Abstract
Clostridium perfringens type A food poisoning is the second most commonly identified bacterial food-borne illness. Sporulation contributes to this disease in two ways: (i) most food-poisoning strains form exceptionally resistant spores to facilitate their survival of food-associated stresses, and (ii) the enterotoxin (CPE) responsible for the symptoms of this food poisoning is synthesized only during sporulation. In Bacillus subtilis, four alternative sigma factors mediate sporulation. The same four sigma factors are encoded by C. perfringens genomes, and two (SigE and SigK) have previously been shown to be necessary for sporulation and CPE production by SM101, a transformable derivative of a C. perfringens food-poisoning strain (K. H. Harry, R. Zhou, L. Kroos, and S. B. Melville, J. Bacteriol. 2009, 191:2728-2742). However, the importance of SigF and SigG for C. perfringens sporulation or CPE production had not yet been assessed. In the current study, after confirming that sporulating wild-type SM101 cultures produce SigF (from a tricistronic operon) and SigG, we prepared isogenic sigF- or sigG-null mutants. Whereas SM101 formed heat-resistant, phase-refractile spores, spore formation was blocked in the sigF- and sigG-null mutants. Complementation fully restored sporulation by both mutants. By use of these mutants and complementing strains, CPE production was shown to be SigF dependent but SigG independent. This finding apparently involved regulation of the production of SigE and SigK, which Harry et al. showed to be necessary for CPE synthesis, by SigF. By combining these findings with those previous results, it is now apparent that all four alternative sigma factors are necessary for C. perfringens sporulation, but only SigE, SigF, and SigK are needed for CPE synthesis.
Clostridium perfringens type A isolates producing enterotoxin (CPE) are the second most commonly identified cause of bacterial food-borne disease in the United States, where an estimated 250,000 cases of C. perfringens type A food poisoning occur annually (18, 19). This food poisoning begins with the consumption of foods contaminated with vegetative cells of an enterotoxigenic type A strain. Those ingested bacteria then sporulate in the small intestine and produce CPE, which has been shown to cause the diarrheal and abdominal-cramping symptoms that characterize C. perfringens type A food poisoning (18, 24). CPE-producing type A strains are also an important cause of human non-food-borne gastrointestinal diseases (2, 24). For example, it has been estimated that these bacteria account for 5 to 15% of all cases of antibiotic-associated diarrhea (2).
Enterotoxigenic type A isolates produce CPE only during sporulation (18). The sporulation-associated nature of CPE expression is controlled at the transcriptional level, since both Northern blotting and reporter construct studies detected enterotoxin gene (cpe) transcription in sporulating cultures, but not in vegetative cultures, of CPE-positive C. perfringens strains (4, 30). CPE can account for as much as 20% of the total protein present in a sporulating C. perfringens cell (5). Three strong promoters controlling cpe transcription have been identified upstream of the cpe open reading frame (ORF) (30), probably explaining why type A isolates often produce such high levels of CPE during sporulation.
Interestingly, the sequence of one cpe promoter (named P1) resembles a Bacillus subtilis SigK-dependent promoter, while the sequences of the other two cpe promoters (named P2 and P3) show similarity to B. subtilis SigE-dependent promoters (30). SigE and SigK are two of the four sporulation-associated sigma factors produced by Bacillus subtilis, which has traditionally been the prototype for the study of sporulation in Gram-positive bacteria (6, 11). These four sporulation-associated sigma factors are now well established as major regulators of B. subtilis sporulation, where SigF and SigG regulate gene expression in the forespore, while SigE and SigK control gene expression in the mother cell (6, 11). In B. subtilis, expression of these sporulation-associated sigma factors occurs in an ordered temporal cascade, with SigF appearing first, followed sequentially by SigE, SigG, and SigK (6, 11).
Sporulation has been much less studied in C. perfringens than in B. subtilis, but homologues of genes encoding SigE, SigF, SigG, and SigK have been identified in C. perfringens genomes (22, 27). In addition, a recent study by Harry et al. (7) used reverse transcription-PCR (RT-PCR) to detect transcripts of all four sigma factors in sporulating cultures of the CPE-positive C. perfringens type A isolate SM101, a transformable derivative of a food-poisoning isolate. Furthermore, Western blot analyses performed in that study confirmed SigE and SigK production by SM101 growing in Duncan-Strong (DS) sporulation medium (7). Since putative SigK- and SigE-dependent promoters had previously been identified upstream of the cpe gene (30), Harry et al. constructed sigK- and sigE-null mutants of SM101 in order to evaluate the importance of SigK and SigE for sporulation and CPE production by SM101. Consistent with the dependence of cpe transcription on these two sporulation-associated sigma factors, both mutants failed to drive β-glucuronidase production when transformed with a plasmid construct carrying the cpe promoter region fused to the Escherichia coli reporter gene gusA. In addition, neither the sigK- nor the sigE-null mutant of SM101 could sporulate.
The recent study by Harry et al. (7) also suggested differences in some developmental events during the sporulation of C. perfringens strain SM101 versus B. subtilis. For example, that recent study suggested that the normal accumulation of sigF and sigE transcripts in sporulating SM101 cells is dependent on SigK, whereas SigK is the last sigma factor expressed during B. subtilis sporulation. In addition, sigF transcript accumulation was reportedly delayed in a SM101 sigE-null mutant, even though sigF is the first transcript produced during B. subtilis sporulation. Harry et al. also suggested that C. perfringens and B. subtilis regulate SigG production differently, since (i) sigG transcripts were detected in SM101 sigE and sigK mutants reportedly containing little or no sigE or sigF transcript, while (ii) sigG transcription by B. subtilis requires both SigE and SigF. Finally, transcripts of all four sigma factor genes were detected much earlier in SM101 than has been reported for B. subtilis.
Given those apparent differences between sporulation in C. perfringens and that in B. subtilis, the current study constructed isogenic SM101 sigF- and sigG-null mutants and complementing strains. Those strains were then employed to explore whether these two sigma factors might also be involved in sporulation and CPE production by SM101.
MATERIALS AND METHODS
Bacterial strains, media, and reagents.
C. perfringens type A isolate SM101, a derivative of food-poisoning isolate NCTC8798 (Hobbs serotype 9), is transformable by electroporation (30). ATCC 3624 is a transformable, CPE-negative type A isolate (5).
Isolates were stored in cooked-meat medium (Oxoid) at −20°C. FTG (fluid thioglycolate; Difco Laboratories) and TGY broth (3% tryptic soy broth [Becton Dickinson and Company], 2% glucose [Fisher Scientific], 1% yeast extract [Difco], and 0.1% sodium thioglycolate [Sigma Chemical]) were used for growing vegetative cultures. Brain heart infusion (BHI) agar (Difco) supplemented with 15 μg/ml of chloramphenicol (Fisher Scientific) was used in screening to identify mutant clones. DS sporulation medium was used to induce the sporulation of C. perfringens strains (4). E. coli DH5α was grown at 37°C in LB broth, with shaking, or on LB agar containing 30 μg/ml chloramphenicol.
