Abstract
The amino sugar N-acetylglucosamine (GlcNAc) is known to be an important structural component of cells from bacteria to humans, but its roles in cell signaling are less well understood. GlcNAc induces two pathways in the human fungal pathogen Candida albicans. One activates cyclic AMP (cAMP) signaling, which stimulates the formation of hyphal cells and the expression of virulence genes, and the other pathway induces genes needed to catabolize GlcNAc. Microarray analysis of gene expression was carried out under four different conditions in order to characterize the transcriptional changes induced by GlcNAc. The most highly induced genes include those that encode a GlcNAc transporter (NGT1) and the GlcNAc catabolic enzymes (HXK1, DAC1, and NAG1). GlcNAc also activated most of the genes whose expression is increased when cells are triggered with other stimuli to form hyphae. Surprisingly, GlcNAc also induced a subset of genes that are regulated by galactose (GAL1, GAL7, and GAL10), which may be due to cross talk between signaling pathways. A novel GlcNAc-induced gene, GIG1, which is not essential for GlcNAc catabolism or the induction of hyphae, was identified. However, a Gig1-green fluorescent protein (GFP) fusion protein was specifically induced by GlcNAc, and not by other sugars. Gig1-GFP localized to the cytoplasm, where GlcNAc metabolism occurs. Significantly, a gig1Δ mutant displayed increased resistance to nikkomycin Z, which inhibits chitin synthase from converting UDP-GlcNAc into cell wall chitin. Gig1 is highly conserved in fungi, especially those that contain GlcNAc catabolic genes. These results implicate Gig1 in GlcNAc metabolism.
The amino sugar N-acetylglucosamine (GlcNAc) plays important roles in a wide range of organisms from bacteria to humans. One major role for GlcNAc is in the structure of the extracellular surfaces of cells. GlcNAc is converted to UDP-GlcNAc, which is a substrate for the transfer of the GlcNAc moiety to other macromolecules. For example, GlcNAc is an important component of the bacterial cell wall peptidoglycan layer and the extracellular matrix glycosaminoglycans in animal cells. In fungi, UDP-GlcNAc is also a substrate for the chitin synthases that form the cell wall chitin, which is a polymer of GlcNAc. UDP-GlcNAc is also used in eukaryotes to initiate N-linked glycosylation and for the synthesis of glycosylphosphatidylinositol (GPI) anchors on membrane proteins.
In addition to its structural roles, GlcNAc is also an important signaling molecule in bacteria, fungi, and animal cells. Bacterial cells respond to GlcNAc by modulating the production of curli fibers, which are important in biofilm formation (3). Studies with animal cells have demonstrated that many critical transcription factors, including c-myc, p53, and NF-κB, are regulated by O-linked attachment of GlcNAc (13). This reversible modification is mediated by the O-GlcNAc transferase enzyme, which uses UDP-GlcNAc as a substrate to modify Ser and Thr residues with GlcNAc. GlcNAc also signals the human fungal pathogen Candida albicans to induce two signaling pathways. One induces the cyclic AMP (cAMP) signal pathway, which promotes the switch from budding to hyphal growth (2, 8, 20, 29). Activation of the cAMP pathway by GlcNAc was also recently shown to promote the switching of C. albicans cells from white to opaque phase (15). The second pathway induces the expression of the GlcNAc transporter NGT1 and the genes needed to catabolize GlcNAc (18, 41). The fungal GlcNAc signaling pathways are not well understood. In contrast to the situation in metazoans, O-GlcNAc modification is not known to occur in C. albicans, and the commonly studied model yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe lack the genes needed to catabolize GlcNAc.
The ability of the human fungal pathogen C. albicans to respond to GlcNAc has significance for the understanding of the mechanisms of pathogenesis. C. albicans grows as a commensal organism on most humans, but under certain conditions it can cause life-threatening infections in susceptible individuals (14, 25). One characteristic feature of C. albicans is its ability to grow in different morphologies, including rounded buds, chains of elongated cells known as pseudohyphae, and long, filamentous hyphal cells (34). The switch to hyphal morphology is significant because it correlates with increased expression of virulence genes and is important for biofilm formation (4, 7, 17, 37). Interestingly, GlcNAc is a potent inducer of hyphal growth, whereas other sugars are not. GlcNAc also induces C. albicans to express genes that encode proteins needed to catabolize GlcNAc, including Ngt1, a GlcNAc transporter; Hxk1, a kinase that converts GlcNAc to GlcNAc-6-PO4; Dac1, a deacetylase that converts it to glucosamine-6-PO4; and Nag1, a deamidase that converts GlcNAc to fructose-6-PO4 (2, 18, 36). Therefore, microarray analysis of gene expression was carried out to comprehensively identify the transcriptional response of C. albicans to GlcNAc. As part of these studies, we identified a novel gene, GIG1, that is important for normal sensitivity to nikkomycin Z, an inhibitor of chitin synthase.
MATERIALS AND METHODS
Strains and media.
The C. albicans strains used in this study, described in Table 1, were propagated on rich yeast extract-peptone-dextrose (YPD) medium or on synthetic medium essentially as described previously (28), except that in some cases 80 mg/liter uridine was added to enable ura3 mutants to grow.
Table 1.
C. albicans strains used in this study
| Strain | Parent | Genotype |
|---|---|---|
| BWP17 | Sc5314 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| DIC185 | BWP17 | ura3Δ::λimm434/URA3 his1::hisG/HIS1 arg4::hisG/ARG4 |
| YJA21 | BWP17 | GIG1/gig1::ARG4 ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| YJA22 | YJA21 | GIG1/gig1::ARG4 URA3/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| YJA23 | YJA21 | GIG1/gig1::ARG4 GIG1::URA3 ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| YJA24 | YJA21 | gig1::HIS1/gig1::ARG4 ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| YJA25 | YJA24 | gig1::HIS1/gig1::ARG4 URA3/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| YJA26 | YJA24 | gig1::HIS1/gig1::ARG4 GIG1::URA3 ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| YLD23 | BWP17 | GIG1-GFP::URA3 ura3Δ::λimm434/URA3 his1::hisG/HIS1 arg4::hisG/ARG4 |
| CR323 | CAI4 | ura3::λ imm434/ura3::λ imm434 cdc35Δ::hisG/cdc35Δ::hisG pVec-URA3 |
A homozygous gig1Δ/gig1Δ mutant (YJA25) lacking the open reading frame orf19.1066 was constructed in C. albicans strain BWP17 using methods described previously (39). In brief, PCR primers containing ∼70 bp of sequence homologous to the sequences flanking the open reading frame of GIG1 were used to amplify either the ARG4 or the HIS1 selectable marker gene. Integration of the deletion cassettes at the appropriate sites was verified by PCR using combinations of primers that flanked the integration as well as primers that annealed within the cassettes that had been introduced. Complemented strains were constructed by introducing a plasmid carrying one wild-type copy of GIG into the genome. The GIG1 plasmid was constructed by PCR amplification of the genomic DNA from 1,000 bp upstream of the initiator ATG codon to 300 bases downstream of the terminator codon of GIG1. This DNA fragment was then inserted between the SacI and SacII restriction sites of the URA3 plasmid pDDB57 (38). The resulting plasmid was linearized in the promoter region by digestion with AgeI and was integrated into the GIG1/gig1Δ strain YJA21 and the gig1Δ/gig1Δ strain YJA24 by using URA3 selection to create complemented strains YJA23 and YJA26.
