepacia promoters regulated by CepR (157), and evidence also suggests that the cepRI system could potentially be involved in cross-species communication with P. aeruginosa (95). One molecule that is controlled by the cepRI system and that has been implicated in virulence is AidA, a surface protein required for the killing of C. elegans by an unknown mechanism (66).
B. cenocepacia also possesses genes for a second homoserine lactone-producing quorum-sensing system, designated cciRI, and an orphan regulator, designated cepR2, that is not encoded by a homoserine lactone synthase gene (108, 109). These systems also contribute to the regulation of many of the same functions as cepRI, though often in reverse to the regulation by cepRI, suggesting a complex network of gene regulation in response to bacterial cell density (108, 109, 119, 157).
Bacterial quorum-sensing systems can also utilize nonhomoserine lactone compounds, an example of which is the diffusible signal factor of Xanthomonas campestris (63). B. cenocepacia has one such system and produces cis-2-dodecenoic acid in a cell density-dependent manner (19). This diffusible signal molecule alters many of the same functions controlled by cepRI, cciRI, and cepR2; mutants unable to synthesize cis-2-dodecenoic acid have decreased motility, biofilm synthesis, and virulence in the G. mellonella moth larvae and zebra fish infection models (19, 44, 136). This molecule may also be involved in communication between B. cenocepacia and other bacteria as well as in interkingdom communication with Candida albicans (19).
Biofilms.
Biofilms are complex, multicellular bacterial communities that can protect bacteria from antibiotics and the host immune system (50). Bcc bacteria are thought to live in CF lungs in biofilms, including mixed biofilms with P. aeruginosa, where they may even communicate with P. aeruginosa via quorum-sensing systems (134, 161). B. cenocepacia can also form biofilms in vitro (39), and the various genes required for the formation of these multicellular bacterial aggregates have been identified (67). Biofilm formation can be affected by multiple gene regulation systems, including quorum sensing (162), the alternative sigma factor RpoN (141), ShvR, a LysR-type regulator (14), and AtsR, a hybrid sensor kinase-response regulator that acts as a negative regulator of multiple virulence properties (6). Biofilm formation is also affected by exopolysaccharide synthesis (41), motility (67), and iron availability (13). There are conflicting reports concerning the increased antibiotic resistance in B. cenocepacia growing in biofilms compared to that of planktonic cells, with some studies showing increases in antibiotic resistance in biofilms (27, 45). However, another study showed little difference in antibiotic resistance between bacteria grown planktonically and those grown in biofilms (126). Together, these attributes indicate that biofilm formation is a complex process involving numerous B. cenocepacia virulence determinants. This makes the disruption of biofilm growth an attractive target for the development of new antibiotics, and many quorum-sensing inhibitors that are being developed also inhibit biofilm formation (124).
Secretion systems.
Many pathogenic bacteria employ specialized systems for the secretion of effector molecules that contribute to cause disease by disrupting host cellular processes (55). Various secretion systems have been disrupted in B. cenocepacia to investigate their relevance to virulence. The type III secretion system (T3SS) is required by B. cenocepacia for survival during murine agar bead infections (159). In this study, the average number of bacteria in the spleens and lungs of mice infected with a T3SS mutant was reduced by 2 and 3 log units, respectively, compared to the number of bacteria in mice infected with the wild-type strain (159). One of the two predicted type IV secretion systems of B. cenocepacia is necessary for causing disease in onions (48) and for intracellular survival in epithelial cells and macrophages (138), while the other is involved in plasmid mobilization (181). The type VI secretion system (T6SS) was recently identified in a number of organisms, including B. cenocepacia, where its expression is negatively regulated by the sensor kinase-response regulator AtsR (6). The T6SS is also required for protection from predation by the amoeba Dictyostelium discoideum (6). Furthermore, macrophages infected with B. cenocepacia cells overexpressing the T6SS make actin-rich cellular projections that can harbor bacterium-containing vacuoles (6). Finally, the T6SS also plays a role in infection, as three independent mutants in the T6SS gene cluster were attenuated in the rat agar bead model (68).
Secreted proteins.
B. cenocepacia produces two distinct extracellular zinc metalloproteases, named ZmpA and ZmpB (40, 81). Disruption of zmpA in one of two strains of B. cenocepacia resulted in a mutant that was much less persistent in the rat agar bead model, and many rats cleared the mutant by day 14 after infection (40). However, the second strain of B. cenocepacia did not require ZmpA for virulence in this model, indicating strain differences in terms of the requirement for virulence determinants (40). A B. cenocepacia zmpB mutant persisted in the rat agar bead model at numbers similar to those of the wild-type strain. However, rats infected with the zmpB mutant have decreased lung tissue inflammation, based on the histopathology at 14 days postinfection (81). Purified, recombinant versions of both of these proteins can degrade lactoferrin, type IV collagen, immunoglobulins, and antimicrobial peptides, such as human β-defensin-1 and LL-37 in vitro (79-81).
Secreted lipases have also been implicated in the ability of B. cenocepacia to cause disease. Mullen et al. (116) showed that Bcc species, including B. cenocepacia, secrete lipases and that pretreatment of epithelial cells with commercially available B. cepacia lipase resulted in roughly 50% more invasion of epithelial cells by B. cenocepacia, while pretreatment of B. cenocepacia with a lipase inhibitor decreased the level of invasion.
Colony variants.
B. cenocepacia can produce different colony morphologies, at least one of which, the shiny colony morphology variant (or shv), results in bacteria that are avirulent in the alfalfa model of infection (14). These isolates can still establish chronic infections in the rat agar bead model, but the average number of viable bacteria per lung in the infected rats decreased by approximately 1 log unit compared to the parental strain number, and infection with the shv isolate resulted in decreased inflammation. Switching to the shv morphology has pleiotropic effects on the bacteria, with decreases in biofilm formation, motility, and production of extracellular matrices and siderophores being found. The extracellular matrix made by the virulent, rough colony morphology isolates is shown in Fig. 1A. Some of the shv isolates arise due to a spontaneous mutation in a LysR-type regulator (ShvR), while others do not have this spontaneous mutation, suggesting that there are multiple pathways that B. cenocepacia can take to arrive at the shv phenotype (14).
Resistance to oxidative stress.
Phagocytic cells produce reactive oxygen species to help eliminate bacteria, and B. cenocepacia has numerous mechanisms to resist oxidative stress. Multiple strains of B. cenocepacia possess in vitro catalase, peroxidase, and superoxide dismutase activities (92). Two catalase/peroxidases with different functions were identified in B. cenocepacia: KatB, the major catalase/peroxidase of B. cenocepacia, and KatA, a secondary catalase/peroxidase required for resistance to hydrogen peroxide under conditions of iron limitation and growth in the presence of carbon sources metabolized through the tricarboxylic acid cycle (91). Also, B. cenocepacia possesses a periplasmic superoxide dismutase, SodC, required for resistance to extracellular superoxide, and a B. cenocepacia sodC mutant is more rapidly killed, in an NADPH oxidase-dependent fashion, by murine macrophages than the wild-type strain (76). Similar results were observed for a melanin-like pigment expressed by strains of B. cenocepacia that belong to the IIIA lineage (75). Disruption of hppD, a gene encoding an enzyme required for production of the pigment, resulted in a nonpigmented strain with increased sensitivity to extracellular hydrogen peroxide and superoxide and decreased survival in murine macrophages (75). Inhibition of either NAPDH oxidase or inducible nitric oxide synthase resulted in increased survival of the mutant during macrophage infections (75), suggesting that this pigment protects B. cenocepacia from both reactive oxygen and reactive nitrogen species.
Iron acquisition.
The ability to grow under conditions of iron limitation is an important characteristic for pathogens because the host is extremely limited in the amount of freely available iron that it has (115). Members of the Bcc can synthesize four different siderophores (pyochelin, ornibactin, cepaciachelin, and cepabactin) for iron chelation and uptake (158). The predominant siderophore produced by most strains of B. cenocepacia appears to be ornibactin, while some strains also synthesize small amounts of pyochelin (42, 158). The synthesis and uptake pathways of ornibactin and their regulation have been well characterized in B. cenocepacia (3, 94, 149, 150), and this siderophore is required for virulence in the rat agar bead, G. mellonella, and C. elegans infection models (150, 164, 172). B. cenocepacia can also use iron obtained from the iron-binding protein ferritin, likely through the proteolytic degradation of ferritin (174). Since ferritin is found at up to 100-fold higher concentrations in the bronchoalveolar lavage fluid of CF patients than in that of healthy individuals (156), it has been suggested that ferritin might serve as an important iron source to B. cenocepacia during CF lung infections (174), although this has not yet been tested experimentally. B. cenocepacia can also use heme as a source of iron (174). Together, these results demonstrate that like many other bacterial pathogens, B. cenocepacia has specialized mechanisms to acquire iron during infections.
Lipopolysaccharide and other cell envelope structures.