Construction of sigF- and sigG-null mutants of C. perfringens isolate SM101.
The sigF and sigG genes of SM101 were inactivated by insertion of a group II intron using the Clostridium-modified TargeTron (Sigma) insertional mutagenesis system (3). Utilizing optimal intron insertion sites identified in the SM101 genome sequence (22) and the Sigma TargeTron website, an intron was targeted, in the sense orientation, between nucleotides 86 and 87 of the sigF ORF. The primers used for PCR targeting the intron to the sigF ORF were 86/87s-IBS (5′-AAAAAAGCTTATAATTATCCTTAGGAAACAAGGAAGTGCGCCCAGATAGGGTG-3′), 86/87s-EBS1d (5′-CAGATTGTACAAATGTGGTGATAACAGATAAGTCAAGGAAGCTAACTTACCTTTCTTTGT-3′), and 86/87s-EBS2 (5′-TGAACGCAAGTTTCTAATTTCGATTTTTCCTCGATAGAGGAAAGTGTCT-3′). Another intron was inserted, in the sense orientation, between nucleotides 159 and 160 of the sigG ORF. This intron was prepared by PCR using primers 159/160s-IBS (5′-AAAAAAGCTTATAATTATCCTTAAGAAACAAGAAGTGCGCCCAGATAGGGTG-3′), 159/160s-EBS1d (5′-CAGATTGTACAAATGTGGTGATAACAGATAAGTCAAGAAGATTAACTTACCTTTCTTTGT-3′), and 159/160s-EBS2 (5′-TGAACGCAAGTTTCTAATTTCGATTTTTCTTCGATAGAGGAAAGTGTCT-3′). The 350-bp PCR products were inserted into pJIR750ai in order to construct sigF- and sigG-specific TargeTron plasmids. The resultant plasmids, named pJIR750sigFi and pJIR750sigGi, respectively, were electroporated into wild-type SM101. The transformation efficiency for SM101 was about 5 × 104 transformants/μg plasmid DNA. Transformants were selected on BHI agar plates containing 15 μg/ml of chloramphenicol, and cells carrying an intron insertion were screened by PCR using primers sigFKOF (5′-AACTCTCATAGTCATGGCTAAAG-3′) and sigFKOR (5′-AGTTCTTCCTTATTGTAA-TGAAGC-3′) for sigF-null mutants (SM101::sigF) or primers sigGKOF (5′-TGTATAGGACTTATGAAATCTATAG-3′) and sigGKOR (5′-ACATATATTGCATCTCCACCATC) for sigG-null mutants (SM101::sigG).
Each reaction mixture was subjected to the following PCR amplification conditions: cycle 1, 95°C for 2 min; cycles 2 through 35, 95°C for 30 s, 55°C for 40 s, and 68°C for 80 s; and a final extension for 8 min at 68°C. An aliquot (20 μl) of each PCR sample was electrophoresed on a 1.5% agarose gel and was then visualized by staining with ethidium bromide.
A digoxigenin (DIG)-labeled, intron sequence-specific probe was prepared, as described previously (3), using primers IBS and EBS1d and a DIG-labeling kit (Roche). That probe was then employed for Southern blotting to confirm the presence of a single intron insertion in the SM101::sigF and SM101::sigG mutants. Briefly, DNA from wild-type SM101, the sigF-null mutant, or the sigG-null mutant was isolated using the MasterPure Gram-positive DNA purification kit (Epicentre, Wisconsin). A 2.5-μg aliquot of each isolated DNA sample was digested overnight with EcoRI according to the manufacturer's (New England Biolabs) instructions. The digested DNA samples were then electrophoresed on a conventional 1% agarose gel. The separated DNA digestion products were transferred onto a nylon membrane (Roche) for hybridization with the intron probe, as described previously (3).
Construction of complementing strains for sigF- and sigG-null mutants.
The SM101 sigF-null mutant was complemented by cloning the entire sigF operon (see Results), including two upstream ORFs and the sigF ORF, along with 500 bp of upstream sequence and 500 bp of downstream sequence, into the pJIR750 E. coli/C. perfringens shuttle vector (1) and then transforming that plasmid into SM101::sigF. Briefly, DNA was isolated from wild-type SM101 as described above. PCR was then performed with that DNA using the Long-Range Taq DNA polymerase from New England Biolabs and primers sigFCOMN2F (5′-ttacgaattcGTGTAAATGGATTGTCTGTTATAG-3′ with an added EcoRI site [lowercase]) and SigFCOMR (5′-atgcctgcagCATTTATAATCAATCCTCTTCTAG-3′ with an added PstI site [lowercase]), designed according to sequencing results from a previous study (22). The PCR was performed in a Techne thermocycler using the following amplification conditions: 95°C for 2 min; 35 cycles of 95°C for 30 s, 55°C for 40 s, and 65°C for 3 min; and finally a single extension at 65°C for 10 min. The resultant 2.5-kb PCR product was cloned into the Topo 2.1 vector (Invitrogen). The Topo plasmid was then digested with EcoRI and PstI. The excised insert containing the sigF operon was gel purified and ligated into shuttle plasmid pJIR750, which had been digested with EcoRI and PstI, to produce plasmid pJIR750sigFcomp. The SM101 sigF-null mutant was transformed by electroporation with pJIR750sigFcomp; a complementing strain named SM101sigFcomp was then selected from those transformants by growth on a BHI agar plate containing 15 μg/ml of chloramphenicol.
The SM101 sigG-null mutant was complemented by cloning the sigG ORF and 500 bp of upstream sequence into pJIR750 and then transforming that new plasmid into SM101::sigG. Briefly, DNA was isolated from wild-type SM101 as described above. PCR was then performed using the Long-Range Taq DNA polymerase from New England Biolabs and primers SigGCOMF (5′-ttacgaattcTTGCAACCTATGCATCAAGATG-3′ with an added EcoRI site [lowercase]) and SigGCOMR (5′-atgcctgcagAGAGCTCTATACATACTTCCTC-3′ with an added PstI site [lowercase]), designed according to sequence results from a previous study (22). The PCR was performed in a Techne thermocycler and used the following amplification conditions: 95°C for 2 min; 35 cycles of 95°C for 30 s, 55°C for 40 s, and 65°C for 90 s; and finally a single extension at 65°C for 10 min. The resultant 1.2-kb PCR product was cloned into the Topo 2.1 vector (Invitrogen). The Topo plasmid was then digested with EcoRI and PstI. The excised insert containing sigG was gel purified and ligated into shuttle plasmid pJIR750, which had been digested with EcoRI and PstI, producing plasmid pJIR750sigGcomp. The SM101 sigG-null mutant was transformed by electroporation with pJIR750sigGcomp; the complementing strain SM101sigGcomp was then selected on BHI agar plates containing 15 μg/ml of chloramphenicol.