A C. albicans GIG1-GFP strain was created by homologous recombination of GFP sequences into the 3′ end of the GIG open reading frame. The DNA used for the transformation was created by PCR using primers that contain ∼70 bp of sequence homologous to the 3′ end of the GIG1 open reading frame to amplify a cassette containing green fluorescent protein (GFP) and a URA3 selectable marker (11). The colonies resulting from transformation into C. albicans were then screened by PCR to identify a GIG1-GFP fusion strain, which was named YLD23.
Broth dilution assays were used to examine sensitivity to tunicamycin (Sigma-Aldrich Corp.) and nikkomycin Z (Calbiochem). Cell cultures were washed and were then adjusted to a density of 1 × 104/ml in synthetic medium containing amino acids and either 2.5 mM dextrose or 2.5 mM GlcNAc. Aliquots were placed in the wells of a 96-well plate, and then serial dilutions of drug were applied. The plates were covered with an oxygen-permeable AeraSeal membrane (Research Products International Corp.) and were incubated at 37°C for 2 days.
Microscopy.
YLD23 cells carrying the GIG1-GFP fusion gene and control strain DIC185 cells were grown overnight to log phase in synthetic medium containing dextrose. The cells were then collected by centrifugation, resuspended in synthetic medium containing either 100 mM dextrose or 50 mM GlcNAc, and then grown for 6 h at 30°C. Gig1-GFP was detected by fluorescence microscopy, and cell morphology was detected using differential interference contrast (DIC) optics. The bud and hyphal morphogenesis of the gig1Δ mutant was examined using cells grown overnight to log phase in synthetic medium with dextrose. The cells were then pelleted by centrifugation, washed in synthetic medium without sugar, and resuspended in synthetic medium containing either 50 mM glucose or 50 mM GlcNAc. Images were captured using an Olympus BH2 microscope equipped with a Zeiss AxioCam digital camera.
Microarray analysis of gene expression.
A fresh overnight culture of the wild type control strain DIC185, the gig1Δ strain YJA25, or the cdc35Δ strain CR323 (26) was inoculated into synthetic medium containing either glucose or galactose. The medium also contained 10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) to maintain the cultures at about pH 6.8. The cell cultures were periodically adjusted to keep the cells in log phase for about 24 h prior to a shift to GlcNAc medium. For comparisons of dextrose with GlcNAc, the cells were harvested by centrifugation and were then resuspended at a density of 2 × 106/ml in prewarmed synthetic medium containing dextrose or GlcNAc (5 mM). For comparisons of galactose alone with galactose plus GlcNAc, cells were grown overnight in a medium containing 2% galactose, harvested by centrifugation, and resuspended at 2 × 106 cells/ml in a synthetic medium containing 2% galactose; then GlcNAc was added to one sample to a final concentration of 5 mM. The cultures were grown for 2 h, and then cell pellets were harvested for analysis.
Microarray analysis of gene expression was carried out by the Spotted Array Facility at Stony Brook University. RNA was extracted from 3 × 108 cells using a RiboPure-Yeast RNA isolation kit (Ambion). cDNA was then synthesized from total RNA using a SuperScript II kit (Invitrogen) for a poly(T)-primed reaction with a mixture of aminoallyl-dUTP and dTTP in a 3:2 ratio plus dATP, dCTP, and dGTP. Samples were treated with RNase A and were purified using a PCR cleanup column (Qiagen), and then the cDNA was coupled to N-hydroxysuccinimide (NHS)-functionalized Cy3 or Cy5 dyes (GE Biosciences). The yield and extent of dye incorporation were measured spectrophotometrically. Microarrays containing 70-mer oligonucleotides representing the open reading frames in the C. albicans ORF19 database spotted in triplicate were obtained from the Washington University Genome Sequencing Center (6). Control and experimental samples were hybridized to the microarrays using a Tecan HS 4300 Pro hybridization station. After a wash, the microarrays were scanned using an Agilent G2505B microarray scanner.
Western blot analysis.
Cells were grown overnight in synthetic medium containing dextrose, washed, resuspended in synthetic medium containing either dextrose, GlcNAc, glucosamine, fructose, or galactose, and grown for 3 h at 30°C. Approximately 3 × 108 cells were harvested and resuspended in 400 μl cold lysis buffer (10 mM Tris, 1 mM EDTA, 150 mM NaCl) with a protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN). The cells were mixed with glass beads and were then lysed by shaking in a bead beater for 3 cycles of agitation for 45 s each, with cooling on ice between cycles. The cell extract was mixed with 2× sodium dodecyl sulfate (SDS) gel sample buffer (8 M urea, 4% SDS, 50 mM Tris [pH 6.8]) and was heated to 90°C for 3 min; the proteins were separated by electrophoresis on a 9% SDS-polyacrylamide gel and were then electrophoretically transferred to a nitrocellulose membrane. The membrane was probed with an anti-GFP monoclonal antibody (Roche Applied Science, Indianapolis, IN) and was then incubated with IRDye 800CW-conjugated goat anti-mouse IgG (Li-Cor Biosciences, Lincoln, NE) as a secondary antibody. As a control, the blots were also probed either with a rabbit anti-actin antibody or with rabbit anti-glucose-6-phosphate dehydrogenase (Sigma-Aldrich Corp., St. Louis, MO) and were then incubated with an IRDye 680-conjugated goat anti-rabbit antibody (Li-Cor Biosciences, Lincoln, NE). Images were acquired using an Odyssey digital infrared imaging system (Li-Cor Biosciences, Lincoln, NE).
Analysis of Gig1 orthologs.