LPS is a complex glycolipid located in the outer leaflet of the outer membrane of Gram-negative bacteria, which plays a significant role in bacterial pathogenesis (129). B. cenocepacia LPS can activate immune cells through Toll-like receptor 4-mediated signaling (11). The O-antigen portion of the LPS molecule is important for resistance to serum-mediated killing (120). It is also prevents both bacterial binding to epithelial cells and bacterial phagocytosis by macrophages (142). However, not all strains of B. cenocepacia, including epidemic strain J2315, make polymeric O antigen, so it is not required for virulence (120). Mutant strains of B. cenocepacia unable to synthesize the LPS inner core oligosaccharide (deep-rough LPS mutants) have impaired virulence in the rat agar bead, C. elegans, and G. mellonella infection models and fail to survive in murine macrophages (99, 121, 164). B. cenocepacia deep-rough LPS mutants are also more sensitive to cationic antimicrobial peptides, including human neutrophil peptide 1 (99).
Another interesting and unique aspect of the LPS biology of B. cenocepacia and other Burkholderia spp. is the constitutive presence of 4-amino-4-deoxy-l-arabinose (Ara4N) residues in the lipid A and inner core oligosaccharide. Many Gram-negative bacteria replace lipid A phosphate residues with molecules of Ara4N in vivo as a regulated mechanism of cationic antimicrobial peptide resistance, and this pathway is usually dispensable under most growth conditions (49, 117, 166, 175). Remarkably, this pathway is essential for B. cenocepacia survival, and depletion of the proteins of this pathway in conditional mutants leads to increased susceptibility to antimicrobial agents and accumulation of membranous material inside the cell (122) in a fashion that is similar to depletion of the machinery responsible for LPS transport to the outer membrane (178).
Other cell envelope structures required for virulence include the flagellum (Fig. 1B), disruption of which results in a nonmotile strain that is avirulent in the mouse agar bead model of infection (165), and cable pili, which are required for binding to cytokeratin 13 on epithelial cells (139) and killing of human airway epithelial cells ex vivo (32). Additionally, Sousa et al. (152) have shown that a B. cenocepacia mutant strain lacking an acyl carrier protein (ACP) has alterations in fatty acid content and increased cell surface hydrophobicity. This mutant strain has a greater ability to form biofilms in vitro but has decreased pathogenicity in the C. elegans infection model (152).
MgtC.
MgtC is a virulence protein that is required by distantly related bacterial pathogens, such as Salmonella enterica, Mycobacterium tuberculosis, and B. cenocepacia, for intraphagosomal survival in macrophages and growth under conditions of low magnesium but whose function is currently unknown (18, 20, 89, 107). Data suggest that these are two separate roles for MgtC (132). B. cenocepacia mgtC mutant strains fail to survive in murine macrophages and in the rat agar bead model of chronic lung infection (68, 107), but MgtC is not required for resistance in vitro to conditions that are encountered in macrophages, such as reactive oxygen and nitrogen species, low pH, and cationic antimicrobial peptides (107), so the function of this protein in virulence remains elusive.
Phenylacetic acid catabolic pathway.
As noted above, transposon mutants in numerous metabolic pathways were attenuated in the rat agar bead model, including mutants in the phenylacetic acid catabolic pathway (68). This pathway is the point at which the catabolism of many aromatic compounds converges (100). In B. cenocepacia, this pathway is also required for virulence in the C. elegans model (90) and is upregulated in vitro when bacteria are grown in synthetic cystic fibrosis medium (62), a defined medium developed by Palmer et al. (123) that is based on the contents of CF sputum and that contains significant amounts of aromatic amino acids. Since phenylacetic acid catabolism is linked to the catabolism of aromatic amino acids, it is conceivable that this pathway may be important for nutrient acquisition or the metabolism of infecting bacteria in the host environment.
Exopolysaccharide.
Exopolysaccharides (EPSs) are branched, repeating polysaccharide subunits that are secreted by bacteria into the extracellular milieu (173). The synthesis of EPS occurs during CF infections with Bcc bacteria (38, 64), although it is not always seen, and commonly studied B. cenocepacia clinical isolates such as J2315 and K56-2 produce little or no exopolysaccharide (182). EPS from EPS-producing B. cenocepacia clinical isolates may contribute to the virulence of the organism through inhibiting both neutrophil chemotaxis and neutrophil generation of H2O2 and O2− (23), and production of EPS results in slower clearance of bacteria from murine lungs (38).
Genomic islands.
The genome of B. cenocepacia strain J2315 contains more than a dozen genomic islands, most of which are still uncharacterized (65). One genomic island, the Burkholderia cepacia epidemic strain marker (BCESM), contains putative virulence-enhancing factors, including an amidase (AmiI), an outer membrane porin (OpcI), and the cciRI quorum-sensing system (10). Recently, a genomic island identified in B. cenocepacia strain K56-2 that is absent from strain J2315 was shown to contain a gene that encodes a predicted regulatory protein, Pbr (131). Disruption of pbr resulted in a strain with a number of defects, including increased susceptibility to oxidative and temperature stress, decreased virulence in C. elegans, and an inability to synthesize phenazines, which are antibiotics that are thought to interfere with electron transport and impair respiration (131).
LIMITATIONS
Despite the abundance of studies described above, there are some important limitations to the currently available data on virulence determinants in B. cenocepacia. First, for many of the determinants described above, experiments have been limited to only one or two infection models. This is particularly relevant if the infection models chosen are limited to only tissue-cultured cell-based assays, and many promising mutant strains must be characterized in additional infection models in order to determine the scope to which they are required for virulence.
Uehlinger et al. (164) have made a first attempt at addressing this issue. The authors of that study tested over a dozen B. cenocepacia mutant strains in up to four different infection models and found that the LPS core oligosaccharide, the siderophore ornibactin, and the cepRI quorum-sensing system were required in at least three of the models tested (Table 2). The remaining virulence determinants tested in that study were required in only one or two of the infection models. To date there is no mutant strain that has been tested and shown to be avirulent in all the different types of infection models (vertebrate, invertebrate, plant, and tissue culture models).
Second, there appears to be significant disparity between results for a given virulence determinant when it is tested across many virulence models. For example, mutants lacking ornibactin or the LPS core oligosaccharide have significant impairments in virulence in multiple animal models but have wild-type levels of virulence in the alfalfa seedling model (Table 2). The infection models used represent different environmental and clinical niches that Bcc bacteria can be found in, none of which accurately represent the environment of the CF lung. The rat agar bead model does replicate some of the characteristics of the CF lung, such as increased cytokine levels and neutrophil influx, and the agar mimics bacterial biofilms. However, instillation of the bacteria into the lungs circumvents innate host defenses and initial steps in bacterial colonization (85). Perhaps new models that appear to more closely replicate human CF disease, such as the cftr−/− pig (135), will be helpful in identifying determinants of B. cenocepacia virulence that also contribute to disease in CF patients.
Third, there is great strain-to-strain variability within the species B. cenocepacia. For example, three well-studied strains, strains H111, K56-2, and J2315 (the last two being clonally related isolates, as described above), all behave quite differently in the G. mellonella and C. elegans infection models (164). Additionally there are a number of examples of strain-to-strain variation in the production of, requirement for, or even presence of certain virulence determinants. A melanin-like pigment required by B. cenocepacia strain C5424 for survival in macrophages is made in much larger amounts in this strain than in clonally related strain J2315 or K56-2 (75). The extracellular protease ZmpA is required for virulence in the rat agar bead model in a strain-dependent fashion (40). The regulatory protein Pbr that contributes to virulence of strain K56-2 in C. elegans is found on a genomic island that is absent from strain J2315 (131). Virulence determinants required in a range of infection models should be tested to see if they are required for virulence in a variety of B. cenocepacia strains as well as in other Bcc bacteria.
CONCLUDING REMARKS
Since Bcc bacteria were initially recognized as causing severe lung infections in CF patients (69), much progress in the characterization of virulence determinants of B. cenocepacia has been made, particularly in the last decade. These include extracellular and cell surface polysaccharides, systems that regulate gene expression, secretion systems, metabolic and nutrient acquisition pathways, molecules required for resistance to host antimicrobial compounds, and proteins whose functions are not well understood. To aid in these studies, a number of genomes have been sequenced, sophisticated tools for genetic manipulation have been developed, and numerous in vivo models have been established.
These studies should form the base for the development of novel antimicrobial agents that can target B. cenocepacia in vivo. Some of these types of studies are under way, such as the development of molecules to inhibit the addition of Ara4N residues to lipid A (78) or the synthesis of l-glycero-d-manno-heptose sugars required for the LPS inner core oligosaccharide (43), as well as the manipulation of siderophores to act as “Trojan horses” for the delivery of antibiotics to bacteria (115). Furthermore a vaccine that prevents B. cenocepacia colonization in CF patients could provide additional benefit, and mucosal vaccines for this purpose are being developed (17, 106).
B. cenocepacia is a member of a highly adaptable genus of bacteria that live under diverse circumstances in nature (36) and that can rapidly evolve under in vitro stress conditions or during infections (46, 125, 128). This adaptability may be key to the pathogenesis of B. cenocepacia, especially to the ability of these bacteria to establish opportunistic chronic infections (167). Recently identified factors required for B. cenocepacia virulence, such as metabolic pathways, alternative sigma factors and other regulatory molecules, and mechanisms of resistance to oxidative stress and host antimicrobial proteins and peptides, support this idea. Finally, several other bacterial species phenotypically similar to Bcc bacteria and often even misidentified at hospitals as Bcc bacteria have also been isolated from CF lung infections (34, 155), and it is possible that the study of B. cenocepacia pathogenesis may serve as a model for these other organisms.