RNA extraction and RT-PCR.
To conduct a time course study of cpe gene expression during sporulation, a starter culture of wild-type SM101 was first grown overnight at 37°C in FTG. A 0.1-ml aliquot of that culture was then transferred to 10 ml of DS medium, which was incubated at 37°C for 1 to 8 h, with sample aliquots removed each hour. After centrifugation of each removed culture aliquot, total C. perfringens RNA was extracted from the pelleted cells by using saturated phenol (Fisher Scientific), as described in a previous study (29). All phenol-extracted samples were then treated with 2 U of DNase I (Ambion) at 37°C for 30 min. To stop this DNase I activity, a DNase I inhibitor (Ambion) was added to each reaction tube. RNA was quantified by absorbance at 260 nm and was stored at −80°C for no more than 1 month.
RT-PCR analysis for cpe gene transcription was then performed with these DNase-treated RNA samples, using the AccessQuick RT-PCR kit from Promega. Briefly, each RNA sample (100 ng) was reverse transcribed to cDNA at 45°C for 1 h, and the cDNA was then used as a template for PCR with primers targeting cpe sequences or spo0A (as a control housekeeping gene). Control RT-PCRs were performed similarly, except for the omission of reverse transcriptase. As an additional control, a PCR amplifying cpe or spo0A sequences was performed using DNA extracted from the MasterPure Gram-positive DNA purification kit as described above. The PCR primers used to amplify the cpe gene were 3F and 4R (21), and the spo0A primers were Spo0A-F (5′-AACAACCAGATTTAGTTGTATTAG-3′) and Spo0A-R (5′-CTCTATTTGTCCTCT-TCCCCAAGC-3′). The RT-PCR conditions used were as follows: 95°C for 2 min; 45°C for 1 h; 35 cycles of 95°C for 30 s, 55°C for 40 s, and 72°C for 40 s; and finally a single extension at 72°C for 5 min.
Overlapping RT-PCR analyses of sigF operon expression were performed using 100 ng of RNA extracted from SM101 grown for 5 h in DS medium as described above. The primers used (Table 1) spanned the sigF ORF, three upstream ORFs, and one downstream ORF. Each RT-PCR mixture contained 1 μl of template RNA (100 ng), 10 μl of 2× TAQ Complete mix, 0.5 μl transcriptase, and 1 μl of each primer pair (final concentration, 1 μM). The RT-PCR conditions used were as follows: 95°C for 2 min; 45°C for 1 h; 35 cycles of 95°C for 30 s, 55°C for 40 s, and 72°C for 1 min 40 s; and finally a single extension at 72°C for 5 min. One control RT-PCR was performed similarly using spo0A primers, except for the omission of reverse transcriptase. Another control reaction used these primers and DNA extracted from the MasterPure Gram-positive DNA purification kit as described above.
TABLE 1.
Primers using sigF operon overlap PCR
| Primer | Sequence | Product size (bp) |
|---|---|---|
| sigFCOMN2F | 5′-GTGTAAATGGATTGTCTGTTATAG-3′ | 853 |
| sigFNR | 5′-TTCTGCACTATGATGATCTAATTC-3′ | |
| sigFCOMpNF | 5′-AATTAGATCATCATAGTGCAGAAG-3′ | 400 |
| sigFR1 | 5′-CTTCAATAGTTGGATCTAACTGAG-3′ | |
| sigFCOMpF | 5′-CTCAGTTAGATCCAACTATTGAAG-3′ | 729 |
| sigFKOR | 5′-AGTTCTTCCTTATTGTAATGAAGC-3′ | |
| sigFF2 | 5′-GCTTCATTACAATAAGGAAGAACT-3′ | 364 |
| sigFR | 5′-CTCTATTCTAGAAACTTGAACTTG-3′ | |
| sigFFN | 5′-CAAGTTCAAGTTTCTAGAATAGAG-3′ | 460 |
| sigFR2 | 5′-CTATTTTATCATATAATCCAAGTCC-3′ |
Northern blot analysis of sigF transcription by wild-type SM101, a sigF-null mutant, and a complementing strain.
Total RNA was phenol extracted from SM101, SM101::sigF, and SM101sigFcomp using pelleted cells from a 5-h DS culture, as described above. Northern blot analysis was then performed using the Northern Max kit (a formaldehyde-based system for Northern blotting) from Ambion. Briefly, gel preparation, electrophoresis, transfer of RNA to a nylon membrane (Roche), prehybridization, and hybridization were each performed according to the manufacturer's instructions. A digoxigenin-labeled, sigF internal sequence-specific probe was prepared, as described previously (16, 26), using primers sigFKOF and sigFKOR and the DIG-labeling kit from Roche.
Western blotting.
For Western blot analysis of CPE production, a 0.2-ml aliquot of an FTG culture of wild-type SM101, SM101::sigF, SM101sigFcomp, SM101::sigG, or SM101sigGcomp was inoculated into 10 ml of DS sporulation medium. After incubation at 37°C for 8 h, each DS culture was visually examined by phase-contrast microscopy to assess the presence of spores. At that time point, aliquots of each DS culture were sonicated until more than 95% of cells had lysed. The sonicated samples were then centrifuged to remove debris and unlysed cells. The supernatant from each centrifuged DS culture lysate was analyzed for the presence of CPE by using a previously described CPE Western immunoblot procedure (10).
For Western immunoblot analysis of alternative sigma factor production, a 1.5-ml aliquot of a 5-h DS culture was centrifuged. The pellets were then resuspended in Tris-EDTA (TE) buffer, and 1 μl of Ready to Use Lysozyme (Epicentre) was added. After 1 h of incubation at 37°C, SDS loading dye was added, and the sample was boiled for 5 min. Western blot analysis of these samples to detect the presence of SigE, SigK, SigF, and SigG used a 1:1,000 dilution of rabbit antibodies raised specifically against purified recombinant B. subtilis sigma factor E, F, G, or K produced by E. coli. These antibodies were kindly supplied by Richard Losick and Masaya Fujita.
Quantitative spore counts.
Quantitative spore counts were determined as described previously (15). Briefly, an overnight DS culture of wild-type SM101, SM101::sigF, SM101::sigG, SM101sigFcomp, or SM101sigGcomp was heated at 75°C for 20 min to kill the remaining vegetative cells and to promote the germination of mature spores. Each heat-shocked suspension was then serially diluted from 10−2 to 10−7 with sterile water and was plated onto BHI agar plates, which were incubated anaerobically overnight at 37°C prior to colony counting. Quantitative spore counting was performed three times each for wild-type SM101, isogenic sigma factor mutants, and complementing strains.