BLAST searches (1) were carried out to identify Gig1 homologs in the genome sequences of other organisms available at the websites of the Broad Institute (http://www.broadinstitute.org/) and the National Center for Biotechnology (http://www.ncbi.nlm.nih.gov/BLAST/). The Gig1 protein sequences were aligned using ClustalW (35).
RESULTS
Effects of GlcNAc on gene expression in C. albicans.
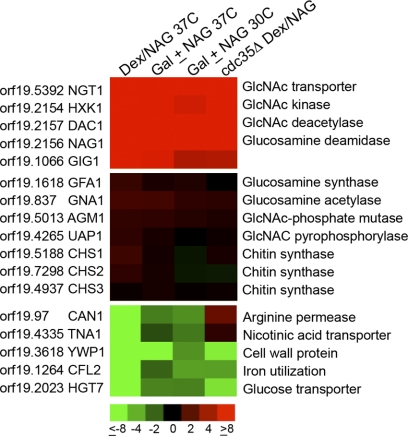
The transcriptional response to GlcNAc was analyzed using microarrays comprising 70-mer oligonucleotides corresponding to the open reading frames in the ORF19 version of the C. albicans genome sequence (6). For the initial analysis, cells were grown in synthetic medium containing dextrose at 37°C and were then shifted to a fresh medium containing either dextrose or GlcNAc and allowed to grow for 2 more hours. Comparison of cells grown in dextrose and GlcNAc media revealed that most of the genes induced to high levels, including NGT1, HXK1, DAC1, and NAG1, are known to be involved in GlcNAc catabolism (Fig. 1A; see also the supplemental material). Another gene induced to high levels corresponded to an unstudied open reading frame, orf19.1066, that will be described in more detail below.
Fig. 1.
Microarray analysis of GlcNAc-regulated genes. The effects of GlcNAc on gene expression were analyzed in four different types of microarray experiments, each carried out in duplicate. (Top) The genes induced most consistently and to the highest levels; (center) genes encoding proteins that synthesize GlcNAc and chitin synthase genes, which show essentially no change in expression; (bottom) the most highly repressed genes. The color code for the fold change in gene expression is given at the bottom (red, induced; green, repressed). Columns show results for cells of the wild-type strain DIC185 grown in dextrose or switched to GlcNAc for 2 h at 37°C (Dex/Nag 37C), strain DIC185 cells grown at 37°C in 2% galactose alone or with the addition of 5 mM GlcNAc for 2 h (Gal ± NAG 37C), cells grown in galactose with or without GlcNAc at 30°C (Gal ± NAG 30C), and a cdc35Δ strain that lacks adenylyl cyclase grown in dextrose or switched to GlcNAc for 2 h (cdc35Δ Dex/NAG).
Surprisingly, the expression of the genes that contribute to the synthesis of GlcNAc, GFA1 (encoding glutamine synthase) and GNA1 (encoding glucosamine acetyltransferase), did not change in the presence of GlcNAc (Fig. 1B). Similarly, no significant changes were detected in the expression of the genes needed to convert GlcNAc to UDP-GlcNAc (AGM1 and UAP1) or the genes that encode the chitin synthases (CHS1, CHS2, and CHS3), which convert UDP-GlcNAc to chitin. This indicates that exogenous levels of GlcNAc do not affect the transcript abundance of genes involved in GlcNAc synthesis in C. albicans.
In contrast to the induced genes, the most highly repressed genes did not fit a simple pattern. Among these genes, it was not surprising that HGT7 showed reduced expression in GlcNAc medium, since it encodes a putative glucose transporter and is typically expressed under high-glucose conditions (6).
Hyphal genes induced by GlcNAc.
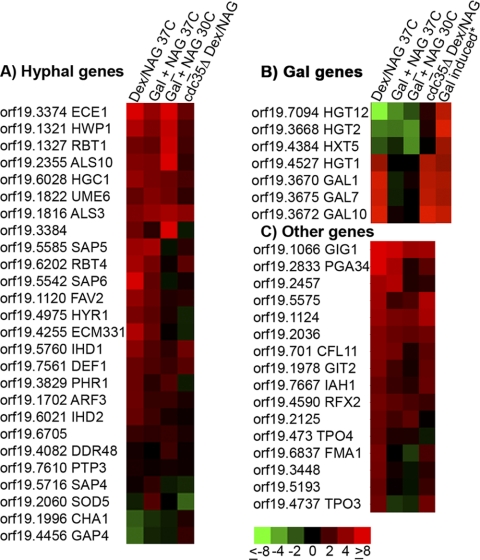
In addition to the GlcNAc catabolic genes, microarrays identified 88 other genes with ≥2-fold-increased expression that could be hypha-induced genes or novel GlcNAc-induced genes (Fig. 2; see also the supplemental material). Some of these genes may also have been derepressed by a shift out of glucose medium rather than induced by GlcNAc (27). To help distinguish between these categories, we examined cells grown in different carbon sources. In one set of experiments, glucose derepression was avoided by growing cells in galactose medium at 37°C and then adding GlcNAc as an inducer. Galactose medium was selected because our previous studies demonstrated that galactose does not prevent GlcNAc induction of hyphae or of the GlcNAc transporter gene NGT1 (2). Another set of microarrays compared cells grown in galactose with or without GlcNAc at 30°C, since most hyphal genes are poorly induced at this lower temperature (16). In addition, we also examined a cdc35Δ strain lacking adenylyl cyclase, which is deficient in inducing hyphal genes (26).
Fig. 2.
GlcNAc induces hyphal genes, a subset of galactose-induced genes, and a set of novel genes. The growth conditions for the cells used in the microarray analyses of gene expression are indicated at the top of each column and explained in the legend to Fig. 1. (A) Analysis of the effects of GlcNAc on genes that have previously been reported to be induced when cells are treated with serum to form hyphae (12, 16). (B) GlcNAc induced a subset of galactose-induced genes. (C) A novel set of genes induced by GlcNAc, of which the orf19.1066 gene is induced to the highest levels and most consistently under all four conditions.
Comparison of the GlcNAc-induced genes with previously published analyses of cells induced with serum to form hyphae revealed a high degree of overlap (Fig. 2A). Despite this similarity, we did not observe induction of DDR48, PTP3, SOD5, CHA1, or GAP4, which had previously been reported to be hypha-induced genes (12, 16). Possibly these genes are induced by factors in serum that stimulate other pathways not activated by GlcNAc. Alternatively, since serum is the strongest known inducer of hyphal growth, perhaps upregulation of these genes requires a stronger hyphal induction signal than that provided by GlcNAc.