Acknowledgments
We thank past and present members of the Miguel A. Valvano laboratory for ongoing discussions and comments regarding B. cenocepacia virulence and building of genetic tools. We also thank Marta Eva Llamera for providing M.A.V.'s photograph accompanying the author's bio.
S.A.L. is supported by a studentship from the Canadian Cystic Fibrosis Foundation. Research in the authors' laboratory was supported by grants from the Canadian Cystic Fibrosis Foundation, the Canadian Institutes of Health Research Special Initiative on Novel Antimicrobials, and the B. cepacia microarray initiative from the U.S. Cystic Fibrosis Foundation. M.A.V. holds a Canada Research Chair in Infectious Diseases and Microbial Pathogenesis and received a Senior Research Training Award from the Canadian Cystic Fibrosis Foundation.
Biography
 Slade A. Loutet obtained his B.Sc. in microbiology and immunology from the University of British Columbia. During his undergraduate training he spent time as a research assistant in the laboratory of Dr. Connie J. Eaves at the British Columbia Cancer Research Centre and at Vancouver Biotech, Ltd. After a sojourn teaching English overseas, he entered the world of bacterial pathogenesis in 2003 when he joined the laboratory of Dr. Miguel A. Valvano at the University of Western Ontario as a lab technician. He quickly upgraded to the level of graduate student and recently (April 2010) completed his Ph.D. dissertation on antimicrobial peptide resistance mechanisms of Burkholderia cenocepacia under the supervision of Dr. Valvano. He remains a member of the Valvano laboratory while completing additional publications based on his Ph.D. dissertation and exploring options for future postdoctoral fellowship training.
Slade A. Loutet obtained his B.Sc. in microbiology and immunology from the University of British Columbia. During his undergraduate training he spent time as a research assistant in the laboratory of Dr. Connie J. Eaves at the British Columbia Cancer Research Centre and at Vancouver Biotech, Ltd. After a sojourn teaching English overseas, he entered the world of bacterial pathogenesis in 2003 when he joined the laboratory of Dr. Miguel A. Valvano at the University of Western Ontario as a lab technician. He quickly upgraded to the level of graduate student and recently (April 2010) completed his Ph.D. dissertation on antimicrobial peptide resistance mechanisms of Burkholderia cenocepacia under the supervision of Dr. Valvano. He remains a member of the Valvano laboratory while completing additional publications based on his Ph.D. dissertation and exploring options for future postdoctoral fellowship training.
 Miguel A. Valvano obtained his M.D. from the University of Buenos Aires, Buenos Aires, Argentina, and acquired further specialization in clinical pediatrics and infectious diseases. He trained in molecular biology and microbial genetics as a research fellow under the supervision of J. H. Crosa in the Department of Microbiology and Immunology, Oregon Health Sciences University, before becoming a faculty member in the Department of Microbiology and Immunology of the University of Western Ontario. Dr. Valvano is currently department chair and holds a Canada Research Chair in Infectious Diseases and Microbial Pathogenesis. His laboratory conducts research on lipopolysaccharide genetics and biosynthesis, as well as on the pathogenesis of Burkholderia cepacia, with special emphasis on bacteria-macrophage interactions.
Miguel A. Valvano obtained his M.D. from the University of Buenos Aires, Buenos Aires, Argentina, and acquired further specialization in clinical pediatrics and infectious diseases. He trained in molecular biology and microbial genetics as a research fellow under the supervision of J. H. Crosa in the Department of Microbiology and Immunology, Oregon Health Sciences University, before becoming a faculty member in the Department of Microbiology and Immunology of the University of Western Ontario. Dr. Valvano is currently department chair and holds a Canada Research Chair in Infectious Diseases and Microbial Pathogenesis. His laboratory conducts research on lipopolysaccharide genetics and biosynthesis, as well as on the pathogenesis of Burkholderia cepacia, with special emphasis on bacteria-macrophage interactions.
Editor: H. L. Andrews-Polymenis
Footnotes
Published ahead of print on 19 July 2010.
REFERENCES
- 1.Aaron, S. D., W. Ferris, D. A. Henry, D. P. Speert, and N. E. Macdonald. 2000. Multiple combination bactericidal antibiotic testing for patients with cystic fibrosis infected with Burkholderia cepacia. Am. J. Respir. Crit. Care Med. 161:1206-1212. [DOI] [PubMed] [Google Scholar]
- 2.Ades, S. E. 2004. Control of the alternative sigma factor σE in Escherichia coli. Curr. Opin. Microbiol. 7:157-162. [DOI] [PubMed] [Google Scholar]
- 3.Agnoli, K., C. A. Lowe, K. L. Farmer, S. I. Husnain, and M. S. Thomas. 2006. The ornibactin biosynthesis and transport genes of Burkholderia cenocepacia are regulated by an extracytoplasmic function σ factor which is a part of the Fur regulon. J. Bacteriol. 188:3631-3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agodi, A., E. Mahenthiralingam, M. Barchitta, V. Gianninò, A. Sciacca, and S. Stefani. 2001. Burkholderia cepacia complex infection in Italian patients with cystic fibrosis: prevalence, epidemiology, and genomovar status. J. Clin. Microbiol. 39:2891-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrade, G., K. L. Mihara, R. G. Linderman, and G. J. Bethlenfalvay. 1997. Bacteria from rhizosphere and hyphosphere soils of different arbuscular-mycorrhizal fungi. Plant Soil 192:71-79. [Google Scholar]
- 6.Aubert, D. F., R. S. Flannagan, and M. A. Valvano. 2008. A novel sensor kinase-response regulator hybrid controls biofilm formation and type VI secretion system activity in Burkholderia cenocepacia. Infect. Immun. 76:1979-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Azegami, K., K. Nishiyama, Y. Watanabe, I. Kadota, A. Ohuchi, and C. Fukazawa. 1987. Pseudomonas plantarii sp. nov., the causal agent of rice seedling blight. Int. J. Syst. Bacteriol. 37:144-152. [Google Scholar]
- 8.Balandreau, J., V. Viallard, B. Cournoyer, T. Coenye, S. Laevens, and P. Vandamme. 2001. Burkholderia cepacia genomovar III is a common plant-associated bacterium. Appl. Environ. Microbiol. 67:982-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baldwin, A., E. Mahenthiralingam, P. Drevinek, C. Pope, D. J. Waine, D. A. Henry, D. P. Speert, P. Carter, P. Vandamme, J. J. LiPuma, and C. G. Dowson. 2008. Elucidating global epidemiology of Burkholderia multivorans in cases of cystic fibrosis by multilocus sequence typing. J. Clin. Microbiol. 46:290-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baldwin, A., P. A. Sokol, J. Parkhill, and E. Mahenthiralingam. 2004. The Burkholderia cepacia epidemic strain marker is part of a novel genomic island encoding both virulence and metabolism-associated genes in Burkholderia cenocepacia. Infect. Immun. 72:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bamford, S., H. Ryley, and S. K. Jackson. 2007. Highly purified lipopolysaccharides from Burkholderia cepacia complex clinical isolates induce inflammatory cytokine responses via TLR4-mediated MAPK signalling pathways and activation of NFΚB. Cell. Microbiol. 9:532-543. [DOI] [PubMed] [Google Scholar]
- 12.Barrett, A. R., Y. Kang, K. S. Inamasu, M. S. Son, J. M. Vukovich, and T. T. Hoang. 2008. Genetic tools for allelic replacement in Burkholderia species. Appl. Environ. Microbiol. 74:4498-4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berlutti, F., C. Morea, A. Battistoni, S. Sarli, P. Cipriani, F. Superti, M. G. Ammendolia, and P. Valenti. 2005. Iron availability influences aggregation, biofilm, adhesion and invasion of Pseudomonas aeruginosa and Burkholderia cenocepacia. Int. J. Immunopathol. Pharmacol. 18:661-670. [DOI] [PubMed] [Google Scholar]