Photomicroscopy.
Spore formation in 8-h DS cultures was documented using a Zeiss phase-contrast microscope (total magnification, ×1,000).
RESULTS
Construction of SM101 sigF- and sigG-null mutants.
As shown in Fig. 1, inspection of the published C. perfringens strain SM101 genomic sequence (22) revealed that the sigE, sigG, and sigK ORFs cluster together, while the sigF ORF is separated from those ORFs by ∼200 kb. While (i) a previous study had demonstrated that sigE and sigK are essential for CPE expression and sporulation by SM101 (7) and (ii) both SigF and SigG are necessary for B. subtilis sporulation (6), the importance (if any) of sigF or sigG for either C. perfringens sporulation or CPE expression had not yet been experimentally evaluated. Since significant differences between B. subtilis and C. perfringens sporulation have been reported (7), the current study used sigF- and sigG-null mutants to directly evaluate whether sigF and sigG are involved in CPE synthesis by, and sporulation of, SM101.
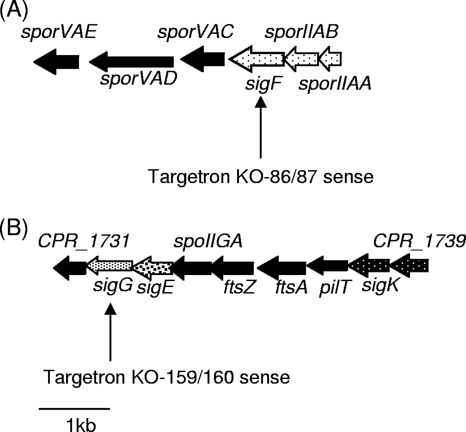
FIG. 1.
Arrangement of the sigma factor ORFs in C. perfringens SM101 based on previous genome-sequencing results (22). (A) Arrangement of the sigF locus in SM101. The results of the current study (see the text) indicate that sigF, sporIIAB, and sporIIAA are transcribed in one operon (dotted arrows). (B) Arrangement of the sigG, sigE, and sigK loci in SM101. As indicated by the black arrows with white dots, sigK and CPR_1739 are also transcribed in one operon, according to reference 7. A 1-kb scale marker is included for size comparisons.
The SM101 sigF- and sigG-null mutants were constructed by a TargeTron-mediated insertional mutagenesis approach (3). The identities of the putative mutants obtained from this mutagenesis procedure were first evaluated by PCR using primers corresponding to internal sigF or sigG ORF sequences (Fig. 2A). In wild-type SM101, these internal PCR primers amplified sigF- or sigG-specific products of ∼750 bp or ∼550 bp, respectively. However, with DNA from the intron-inactivated mutants, the same primers supported PCR amplification of products that were ∼900 bp larger than the products amplified using wild-type SM101 template DNA. The larger sizes of these PCR products amplified from both putative mutants matched the expected size change resulting from an intron insertion into the sigF or sigG ORF.
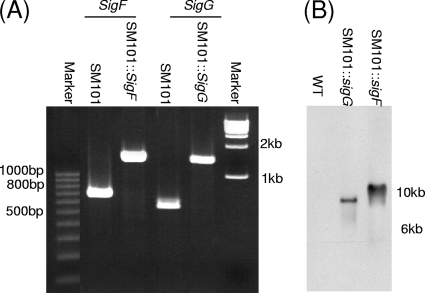
FIG. 2.
Intron-based insertional mutagenesis to create SM101 sigF- and sigG-null mutants. (A) Internal sigF- and sigG-specific PCR results for wild-type SM101, SM101::sigF, and SM101::sigG. The migration of 100-bp DNA markers is shown on the left, and the migration of 1-kb DNA markers is shown on the right. (B) Southern blot analysis of wild-type SM101 and SM101::sigF and SM101::sigG null mutant strains with a DIG-labeled, intron-specific probe. DNA size markers are shown on the right.
To specifically demonstrate the presence of an intron insertion in the sigF- and sigG-null mutants, and also to show that these mutants carried only a single intron insertion, Southern blot analysis was performed using an intron-specific probe (Fig. 2B). As expected, this experiment detected no hybridization of the intron-specific probe to wild-type SM101 DNA. In contrast, these Southern blot results showed the presence of a single intron insertion in the SM101::sigF and SM101::sigG mutants.
Disruption of sigF or sigG expression by the two mutants was then assessed directly using Western blotting (Fig. 3A and B). These analyses first demonstrated that wild-type SM101 produces both SigF and SigG during sporulation. Furthermore, these Western blots also showed the complete loss of SigF or SigG production by sporulating cultures of the sigF- or sigG-null mutant, respectively.
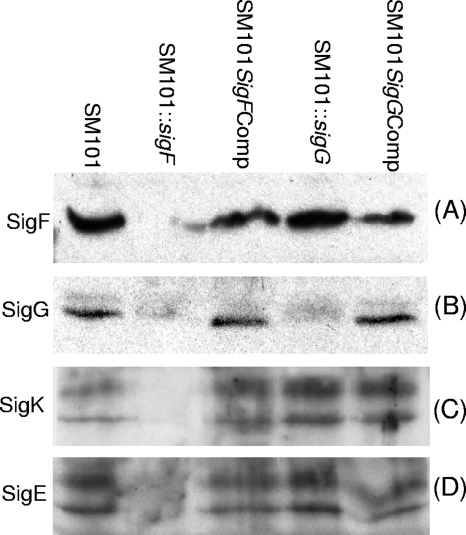
FIG. 3.
Western blot analyses of alternative sigma factor production by SM101. Shown are Western blot results for the production of SigF (A), SigG (B), SigK (C), and SigE (D) by DS cultures of wild-type, SM101::sigF, SM101sigFcomp, SM101::sigG, and SM101sigGcomp strains grown for 5 h at 37°C. See Results for discussion of the two SigE or SigK bands present in some lanes.
Complementation of the SM101 sigF- and sigG-null mutants.
The SM101::sigG mutant was complemented by PCR cloning, from wild-type SM101, of DNA corresponding to the sigG ORF and 500 bp of upstream sequence. This PCR product was cloned into the pJIR750 shuttle plasmid and was then transformed into the sigG-null mutant to create SM101::sigGcomp. The presence of a wild-type sigG gene in this complementing strain was confirmed by PCR (data not shown). SigG production by SM101::sigGcomp was then demonstrated directly by Western blotting (Fig. 3B).