Galactose-induced genes.
After removal of the known GlcNAc-induced and hypha-induced genes, the remaining upregulated transcripts unexpectedly included three genes needed for galactose catabolism: GAL1, GAL7, and GAL10 (Fig. 2B). Comparison with previous microarray studies of the effects of galactose (5, 23) showed that GlcNAc stimulated only a subset of galactose-induced genes (Fig. 2B). For example, the putative galactose transporter HGT2/orf19.3668 (23) was not induced by GlcNAc, and neither were HGT12 and HXT5. Although the relative expression of the galactose-regulated genes was unchanged in the comparison of galactose with or without GlcNAc, since galactose was present under both conditions, the microarray signal intensities confirmed that HGT2/orf19.3668 and the GAL genes were all expressed at high levels. One interesting possibility, addressed further in the Discussion, is that these results are due to activation of Cph1, a transcription factor that induces hyphal genes and is also activated by galactose to stimulate expression of the GAL1, GAL7, and GAL10 genes (23).
Identification of GIG1/orf19.1066 as a novel GlcNAc-induced gene.
Genes that were not stimulated by serum or galactose were analyzed in order to identify novel GlcNAc-induced transcripts. In this grouping (Fig. 2C), orf19.1066 stood out as the most highly induced gene under all four microarray conditions, and it was therefore named GIG1 (GlcNAc-induced gene). The GIG1 open reading frame is relatively short—254 amino acids—and does not contain any previously described functional motifs. However, GIG1 is highly conserved in the fungal kingdom and in some other unicellular eukaryotes (see Discussion).
To confirm that GlcNAc induces the expression of the GIG1 gene and protein, Western blot analysis was carried out with a C. albicans strain engineered to express a GFP-tagged version of GIG1 (Fig. 3). Gig1-GFP was induced by GlcNAc but not by the GlcNAc catabolic breakdown products glucosamine and fructose or by the unrelated sugar galactose. These data confirm the microarray studies and further demonstrate that GIG1 is specifically induced by GlcNAc.
Fig. 3.
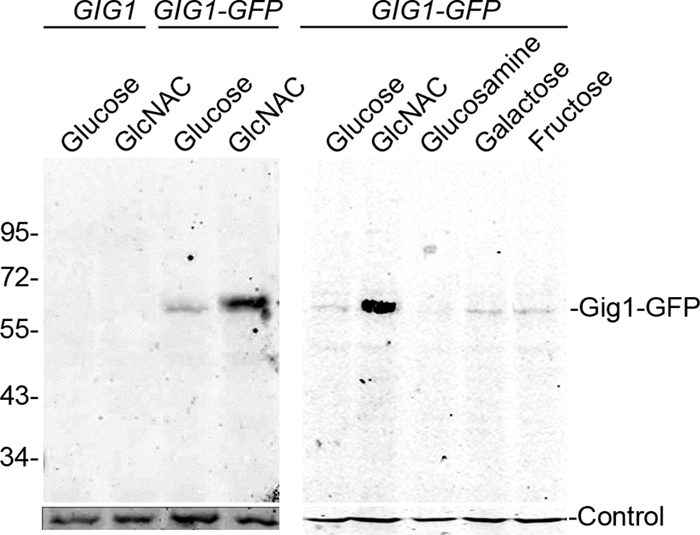
Gig1-GFP is specifically induced by GlcNAc. The wild-type control strain DIC185 or the GIG1-GFP strain YLD23 was grown in synthetic medium containing the indicated sugar. Cell extracts were then analyzed on a Western blot probed with an anti-GFP antibody. Control blots were probed with rabbit anti-glucose-6-phosphate dehydrogenase (left) or rabbit anti-actin (right).
The subcellular localization of Gig1-GFP was analyzed by fluorescence microscopy. Gig1-GFP fluorescence was elevated in the cells carrying the fusion gene, but not in the untagged control cells, after they were shifted to GlcNAc medium (Fig. 4A). The GIG1-GFP cells did not show higher fluorescence than the control cells when the cells were grown in a repressing medium containing glucose (Fig. 4B). The Gig1-GFP fluorescence appeared to be spread throughout the cell, indicating that Gig1-GFP is a cytoplasmic protein. This localization was not due to proteolytic cleavage of the GFP from the fusion protein, since Gig1-GFP migrated at its expected position on a Western blot (Fig. 3).
Fig. 4.
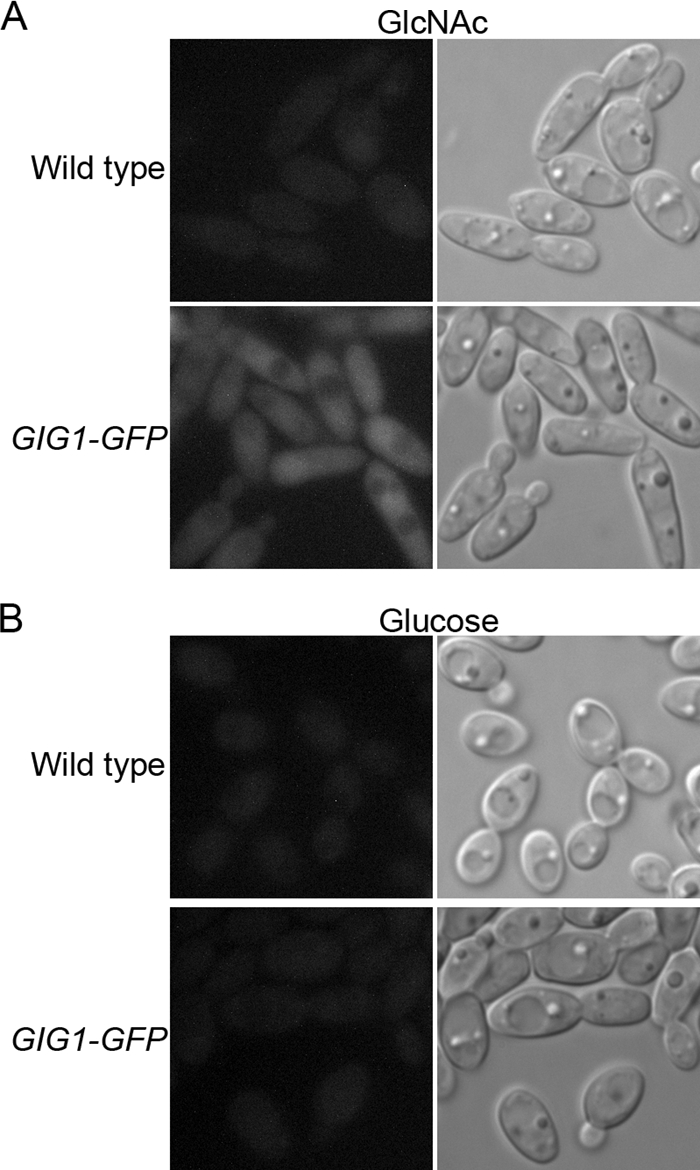
Gig1-GFP localizes to the cytoplasm. The wild-type control strain DIC185 or the GIG1-GFP strain YLD23 was grown in synthetic medium containing GlcNAc (A) or glucose (B). Gig1-GFP was then visualized by fluorescence microscopy (left). Cell morphology was visualized by light microscopy (right).