- 14.Bernier, S. P., D. T. Nguyen, and P. A. Sokol. 2008. A LysR-type transcriptional regulator in Burkholderia cenocepacia influences colony morphology and virulence. Infect. Immun. 76:38-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernier, S. P., L. Silo-Suh, D. E. Woods, D. E. Ohman, and P. A. Sokol. 2003. Comparative analysis of plant and animal models for characterization of Burkholderia cepacia virulence. Infect. Immun. 71:5306-5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernier, S. P., and P. A. Sokol. 2005. Use of suppression-subtractive hybridization to identify genes in the Burkholderia cepacia complex that are unique to Burkholderia cenocepacia. J. Bacteriol. 187:5278-5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertot, G. M., M. A. Restelli, L. Galanternik, R. C. Aranibar Urey, M. A. Valvano, and S. Grinstein. 2007. Nasal immunization with Burkholderia multivorans outer membrane proteins and the mucosal adjuvant adamantylamide dipeptide confers efficient protection against experimental lung infections with B. multivorans and B. cenocepacia. Infect. Immun. 75:2740-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blanc-Potard, A.-B., and E. A. Groisman. 1997. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J. 16:5376-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boon, C., Y. Deng, L. H. Wang, Y. He, J. L. Xu, Y. Fan, S. Q. Pan, and L. H. Zhang. 2008. A novel DSF-like signal from Burkholderia cenocepacia interferes with Candida albicans morphological transition. ISME J. 2:27-36. [DOI] [PubMed] [Google Scholar]
- 20.Buchmeier, N., A.-B. Blanc-Potard, S. Ehrt, D. Piddington, L. Riley, and E. A. Groisman. 2000. A parallel intraphagosomal survival strategy shared by Mycobacterium tuberculosis and Salmonella enterica. Mol. Microbiol. 35:1375-1382. [DOI] [PubMed] [Google Scholar]
- 21.Burkholder, W. H. 1950. Sour skin, a bacterial rot of onion bulbs. Phytopathology 40:115-117. [Google Scholar]
- 22.Burns, J. L., C. D. Wadsworth, J. J. Barry, and C. P. Goodall. 1996. Nucleotide sequence analysis of a gene from Burkholderia (Pseudomonas) cepacia encoding an outer membrane lipoprotein involved in multiple antibiotic resistance. Antimicrob. Agents Chemother. 40:307-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bylund, J., L. A. Burgess, P. Cescutti, R. K. Ernst, and D. P. Speert. 2006. Exopolysaccharides from Burkholderia cenocepacia inhibit neutrophil chemotaxis and scavenge reactive oxygen species. J. Biol. Chem. 281:2526-2532. [DOI] [PubMed] [Google Scholar]
- 24.Bylund, J., P. A. Campsall, R. C. Ma, B. A. Conway, and D. P. Speert. 2005. Burkholderia cenocepacia induces neutrophil necrosis in chronic granulomatous disease. J. Immunol. 174:3562-3569. [DOI] [PubMed] [Google Scholar]
- 25.Caballero-Mellado, J., J. Onofre-Lemus, P. Estrada-de los Santos, and L. Martinez-Aguilar. 2007. The tomato rhizosphere, an environment rich in nitrogen-fixing Burkholderia species with capabilities of interest for agriculture and bioremediation. Appl. Environ. Microbiol. 73:5308-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cain, C. C., A. T. Henry, R. H. Waldo III, L. J. Casida, Jr., and J. O. Falkinham III. 2000. Identification and characteristics of a novel Burkholderia strain with broad-spectrum antimicrobial activity. Appl. Environ. Microbiol. 66:4139-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caraher, E., G. Reynolds, P. Murphy, S. McClean, and M. Callaghan. 2007. Comparison of antibiotic susceptibility of Burkholderia cepacia complex organisms when grown planktonically or as biofilm in vitro. Eur. J. Clin. Microbiol. Infect. Dis. 26:213-216. [DOI] [PubMed] [Google Scholar]
- 28.Cardona, S. T., C. L. Mueller, and M. A. Valvano. 2006. Identification of essential operons with a rhamnose-inducible promoter in Burkholderia cenocepacia. Appl. Environ. Microbiol. 72:2547-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cash, H. A., D. E. Woods, B. McCullough, W. G. Johanson, Jr., and J. A. Bass. 1979. A rat model of chronic respiratory infection with Pseudomonas aeruginosa. Am. Rev. Respir. Dis. 119:453-459. [DOI] [PubMed] [Google Scholar]
- 30.Chavan, A., and S. Mukherji. 2008. Treatment of hydrocarbon-rich wastewater using oil degrading bacteria and phototrophic microorganisms in rotating biological contactor: effect of N:P ratio. J. Haz. Mat. 154:63-72. [DOI] [PubMed] [Google Scholar]
- 31.Cheng, A. C., and B. J. Currie. 2005. Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 18:383-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheung, K. J., Jr., G. Li, T. A. Urban, J. B. Goldberg, A. Griffith, F. Lu, and J. L. Burns. 2007. Pilus-mediated epithelial cell death in response to infection with Burkholderia cenocepacia. Microbes Infect. 9:829-837. [DOI] [PubMed] [Google Scholar]
- 33.Choi, K. H., T. Mima, Y. Casart, D. Rholl, A. Kumar, I. R. Beacham, and H. P. Schweizer. 2008. Genetic tools for select-agent-compliant manipulation of Burkholderia pseudomallei. Appl. Environ. Microbiol. 74:1064-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coenye, T., J. Goris, T. Spilker, P. Vandamme, and J. J. LiPuma. 2002. Characterization of unusual bacteria isolated from respiratory secretions of cystic fibrosis patients and description of Inquilinus limosus gen. nov., sp. nov. J. Clin. Microbiol. 40:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Compant, S., H. Kaplan, A. Sessitsch, J. Nowak, E. A. Barka, and C. Clément. 2008. Endophytic colonization of Vitis vinifera L. by Burkholderia phytofirmans strain PsJN: from the rhizosphere to inflorescence tissues. FEMS Microbiol. Ecol. 63:84-93. [DOI] [PubMed] [Google Scholar]
- 36.Compant, S., J. Nowak, T. Coenye, C. Clément, and E. A. Barka. 2008. Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol. Rev. 32:607-626. [DOI] [PubMed] [Google Scholar]
- 37.Compant, S., B. Reiter, A. Sessitsch, J. Nowak, C. Clement, and E. Ait Barka. 2005. Endophytic colonization of Vitis vinifera L. by plant growth-promoting bacterium Burkholderia sp. strain PsJN. Appl. Environ. Microbiol. 71:1685-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conway, B.-A. D., K. K. Chu, J. Bylund, E. Altman, and D. P. Speert. 2004. Production of exopolysaccharide by Burkholderia cenocepacia results in altered cell-surface interactions and altered bacterial clearance in mice. J. Infect. Dis. 190:957-966. [DOI] [PubMed] [Google Scholar]
- 39.Conway, B.-A. D., V. Venu, and D. P. Speert. 2002. Biofilm formation and acyl homoserine lactone production in the Burkholderia cepacia complex. J. Bacteriol. 184:5678-5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Corbett, C. R., M. N. Burtnick, C. Kooi, D. E. Woods, and P. A. Sokol. 2003. An extracellular zinc metalloprotease gene of Burkholderia cepacia. Microbiology 149:2263-2271. [DOI] [PubMed] [Google Scholar]
- 41.Cunha, M. V., S. A. Sousa, J. H. Leitão, L. M. Moreira, P. A. Videira, and I. Sá-Correia. 2004. Studies on the involvement of the exopolysaccharide produced by cystic fibrosis-associated isolates of the Burkholderia cepacia complex in biofilm formation and in persistence of respiratory infections. J. Clin. Microbiol. 42:3052-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Darling, P., M. Chan, A. D. Cox, and P. A. Sokol. 1998. Siderophore production by cystic fibrosis isolates of Burkholderia cepacia. Infect. Immun. 66:874-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Leon, G. P., N. H. Elowe, K. P. Koteva, M. A. Valvano, and G. D. Wright. 2006. An in vitro screen of bacterial lipopolysaccharide biosynthetic enzymes identifies an inhibitor of ADP-heptose biosynthesis. Chem. Biol. 13:437-441. [DOI] [PubMed] [Google Scholar]
- 44.Deng, Y., C. Boon, L. Eberl, and L.-H. Zhang. 2009. Differential modulation of Burkholderia cenocepacia virulence and energy metabolism by the quorum-sensing signal BDSF and its synthase. J. Bacteriol. 191:7270-7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Desai, M., T. Buhler, P. H. Weller, and M. R. Brown. 1998. Increasing resistance of planktonic and biofilm cultures of Burkholderia cepacia to ciprofloxacin and ceftazidime during exponential growth. J. Antimicrob. Chemother. 42:153-160. [DOI] [PubMed] [Google Scholar]
- 46.Drevinek, P., A. Baldwin, L. Lindenburg, L. T. Joshi, A. Marchbank, S. Vosahlikova, C. G. Dowson, and E. Mahenthiralingam. 2010. Oxidative stress of Burkholderia cenocepacia induces insertion sequence-mediated genomic rearrangements that interfere with macrorestriction-based genotyping. J. Clin. Microbiol. 48:34-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Drevinek, P., M. Holden, Z. Ge, A. Jones, I. Ketchell, R. Gill, and E. Mahenthiralingam. 2008. Gene expression changes linked to antimicrobial resistance, oxidative stress, iron depletion and retained motility are observed when Burkholderia cenocepacia grows in cystic fibrosis sputum. BMC Infect. Dis. 8:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Engledow, A. S., E. G. Medrano, E. Mahenthiralingam, J. J. LiPuma, and C. F. Gonzalez. 2004. Involvement of a plasmid-encoded type IV secretion system in the plant tissue watersoaking phenotype of Burkholderia cenocepacia. J. Bacteriol. 186:6015-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ernst, R. K., E. C. Yi, L. Guo, K. B. Lim, J. L. Burns, M. Hackett, and S. I. Miller. 1999. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science 286:1561-1565. [DOI] [PubMed] [Google Scholar]