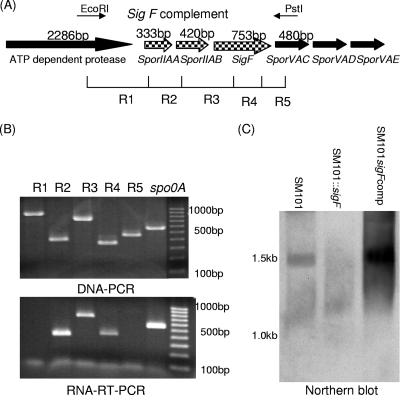
Complementation of the sigF mutant proved more complicated. Initial experiments indicated that transformation of a plasmid carrying a PCR product corresponding to the sigF ORF and ∼500 bp of upstream sequence failed to restore SigF expression to SM101::sigF (data not shown). Therefore, it was considered that the sigF ORF might be transcribed as part of an operon. To test this hypothesis, overlapping RT-PCR and PCR were first performed. The overlapping PCR primers for these analyses were designed to link sigF with three upstream genes, i.e., the ATP-dependent protease gene, sporIIAA, and sporIIAB, along with one downstream gene, i.e., sporVAC (Fig. 4A). Using wild-type SM101 DNA as the template for this overlap PCR assay, each of the five expected products was amplified (Fig. 4B), confirming the validity of the primers. However, using an SM101 RNA template, the RT-PCR amplified products only from reactions R2 to R4, indicating that the sporIIAA, sporIIAB, and sigF ORFs are cotranscribed as a single mRNA.
FIG. 4.
The sigF ORF is transcribed as part of a tricistronic operon. (A) Arrangement of the sigF locus in SM101, based on sequencing results (22). The locations of the primers used in panel B for overlap PCR are indicated by R1 to R5. (B) Overlap PCR analysis of the sigF locus region in five reactions (R1 to R5). The results of a DNA PCR (top) and an RNA RT-PCR (bottom) using reaction R1 to R5 primers (Table 1) are shown. Size markers are shown on the right. (C) Northern blot analysis using a DIG-labeled sigF probe. Results for wild-type, SM101::sigF, and SM101sigFcomp strains are shown. Size markers are shown on the left.
Northern blot analysis (Fig. 4C) supported this finding by indicating that the sigF gene is transcribed as a ∼1.5-kb mRNA, matching the expected size of a single mRNA encoding SporIIAA, SporIIAB, and SigF. In addition, these Northern blot analyses also failed to detect the presence of any sigF mRNA in the sigF-null mutant, consistent with the Western blot results in Fig. 3A.
Since both the overlap RT-PCR and Northern blot results indicated expression of the sigF gene as part of a tricistronic operon containing two upstream genes, a PCR product containing those three genes and 500 bp of upstream and downstream sequence was ligated into shuttle vector pJIR750, followed by transformation of the resultant plasmid (pJIR750sigFcomp) into the sigF-null mutant. Both Western blot and Northern blot analyses demonstrated SigF expression by this complementing strain (Fig. 3A and 4C), named SM101::sigFcomp.
Regulation of sigma factor production by SigF and SigG.
The sigF- and sigG-null mutants were first used to assess whether SigF or SigG might regulate each other's production. Western blotting (Fig. 3B) showed that the sigF mutant was impaired for production of SigG, while the SM101::sigFcomp complementing strain produced near-wild-type levels of SigG. In contrast, the sigG mutant was still able to produce wild-type levels of SigF (Fig. 3A).
The sigF- and sigG-null mutants were next employed to test whether SigF or SigG might regulate the production of SigE or SigK, which have previously been tied to C. perfringens sporulation and the regulation of CPE production (7). Western blot analyses comparing SigE and SigK production by sporulating cultures of wild-type SM101 revealed that, as expected, 5-h DS cultures grown at 37°C produce both SigE and SigK (Fig. 3C and D). The presence of two visible bands for SigE and SigK on Western blots has been observed previously for C. perfringens (7), and those bands were suggested to correspond to proprotein or mature, active forms of these two sigma factors.
Compared to the results for wild-type SM101 in Fig. 3, Western blotting detected sharply reduced or no production of SigE or SigK by the SM101 sigF-null mutant grown under the same incubation conditions for 5 h at 37°C in DS medium (Fig. 3C and D). Furthermore, complementation of the sigF-null mutant fully restored SigE and SigK production after a similar 5 h of incubation at 37°C in DS medium (Fig. 3C and D).
In contrast to these results indicating that SigF regulates the production of SigE and SigK, inactivation of the sigG gene had no effect on the production of either SigE or SigK in cultures grown for 5 h in DS medium at 37°C (Fig. 3).
Evidence that both SigF and SigG are essential for C. perfringens sporulation.
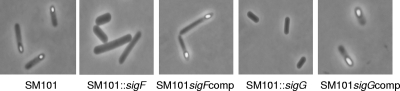
To address whether SigF or SigG is required for sporulation by wild-type SM101, the sigF- and sigG-null mutants (or complementing strains of those mutants) were first examined using phase-contrast microscopy (Fig. 5). This analysis showed that, by 8 h of incubation at 37°C in DS sporulation medium, wild-type SM101 had already formed refractile spores, with an efficiency of >95%. In contrast, the sigG- and sigF-null mutants failed to form any refractile spores after a similar 8 h of incubation in DS medium at 37°C. Furthermore, both mutants remained unable to form refractile spores, even when incubated for 24 or 48 h at 37°C in DS medium (not shown). However, complementation of either mutant restored sporulation after 8 h of incubation at 37°C in DS medium, by which time >95% of the complementing strain cells had formed refractile spores (Fig. 5).
FIG. 5.
Formation of phase-refractile spores by wild-type SM101, the sigF- and sigG-null mutants, and complementing strains. Shown are results for DS cultures of each strain after 8 h of growth at 37°C in DS. Photographs were taken using a phase-contrast microscope (magnification, ×1,000).
The sporulating abilities of SM101, the sigG- and sigF-null mutants, and complementing strains of those mutants were also directly compared by measuring the formation of heat-resistant spores after 24 h of incubation at 37°C in DS medium. Under these conditions, wild-type SM101 formed ∼ 7.7 × 108 ± 1.7 × 108 heat-resistant spores/ml. However, under the same incubation conditions, no heat-resistant spores were detected for either the SM101 sigG-null mutant or the SM101 sigF-null mutant. This inability of the sigG and sigF mutants to form heat-resistant spores was not due simply to slower spore production, since no heat-resistant spores were detected even when these mutants were incubated at 37°C in DS sporulation medium for 2, 3, or 7 days. Complementation of either sigma factor mutant restored the ability to form heat-resistant spores to near-wild-type levels following overnight incubation in DS medium at 37°C. Specifically, SM101sigFcomp formed 3.2 × 108 ± 1.5 ×108 heat-resistant spores/ml, and SM101sigGcomp formed 9.2 ×107 ± 0.4 ×107 heat-resistant spores/ml. Collectively, these results indicated that SigF and SigG are essential for the formation of heat-resistant mature spores by SM101.
CPE expression during sporulation is SigF dependent but does not require SigG.