Analysis of gig1Δ mutant cells.
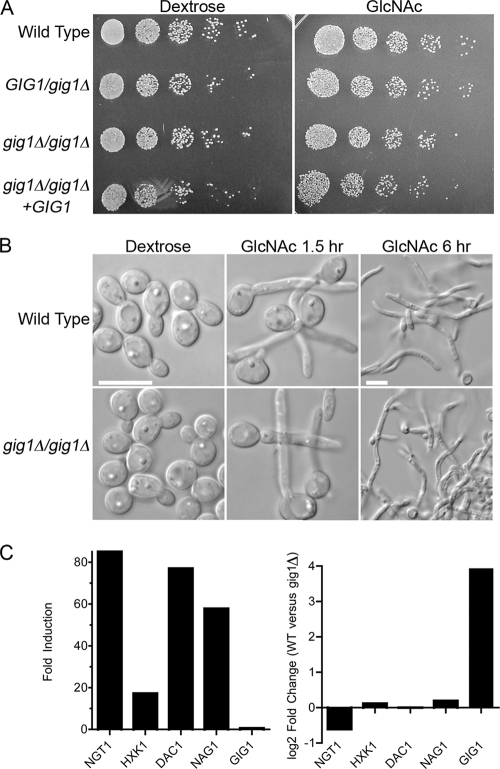
A gig1Δ mutant strain in which both copies of GIG1 were deleted from the diploid C. albicans genome was constructed. The gig1Δ cells were then examined for their ability to catabolize GlcNAc by spotting dilutions of cells onto a solid agar medium containing either GlcNAc or dextrose as the carbon source (Fig. 5A). No obvious difference in the growth rate was observed. Additional studies carried out with higher or lower GlcNAc concentrations also failed to reveal a difference in the growth rate (data not shown). The gig1Δ mutant was also able to form hyphae in response to GlcNAc (Fig. 5B). Thus, GIG1 is not essential for growth on GlcNAc or for the induction of hyphae.
Fig. 5.
Growth and morphogenesis of gig1Δ mutant cells. (A) A 10-fold dilution series of the indicated C. albicans strains was spotted onto synthetic medium agar plates containing either glucose or GlcNAc at 25 mM. The cells were grown for 2 days at 30°C and were then photographed. The strains used included the wild-type control DIC185, YJA22 (GIG1/gig1Δ), YJA25 (gig1Δ/gig1Δ), and YJA26 (gig1Δ/gig1Δ + GIG1). (B) Cells were grown overnight in synthetic dextrose medium, washed, resuspended in synthetic medium containing either dextrose or GlcNAc at 50 mM, and then grown at 37°C for the indicated times. The GlcNAc 6-h samples are shown at a lower magnification to allow the visualization of long hyphal structures. Bars, 10 μm. (C) (Left) Fold induction of the indicated genes after a 2-h incubation of the gig1Δ strain in GlcNAc, as determined by a microarray experiment. (Right) Direct comparison on the same microarray of wild-type control cells and gig1Δ cells that were shifted to GlcNAc for 2 h.
Microarray analysis of gene expression revealed that the gig1Δ mutant showed essentially wild-type gene induction in response to GlcNAc, including the expected increase in expression of the catabolic genes (Fig. 5C; see also the supplemental material). To examine differences in gene expression in a more sensitive manner, RNA samples from GlcNAc-induced wild-type and gig1Δ mutant cells were mixed and compared on the same microarray, but this also failed to detect interesting differences in gene expression (Fig. 5C; see also the supplemental material).
Deletion of GIG1 causes increased resistance to the chitin synthase inhibitor nikkomycin Z.
Two of the pathways that make use of UDP-GlcNAc in fungal cells are N-linked glycosylation and synthesis of the chitin layer of the cell wall. The potential role of Gig1 in these pathways was probed using broth dilution assays in 96-well plates to test sensitivity to tunicamycin and nikkomycin Z. Tunicamycin inhibits the first step of N-linked glycosylation by blocking the transfer of GlcNAc-1-PO4 from UDP-GlcNAc to dolichol phosphate. Mutation of GIG1 did not detectably alter sensitivity to tunicamycin (data not shown).
Nikkomycin Z inhibits the conversion of UDP-GlcNAc into chitin by chitin synthase. Sensitivity to nikkomycin Z did not show a sharp cutoff, so the concentrations at which ≥50% growth occurred and the concentration at which no growth occurred were recorded. Interestingly, wild-type cells grown in GlcNAc medium were ∼3-fold more resistant to nikkomycin Z than wild-type cells grown in glucose medium (Fig. 6). In dextrose medium, gig1Δ cells showed strong growth at a 3-fold-higher dose of nikkomycin Z than that for the wild type, as well as a partial ability to grow at 9-fold-higher concentrations of the drug. When grown in GlcNAc medium under conditions that would ordinarily induce GIG1 expression, the gig1Δ cells showed even higher levels of resistance to nikkomycin Z. Growth of gig1Δ cells was detectable at nikkomycin Z doses more than 25-fold higher than a fully lethal dose for the wild-type cells.
Fig. 6.