- 50.Estrela, A. B., M. G. Heck, and W. R. Abraham. 2009. Novel approaches to control biofilm infections. Curr. Med. Chem. 16:1512-1530. [DOI] [PubMed] [Google Scholar]
- 51.Flannagan, R. S., D. Aubert, C. Kooi, P. A. Sokol, and M. A. Valvano. 2007. Burkholderia cenocepacia requires a periplasmic HtrA protease for growth under thermal and osmotic stress and for survival in vivo. Infect. Immun. 75:1679-1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Flannagan, R. S., T. Linn, and M. A. Valvano. 2008. A system for the construction of targeted unmarked gene deletions in the genus Burkholderia. Environ. Microbiol. 10:1652-1660. [DOI] [PubMed] [Google Scholar]
- 53.Flannagan, R. S., and M. A. Valvano. 2008. Burkholderia cenocepacia requires RpoE for growth under stress conditions and delay of phagolysosomal fusion in macrophages. Microbiology 154:643-653. [DOI] [PubMed] [Google Scholar]
- 54.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 55.Galán, J. E. 2009. Common themes in the design and function of bacterial effectors. Cell Host Microbe 5:571-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garbaye, J., and G. D. Bowen. 1989. Stimulation of ectomycorrhizal infection of Pinus radiata by some microorganisms associated with the mantle of ectomycorrhizas. New Phytol. 112:383-388. [Google Scholar]
- 57.Gold, R., E. Jin, H. Levison, A. Isles, and P. C. Fleming. 1983. Ceftazidime alone and in combination in patients with cystic fibrosis: lack of efficacy in treatment of severe respiratory infections caused by Pseudomonas cepacia. J. Antimicrob. Chemother. 12(Suppl. A):331-336. [DOI] [PubMed] [Google Scholar]
- 58.Govan, J. R., P. H. Brown, J. Maddison, C. J. Doherty, J. W. Nelson, M. Dodd, A. P. Greening, and A. K. Webb. 1993. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet 342:15-19. [DOI] [PubMed] [Google Scholar]
- 59.Gregory, C. W., D. M. Estes, and G. T. Alfredo. 2007. Glanders: off to the races with Burkholderia mallei. FEMS Microbiol. Lett. 277:115-122. [DOI] [PubMed] [Google Scholar]
- 60.Hamad, M. A., A. M. Skeldon, and M. A. Valvano. 2010. Construction of aminoglycoside-sensitive Burkholderia cenocepacia strains for studying intracellular bacteria by the gentamicin protection assay. Appl. Environ. Microbiol. 76:3170-3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hamad, M. A., S. L. Zajdowicz, R. K. Holmes, and M. I. Voskuil. 2009. An allelic exchange system for compliant genetic manipulation of the select agents Burkholderia pseudomallei and Burkholderia mallei. Gene 430:123-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hamlin, J., R. Bloodworth, and S. Cardona. 2009. Regulation of phenylacetic acid degradation genes of Burkholderia cenocepacia K56-2. BMC Microbiol. 9:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He, Y.-W., and L.-H. Zhang. 2008. Quorum sensing and virulence regulation in Xanthomonas campestris. FEMS Microbiol. Rev. 32:842-857. [DOI] [PubMed] [Google Scholar]
- 64.Herasimenka, Y., P. Cescutti, G. Impallomeni, S. Campana, G. Taccetti, N. Ravenni, F. Zanetti, and R. Rizzo. 2007. Exopolysaccharides produced by clinical strains belonging to the Burkholderia cepacia complex. J. Cyst. Fibros. 6:145-152. [DOI] [PubMed] [Google Scholar]
- 65.Holden, M. T., H. M. Seth-Smith, L. C. Crossman, M. Sebaihia, S. D. Bentley, A. M. Cerdeño-Tárraga, N. R. Thomson, N. Bason, M. A. Quail, S. Sharp, I. Cherevach, C. Churcher, I. Goodhead, H. Hauser, N. Holroyd, K. Mungall, P. Scott, D. Walker, B. White, H. Rose, P. Iversen, D. Mil-Homens, E. P. Rocha, A. M. Fialho, A. Baldwin, C. Dowson, B. G. Barrell, J. R. Govan, P. Vandamme, C. A. Hart, E. Mahenthiralingam, and J. Parkhill. 2009. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J. Bacteriol. 191:261-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huber, B., F. Feldmann, M. Köthe, P. Vandamme, J. Wopperer, K. Riedel, and L. Eberl. 2004. Identification of a novel virulence factor in Burkholderia cenocepacia H111 required for efficient slow killing of Caenorhabditis elegans. Infect. Immun. 72:7220-7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huber, B., K. Riedel, M. Köthe, M. Givskov, S. Molin, and L. Eberl. 2002. Genetic analysis of functions involved in the late stages of biofilm development in Burkholderia cepacia H111. Mol. Microbiol. 46:411-426. [DOI] [PubMed] [Google Scholar]
- 68.Hunt, T. A., C. Kooi, P. A. Sokol, and M. A. Valvano. 2004. Identification of Burkholderia cenocepacia genes required for bacterial survival in vivo. Infect. Immun. 72:4010-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Isles, A., I. Maclusky, M. Corey, R. Gold, C. Prober, P. Fleming, and H. Levison. 1984. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J. Pediatr. 104:206-210. [DOI] [PubMed] [Google Scholar]
- 70.Izumi, H., E. R. B. Moore, K. Killham, I. J. Alexander, and I. G. Anderson. 2007. Characterisation of endobacterial communities in ectomycorrhizas by DNA- and RNA-based molecular methods. Soil Biol. Biochem. 39:891-899. [Google Scholar]
- 71.Jacobs, J. L., A. C. Fasi, A. Ramette, J. J. Smith, R. Hammerschmidt, and G. W. Sundin. 2008. Identification and onion pathogenicity of Burkholderia cepacia complex isolates from the onion rhizosphere and onion field soil. Appl. Environ. Microbiol. 74:3121-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jeong, Y., J. Kim, S. Kim, Y. Kang, T. Nagamatsu, and I. Hwang. 2003. Toxoflavin produced by Burkholderia glumae causing rice grain rot is responsible for inducing bacterial wilt in many field crops. Plant Dis. 87:890-895. [DOI] [PubMed] [Google Scholar]
- 73.Jiang, C., X. Sheng, M. Qian, and Q. Wang. 2008. Isolation and characterization of a heavy metal-resistant Burkholderia sp. from heavy metal-contaminated paddy field soil and its potential in promoting plant growth and heavy metal accumulation in metal-polluted soil. Chemosphere 72:157-164. [DOI] [PubMed] [Google Scholar]
- 74.Keith, K. E., D. W. Hynes, J. E. Sholdice, and M. A. Valvano. 2009. Delayed association of the NADPH oxidase complex with macrophage vacuoles containing the opportunistic pathogen Burkholderia cenocepacia. Microbiology 155:1004-1015. [DOI] [PubMed] [Google Scholar]
- 75.Keith, K. E., L. Killip, P. He, G. R. Moran, and M. A. Valvano. 2007. Burkholderia cenocepacia C5424 produces a pigment with antioxidant properties using a homogentisate intermediate. J. Bacteriol. 189:9057-9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Keith, K. E., and M. A. Valvano. 2007. Characterization of SodC, a periplasmic superoxide dismutase from Burkholderia cenocepacia. Infect. Immun. 75:2451-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kikuchi, Y., X.-Y. Meng, and T. Fukatsu. 2005. Gut symbiotic bacteria of the genus Burkholderia in the broad-headed bugs Riptortus clavatus and Leptocorisa chinensis (Heteroptera: Alydidae). Appl. Environ. Microbiol. 71:4035-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kline, T., M. S. Trent, C. M. Stead, M. S. Lee, M. C. Sousa, H. B. Felise, H. V. Nguyen, and S. I. Miller. 2008. Synthesis of and evaluation of lipid A modification by 4-substituted 4-deoxy arabinose analogs as potential inhibitors of bacterial polymyxin resistance. Bioorg. Med. Chem. Lett. 18:1507-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kooi, C., C. R. Corbett, and P. A. Sokol. 2005. Functional analysis of the Burkholderia cenocepacia ZmpA metalloprotease. J. Bacteriol. 187:4421-4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kooi, C., and P. A. Sokol. 2009. Burkholderia cenocepacia zinc metalloproteases influence resistance to antimicrobial peptides. Microbiology 155:2818-2825. [DOI] [PubMed] [Google Scholar]
- 81.Kooi, C., B. Subsin, R. Chen, B. Pohorelic, and P. A. Sokol. 2006. Burkholderia cenocepacia ZmpB is a broad-specificity zinc metalloprotease involved in virulence. Infect. Immun. 74:4083-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Köthe, M., M. Antl, B. Huber, K. Stoecker, D. Ebrecht, I. Steinmetz, and L. Eberl. 2003. Killing of Caenorhabditis elegans by Burkholderia cepacia is controlled by the cep quorum-sensing system. Cell. Microbiol. 5:343-351. [DOI] [PubMed] [Google Scholar]
- 83.Kroon, A. G. M., and C. G. van Ginkel. 2001. Complete mineralization of dodecyldimethylamine using a two-membered bacterial culture. Environ. Microbiol. 3:131-136. [DOI] [PubMed] [Google Scholar]