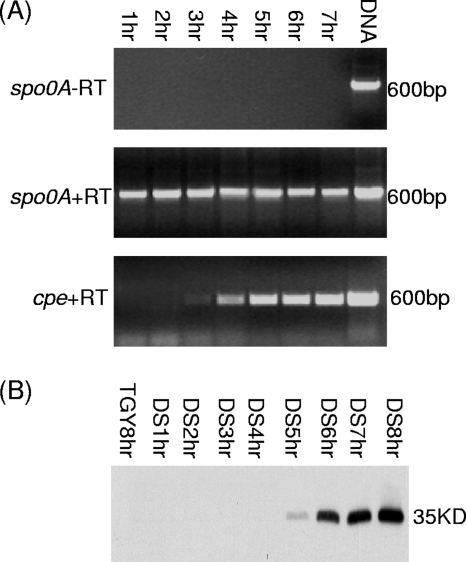
RT-PCR analyses showed that, for wild-type SM101 growing at 37°C in DS medium, cpe gene transcription starts at ∼3 to 4 h and then increases over the next several hours (Fig. 6). In addition, Western blot analyses demonstrated that, under the same incubation conditions, CPE production by SM101 became detectable within 5 h in DS cultures incubated at 37°C and then increased markedly by 6 to 8 h (Fig. 6 and 7). In contrast, no cpe transcription or CPE production was observed in 8-h cultures of SM101 grown at 37°C in TGY medium, a condition that does not trigger C. perfringens sporulation (data not shown). As another control, no immunoreactivity with an anti-CPE antibody was observed using 8-h DS cultures of the cpe-negative strain ATCC 3624.
FIG. 6.
Kinetics of cpe gene expression and CPE production by DS cultures of SM101. (A) RT-PCR analysis of cpe gene transcription in cultures growing from 1 to 7 h in DS medium at 37°C. The spo0A gene was used as a housekeeping gene, and a sample lacking reverse transcriptase (−RT) was run to show the absence of DNA contamination in these samples. The sizes of products, based on comparison against DNA size markers (not shown), are shown to the right of each gel. (B) Western blot analyses of CPE production after 1 to 7 h of growth in DS medium at 37°C. To show that CPE production is dependent on sporulation, this figure shows the absence of CPE production by an 8-h culture of SM101 grown at 37°C in TGY, a medium that does not induce sporulation. The size of the immunoreactive protein, based on the migration of prestained markers (not shown), is indicated on the right.
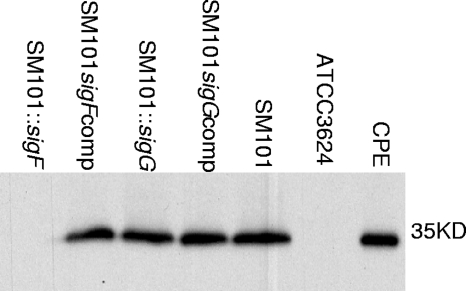
FIG. 7.
Comparison of CPE production by wild-type SM101, sigF- and sigG-null mutants, and complementing strains. Western blot results for CPE production are shown for sporulating 8-h DS cultures incubated at 37°C. As controls, similar analyses are shown for an 8-h DS culture of the CPE-negative strain ATCC 3624 grown at 37°C and for purified CPE. The size of the immunoreactive protein, based on the migration of prestained markers (not shown), is given on the right.
Similar Western blot analyses of 8-h DS cultures incubated at 37°C were used to evaluate CPE production by the sigF- and sigG-null mutants. Those Western blot studies detected no CPE production by the SM101::sigF strain (Fig. 7). However, under the same incubation conditions, complementation of the sigF mutant to restore SigF expression also restored CPE production to near-wild-type SM101 levels. Collectively, these results demonstrated that SigF is required for CPE expression.
In contrast, similar Western blot experiments revealed that 8-h DS cultures of the sigG-null mutant still produced normal levels of CPE, as did similar cultures of the complementing strain for that mutant. Therefore, SigG is necessary for the production of spores by SM101 but is not required for CPE production.
DISCUSSION
The ability to sporulate is an integral contributor to several important C. perfringens diseases. For example, contamination of wounds with spores can lead to gas gangrene (28). Spores also contribute to the transmission of C. perfringens type A food poisoning, particularly because the survival of food-poisoning strains is often facilitated by the formation of spores that are exceptionally resistant to food environment stresses, such as heat, cold, and preservatives (13-15, 17, 23, 25). Furthermore, the sporulation process itself is essential for C. perfringens type A food poisoning, i.e., the enterotoxin (CPE) causing the gastrointestinal symptoms of this food-borne illness is produced only by sporulating cells (5, 18, 19). Therefore, knowledge of C. perfringens sporulation is important for full understanding of the pathogenesis of C. perfringens type A food poisoning and other C. perfringens diseases.
While C. perfringens sporulation is still understood only at a rudimentary level, both similarities to and differences from B. subtilis sporulation have been reported (7). Regarding sporulation similarities, both bacteria were found to utilize the transcription factor Spo0A (6, 8) and to express four alternative sigma factors that, for B. subtilis, have been shown to regulate sporulation-associated gene expression (6, 7). Among sporulation differences, C. perfringens lacks the phosphorelay that helps signal the start of B. subtilis sporulation (22, 27). Also, it has been suggested that the production of the four alternative sigma factors may be regulated differently in these two bacteria (7). Given these apparent differences between B. subtilis versus C. perfringens sporulation, it has been important to establish whether all four alternative sigma factors are required for C. perfringens sporulation. To begin addressing that question, Harry et al. showed that SigE and SigK are necessary for the sporulation of C. perfringens strain SM101 (7).
The current results have demonstrated that SigF and SigG are also required for C. perfringens sporulation, i.e., mutants unable to produce SigF or SigG were completely blocked for spore formation, and complementation restored sporulation to near-wild-type levels. This complementation of sigF and sigG mutants back to near-wild-type sporulation levels contrasts notably with results from the study by Harry et al. (7), where complementation restored only 1 to 2% of wild-type sporulation ability to sigE- and sigK-null mutants. Harry et al. suggested that their more limited complementation might be attributable to substantial wild-type sigE and sigK transcription from upstream genes, an effect not encoded by their complementing plasmid. This would be similar to our finding that, as in B. subtilis (20), SM101 transcribes sigF as part of a spoIIA tricistronic operon containing the sigF (spoIIAC), spoIIAA, and spoIIAB genes. This genetic similarity could suggest that, as in B. subtilis (20), SpoIIAA and SpoIIAB regulate SigF activity in SM101.
The current results add important support for another proposed similarity between the sporulation of SM101 and that of B. subtilis, where SigE and SigK, but not SigF or SigG, are initially produced as inactive proproteins that are then proteolytically processed to an active, mature form (6, 7). Specifically, our Western blot analyses showed that antibodies raised against SigF or SigG reacted only with single proteins that corresponded to the appropriate sigma factor based on size, and this immunoreactive band was absent from Western blot lanes containing lysates of sigF- or sigG-null mutants.