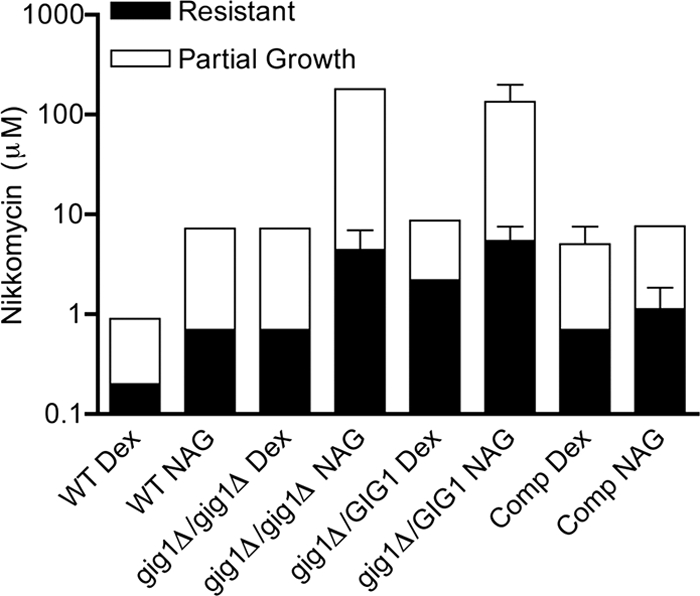
Sensitivity to nikkomycin Z. Cells were grown in synthetic medium containing either dextrose (Dex) or GlcNAc (NAG) and different concentrations of nikkomycin, an inhibitor of chitin synthase. The cells were incubated for 2 days at 37°C, and then growth was monitored. The filled portions of the bars indicate extensive resistance to nikkomycin Z. The open portions of the bars indicate partial resistance, as evidenced by weaker growth of the cells that tapered off with an increased dose. Results, expressed as the concentrations of nikkomycin Z at which cells were extensively or partially resistant, are averages for at least three independent experiments. Error bars indicate standard deviations The strains used were the wild-type control strain DIC185, YJA25 (gig1Δ/gig1Δ), YJA22 (GIG1/gig1Δ), and the complemented strain YJA23 (GIG1/gig1Δ + GIG1) (Comp).
The GIG1/gig1Δ heterozygous mutant also showed increased resistance to nikkomycin Z, indicating that the loss of one copy is sufficient to alter sensitivity to this drug (Fig. 6). The reintroduction of a second copy of GIG1 into the heterozygous mutant restored normal sensitivity to nikkomycin Z. Thus, cells are sensitive to the gene dosage of GIG1, consistent with regulation of the expression of GIG1 by GlcNAc.
DISCUSSION
GIG1 is a novel GlcNAc-induced gene linked to GlcNAc metabolism.
GlcNAc plays important roles in a wide range of organisms from bacteria to humans. It is a major structural component of bacterial cell wall peptidoglycan, fungal cell wall chitin, and the extracellular matrix of animal cells. GlcNAc also has functions in cell signaling that have only recently begun to be appreciated, so the mechanisms are not well understood. Therefore, we used microarray analysis to examine the changes in transcription caused by GlcNAc in the human fungal pathogen C. albicans. GlcNAc stimulates two pathways in C. albicans: hyphal growth and induction of the genes needed to catabolize GlcNAc. Four different microarray conditions were used to help classify the genes as hyphally induced or specifically induced by GlcNAc. These experiments also showed that GlcNAc did not repress the GlcNAc synthesis genes (Fig. 1). This finding suggests that the proposed mechanism of feedback inhibition of glucosamine synthase (Gfa1) by UDP-GlcNAc may be the primary mode for regulating GlcNAc synthesis (24).
In addition to inducing the expected hyphal and catabolic genes, GlcNAc also induced genes that have not been characterized previously. The most highly and consistently induced gene under all of the different microarray conditions was GIG1/orf19.1066. The GIG1 gene resides on chromosome 1 and therefore is not linked to the cluster of GlcNAc catabolic genes (HXK1, NAG1, and DAC1) on chromosome 6 or the GlcNAc transporter NGT1 on chromosome 3. GIG1 was induced ∼12-fold after a 2-h shift from dextrose to GlcNAc. A Gig1-GFP fusion protein was also highly induced by GlcNAc, confirming that both the GIG1 gene and the Gig1 protein are responsive to GlcNAc. Further analysis showed that Gig1-GFP was specifically induced by GlcNAc, and not by the catabolic degradation products glucosamine and fructose or by the unrelated sugar galactose (Fig. 3). This pattern of expression implicates GIG1 in GlcNAc metabolism. Consistent with this, Gig1-GFP was detected in the cytoplasm (Fig. 4).
GIG1 was further linked to GlcNAc metabolism by analysis of the gig1Δ mutant phenotypes. The gig1Δ mutant was not obviously defective in growth on GlcNAc or in the induction of hyphal morphogenesis (Fig. 5), so it was examined under more stressful conditions by testing sensitivity to inhibitors of enzymes that utilize UDP-GlcNAc as a substrate. The gig1Δ mutant showed normal sensitivity to tunicamycin, which blocks an early enzymatic step in N-linked glycosylation. However, the gig1Δ mutant was more resistant to nikkomycin Z, which inhibits chitin synthase from using UDP-GlcNAc to form chitin (Fig. 6). A gig1Δ/GIG1 heterozygote also showed increased resistance to GlcNAc, indicating that the cells are sensitive to the level of GIG1 expression, consistent with the induction of GIG1 expression by GlcNAc. These results are also significant in that they suggest that at least some types of chitin synthesis inhibitors may have limited therapeutic value because of the potential for resistant strains to develop via mutation of GIG1. (Chitin synthase has been proposed as an attractive drug target, because humans do not synthesize chitin.)
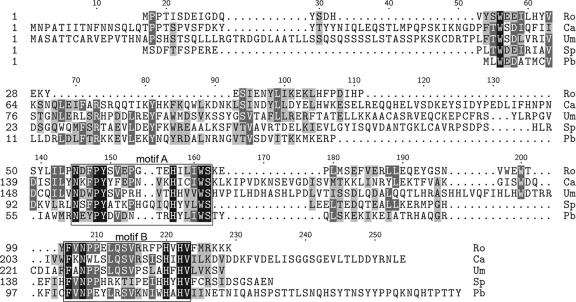
Comparison of the presence or absence of Gig1 orthologs in other species also implicates Gig1 in GlcNAc metabolism. Gig belongs a conserved family of proteins (DUF3605 domain) present in a wide range of fungi and some unicellular eukaryotes, including Dictyostelium discoideum and Chlamydomonas reinhardtii. Gig1 orthologs were defined on the basis of overall sequence identity and the presence of two highly conserved motifs (Fig. 7). The detection of a Gig1 ortholog in S. cerevisiae (YPL067c) was interesting, since this yeast lacks GlcNAc catabolic genes. Although this could suggest that the function of Gig1 is related to GlcNAc synthesis rather than to catabolism, it is noteworthy that no Gig1 ortholog is present in Ashbya gossypii, a species related to S. cerevisiae that lacks GlcNAc catabolic genes. Furthermore, BLAST searches carried out with Broad Institute genome databases failed to detect Gig1 homologs in the more distantly related fission yeasts Schizosaccharomyces pombe, Schizosaccharomyces japonicus, Schizosaccharomyces octosporus, and Schizosaccharomyces cryophilus, all of which lack GlcNAc catabolic genes. In contrast, GIG1 and GlcNAc catabolic genes were detected in the Broad Institute databases for seven species closely related to Candida albicans and for eight Aspergillus species, which are more distantly related filamentous fungi. GIG1 and the GlcNAc catabolic gene orthologs were also identified in members of four different fungal phyla (Ascomycota, Basidiomycota, Mucormycotina, and Chytridiomycota), but both were absent from the genomes of 4 fungi in the phylum Microsporidia. The GIG1 gene and GlcNAc catabolic genes were also missing from Batrachochytrium dendrobatidis in the phylum Chytridiomycotina. This overall pattern of gene conservation in different organisms is analogous to observations for the galactose genes, which have been lost in various fungal species (9, 31).