- 84.Krumme, M. L., K. N. Timmis, and D. F. Dwyer. 1993. Degradation of trichloroethylene by Pseudomonas cepacia G4 and the constitutive mutant strain G4 5223 PR1 in aquifer microcosms. Appl. Environ. Microbiol. 59:2746-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kukavica-Ibrulj, I., and R. C. Levesque. 2008. Animal models of chronic lung infection with Pseudomonas aeruginosa: useful tools for cystic fibrosis studies. Lab. Anim. 42:389-412. [DOI] [PubMed] [Google Scholar]
- 86.Lamothe, J., K. K. Huynh, S. Grinstein, and M. A. Valvano. 2007. Intracellular survival of Burkholderia cenocepacia in macrophages is associated with a delay in the maturation of bacteria-containing vacuoles. Cell. Microbiol. 9:40-53. [DOI] [PubMed] [Google Scholar]
- 87.Lamothe, J., S. Thyssen, and M. A. Valvano. 2004. Burkholderia cepacia complex isolates survive intracellularly without replication within acidic vacuoles of Acanthamoeba polyphaga. Cell. Microbiol. 6:1127-1138. [DOI] [PubMed] [Google Scholar]
- 88.Lamothe, J., and M. A. Valvano. 2008. Burkholderia cenocepacia-induced delay of acidification and phagolysosomal fusion in cystic fibrosis transmembrane conductance regulator (CFTR)-defective macrophages. Microbiology 154:3825-3834. [DOI] [PubMed] [Google Scholar]
- 89.Lavigne, J.-P., D. O'Callaghan, and A.-B. Blanc-Potard. 2005. Requirement of MgtC for Brucella suis intramacrophage growth: a potential mechanism shared by Salmonella enterica and Mycobacterium tuberculosis for adaptation to a low-Mg2+ environment. Infect. Immun. 73:3160-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Law, R. J., J. N. R. Hamlin, A. Sivro, S. J. McCorrister, G. A. Cardama, and S. T. Cardona. 2008. A functional phenylacetic acid catabolic pathway is required for full pathogenicity of Burkholderia cenocepacia in the Caenorhabditis elegans host model. J. Bacteriol. 190:7209-7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lefebre, M. D., R. S. Flannagan, and M. A. Valvano. 2005. A minor catalase/peroxidase from Burkholderia cenocepacia is required for normal aconitase activity. Microbiology 151:1975-1985. [DOI] [PubMed] [Google Scholar]
- 92.Lefebre, M. D., and M. A. Valvano. 2001. In vitro resistance of Burkholderia cepacia complex isolates to reactive oxygen species in relation to catalase and superoxide dismutase production. Microbiology 147:97-109. [DOI] [PubMed] [Google Scholar]
- 93.Lessie, T. G., W. Hendrickson, B. D. Manning, and R. Devereux. 1996. Genomic complexity and plasticity of Burkholderia cepacia. FEMS Microbiol. Lett. 144:117-128. [DOI] [PubMed] [Google Scholar]
- 94.Lewenza, S., B. Conway, E. P. Greenberg, and P. A. Sokol. 1999. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J. Bacteriol. 181:748-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lewenza, S., M. B. Visser, and P. A. Sokol. 2002. Interspecies communication between Burkholderia cepacia and Pseudomonas aeruginosa. Can. J. Microbiol. 48:707-716. [DOI] [PubMed] [Google Scholar]
- 96.LiPuma, J. J., T. Spilker, T. Coenye, and C. F. Gonzalez. 2002. An epidemic Burkholderia cepacia complex strain identified in soil. Lancet 359:2002. [DOI] [PubMed] [Google Scholar]
- 97.LiPuma, J. J., T. Spilker, L. H. Gill, P. W. Campbell III, L. Liu, and E. Mahenthiralingam. 2001. Disproportionate distribution of Burkholderia cepacia complex species and transmissibility markers in cystic fibrosis. Am. J. Respir. Crit. Care Med. 164:92-96. [DOI] [PubMed] [Google Scholar]
- 98.López, C. M., D. A. Rholl, L. A. Trunck, and H. P. Schweizer. 2009. Versatile dual-technology system for markerless allele replacement in Burkholderia pseudomallei. Appl. Environ. Microbiol. 75:6496-6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Loutet, S. A., R. S. Flannagan, C. Kooi, P. A. Sokol, and M. A. Valvano. 2006. A complete lipopolysaccharide inner core oligosaccharide is required for resistance of Burkholderia cenocepacia to antimicrobial peptides and bacterial survival in vivo. J. Bacteriol. 188:2073-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Luengo, J. M., J. L. Garcia, and E. R. Olivera. 2001. The phenylacetyl-CoA catabolon: a complex catabolic unit with broad biotechnological applications. Mol. Microbiol. 39:1434-1442. [DOI] [PubMed] [Google Scholar]
- 101.MacDonald, K. L., and D. P. Speert. 2008. Differential modulation of innate immune cell functions by the Burkholderia cepacia complex: Burkholderia cenocepacia but not Burkholderia multivorans disrupts maturation and induces necrosis in human dendritic cells. Cell. Microbiol. 10:2138-2149. [DOI] [PubMed] [Google Scholar]
- 102.Mahenthiralingam, E., A. Baldwin, and C. G. Dowson. 2008. Burkholderia cepacia complex bacteria: opportunistic pathogens with important natural biology. J. Appl. Microbiol. 104:1539-1551. [DOI] [PubMed] [Google Scholar]
- 103.Mahenthiralingam, E., M. E. Campbell, D. A. Henry, and D. P. Speert. 1996. Epidemiology of Burkholderia cepacia infection in patients with cystic fibrosis: analysis by randomly amplified polymorphic DNA fingerprinting. J. Clin. Microbiol. 34:2914-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mahenthiralingam, E., T. Coenye, J. W. Chung, D. P. Speert, J. R. Govan, P. Taylor, and P. Vandamme. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38:910-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mahenthiralingam, E., T. A. Urban, and J. B. Goldberg. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3:144-156. [DOI] [PubMed] [Google Scholar]
- 106.Makidon, P. E., J. Knowlton, J. V. Groom II, L. P. Blanco, J. J. LiPuma, A. U. Bielinska, and J. R. Baker, Jr. 2010. Induction of immune response to the 17 kDa OMPA Burkholderia cenocepacia polypeptide and protection against pulmonary infection in mice after nasal vaccination with an OMP nanoemulsion-based vaccine. Med. Microbiol. Immunol. 199:81-92. [DOI] [PubMed] [Google Scholar]
- 107.Maloney, K. E., and M. A. Valvano. 2006. The mgtC gene of Burkholderia cenocepacia is required for growth under magnesium limitation conditions and intracellular survival in macrophages. Infect. Immun. 74:5477-5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Malott, R. J., A. Baldwin, E. Mahenthiralingam, and P. A. Sokol. 2005. Characterization of the cciIR quorum-sensing system in Burkholderia cenocepacia. Infect. Immun. 73:4982-4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Malott, R. J., E. P. O'Grady, J. Toller, S. Inhulsen, L. Eberl, and P. A. Sokol. 2009. A Burkholderia cenocepacia orphan LuxR homolog is involved in quorum-sensing regulation. J. Bacteriol. 191:2447-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Manno, G., C. Dalmastri, S. Tabacchioni, P. Vandamme, R. Lorini, L. Minicucci, L. Romano, A. Giannattasio, L. Chiarini, and A. Bevivino. 2004. Epidemiology and clinical course of Burkholderia cepacia complex infections, particularly those caused by different Burkholderia cenocepacia strains, among patients attending an Italian cystic fibrosis center. J. Clin. Microbiol. 42:1491-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McLoughlin, T. J., J. P. Quinn, A. Bettermann, and R. Bookland. 1992. Pseudomonas cepacia suppression of sunflower wilt fungus and role of antifungal compounds in controlling the disease. Appl. Environ. Microbiol. 58:1760-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mendes, R., A. A. Pizzirani-Kleiner, W. L. Araujo, and J. M. Raaijmakers. 2007. Diversity of cultivated endophytic bacteria from sugarcane: genetic and biochemical characterization of Burkholderia cepacia complex isolates. Appl. Environ. Microbiol. 73:7259-7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Merrick, M. J. 1993. In a class of its own—the RNA polymerase sigma factor σ54 (σN). Mol. Microbiol. 10:903-909. [DOI] [PubMed] [Google Scholar]
- 114.Meyer, S. L. F., S. I. Massoud, D. J. Chitwood, and D. P. Roberts. 2000. Evaluation of Trichoderma virens and Burkholderia cepacia for antagonistic activity against root-knot nematode, Meloidogyne incognita. Nematology 2:871-879. [Google Scholar]
- 115.Miethke, M., and M. A. Marahiel. 2007. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71:413-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mullen, T., K. Markey, P. Murphy, S. McClean, and M. Callaghan. 2007. Role of lipase in Burkholderia cepacia complex (Bcc) invasion of lung epithelial cells. Eur. J. Clin. Microbiol. Infect. Dis. 26:869-877. [DOI] [PubMed] [Google Scholar]