In B. subtilis, synthesis of the four alternative sigma factors begins during the initiation of sporulation and follows the temporal pattern of SigF appearing first, followed sequentially by SigE, SigG, and SigK (6). In contrast, Harry et al. (7) found that all four sigma factors accumulated rapidly after SM101 was inoculated into sporulation medium and that early accumulation of sigF transcript appeared to be SigK dependent, suggesting that SigK might act prior to SigF in the C. perfringens sporulation regulatory network. Therefore, it was interesting that the current study detected little or no or SigK production by the sigF-null mutant, although production of that sigma factor was observed after complementation to restore SigF production. As expanded on below, our current results are most consistent with SM101 sporulation involving a cascade of alternative sigma factors reminiscent of the process in B. subtilis, where SigF regulates the production of SigG, SigE, and SigK. Notably, the same classical sporulation-associated sigma factor regulatory cascade has also been described for the nonpathogenic organism Clostridium acetobutylicum (9).
Harry et al. (7) also concluded that SigG regulation substantially differs between B. subtilis and C. perfringens. Whereas SigG production in B. subtilis is dependent on both SigF and SigE, the study by Harry et al. (7) had reported that a SM101 sigE-null mutant still produced SigG, even though it produced little or no SigE or SigF. However, our current study found that a SM101 sigF-null mutant no longer produced SigG. Consistent with that finding, bioinformatics analysis identified a consensus sigF promoter sequence upstream of sigG. These findings would suggest that, as in B. subtilis, SigG production by C. perfringens requires SigF.
As mentioned in the introduction, the P1 cpe promoter possesses sequence similarity to a SigK-dependent promoter, while the P2 and P3 cpe promoter sequences resemble sequences found in SigE-dependent promoters (30). Consistent with those homologies, Harry et al. recently used gusA fusion assays to conclude that cpe transcription is blocked in sigE- and sigK-null mutants of SM101 (7). The results from our current study now establish that cpe transcription and CPE production are also blocked in an SM101 sigF-null mutant. However, we also found that, despite the previously reported early appearance of sigG transcripts in sporulating SM101 cultures (7), a sigG-null mutant still shows normal levels of cpe transcription and CPE production, thus documenting that not all sporulation-associated sigma factors are required for cpe transcription and CPE production.
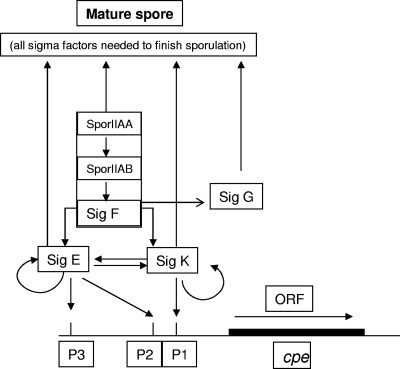
At least two possibilities could explain how SigF regulates cpe transcription. First, a cpe promoter might be SigF dependent. This possibility seems less likely, since promoter-mapping studies identified only SigE- and SigK-dependent cpe promoter sequences (30). A second possible explanation, shown in the Fig. 8 model, might be that (as with B. subtilis and C. acetobutylicum) SM101 uses SigF to regulate the production of other sporulation-associated sigma factors. This model is supported by the current Western blot results showing little or no production of SigE and SigK by the sigF-null mutant. This model also indicates that (as with B. subtilis and C. acetobutylicum [6, 9, 11]) SigG production by SM101 apparently requires SigF, consistent with the Western blot results of our current study.
FIG. 8.
Model describing sigma factor regulation of C. perfringens CPE production and sporulation. The model incorporates findings from the current work and research by Harry et al. (7). Not shown is the proven involvement of Spo0A in C. perfringens sporulation (8), since it is not yet clear how that protein leads to sporulation-associated sigma factor production by C. perfringens.
Because older studies (12) had suggested that CPE production levels may involve posttranscriptional regulatory effects not modeled by transcriptional fusion assays, the current study directly compared the timing of cpe transcription and CPE production in sporulating SM101 cultures. These analyses detected cpe transcription beginning ∼3 h after inoculation into DS medium. The appearance of CPE protein lagged slightly; it became identifiable by Western blotting at 4 to 5 h postinoculation. These results largely agree with the transcriptional fusion assay results of Harry et al. (7), who noted cpe promoter-driven GusA activity ∼3 to 4 h after inoculation into a slightly different sporulation medium. However, our results do not necessarily eliminate posttranscriptional contributions to CPE production, a topic worthy of further evaluation.
In summary, there is now increasing evidence for both similarities and differences between the sporulation of C. perfringens and that of B. subtilis. Sporulation similarities apparently include the involvement of Spo0A and SigF-mediated control of the other sporulation-associated sigma factors, while differences include the absence of a B. subtilis-like phosphorelay from C. perfringens. Future transcriptomic studies should identify transcripts regulated by each alternative sigma factor in C. perfringens. Another largely unstudied topic for C. perfringens sporulation concerns the initial signaling cascade that leads to Spo0A phosphorylation and triggers sporulation. Finally, the results of this study indicate a previously unappreciated level of complexity for the regulation of cpe transcription (i.e., the involvement of SigF). However, in combination with the earlier results of Harry et al., the now demonstrated involvement of three sporulation-associated sigma factors in the regulation of cpe transcription offers an explanation for the ∼40-year-old observation of a strong linkage between CPE production and sporulation.
Acknowledgments
This research was generously supported by grant R37 AI19844-27 from the National Institute of Allergy and Infectious Diseases.
We thank Richard Losick and Masaya Fujita for supplying the sigma factor antibodies used in this study.
Editor: A. Camilli
Footnotes
Published ahead of print on 19 July 2010.