Fig. 7.
Alignment of conserved amino acids in Gig1 proteins. Gig1-homologous proteins in different species were identified by BLAST searches and were then aligned using ClustalW. Two very highly conserved sequence motifs, labeled motif A and motif B, are boxed. The Gig1 orthologs are labeled by the initials of the organisms from which they come and include representatives from four different fungal phyla and an algal virus. Ro, Rhizopus oryzae; Ca, Candida albicans; Um, Ustilago maydis; Sp, Spizellomyces punctatus; Pb, Paramecium bursaria chlorella virus.
Further support for linking Gig1 to GlcNAc metabolism comes from the presence of GIG1 orthologs in Paramecium bursaria chlorella viruses (PBCVs), which infect unicellular green algae of the genus Chlorella (40). PBCV genomes are unusual in that they encode many proteins involved in glycosylation and the synthesis of GlcNAc polymers (40). For example, PBCVs carry a gene for glucosamine synthase, which mediates the first committed step in GlcNAc synthesis. They also encode enzymes to synthesize hyaluronan, a polymer of glucose and GlcNAc, or, in some cases, chitin. These GlcNAc-containing polymers coat the outer surfaces of the infected algae. Taken together, the regulation of GIG1 expression, the cytoplasmic localization of Gig1-GFP, the resistance of a gig1Δ mutant to nikkomycin Z, and the pattern of Gig1 evolutionary conservation suggest that Gig1 functions in GlcNAc metabolism.
Galactose genes.
It was surprising that GlcNAc induced the expression of the GAL1, GAL7, and GAL10 genes, which mediate galactose catabolism (Fig. 2B). There is no obvious link with galactose metabolism that would account for the induction of these genes by GlcNAc. An interesting possibility is that these GAL genes are induced by activation of Cph1 in response to GlcNAc. Cph1 is a transcription factor involved in inducing both hyphal genes and galactose-regulated genes (21, 23). GAL1, GAL7, and GAL10 are the only C. albicans genes that have been reported to contain a putative Cph1 recognition motif in their upstream regulatory regions (23). The predicted galactose transporter HGT2/orf19.3668, which was induced by galactose but not by GlcNAc, contains a divergent recognition motif for Cph1 (23). Other galactose-induced genes that lack the Cph1 recognition site were not induced by GlcNAc (Fig. 2B).
GlcNAc and virulence.
The ability of GlcNAc to induce hyphae and the expression of virulence genes is significant for an understanding of the molecular mechanisms of C. albicans pathogenesis (Fig. 2A). However, the role of GlcNAc during infection is not understood. Previous studies suggested that the GlcNAc catabolic genes NAG1, DAC1, and HXK1 are important for virulence (30, 41). Unfortunately, the gene deletion methods used in these studies are now considered to be unreliable for assessing virulence (19). (The URA3 gene used to select for gene deletions was present in different chromosomal contexts in the mutant and control strains, and URA3 expression levels have a significant impact on virulence.) Furthermore, in a systemic infection, C. albicans cells are likely to be exposed to serum, which does not contain significant amounts of GlcNAc. Serum is also high in glucose, which represses the expression of the GlcNAc transporter NGT1, even in the presence of GlcNAc (2). It was also interesting that GlcNAc failed to detectably induce several genes that are stimulated to form hyphae when C. albicans is treated with serum (Fig. 2A), including some genes, such as the SOD5 superoxide dismutase gene, that are important for virulence in a mouse model of systemic infection (10, 22).
Another possibility is that GlcNAc may not be important for systemic infection but could be important for the growth of C. albicans on the skin or mucosa. Extracellular matrix glycosaminoglycans, present in these niches, are rich in GlcNAc. GlcNAc may also be released by bacteria in the gastrointestinal (GI) tract. In this regard, it is interesting that activation of the cAMP signal pathway by GlcNAc also stimulates white-phase C. albicans cells to switch to the opaque phase (15), which is thought to be better suited for skin and mucosal infections (33). Hyphal induction is also important for biofilm formation by C. albicans, so it is possible that GlcNAc may play a role in regulating this process. Interestingly, Escherichia coli appears to respond in the opposite manner. GlcNAc reduces the production by bacteria of curli fibers, which are important for biofilm formation, and also inhibits the expression of the fimbrial adhesin fimB (3, 32). Thus, further studies will be required to determine the roles of GlcNAc in the virulence of C. albicans.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of our lab for helpful advice and comments on the manuscript and Cristina Commesso for assistance. We also thank Malcolm Whiteway for providing C. albicans strains.
This research was supported by a grant awarded to J.B.K. from the National Institutes of Health (R01 GM087368).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 30 July 2010.