- 117.Nummila, K., I. Kilpeläinen, U. Zähringer, M. Vaara, and I. M. Helander. 1995. Lipopolysaccharides of polymyxin B-resistant mutants of Escherichia coli are extensively substituted by 2-aminoethyl pyrophosphate and contain aminoarabinose in lipid A. Mol. Microbiol. 16:271-278. [DOI] [PubMed] [Google Scholar]
- 118.Nzula, S., P. Vandamme, and J. R. Govan. 2002. Influence of taxonomic status on the in vitro antimicrobial susceptibility of the Burkholderia cepacia complex. J. Antimicrob. Chemother. 50:265-269. [DOI] [PubMed] [Google Scholar]
- 119.O'Grady, E. P., D. F. Viteri, R. J. Malott, and P. A. Sokol. 2009. Reciprocal regulation by the CepIR and CciIR quorum sensing systems in Burkholderia cenocepacia. BMC Genomics 10:441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ortega, X., T. A. Hunt, S. Loutet, A. D. Vinion-Dubiel, A. Datta, B. Choudhury, J. B. Goldberg, R. Carlson, and M. A. Valvano. 2005. Reconstitution of O-specific lipopolysaccharide expression in Burkholderia cenocepacia strain J2315, which is associated with transmissible infections in patients with cystic fibrosis. J. Bacteriol. 187:1324-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ortega, X., A. Silipo, M. S. Saldías, C. C. Bates, A. Molinaro, and M. A. Valvano. 2009. Biosynthesis and structure of the Burkholderia cenocepacia K56-2 lipopolysaccharide core oligosaccharide: truncation of the core oligosaccharide leads to increased binding and sensitivity to polymyxin B. J. Biol. Chem. 284:21738-21751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ortega, X. P., S. T. Cardona, A. R. Brown, S. A. Loutet, R. S. Flannagan, D. J. Campopiano, J. R. Govan, and M. A. Valvano. 2007. A putative gene cluster for aminoarabinose biosynthesis is essential for Burkholderia cenocepacia viability. J. Bacteriol. 189:3639-3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Palmer, K. L., L. M. Aye, and M. Whiteley. 2007. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 189:8079-8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pan, J., and D. Ren. 2009. Quorum sensing inhibitors: a patent overview. Expert Opin. Ther. Pat. 19:1581-1601. [DOI] [PubMed] [Google Scholar]
- 125.Pearson, T., J. M. U'Ren, J. M. Schupp, G. J. Allan, P. G. Foster, M. J. Mayo, D. Gal, J. L. Choy, R. L. Daugherty, S. Kachur, C. L. Friedman, B. Leadem, S. Georgia, H. Hornstra, A. J. Vogler, D. M. Wagner, P. Keim, and B. J. Currie. 2007. VNTR analysis of selected outbreaks of Burkholderia pseudomallei in Australia. Infect. Genet. Evol. 7:416-423. [DOI] [PubMed] [Google Scholar]
- 126.Peeters, E., H. J. Nelis, and T. Coenye. 2009. In vitro activity of ceftazidime, ciprofloxacin, meropenem, minocycline, tobramycin and trimethoprim/sulfamethoxazole against planktonic and sessile Burkholderia cepacia complex bacteria. J. Antimicrob. Chemother. 64:801-809. [DOI] [PubMed] [Google Scholar]
- 127.Pirone, L., L. Chiarini, C. Dalmastri, A. Bevivino, and S. Tabacchioni. 2005. Detection of cultured and uncultured Burkholderia cepacia complex bacteria naturally occurring in the maize rhizosphere. Environ. Microbiol. 7:1734-1742. [DOI] [PubMed] [Google Scholar]
- 128.Price, E. P., H. M. Hornstra, D. Limmathurotsakul, T. L. Max, D. S. Sarovich, A. J. Vogler, J. L. Dale, J. L. Ginther, B. Leadem, R. E. Colman, J. T. Foster, A. Tuanyok, D. M. Wagner, S. J. Peacock, T. Pearson, and P. Keim. 2010. Within-host evolution of Burkholderia pseudomallei in four cases of acute melioidosis. PLoS Pathog. 6:e1000725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ramette, A., J. J. LiPuma, and J. M. Tiedje. 2005. Species abundance and diversity of Burkholderia cepacia complex in the environment. Appl. Environ. Microbiol. 71:1193-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ramos, C. G., S. A. Sousa, A. M. Grilo, L. Eberl, and J. H. Leitão. 2010. The Burkholderia cenocepacia K56-2 pleiotropic regulator Pbr, is required for stress resistance and virulence. Microb. Pathog. 48:168-177. [DOI] [PubMed] [Google Scholar]
- 132.Rang, C., E. Alix, C. Felix, A. Heitz, L. Tasse, and A.-B. Blanc-Potard. 2007. Dual role of the MgtC virulence factor in host and non-host environments. Mol. Microbiol. 63:605-622. [DOI] [PubMed] [Google Scholar]
- 133.Reik, R., T. Spilker, and J. J. Lipuma. 2005. Distribution of Burkholderia cepacia complex species among isolates recovered from persons with or without cystic fibrosis. J. Clin. Microbiol. 43:2926-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Riedel, K., M. Hentzer, O. Geisenberger, B. Huber, A. Steidle, H. Wu, N. Høiby, M. Givskov, S. Molin, and L. Eberl. 2001. N-Acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology 147:3249-3262. [DOI] [PubMed] [Google Scholar]
- 135.Rogers, C. S., D. A. Stoltz, D. K. Meyerholz, L. S. Ostedgaard, T. Rokhlina, P. J. Taft, M. P. Rogan, A. A. Pezzulo, P. H. Karp, O. A. Itani, A. C. Kabel, C. L. Wohlford-Lenane, G. J. Davis, R. A. Hanfland, T. L. Smith, M. Samuel, D. Wax, C. N. Murphy, A. Rieke, K. Whitworth, A. Uc, T. D. Starner, K. A. Brogden, J. Shilyansky, P. B. McCray, Jr., J. Zabner, R. S. Prather, and M. J. Welsh. 2008. Disruption of the CFTR gene produces a model of cystic fibrosis in newborn pigs. Science 321:1837-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ryan, R. P., Y. McCarthy, S. A. Watt, K. Niehaus, and J. M. Dow. 2009. Intraspecies signaling involving the diffusible signal factor BDSF (cis-2-dodecenoic acid) influences virulence in Burkholderia cenocepacia. J. Bacteriol. 191:5013-5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Saini, L. S., S. B. Galsworthy, M. A. John, and M. A. Valvano. 1999. Intracellular survival of Burkholderia cepacia complex isolates in the presence of macrophage cell activation. Microbiology 145:3465-3475. [DOI] [PubMed] [Google Scholar]
- 138.Sajjan, S. U., L. A. Carmody, C. F. Gonzalez, and J. J. LiPuma. 2008. A type IV secretion system contributes to intracellular survival and replication of Burkholderia cenocepacia. Infect. Immun. 76:5447-5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sajjan, U. S., F. A. Sylvester, and J. F. Forstner. 2000. Cable-piliated Burkholderia cepacia binds to cytokeratin 13 of epithelial cells. Infect. Immun. 68:1787-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sajjan, U. S., J. H. Yang, M. B. Hershenson, and J. J. LiPuma. 2006. Intracellular trafficking and replication of Burkholderia cenocepacia in human cystic fibrosis airway epithelial cells. Cell. Microbiol. 8:1456-1466. [DOI] [PubMed] [Google Scholar]
- 141.Saldías, M. S., J. Lamothe, R. Wu, and M. A. Valvano. 2008. Burkholderia cenocepacia requires the RpoN sigma factor for biofilm formation and intracellular trafficking within macrophages. Infect. Immun. 76:1059-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Saldías, M. S., X. Ortega, and M. A. Valvano. 2009. Burkholderia cenocepacia O antigen lipopolysaccharide prevents phagocytosis by macrophages and adhesion to epithelial cells. J. Med. Microbiol. 58:1542-1548. [DOI] [PubMed] [Google Scholar]
- 143.Saldías, M. S., and M. A. Valvano. 2009. Interactions of Burkholderia cenocepacia and other Burkholderia cepacia complex bacteria with epithelial and phagocytic cells. Microbiology 155:2809-2817. [DOI] [PubMed] [Google Scholar]
- 144.Santos, A. V., R. J. Dillon, V. M. Dillon, S. E. Reynolds, and R. I. Samuels. 2004. Occurrence of the antibiotic producing bacterium Burkholderia sp. in colonies of the leaf-cutting ant Atta sexdens rubropilosa. FEMS Microbiol. Lett. 239:319-323. [DOI] [PubMed] [Google Scholar]
- 145.Schwab, U., M. Leigh, C. Ribeiro, J. Yankaskas, K. Burns, P. Gilligan, P. Sokol, and R. Boucher. 2002. Patterns of epithelial cell invasion by different species of the Burkholderia cepacia complex in well-differentiated human airway epithelia. Infect. Immun. 70:4547-4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Seed, K. D., and J. J. Dennis. 2008. Development of Galleria mellonella as an alternative infection model for the Burkholderia cepacia complex. Infect. Immun. 76:1267-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Seeger, M., M. Zielinski, K. N. Timmis, and B. Hofer. 1999. Regiospecificity of dioxygenation of di- to pentachlorobiphenyls and their degradation to chlorobenzoates by the bph-encoded catabolic pathway of Burkholderia sp. strain LB400. Appl. Environ. Microbiol. 65:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Silvia, A. S., U. Martina, B. Alessandra, B. Margaret, W. Dieter, H. L. Jorge, M. Christoph, E. Leo, S.-C. Isabel, and D. Gerd. 2007. Virulence of Burkholderia cepacia complex strains in gp91phox−/− mice. Cell. Microbiol. 9:2817-2825. [DOI] [PubMed] [Google Scholar]