REFERENCES
- 1.Bannam, T. L., and J. I. Rood. 1993. Clostridium perfringens-Escherichia coli shuttle vectors that carry single antibiotic resistance determinants. Plasmid 29:233-235. [DOI] [PubMed] [Google Scholar]
- 2.Carman, R. J. 1997. Clostridium perfringens in spontaneous and antibiotic-associated diarrhoea of man and other animals. Rev. Med. Microbiol. 8(Suppl. 1):S43-S45. [Google Scholar]
- 3.Chen, Y., B. A. McClane, D. J. Fisher, J. I. Rood, and P. Gupta. 2005. Construction of an alpha toxin gene knockout mutant of Clostridium perfringens type A by use of a mobile group II intron. Appl. Environ. Microbiol. 71:7542-7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Czeczulin, J. R., R. E. Collie, and B. A. McClane. 1996. Regulated expression of Clostridium perfringens enterotoxin in naturally cpe-negative type A, B, and C isolates of C. perfringens. Infect. Immun. 64:3301-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Czeczulin, J. R., P. C. Hanna, and B. A. McClane. 1993. Cloning, nucleotide sequencing, and expression of the Clostridium perfringens enterotoxin gene in Escherichia coli. Infect. Immun. 61:3429-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haldenwang, W. G. 1995. The sigma factors of Bacillus subtilis. Microbiol. Rev. 59:1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harry, K. H., R. Zhou, L. Kroos, and S. B. Melville. 2009. Sporulation and enterotoxin (CPE) synthesis are controlled by the sporulation-specific factors SigE and SigK in Clostridium perfringens. J. Bacteriol. 191:2728-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang, I. H., M. Waters, R. R. Grau, and M. R. Sarker. 2004. Disruption of the gene (spo0A) encoding sporulation transcription factor blocks endospore formation and enterotoxin production in enterotoxigenic Clostridium perfringens type A. FEMS Microbiol. Lett. 233:233-240. [DOI] [PubMed] [Google Scholar]
- 9.Jones, S. W., C. J. Paredes, B. Tracy, N. Cheng, R. Sillers, R. S. Senger, and E. T. Papoutsakis. 2008. The transcriptional program underlying the physiology of clostridial sporulation. Genome Biol. 9:R114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kokai-Kun, J. F., J. G. Songer, J. R. Czeczulin, F. Chen, and B. A. McClane. 1994. Comparison of Western immunoblots and gene detection assays for identification of potentially enterotoxigenic isolates of Clostridium perfringens. J. Clin. Microbiol. 32:2533-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kroos, L. 2007. The Bacillus and Myxococcus developmental networks and their transcriptional regulators. Annu. Rev. Genet. 41:13-39. [DOI] [PubMed] [Google Scholar]
- 12.Labbe, R. G., and C. L. Duncan. 1977. Evidence for stable messenger ribonucleic acid during sporulation and enterotoxin synthesis by Clostridium perfringens type A. J. Bacteriol. 129:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, J., and B. A. McClane. 2008. A novel small acid soluble protein variant is important for spore resistance of most Clostridium perfringens food poisoning isolates. PLoS Pathog. 4:e1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, J., and B. A. McClane. 2006. Comparative effects of osmotic, sodium nitrite-induced, and pH-induced stress on growth and survival of Clostridium perfringens type A isolates carrying chromosomal or plasmid-borne enterotoxin genes. Appl. Environ. Microbiol. 72:7620-7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, J., and B. A. McClane. 2006. Further comparison of temperature effects on growth and survival of Clostridium perfringens type A isolates carrying a chromosomal or plasmid-borne enterotoxin gene. Appl. Environ. Microbiol. 72:4561-4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, J., K. Miyamoto, and B. A. McClane. 2007. Comparison of virulence plasmids among Clostridium perfringens type E isolates. Infect. Immun. 75:1811-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, J., D. Paredes-Sabja, M. R. Sarker, and B. A. McClane. 2009. Further characterization of Clostridium perfringens small acid soluble protein-4 (Ssp4) properties and expression. PLoS One 4:e6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McClane, B. A. 2007. Clostridium perfringens, p. 423-444. In M. P. Doyle and L. R. Beuchat (ed.), Food microbiology: fundamentals and frontiers, 3rd ed. ASM Press, Washington, DC.
- 19.McClane, B. A., F. A. Uzal, M. F. Miyakawa, D. Lyerly, and T. Wilkins. 2006. The enterotoxic clostridia, p. 688-752. In M. Dworkin, S. Falkow, E. Rosenburg, K. H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed. Springer, New York, NY.
- 20.Min, K., C. M. Hilditch, B. Diederich, J. Errington, and M. D. Yudkin. 1993. σF, the first compartment-specific transcription factor of Bacillus subtilis, is regulated by an anti-σ factor that is also a protein kinase. Cell 74:735-742. [DOI] [PubMed] [Google Scholar]
- 21.Miyamoto, K., Q. Wen, and B. A. McClane. 2004. Multiplex PCR genotyping assay that distinguishes between isolates of Clostridium perfringens type A carrying a chromosomal enterotoxin gene (cpe) locus, a plasmid cpe locus with an IS1470-like sequence, or a plasmid cpe locus with an IS1151 sequence. J. Clin. Microbiol. 42:1552-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myers, G. S., D. A. Rasko, J. K. Cheung, J. Ravel, R. Seshadri, R. T. DeBoy, Q. Ren, J. Varga, M. M. Awad, L. M. Brinkac, S. C. Daugherty, D. H. Haft, R. J. Dodson, R. Madupu, W. C. Nelson, M. J. Rosovitz, S. A. Sullivan, H. Khouri, G. I. Dimitrov, K. L. Watkins, S. Mulligan, J. Benton, D. Radune, D. J. Fisher, H. S. Atkins, T. Hiscox, B. H. Jost, S. J. Billington, J. G. Songer, B. A. McClane, R. W. Titball, J. I. Rood, S. B. Melville, and I. T. Paulsen. 2006. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 16:1031-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orsburn, B., S. B. Melville, and D. Popham. 2008. Factors contributing to heat resistance of Clostridium perfringens endospores. Appl. Environ. Microbiol. 74:3328-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarker, M. R., R. J. Carman, and B. A. McClane. 1999. Inactivation of the gene (cpe) encoding Clostridium perfringens enterotoxin eliminates the ability of two cpe-positive C. perfringens type A human gastrointestinal disease isolates to affect rabbit ileal loops. Mol. Microbiol. 33:946-958. [DOI] [PubMed] [Google Scholar]
- 25.Sarker, M. R., R. P. Shivers, S. G. Sparks, V. K. Juneja, and B. A. McClane. 2000. Comparative experiments to examine the effects of heating on vegetative cells and spores of Clostridium perfringens isolates carrying plasmid versus chromosomal enterotoxin genes. Appl. Environ. Microbiol. 66:3234-3240. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Sayeed, S., J. Li, and B. A. McClane. 2007. Virulence plasmid diversity in Clostridium perfringens type D isolates. Infect. Immun. 75:2391-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. U. S. A. 99:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stevens, D. L., and J. I. Rood. 2006. Histotoxic clostridia, p. 715-725. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens, 2nd ed. ASM Press, Washington, DC.
- 29.Vidal, J. E., K. Ohtani, T. Shimizu, and B. A. McClane. 2009. Contact with enterocyte-like Caco2 cells induces rapid upregulation of toxin production by Clostridium perfringens type C isolates. Cell. Microbiol. 11:1306-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao, Y., and S. B. Melville. 1998. Identification and characterization of sporulation-dependent promoters upstream of the enterotoxin gene (cpe) of Clostridium perfringens. J. Bacteriol. 180:136-142. [DOI] [PMC free article] [PubMed] [Google Scholar]