REFERENCES
- 1.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2.Alvarez F. J., Konopka J. B. 2007. Identification of an N-acetylglucosamine transporter that mediates hyphal induction in Candida albicans. Mol. Biol. Cell 18:965–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnhart M. M., Lynem J., Chapman M. R. 2006. GlcNAc-6P levels modulate the expression of curli fibers by Escherichia coli. J. Bacteriol. 188:5212–5219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blankenship J. R., Mitchell A. P. 2006. How to build a biofilm: a fungal perspective. Curr. Opin. Microbiol. 9:588–594 [DOI] [PubMed] [Google Scholar]
- 5.Brown V., Sabina J., Johnston M. 2009. Specialized sugar sensing in diverse fungi. Curr. Biol. 19:436–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown V., Sexton J. A., Johnston M. 2006. A glucose sensor in Candida albicans. Eukaryot. Cell 5:1726–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calderone R. A., Fonzi W. A. 2001. Virulence factors of Candida albicans. Trends Microbiol. 9:327–335 [DOI] [PubMed] [Google Scholar]
- 8.Castilla R., Passeron S., Cantore M. L. 1998. N-Acetyl-d-glucosamine induces germination in Candida albicans through a mechanism sensitive to inhibitors of cAMP-dependent protein kinase. Cell. Signal. 10:713–719 [DOI] [PubMed] [Google Scholar]
- 9.Dujon B. 2006. Yeasts illustrate the molecular mechanisms of eukaryotic genome evolution. Trends Genet. 22:375–387 [DOI] [PubMed] [Google Scholar]
- 10.Frohner I. E., Bourgeois C., Yatsyk K., Majer O., Kuchler K. 2009. Candida albicans cell surface superoxide dismutases degrade host-derived reactive oxygen species to escape innate immune surveillance. Mol. Microbiol. 71:240–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerami-Nejad M., Berman J., Gale C. A. 2001. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast 18:859–864 [DOI] [PubMed] [Google Scholar]
- 12.Harcus D., Nantel A., Marcil A., Rigby T., Whiteway M. 2004. Transcription profiling of cyclic AMP signaling in Candida albicans. Mol. Biol. Cell 15:4490–4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hart G. W., Housley M. P., Slawson C. 2007. Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446:1017–1022 [DOI] [PubMed] [Google Scholar]
- 14.Heitman J., Filler S. G., Edwards J. E. J., Mitchell A. P. 2006. Molecular principles of fungal pathogenesis. ASM Press, Washington, DC [Google Scholar]
- 15.Huang G., Yi S., Sahni N., Daniels K. J., Srikantha T., Soll D. R. 2010. N-Acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans. PLoS Pathog. 6:e1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kadosh D., Johnson A. D. 2005. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell 16:2903–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumamoto C. A., Vinces M. D. 2005. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell. Microbiol. 7:1546–1554 [DOI] [PubMed] [Google Scholar]
- 18.Kumar M. J., Jamaluddin M. S., Natarajan K., Kaur D., Datta A. 2000. The inducible N-acetylglucosamine catabolic pathway gene cluster in Candida albicans: discrete N-acetylglucosamine-inducible factors interact at the promoter of NAG1. Proc. Natl. Acad. Sci. U. S. A. 97:14218–14223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lay J., Henry L. K., Clifford J., Koltin Y., Bulawa C. E., Becker J. M. 1998. Altered expression of selectable marker URA3 in gene-disrupted Candida albicans strains complicates interpretation of virulence studies. Infect. Immun. 66:5301–5306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leberer E., Harcus D., Dignard D., Johnson L., Ushinsky S., Thomas D. Y., Schroppel K. 2001. Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol. Microbiol. 42:673–687 [DOI] [PubMed] [Google Scholar]
- 21.Liu H., Kohler J., Fink G. R. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723–1726 [DOI] [PubMed] [Google Scholar]
- 22.Martchenko M., Alarco A. M., Harcus D., Whiteway M. 2004. Superoxide dismutases in Candida albicans: transcriptional regulation and functional characterization of the hyphal-induced SOD5 gene. Mol. Biol. Cell 15:456–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martchenko M., Levitin A., Hogues H., Nantel A., Whiteway M. 2007. Transcriptional rewiring of fungal galactose-metabolism circuitry. Curr. Biol. 17:1007–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milewski S., Gabriel I., Olchowy J. 2006. Enzymes of UDP-GlcNAc biosynthesis in yeast. Yeast 23:1–14 [DOI] [PubMed] [Google Scholar]
- 25.Odds F. C. 1988. Candida and candidosis. Baillière Tindall, Philadelphia, PA [Google Scholar]
- 26.Rocha C. R., Schroppel K., Harcus D., Marcil A., Dignard D., Taylor B. N., Thomas D. Y., Whiteway M., Leberer E. 2001. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol. Cell 12:3631–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sabina J., Brown V. 2009. Glucose sensing network in Candida albicans: a sweet spot for fungal morphogenesis. Eukaryot. Cell 8:1314–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherman F. 1991. Getting started with yeast. Methods Enzymol. 194:3–21 [DOI] [PubMed] [Google Scholar]
- 29.Simonetti N., Strippoli V., Cassone A. 1974. Yeast-mycelial conversion induced by N-acetyl-d-glucosamine in Candida albicans. Nature 250:344–346 [DOI] [PubMed] [Google Scholar]
- 30.Singh P., Ghosh S., Datta A. 2001. Attenuation of virulence and changes in morphology in Candida albicans by disruption of the N-acetylglucosamine catabolic pathway. Infect. Immun. 69:7898–7903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slot J. C., Rokas A. 2010. Multiple GAL pathway gene clusters evolved independently and by different mechanisms in fungi. Proc. Natl. Acad. Sci. U. S. A. 107:10136–10141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sohanpal B. K., El-Labany S., Lahooti M., Plumbridge J. A., Blomfield I. C. 2004. Integrated regulatory responses of fimB to N-acetylneuraminic (sialic) acid and GlcNAc in Escherichia coli K-12. Proc. Natl. Acad. Sci. U. S. A. 101:16322–16327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soll D. R. 2009. Why does Candida albicans switch? FEMS Yeast Res. 9:973–989 [DOI] [PubMed] [Google Scholar]
- 34.Sudbery P., Gow N., Berman J. 2004. The distinct morphogenic states of Candida albicans. Trends Microbiol. 12:317–324 [DOI] [PubMed] [Google Scholar]
- 35.Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wendland J., Schaub Y., Walther A. 2009. N-Acetylglucosamine utilization by Saccharomyces cerevisiae based on expression of Candida albicans NAG genes. Appl. Environ. Microbiol. 75:5840–5845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whiteway M., Oberholzer U. 2004. Candida morphogenesis and host-pathogen interactions. Curr. Opin. Microbiol. 7:350–357 [DOI] [PubMed] [Google Scholar]
- 38.Wilson R. B., Davis D., Enloe B. M., Mitchell A. P. 2000. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast 16:65–70 [DOI] [PubMed] [Google Scholar]
- 39.Wilson R. B., Davis D., Mitchell A. P. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamada T., Onimatsu H., Van Etten J. L. 2006. Chlorella viruses. Adv. Virus Res. 66:293–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamada-Okabe T., Sakamori Y., Mio T., Yamada-Okabe H. 2001. Identification and characterization of the genes for N-acetylglucosamine kinase and N-acetylglucosamine-phosphate deacetylase in the pathogenic fungus Candida albicans. Eur. J. Biochem. 268:2498–2505 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.