- 149.Sokol, P. A., P. Darling, S. Lewenza, C. R. Corbett, and C. D. Kooi. 2000. Identification of a siderophore receptor required for ferric ornibactin uptake in Burkholderia cepacia. Infect. Immun. 68:6554-6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sokol, P. A., P. Darling, D. E. Woods, E. Mahenthiralingam, and C. Kooi. 1999. Role of ornibactin biosynthesis in the virulence of Burkholderia cepacia: characterization of pvdA, the gene encoding l-ornithine N(5)-oxygenase. Infect. Immun. 67:4443-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sokol, P. A., U. Sajjan, M. B. Visser, S. Gingues, J. Forstner, and C. Kooi. 2003. The CepIR quorum-sensing system contributes to the virulence of Burkholderia cenocepacia respiratory infections. Microbiology 149:3649-3658. [DOI] [PubMed] [Google Scholar]
- 152.Sousa, S. A., C. G. Ramos, F. Almeida, L. Meirinhos-Soares, J. Wopperer, S. Schwager, L. Eberl, and J. H. Leitão. 2008. Burkholderia cenocepacia J2315 acyl carrier protein: a potential target for antimicrobials' development? Microb. Pathog. 45:331-336. [DOI] [PubMed] [Google Scholar]
- 153.Speert, D. P., D. Henry, P. Vandamme, M. Corey, and E. Mahenthiralingam. 2002. Epidemiology of Burkholderia cepacia complex in patients with cystic fibrosis, Canada. Emerg. Infect. Dis. 8:181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Spiess, C., A. Beil, and M. Ehrmann. 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97:339-347. [DOI] [PubMed] [Google Scholar]
- 155.Spilker, T., A. Z. Uluer, F. M. Marty, W. W. Yeh, J. H. Levison, P. Vandamme, and J. J. LiPuma. 2008. Recovery of Herbaspirillum species from persons with cystic fibrosis. J. Clin. Microbiol. 46:2774-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Stites, S. W., M. W. Plautz, K. Bailey, A. R. O'Brien-Ladner, and L. J. Wesselius. 1999. Increased concentrations of iron and isoferritins in the lower respiratory tract of patients with stable cystic fibrosis. Am. J. Respir. Crit. Care Med. 160:796-801. [DOI] [PubMed] [Google Scholar]
- 157.Subsin, B., C. E. Chambers, M. B. Visser, and P. A. Sokol. 2007. Identification of genes regulated by the cepIR quorum-sensing system in Burkholderia cenocepacia by high-throughput screening of a random promoter library. J. Bacteriol. 189:968-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Thomas, M. 2007. Iron acquisition mechanisms of the Burkholderia cepacia complex. BioMetals 20:431-452. [DOI] [PubMed] [Google Scholar]
- 159.Tomich, M., A. Griffith, C. A. Herfst, J. L. Burns, and C. D. Mohr. 2003. Attenuated virulence of a Burkholderia cepacia type III secretion mutant in a murine model of infection. Infect. Immun. 71:1405-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tomich, M., C. A. Herfst, J. W. Golden, and C. D. Mohr. 2002. Role of flagella in host cell invasion by Burkholderia cepacia. Infect. Immun. 70:1799-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tomlin, K. L., O. P. Coll, and H. Ceri. 2001. Interspecies biofilms of Pseudomonas aeruginosa and Burkholderia cepacia. Can. J. Microbiol. 47:949-954. [PubMed] [Google Scholar]
- 162.Tomlin, K. L., R. J. Malott, G. Ramage, D. G. Storey, P. A. Sokol, and H. Ceri. 2005. Quorum-sensing mutations affect attachment and stability of Burkholderia cenocepacia biofilms. Appl. Environ. Microbiol. 71:5208-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Turner, J., Y. Cho, N.-N. Dinh, A. J. Waring, and R. I. Lehrer. 1998. Activities of LL-37, a cathelin-associated antimicrobial peptide of human neutrophils. Antimicrob. Agents Chemother. 42:2206-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Uehlinger, S., S. Schwager, S. P. Bernier, K. Riedel, D. T. Nguyen, P. A. Sokol, and L. Eberl. 2009. Identification of specific and universal virulence factors in Burkholderia cenocepacia strains using multiple infection hosts. Infect. Immun. 77:4102-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Urban, T. A., A. Griffith, A. M. Torok, M. E. Smolkin, J. L. Burns, and J. B. Goldberg. 2004. Contribution of Burkholderia cenocepacia flagella to infectivity and inflammation. Infect. Immun. 72:5126-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Vaara, M., T. Vaara, M. Jensen, I. Helander, M. Nurminen, E. T. Rietschel, and P. H. Mäkelä. 1981. Characterization of the lipopolysaccharide from the polymyxin-resistant pmrA mutants of Salmonella typhimurium. FEBS Lett. 129:145-149. [DOI] [PubMed] [Google Scholar]
- 167.Valvano, M. A. 2006. Infections by Burkholderia spp.: the psychodramatic life of an opportunistic pathogen. Future Microbiol. 1:145-149. [DOI] [PubMed] [Google Scholar]
- 168.Vandamme, P., B. Holmes, T. Coenye, J. Goris, E. Mahenthiralingam, J. J. LiPuma, and J. R. Govan. 2003. Burkholderia cenocepacia sp. nov.—a new twist to an old story. Res. Microbiol. 154:91-96. [DOI] [PubMed] [Google Scholar]
- 169.Vandamme, P., B. Holmes, M. Vancanneyt, T. Coenye, B. Hoste, R. Coopman, H. Revets, S. Lauwers, M. Gillis, K. Kersters, and J. R. Govan. 1997. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Evol. Microbiol. 47:1188-1200. [DOI] [PubMed] [Google Scholar]
- 170.Vanlaere, E., A. Baldwin, D. Gevers, D. Henry, E. De Brandt, J. J. Lipuma, E. Mahenthiralingam, D. P. Speert, C. Dowson, and P. Vandamme. 2009. Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. Int. J. Syst. Evol. Microbiol. 59:102-111. [DOI] [PubMed] [Google Scholar]
- 171.Vanlaere, E., J. J. Lipuma, A. Baldwin, D. Henry, E. De Brandt, E. Mahenthiralingam, D. Speert, C. Dowson, and P. Vandamme. 2008. Burkholderia latens sp. nov., Burkholderia diffusa sp. nov., Burkholderia arboris sp. nov., Burkholderia seminalis sp. nov. and Burkholderia metallica sp. nov., novel species within the Burkholderia cepacia complex. Int. J. Syst. Evol. Microbiol. 58:1580-1590. [DOI] [PubMed] [Google Scholar]
- 172.Visser, M. B., S. Majumdar, E. Hani, and P. A. Sokol. 2004. Importance of the ornibactin and pyochelin siderophore transport systems in Burkholderia cenocepacia lung infections. Infect. Immun. 72:2850-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Welman, A. D., and I. S. Maddox. 2003. Exopolysaccharides from lactic acid bacteria: perspectives and challenges. Trends Biotechnol. 21:269-274. [DOI] [PubMed] [Google Scholar]
- 174.Whitby, P. W., T. M. VanWagoner, J. M. Springer, D. J. Morton, T. W. Seale, and T. L. Stull. 2006. Burkholderia cenocepacia utilizes ferritin as an iron source. J. Med. Microbiol. 55:661-668. [DOI] [PubMed] [Google Scholar]
- 175.Winfield, M. D., T. Latifi, and E. A. Groisman. 2005. Transcriptional regulation of the 4-amino-4-deoxy-l-arabinose biosynthetic genes in Yersinia pestis. J. Biol. Chem. 280:14765-14772. [DOI] [PubMed] [Google Scholar]
- 176.Winkelstein, J. A., M. C. Marino, R. B. Johnston, Jr., J. Boyle, J. Curnutte, J. I. Gallin, H. L. Malech, S. M. Holland, H. Ochs, P. Quie, R. H. Buckley, C. B. Foster, S. J. Chanock, and H. Dickler. 2000. Chronic granulomatous disease. Report on a national registry of 368 patients. Medicine (Baltimore) 79:155-169. [DOI] [PubMed] [Google Scholar]
- 177.Winsor, G. L., B. Khaira, T. Van Rossum, R. Lo, M. D. Whiteside, and F. S. L. Brinkman. 2008. The Burkholderia Genome Database: facilitating flexible queries and comparative analyses. Bioinformatics 24:2803-2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wu, T., A. C. McCandlish, L. S. Gronenberg, S. S. Chng, T. J. Silhavy, and D. Kahne. 2006. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 103:11754-11759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yoder-Himes, D. R., P. S. G. Chain, Y. Zhu, O. Wurtzel, E. M. Rubin, J. M. Tiedje, and R. Sorek. 2009. Mapping the Burkholderia cenocepacia niche response via high-throughput sequencing. Proc. Natl. Acad. Sci. U. S. A. 106:3976-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yoder-Himes, D. R., K. T. Konstantinidis, and J. M. Tiedje. 2010. Identification of potential therapeutic targets for Burkholderia cenocepacia by comparative transcriptomics. PLoS One 5:e8724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhang, R., J. J. LiPuma, and C. F. Gonzalez. 2009. Two type IV secretion systems with different functions in Burkholderia cenocepacia K56-2. Microbiology 155:4005-4013. [DOI] [PubMed] [Google Scholar]
- 182.Zlosnik, J. E. A., T. J. Hird, M. C. Fraenkel, L. M. Moreira, D. A. Henry, and D. P. Speert. 2008. Differential mucoid exopolysaccharide production by members of the Burkholderia cepacia complex. J. Clin. Microbiol. 46:1470-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]